Abstract
We evaluated detection of ertapenem (ETP) resistance and Klebsiella pneumoniae carbapenemase (KPC) in 47 Klebsiella pneumoniae isolates using a novel automated microscopy system. Automated microscopy correctly classified 22/23 isolates as ETP resistant and 24/24 as ETP susceptible and correctly classified 21/21 isolates as KPC positive and 26/26 as KPC negative.
TEXT
Carbapenem-resistant Enterobacteriaceae (CRE) are emerging as a global threat; the plasmid-borne carbapenemase gene blaKPC is the predominant mechanism conferring carbapenem resistance in North America (1–5). This resistance gene has been reported in most species of Enterobacteriaceae, but it is most commonly found in Klebsiella pneumoniae. Timely detection of carbapenem resistance is critical for prompt optimization of antimicrobial therapy, but the sensitivity of antimicrobial susceptibility testing methods for CRE detection is variable and turnaround time can be slow (6–10). It has been demonstrated that in vitro detection of K. pneumoniae carbapenemase (KPC) expression can be difficult, varying by bacterial species and level of enzyme expression.
(This study was presented in part at the 113th American Society for Microbiology General Meeting, Denver, CO, May 2013.)
The objective of our study was to evaluate automated microscopy for detection of ertapenem (ETP) resistance in K. pneumoniae and the ability to attribute the mechanism of this resistance to the KPC enzyme. Forty-six K. pneumoniae isolates recovered from clinical specimens obtained at Barnes-Jewish Hospital (St. Louis, MO) and one KPC-negative, extended-spectrum β-lactamase (ESBL)-positive K. pneumoniae quality control strain, ATCC 700603, were tested (Table 1). The ertapenem and meropenem (MEM) susceptibility profiles of the isolates were determined by disk diffusion according to CLSI standards (11), and isolates were characterized for blaKPC using a laboratory-developed real-time PCR assay (6).
TABLE 1.
Characterization of bacterial strains
| Strain type | No. of isolates | No. of isolates with each result as determined by automated microscopy |
Mean (range) zone diameter (mm) witha: |
||||
|---|---|---|---|---|---|---|---|
| KPC |
ETP resistance |
ETP | MEM | ||||
| Positive | Negative | Positive | Negative | ||||
| KPC positive | 21 | 21 | 0 | 21 | 0 | ||
| KPC-2 positive | 13 | 13 | 0 | 13 | 0 | 8 (6–11) | 10 (8–12) |
| KPC-3 positive | 8 | 8 | 0 | 8 | 0 | 12 (8–15) | 13 (12–14) |
| KPC negative | 26 | 0 | 26 | 1 | 25 | ||
| Ertapenem resistant | 2 | 0 | 2 | 1 | 1 | 15 (13–16) | 18 (17–19) |
| Ertapenem susceptible | 24 | 0 | 24 | 0 | 24 | 28 (26–35) | 27 (25–30) |
The CLSI susceptibility breakpoint for ETP is ≥22 mm, and the CLSI susceptibility breakpoint for MEM is ≥23 mm.
For automated microscopy, bacterial suspensions were centrifuged (12,000 × g for 4 min), washed in 1 mM l-histidine buffer at a pH of 7.2, and resuspended in a low-ionic-strength electrokinetic buffer containing 10 mM l-3,4-dihydroxyphenylalanine (l-DOPA) and 1 mM l-histidine at a pH of 7.0 (reagents from Sigma-Aldrich, St. Louis, MO). This created an inoculum of approximately 1 × 106 CFU/ml for testing, and then automated microscopy was performed. Bacterial inocula were pipetted into independent flow cells of a multichannel disposable fluidic cassette (Accelerate Diagnostics Inc., Tucson, AZ). Bacteria were immobilized on the transparent lower surface of each flow cell using electrokinetic concentration (Fig. 1). Mixtures of antibiotics in Mueller-Hinton broth (Becton, Dickinson, Franklin Lakes, NJ) containing 0.85% noble agar (Affymetrix, Santa Clara, CA) were introduced into each flow cell channel. Dark-field images of each flow cell channel were taken at 10-min intervals during a fixed 3-h antibiotic exposure period. An offline image analyzer tracked each immobilized bacterial progenitor cell as it replicated into a clone of daughter cells throughout a series of time-lapse images for each flow cell. The analyzer computed a growth probability score for each growing clone derived from coefficients of a cubic polynomial [f(x) = ax3 + bx2 + cx + d] fitted to the computed log of relative clone mass (integrated pixel intensity) versus time. The growth probability score transformed each clone's growth data into a numerical score ranging between 0 and 1 that represented the probability of the clone continuing to grow (>0.8), to arrest (0.2 to 0.8), or to lyse (<0.2). The slope of a straight line fit by linear regression was used to calculate the division rates for individual clones. The median clonal division rate (div/h), weighted for the number of growing clones and growth probability, was used to calculate a resistance score for each test condition.
FIG 1.
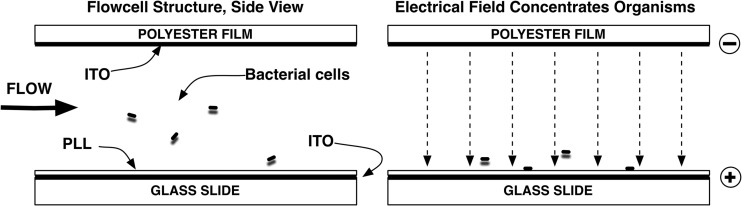
Bacterial cell immobilization by electrokinetic concentration. (Left) Sample suspension flows in and then stops. (Right) Electric field forces bacterial cells to the positively charged poly-l-lysine (PLL) capture coating. PLL binds cells upon contact and immobilizes them after the electric field turns off. ITO, indium tin oxide.
A total of 10 μg/ml of ETP (Merck, Whitehouse Station, NJ) was used for susceptibility testing, and KPC detection used four flow cell channels per sample. The first KPC channel contained 16 μg/ml of MEM (Sigma-Aldrich), the second contained 32 μg/ml of MEM, the third contained 16 μg/ml of MEM plus 32 μg/ml 3-nitrophenylboronic acid (NPBA; Sigma-Aldrich), and the fourth contained 32 μg/ml of MEM plus 32 μg/ml NPBA. NPBA was included because it is an inhibitor of the KPC enzyme (12). Interpretation algorithms in the image analyzer computed the difference between resistance scores in MEM alone and those in MEM plus NPBA to obtain an inhibition score, weighted by the number of growing clones in each condition. The operator interpreting the ETP susceptibility and KPC status of the isolate was blinded to the phenotypic data and the blaKPC PCR result.
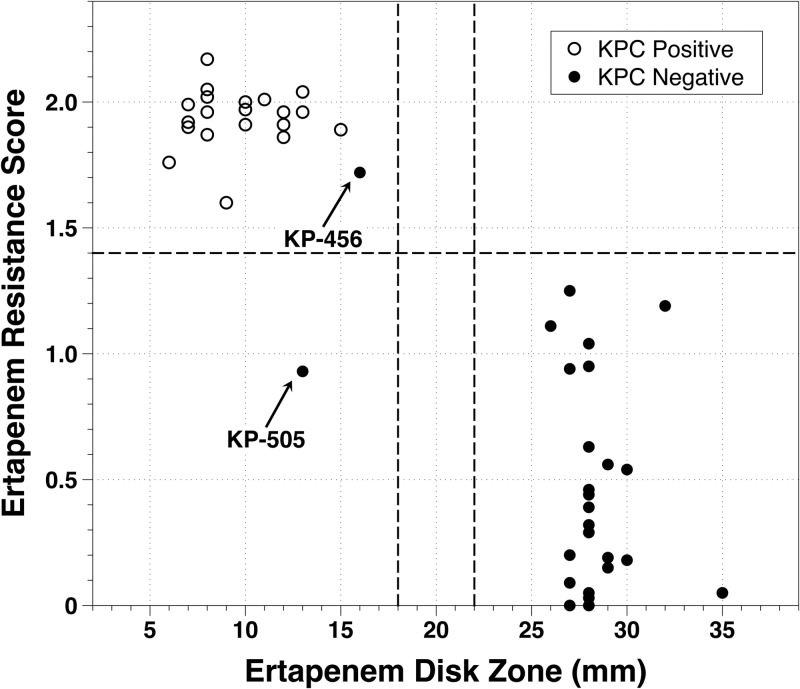
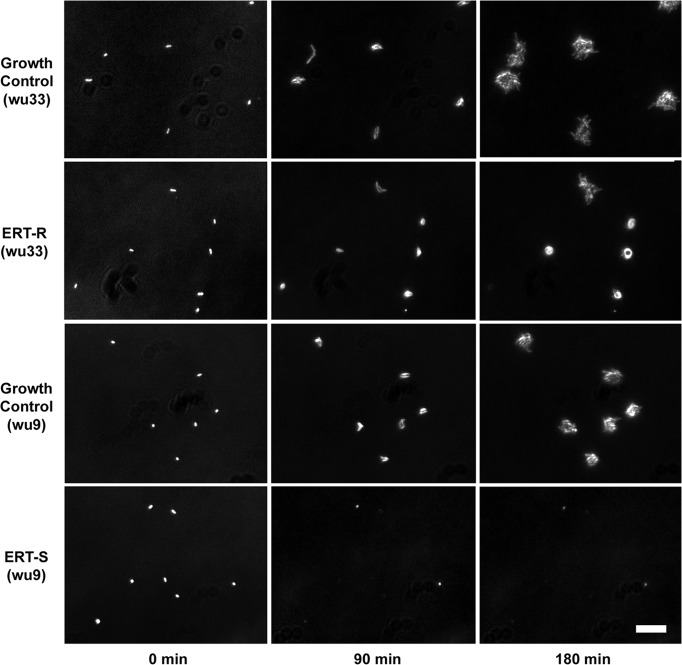
The automated microscopy system classified 22/23 Klebsiella pneumoniae isolates as ETP resistant and 24/24 as ETP susceptible using a resistance score cutoff of 1.4 (Fig. 2) and therefore achieved 96% sensitivity (95% confidence interval [CI], 76% to 100%) and 100% specificity (95% CI, 83% to 100%) for detection of ETP resistance. There was one instance of susceptibility reported by automated microscopy for an isolate that tested as resistant by disk diffusion (KP-505; 13-mm ETP zone diameter, microscopy score of 0.93). Growth controls had an average and standard deviation of 55 ± 27 growing clones per field of view, a number which was therefore the number of viable clones in all channels. The average resistance score and standard deviation was 0.46 ± 0.41 (range, 0.00 to 1.25) for ETP-susceptible isolates and 1.89 ± 0.24 (range, 0.93 to 2.17) for ETP-resistant isolates. Images of ETP-resistant isolate wu33 and ETP-susceptible isolate wu9 at 0, 90, and 180 min after exposure to 10 μg/ml ETP are shown in Fig. 3. Upon exposure to 10 μg/ml ETP, ETP-resistant strains showed robust growth with minimal lysis, with most clones growing in a spheroidal clone morphology. In contrast, ETP-susceptible strains exhibited rapid growth arrest and lysis (Fig. 3).
FIG 2.
Scatter plot of the automated microscopy resistance score versus ertapenem disk diffusion reference. Vertical dashed lines indicate 2013 CLSI breakpoints for susceptible and resistant, and the horizontal dashed line indicates the microscopy score interpretation criterion value of 1.4. Strains KP-456 and KP-505 were KPC negative by PCR but resistant by disk diffusion for ertapenem and meropenem.
FIG 3.
Ertapenem-resistant (ETP-R) (wu33; ETP disk zone = 8 mm) and -susceptible (ETP-S) (wu9; ETP disk zone = 35 mm) K. pneumoniae strains. Rows show clone images at 90-min intervals after exposure to growth medium alone (growth control) or 10 μg/ml ertapenem (ETP-R and ETP-S). Images are zoomed in and contrast enhanced. Scale bar, 20 μm.
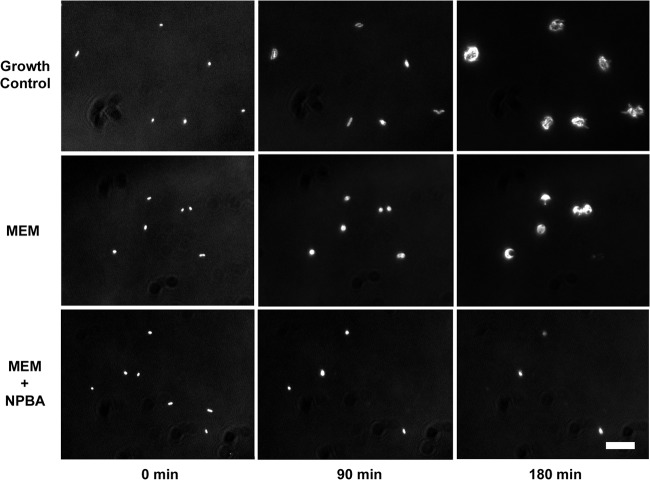
The microscopy system classified 21/21 isolates as KPC positive and 26/26 strains as KPC negative compared to PCR results. Upon exposure to MEM alone, spheroidal clone morphology was observed for KPC-positive clones with minimal lysis (Fig. 4). Rapid growth arrest and lysis were observed for KPC-positive isolates in the presence of MEM plus NPBA as well as for all KPC-negative isolates upon exposure to MEM and MEM plus NPBA. The microscopy method achieved 100% sensitivity (95% CI, 81% to 100%) and 100% specificity (95% CI, 83% to 100%) for KPC characterization of the isolates. The mean inhibition score and standard deviation was 0.54 ± 0.19 (range, 0.19 to 0.91) for KPC-positive isolates and −0.75 ± 0.38 (range, −1.00 to −0.08) for KPC-negative isolates. Images of KPC-positive strain wu15 at 0, 90, and 180 min are shown in Fig. 4. All 21 KPC-positive strains were classified as ETP resistant by microscopy analysis and by disk diffusion.
FIG 4.
KPC-positive K. pneumoniae strain wu15 (disk diffusion zone sizes of 8 mm and 12 mm for ETP and MEM, respectively). Rows show clone images at 90-min intervals after exposure to growth medium alone (top row), 16 μg/ml meropenem (MEM) (middle row), or 16 μg/ml meropenem plus 32 μg/ml of the inhibitor 3-nitrophenylboronic acid (MEM+NPBA) (bottom row). Images are zoomed in and contrast enhanced. Scale bar, 20 μm.
Of note, two strains evaluated in this study (KP-456 and KP-505) were negative for KPC by PCR but resistant to both ETP and MEM using disk diffusion. The microscopy analysis correctly classified both strains as KPC negative. Microscopy analysis correctly classified KP-456 as ETP resistant but incorrectly classified KP-505 as ETP susceptible (Fig. 2). Additional susceptibility testing of isolate KP-505 demonstrated that the isolate was susceptible to piperacillin-tazobactam, ticarcillin-clavulanic acid, cefotetan, amikacin, and tobramycin and resistant to gentamicin; this profile is different from those of the KPC-producing strains in this study. The KPC-producing strains were typically gentamicin susceptible and resistant to piperacillin-tazobactam, ticarcillin-clavulanic acid, amikacin, and tobramycin. Together, these data suggest that in isolate KP-505, carbapenem resistance may be attributed to an ESBL-producing strain with a porin or efflux mutation(s). Visual examination of the time-lapse images for isolate KPC-505 revealed that the microscopy error was due to technical limitations, including loss of focus in two of the six fields of view and very low clone counts in the remaining four fields of view, issues which weighted the scoring algorithm toward a score in the susceptible range. Improvements in instrument focus, setting of strict requirements for the number of clones analyzed prior to reporting, and algorithm optimization may help to reduce or eliminate these sources of error in future analyses. For isolate KP-456, additional susceptibility testing results revealed cefepime susceptibility and cefotetan resistance, suggesting that in this isolate, an acquired AmpC with a porin and/or efflux mutation may be a plausible explanation for the observed carbapenem resistance in the absence of KPC.
The KPC type of each strain was determined (Table 1) as previously described (13). In addition, enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) was used to assess the genetic relatedness of the KPC-producing strains. PCR products were resolved using DiversiLab DNA chips (bioMérieux, Durham, NC) on the Agilent 2100 system (Agilent Technologies, Santa Clara, CA). DiversiLab software was used to compare banding patterns and determine the similarity of the isolates (using a similarity index [SI]), with identical isolates having an SI of 85% or greater (14–17). Molecular typing of the specific KPC variants contained in the isolates identified 13 blaKPC-2- and 8 blaKPC-3-containing strains (Table 1). With ERIC-PCR, the isolates clustered into 4 major strain types: the first cluster with 5 isolates, the second with 3 isolates, the third with 12 isolates, and the fourth with 1 isolate.
State-of-the-art automated antimicrobial susceptibility testing systems vary in their ability to detect carbapenemase production (8, 18). Reliable detection of carbapenem resistance is important to guide selection of appropriate therapy. Automated microscopy, as evaluated in this study, detected ETP resistance and showed promising phenotypic results that were consistent with KPC expression, using data acquired in 3 h. Using ETP as an indicator, the automated microscopy method evaluated in this study was 96% sensitive and 100% specific for detection of carbapenem resistance. This rapid result may be important for guiding antimicrobial therapy in critically ill patients and quickly initiating appropriate infection control measures.
ACKNOWLEDGMENTS
We thank James H. Jorgensen of the University of Texas Health Science Center for assisting with coordination of the KPC typing. We thank Steve Metzger, Alena Shamsheyeva, David Howson, and Christina Chantell of Accelerate Diagnostics, Inc. (Tucson, AZ), for useful discussions on study design, data analysis and interpretation, and assistance with manuscript preparation.
This study was funded through a grant from Accelerate Diagnostics, Inc., to the Washington University School of Medicine.
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Drew RJ, Turton JF, Hill RL, Livermore DM, Woodford N, Paulus S, Cunliffe NA. 2013. Emergence of carbapenem-resistant Enterobacteriaceae in a UK paediatric hospital. J. Hosp. Infect. 84:300–304. 10.1016/j.jhin.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu SK, Wu TL, Chuang YC, Lin JC, Fung CP, Lu PL, Wang JT, Wang LS, Siu LK, Yeh KM. 2013. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One 8:e69428. 10.1371/journal.pone.0069428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel G, Bonomo RA. 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doern CD, Dunne WM, Jr, Burnham CA. 2011. Detection of Klebsiella pneumoniae carbapenemase (KPC) production in non-Klebsiella pneumoniae Enterobacteriaceae isolates by use of the Phoenix, Vitek 2, and disk diffusion methods. J. Clin. Microbiol. 49:1143–1147. 10.1128/JCM.02163-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse EB, Aurbach U, Wisplinghoff H. 2013. Carbapenem-resistant Enterobacteriaceae: laboratory detection and infection control practices. Curr. Infect. Dis. Rep. 15:549–558. 10.1007/s11908-013-0373-x [DOI] [PubMed] [Google Scholar]
- 8.Woodford N, Eastaway AT, Ford M, Leanord A, Keane C, Quayle RM, Steer JA, Zhang J, Livermore DM. 2010. Comparison of BD Phoenix, Vitek 2, and MicroScan automated systems for detection and inference of mechanisms responsible for carbapenem resistance in Enterobacteriaceae. J. Clin. Microbiol. 48:2999–3002. 10.1128/JCM.00341-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J. Antimicrob. Chemother. 65:1119–1125. 10.1093/jac/dkq108 [DOI] [PubMed] [Google Scholar]
- 10.Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE, Jr, Hanna B. 2006. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg. Infect. Dis. 12:1209–1213. 10.3201/eid1208.060291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 12.Ke W, Bethel CR, Papp-Wallace KM, Pagadala SR, Nottingham M, Fernandez D, Buynak JD, Bonomo RA, van den Akker F. 2012. Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226. Antimicrob. Agents Chemother. 56:2713–2718. 10.1128/AAC.06099-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasheed JK, Biddle JW, Anderson KF, Washer L, Chenoweth C, Perrin J, Newton DW, Patel JB. 2008. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 46:2066–2069. 10.1128/JCM.02038-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu PY, Lau YJ, Hu BS, Shyr JM, Shi ZY, Tsai WS, Lin YH, Tseng CY. 1995. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol. 33:1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maslow JN, Mulligan ME, Arbeit RD. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153–164. 10.1093/clinids/17.2.153 [DOI] [PubMed] [Google Scholar]
- 17.Casarez EA, Pillai SD, Di Giovanni GD. 2007. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res. 41:3643–3648. 10.1016/j.watres.2007.03.020 [DOI] [PubMed] [Google Scholar]
- 18.Bulik CC, Fauntleroy KA, Jenkins SG, Abuali M, LaBombardi VJ, Nicolau DP, Kuti JL. 2010. Comparison of meropenem MICs and susceptibilities for carbapenemase-producing Klebsiella pneumoniae isolates by various testing methods. J. Clin. Microbiol. 48:2402–2406. 10.1128/JCM.00267-10 [DOI] [PMC free article] [PubMed] [Google Scholar]