Abstract
HIV-1 superinfection (SI) occurs when an infected individual acquires a distinct new viral strain. The rate of superinfection may be reflective of the underlying HIV risk in a population. The Centre for the AIDS Programme of Research in South Africa (CAPRISA) 004 clinical trial demonstrated that women who used a tenofovir-containing microbicide gel had lower rates of HIV infection than women using a placebo gel. Women who contracted HIV-1 during the trial were screened for the occurrence of superinfection by next-generation sequencing of the viral gag and env genes. There were two cases (one in each trial arm) of subtype C superinfection identified from the 76 women with primary infection screened at two time points (rate of superinfection, 1.5/100 person-years). Both women experienced a >0.5-log increase in viral load during the window when superinfection occurred. The rate of superinfection was significantly lower than the overall primary HIV incidence in the microbicide trial (incidence rate ratio [IRR], 0.20; P = 0.003). The women who seroconverted during the trial reported a significant increase in sexual contact with their stable partner 4 months after seroconversion (P < 0.001), which may have lowered the risk of superinfection in this population. The lower frequency of SI compared to the primary incidence is in contrast to a report from a general heterosexual African population but agrees with a study of high-risk women in Kenya. A better understanding of the rate of HIV superinfection could have important implications for ongoing HIV vaccine research.
INTRODUCTION
HIV superinfection (SI) occurs when an infected individual acquires a new viral strain that is phylogenetically distinct from all previous viral strains (1, 2). This can include both inter- and intrasubtype SI, and it has been reported in a variety of populations around the world (3). The rates at which HIV SI occurs differ between populations and appear to be related, in part, to the underlying primary HIV incidence rate (2–5). Previous analysis of high-risk women in Kenya found a significantly lower rate of SI compared to primary incidence (6). HIV SI can significantly impact HIV disease markers by increasing the HIV viral load temporarily or lastingly, by possibly accelerating disease progression, and by putatively increasing HIV-specific antibody responses (3, 4, 7, 8).
The Centre for the AIDS Programme of Research in South Africa (CAPRISA) 004 clinical trial demonstrated for the first time that an antiretroviral microbicide gel could help to prevent HIV infection in women (9). The rates of primary HIV incidence were relatively high in the full trial population (7.3 per 100 person-years [py]) as well as in both the tenofovir (5.6/100 py) and the placebo (9.1/100 py) arms (9). Women in the CAPRISA 004 trial were followed monthly for 2 years and were provided at each visit with counseling, condoms, and treatment for any other sexually transmitted infections. Women who seroconverted during the CAPRISA 004 trial were initially followed monthly and then every 3 months in the CAPRISA 002 acute infection cohort study, and they were offered treatment through their local AIDS treatment center or the CAPRISA AIDS treatment program once their CD4 count dropped to <350 cells/μl (10). This study examined if HIV SI as identified by next-generation sequencing (NGS) of the viral population occurred at a rate similar to the primary incidence in the trial.
MATERIALS AND METHODS
The CAPRISA 004 study population has been described in detail (9). Briefly, viral genetic sequences were generated for 76 women who seroconverted to HIV and who had plasma samples available within the first 3 months of infection (median time from infection, 10 weeks; interquartile range [IQR], 9 to 11 weeks) and had an available sample at their last visit prior to the initiation of antiretroviral therapy (ART) or at the end of the study (median time from infection, 110.5 weeks; IQR, 70 to 123 weeks). All women who seroconverted during the trial stopped using the vaginal gel regardless of the study arm. The CAPRISA 004 trial and the CAPRISA 002 study were both approved by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal, and all of the subjects provided informed consent to have their samples used for future studies.
Subject samples were examined for SI as previously described (2). Briefly, viral RNA was extracted from the plasma samples, reverse transcribed, and amplified in a nested-PCR format for a region of the viral p24 (∼390 bp) and gp41 (∼324 bp) genes. Samples that amplified for both time points in at least one region were sequenced using the 454 platform (Roche, Branford, CT) (1, 2). The resulting sequencing reads were analyzed using the GS Amplicon Variant Analyzer, version 2.5 (Roche). All sequence reads were compared, and similar sequences were combined into a single consensus sequence. Consensus sequences that were within 10 bases from the ends of the amplicon and comprised a cluster of at least 10 individual near-identical sequence reads were determined (1, 2). These consensus sequences were used for all subsequent phylogenetic analyses (1).
HIV superinfection was defined when an individual's follow-up sample demonstrated two or more distinct consensus sequences forming a phylogenetically distinct cluster that was unlinked from the individual's consensus sequences in the initial sample and was of adequate genetic distance from the baseline sequences to rule out natural evolutionary drift (≥0.55% per year for the p24 region or ≥0.98% per year for the gp41 region) (1). Samples from subjects who initially were identified by their initial and follow-up samples as being superinfected were further analyzed at multiple midpoints (selected from samples between months 3 to 6, 9 to 12, 12 to 24, and 24 to 36) to verify SI and to identify the approximate time of SI. The time of SI was estimated to be the midpoint between the first time point that contained the SI strain and the last sample with amplifiable sequences that did not show a presence of the SI strain. Primary HIV incidence rates were previously calculated as part of the CAPRISA 004 trial (9). Briefly, women were tested monthly with two HIV-1/-2 rapid tests, and positive samples were confirmed with two separate positive viral load tests. The samples from the previous month were then screened for acute HIV infection (9). The SI incidence rate was calculated as the total number of SI events identified during the total follow-up between the closest samples tested.
The NGS consensus sequences for gp41 and p24 are available upon request. The total sequence reads and consensus sequences were each compared for both genomic regions at the baseline and follow-up time points with the Mann-Whitney rank sum test.
Sequences from the initial time point were also analyzed for the presence of initial coinfection (3). The consensus sequences were analyzed for the presence of phylogenetically distinct viral populations with >1% genetic distance between the populations. As recombination can potentially occur rapidly following infection, consensus sequences from women who had these diverse viral populations in either genomic region (n = 7) were further analyzed by scanning subgenomic windows of the sequences for evidence of recombination with a distinct virus. Any regions with >50 bp that were no more similar to the main viral quasispecies than they were to one of a reference panel of 23 region appropriate sequences were flagged as potential recombinant regions. The phylogenies of these regions were then reconstructed in order to verify the presence of phylogenetically unlinked viral sequences.
Sexual partner status (stable versus casual) at the visit immediately prior to the estimated date of infection, near the initial time point screened, and 10 months postinfection (IQR, 9.4 to 10.7 months) for the women screened was calculated to examine the possible behavioral risk factors for SI.
HIV viral loads were measured at each monthly visit as described previously (with the Roche Cobas TaqMan HIV-1 monitor v1.0 or TaqMan version 2.0; Roche Diagnostics, Branchburg, NJ) (9). The rates of SI and primary incidence were calculated as previously described (2, 9). Incidence rate ratios (IRRs) were calculated using STATA 12 (College Station, TX) using a univariate Poisson model.
RESULTS
NGS for at least one region of the viral genome was performed on samples from 76/96 (79.2%) women who seroconverted during the trial from the first 3 months of infection (median time from infection, 10 weeks; IQR, 9 to 11 weeks) and from a later sample prior to the initiation of antiretroviral therapy (ART) (median time from infection, 110.5 weeks; IQR, 70 to 123 weeks) (Table 1). There was no significant difference by trial arm in the percentage of women whose samples were successfully sequenced (tenofovir, 29/35 [82.9%], versus placebo, 47/61 [77.0%]; P = 0.68). The total sequence reads and consensus sequences for the gp41 region were not significantly different (Table 2). However, the sequence reads and consensus sequences for the p24 region were significantly lower for the second time point screened (P < 0.001) (Table 2).
TABLE 1.
Characteristics of women from the CAPRISA 004 trial analyzed for superinfection
| Population characteristics (n = 76) | Median | IQRa |
|---|---|---|
| Age (yr) | 22 | 21–26 |
| HIV viral load (log copies/ml)a | 4.7 | 3.9–5.2 |
| CD4 counta | 476 | 401–647 |
| Follow-up (yr) | 1.9 | 1.1–2.2 |
| % from tenofovir arm | 38.2 |
Interquartile range (IQR), CD4 counts, and viral load were measured at initial time point screened for SI (median, 10 weeks from infection).
TABLE 2.
Sequence reads and consensus sequences
| Time point |
p24 (median [IQR]) |
gp41 (median [IQR]) |
||
|---|---|---|---|---|
| Total reads | Consensus sequences | Total reads | Consensus sequences | |
| Initiala | 7,472b (6,709–8,046) | 50b (37–68) | 9,012 (6,611–11,730) | 77 (40–118) |
| Follow-upc | 6,020b (5,274–7,135) | 35b (27–46) | 8,774 (6,058–12,134) | 58 (39–114) |
Initial time point was a median of 10 weeks postinfection.
Significantly higher reads and consensus sequences at the first time point (P < 0.001, Mann-Whitney rank sum test).
Follow-up time point was a median of 110.5 weeks postinfection.
Except for one woman who was infected with HIV subtype A, all of the seroconverters were infected with HIV subtype C. The sequences from the sample at the first time point for each woman were initially screened phylogenetically for the presence of possible coinfections. The consensus sequences from samples that had distinct phylogenetic populations (>1%) in either genomic region (n = 7) were further analyzed by scanning the sequences for the presence of distinctly different regions. Two cases of possible coinfection were identified, and in these cases, evidence of recombinant regions derived from a phylogenetically unlinked virus was found.
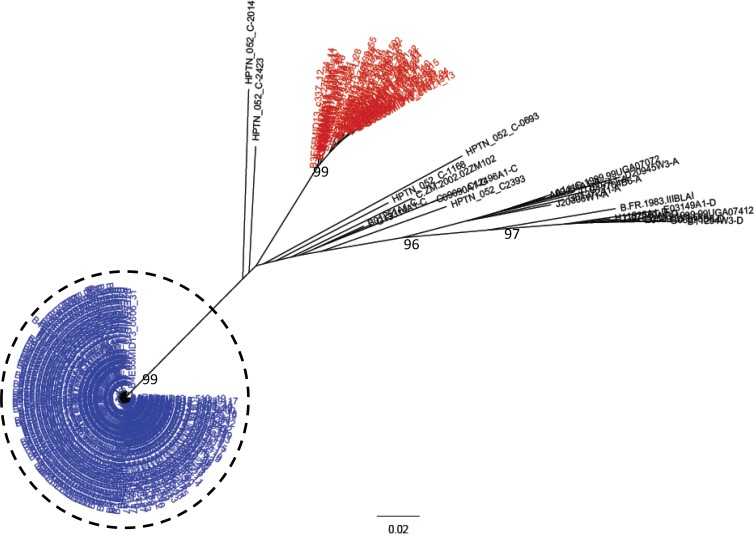
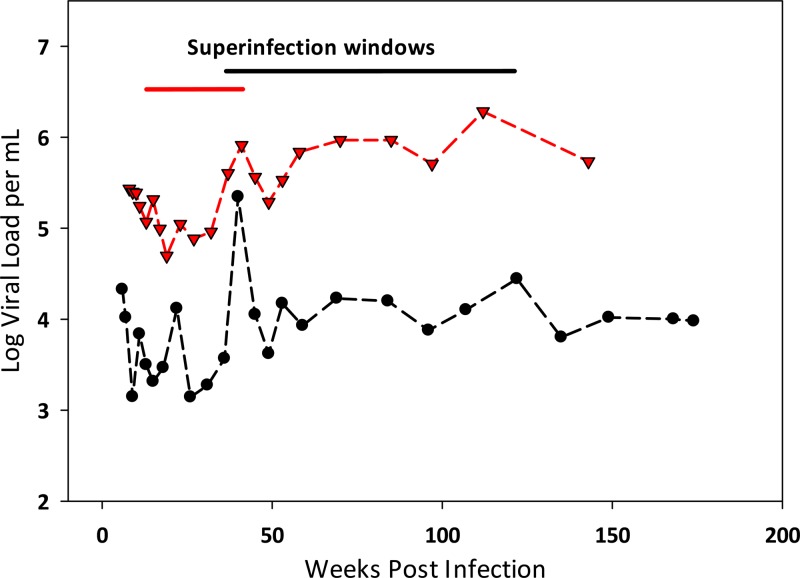
Of all the women screened, two (2.6%) were found to have been superinfected (Fig. 1). There was one case of SI identified in each arm of the study, and in both cases, the initial and superinfecting strains were subtype C. A sample from the midpoint for each SI case was analyzed with NGS to determine the approximate time of superinfection. The SI events occurred between 10 and 41 weeks in one case and 36 and 122 weeks in the other. In each case, the women experienced a spike in their viral loads of at least 0.5 log between visits during this period (Fig. 2). The rate of SI for the seroconverters screened was calculated to be 1.5/100 py (95% confidence interval [CI], 0.2 to 5.4 per 100 py), which was significantly lower than the observed primary infection rate of 7.3/100 py observed for the total trial population (IRR, 0.20, 95% CI, 0.02 to 0.75; P = 0.003). The rate of SI was also significantly lower than the rate of primary infection in the placebo (IRR, 0.16, 95% CI, 0.02 to 0.62; P < 0.001) and treatment arms (IRR, 0.27, 95% CI, 0.03 to 1.03; P = 0.018).
FIG 1.
HIV superinfection cases among CAPRISA 004 seroconverters. Phylogenetic tree of consensus gp41 viral sequences (≥10 reads) derived from 454 pyrosequencing of a seroconversion (red) and a follow-up sample (blue), with the superinfecting strain indicated (dashed circle). The number of repeated sequences represented by each consensus sequence is shown at the end of the consensus identifier. Distance is indicated for the tree by the scale at bottom of the tree, and samples are grouped with a selection of subtype reference sequences (black). Bootstrap values of >80% are indicated (1,000 replicates).
FIG 2.
Viral loads from HIV SI cases. Viral load measurements for the two women (placebo arm, red; tenofovir arm, black) who became superinfected are shown, with the time frame in which SI occurred shown above.
For the 96 women screened, 80 had sexual partner data available directly before seroconversion (median time to HIV infection, 14 days; IQR, 13 to 17 days) and near the first time point screened (median time from HIV infection, 4 months; IQR, 3.5 to 4.6 months). Of these women, 64/80 (80%) reported having had sex in the past 30 days with a stable partner at the preinfection time point, but a significantly higher percentage of these women (79/80 [99%]) reported having had sex with a stable partner in the past 30 days at the time point near the initial SI screening sample (P < 0.001). This significant increase in reported sex with a stable partner was also found at 10 months postinfection (91%; P = 0.049). There was no difference in reported sex with causal partners between the three time points; only one woman reported having had a casual sexual contact preinfection and at 10 months, and only two women reported having had a causal sexual partner at 4 months.
DISCUSSION
HIV SI has been demonstrated to occur at rates that reflect the underlying rate of primary HIV infection in multiple populations spanning a variety of risk levels (2–4, 11, 12). Other studies have found either significantly lower rates or no evidence of SI, but many of these studies utilized less sensitive methods to identify SI (3, 13, 14). The observed frequency of SI in this population enrolled in a microbicide intervention study was significantly lower than the primary HIV incidence, which was relatively high. In both cases, the women experienced an increase in the viral load during the window period when HIV SI occurred. This increase was temporary in one case and lasting in the other, which is similar to the disparity in viral load reactions to SI seen in other studies (3). In addition, two additional women were identified at 3 months postinfection as being likely initially coinfected with two strains of HIV.
This finding of a lower frequency of SI than of primary incidence is in contrast to a previous finding by our group, using identical laboratory methods, where the rate of SI was similar to the rate of primary infection in a general population cohort in the rural Rakai District of Uganda (1, 2). Interestingly, a study of high-risk women in Kenya also found a significantly lower rate of SI than primary incidence, which agrees with these findings (6). To date, these three studies are the largest in Africa to have used deep sequencing to examine for superinfection, and they were distinctly different in terms of rates of primary infections and follow-up. The rate of primary infection in the Rakai study (1.2/100 py), which included men and women, was substantially less than those within the CAPRISA 004 trial (7.3/100 py) and the Kenyan trial (5.75/100 py) (2, 6, 9). The study presented here and the Rakai study calculated IRRs in a similar manner, while the Kenyan study utilized a slightly different hazard ratio analysis (2, 6). The Rakai study was powered to identify rates that were at least half or double the rate of primary incidence, which limited its ability to identify more subtle differences. In all three studies, newly diagnosed individuals were provided with counseling and were referred for care, but the frequency of follow-up was markedly different. While individuals in the Rakai community cohort study were followed annually, seroconverters from the CAPRISA 004 study were followed up at least once per month until 12 months postinfection and every 3 months thereafter until ART initiation, and they continued to receive counseling at each visit. The Kenyan study collected samples quarterly, but the women were seen monthly (6, 15). It is possible that the continuous reinforcement provided by these frequent visits affected the behavior of the women, which lowered their risk of SI.
Interestingly, only 4.3% of the women recruited into the CAPRISA 004 trial reported having had multiple causal sexual partners, and 95.2% reported one stable partner in the past year (16). In the women screened for SI, there was a significant increase in reported sexual contact with a stable partner at both 4 and 10 months after seroconversion. This would suggest that these women may have altered their sexual habits in a way that subsequently lowered their risk of becoming superinfected. Therefore, although these women were at high risk for primary infection from their stable partners, their lack of casual partners may have made SI less likely. It is unknown what risk factors are associated with HIV SI in study cohorts and in the HIV-infected population as a whole or if they are the same as those for primary HIV infection, as would be expected (3). It is difficult to extrapolate what role HIV SI is playing in nonstudy settings and how knowledge of SI affects the behavior of HIV-infected individuals (3). Due to the relatively small number of cases identified here, it was not possible to examine risk factors for HIV SI in more detail. More research in a variety of populations is needed to fully clarify the relationship between HIV SI risk and primary incidence.
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. CAPRISA is part of the Comprehensive International Program of Research on AIDS (CIPRA), which is funded by the National Institute of Allergy and Infectious Disease, National Institutes of Health, the U.S. Department of Health and Human Services (grant AI51794), and the National Research Foundation, South Africa (grant 67385). The TRAPS (Tenofovir gel Research for AIDS Prevention Science) Program is funded in part by CONRAD subproject agreement PPA-09-046, under a Cooperative Agreement (GPO-A-00-08-00005-00) with the U.S. Agency for International Development (USAID). The CAPRISA 004 trial was funded by the USAID and LIFELab, a biotechnology agency of the South African government's Department of Science and Technology.
We do not have any commercial or other associations that might pose a conflict of interest.
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Redd AD, Collinson-Streng A, Martens C, Ricklefs S, Mullis CE, Manucci J, Tobian AA, Selig EJ, Laeyendecker O, Sewankambo N, Gray RH, Serwadda D, Wawer MJ, Porcella SF, Quinn TC. 2011. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda using next generation deep sequencing. J. Clin. Microbiol. 49:2859–2867. 10.1128/JCM.00804-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redd AD, Mullis CE, Serwadda D, Kong X, Martens C, Ricklefs SM, Tobian AA, Xiao C, Grabowski MK, Nalugoda F, Kigozi G, Laeyendecker O, Kagaayi J, Sewankambo N, Gray RH, Porcella SF, Wawer MJ, Quinn TC. 2012. The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J. Infect. Dis. 206:267–274. 10.1093/infdis/jis325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redd AD, Quinn TC, Tobian AA. 2013. Frequency and implications of HIV superinfection. Lancet Infect. Dis. 3:622–628. 10.1016/S1473-3099(13)70066-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DM, Wong JK, Hightower GK, Ignacio CC, Koelsch KK, Daar ES, Richman DD, Little SJ. 2004. Incidence of HIV superinfection following primary infection. JAMA 292:1177–1178. 10.1001/jama.292.10.1177 [DOI] [PubMed] [Google Scholar]
- 5.Wagner GA, Pacold ME, Kosakovsky Pond SL, Caballero G, Chaillon A, Rudolph AE, Morris SR, Little SJ, Richman DD, Smith DM. 22 November 2013. Incidence and prevalence of intrasubtype HIV-1 dual infection in at-risk men in the United States. J. Infect. Dis. 10.1093/infdis/jit633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronen K, McCoy CO, Matsen FA, Boyd DF, Emery S, Odem-Davis K, Jaoko W, Mandaliya K, McClelland RS, Richardson BA, Overbaugh J. 2013. HIV-1 superinfection occurs less frequently than initial infection in a cohort of high-risk Kenyan women. PLoS Pathog. 9:e1003593. 10.1371/journal.ppat.1003593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez V, Odem-Davis K, McClelland RS, Jaoko W, Overbaugh J. 2012. HIV-1 superinfection in women broadens and strengthens the neutralizing antibody response. PLoS Pathog. 8:e1002611. 10.1371/journal.ppat.1002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altfeld M, Allen TM, Yu XG, Johnston MN, Agrawal D, Korber BT, Montefiori DC, O'Connor DH, Davis BT, Lee PK, Maier EL, Harlow J, Goulder PJ, Brander C, Rosenberg ES, Walker BD. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434–439. 10.1038/nature01200 [DOI] [PubMed] [Google Scholar]
- 9.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, Kharsany AB, Sibeko S, Mlisana KP, Omar Z, Gengiah TN, Maarschalk S, Arulappan N, Mlotshwa M, Morris L, Taylor D. 2010. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science 329:1168–1174. 10.1126/science.1193748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, Abdool Karim Q, Grobler A, Barnabas N, Iriogbe I, Abdool Karim SS, Team CAIS. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954. 10.1371/journal.pone.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3:e177. 10.1371/journal.ppat.0030177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piantadosi A, Ngayo MO, Chohan B, Overbaugh J. 2008. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res. Hum. Retroviruses 24:1221. 10.1089/aid.2008.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzales MJ, Delwart E, Rhee SY, Tsui R, Zolopa AR, Taylor J, Shafer RW. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J. Infect. Dis. 188:397–405. 10.1086/376534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rachinger A, Manyenga P, Burger JA, Derks van de Ven TL, Stolte IG, Prins M, van 't Wout AB, Schuitemaker H. 2011. Low incidence of HIV-1 superinfection even after episodes of unsafe sexual behavior of homosexual men in the Amsterdam Cohort Studies on HIV Infection and AIDS. J. Infect. Dis. 203:1621–1628. 10.1093/infdis/jir164 [DOI] [PubMed] [Google Scholar]
- 15.Martin HL, Jr, Nyange PM, Richardson BA, Lavreys L, Mandaliya K, Jackson DJ, Ndinya-Achola JO, Kreiss J. 1998. Hormonal contraception, sexually transmitted diseases, and risk of heterosexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 178:1053–1059. 10.1086/515654 [DOI] [PubMed] [Google Scholar]
- 16.Karim QA, Kharsany AB, Frohlich JA, Baxter C, Yende N, Mansoor LE, Mlisana KP, Maarschalk S, Arulappan N, Grobler A, Sibeko S, Omar Z, Gengiah TN, Mlotshwa M, Samsunder N, Karim SS. 2011. Recruitment of high risk women for HIV prevention trials: baseline HIV prevalence and sexual behavior in the CAPRISA 004 tenofovir gel trial. Trials 12:67. 10.1186/1745-6215-12-67 [DOI] [PMC free article] [PubMed] [Google Scholar]