Abstract
Ceftazidime-avibactam, aztreonam-avibactam, and comparators were tested by reference broth microdilution against 372 nonrepetitive Gram-negative bacilli (346 unselected plus 26 selected meropenem-nonsusceptible Enterobacteriaceae isolates) collected from 11 teaching hospitals in China in 2011 and 2012. Meropenem-nonsusceptible isolates produced extended-spectrum β-lactamases (ESBLs; e.g., CTX-M-14/3), AmpCs (e.g., CMY-2), and/or carbapenemases (e.g., KPC-2 and NDM-1). Avibactam potentiated the activity of ceftazidime against organisms with combinations of ESBLs, AmpCs, and KPC-2. Aztreonam-avibactam was active against all β-lactamase producers (including producers of NDM-1 and IMP-4/8) except blaOXA-containing Acinetobacter baumannii and some Pseudomonas aeruginosa isolates.
TEXT
Avibactam is a member of a class of inhibitors called diazabicyclooctanes (DBOs) that does not contain a β-lactam core but maintains the capacity to covalently acylate its β-lactamase targets (1, 2, 3). Avibactam alone has very little intrinsic antimicrobial activity but has been shown to efficiently restore the in vitro activities of cephalosporins (including ceftazidime and ceftaroline) (4, 5) against Ambler class A, class C, and some class D β-lactamases (6, 7, 8, 9, 10), excluding metallo-β-lactamases (MBLs) and Acinetobacter OXA carbapenemases (2). The combination of aztreonam and avibactam has been proposed as a principal candidate for the treatment of infections with MBL-producing Gram-negative organisms (8, 11). To date, few data have been available in China describing the in vitro activities of ceftazidime-avibactam and aztreonam-avibactam against clinical Gram-negative bacilli, especially Enterobacteriaceae. In this study, we evaluated the in vitro activities of ceftazidime and aztreonam alone and combined with a fixed concentration of 4 mg/liter of avibactam against routinely collected clinical Gram-negative bacilli, including Enterobacteriaceae, Acinetobacter spp., and Pseudomonas aeruginosa, from two national surveillance programs in China, the Chinese Meropenem Susceptibility Surveillance (CMSS) program in 2012 and the Chinese Antimicrobial Resistance Surveillance of Nosocomial Infections (CARES) in 2011.
A total of 372 nonrepetitive, routinely collected isolates (from 2011 and 2012) were obtained from 11 teaching hospitals representing the south, north, northwest, east, and middle districts of mainland China. The 372 organisms included the following. (i) Two hundred ninety-one routinely collected but otherwise unselected Enterobacteriaceae isolates (from CMSS) and 26 carbapenem-nonsusceptible Enterobacteriaceae isolates (meropenem MIC, ≥2 mg/liter; 19 isolates from CARES and the remainder from CMSS) were tested. Of the 26 carbapenem-nonsusceptible Enterobacteriaceae isolates, 10 produced KPC-2, 11 produced IMP β-lactamases (10 isolates produced IMP-4 and 1 isolate produced IMP-8), and 3 produced NDM-1. The remaining 2 isolates possessed TEM-1 and CTX-M, enzymes that are not regarded as carbapenemases. Most of the carbapenemase producers coharbored other β-lactamases (ACT-14/15, CMY-2, DHA-1, and SHV-12/11/107) (Table 1). (ii) Thirty routinely collected but otherwise unselected Acinetobacter isolates (from CMSS), including 11 carbapenem-nonsusceptible isolates in which coexisting genes were detected encoding OXA-23-like, OXA-51-like, and TEM-1 β-lactamases, were tested. (iii) Twenty-five routinely collected but otherwise unselected P. aeruginosa isolates (from CMSS), including 11 carbapenem-nonsusceptible bacteria that harbored genes encoding OXA-50-like and/or TEM-1 β-lactamases, were tested. All the isolates, obtained from intra-abdominal, urinary tract, respiratory tract, or bloodstream infections, were sent to the central laboratory (Laboratory Medicine, Peking University People's Hospital, Beijing, China) for reidentification and antibiotic susceptibility testing. The Vitek GNI system (bioMérieux Vitek Inc., Hazelwood, MO) or API20E or API20NE (bioMérieux, Marcy l'Etoile, France) was used for bacterial identification.
TABLE 1.
In vitro activities of ceftazidime-avibactam, aztreonam-avibactam, and comparators against 48 meropenem-nonsusceptible β-lactamase-producing bacteriaa
| Organism (no. of isolates) | Year of isolation | Cityb | MLSTd | β-Lactamase(s) for which acquired genes were detected | MIC/MIC90 of: |
MIC/MIC90 reduction (fold) | MIC/MIC90 of: |
MIC/MIC90 reduction (fold) | MIC/MIC90 of: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CAZ-AVI | ATM | ATM-AVI | FEP | TZP | MEM | POL | AMK | LVX | |||||||
| Enterobacteriaceae | ||||||||||||||||
| Citrobacter freundii (3) | 2011 | BJ | ND | KPC-2, TEM-1 | 64 | 0.5 | 128 | >128 | 0.5 | >256 | 16 | 128 | 1 | 0.25 | 256 | ND |
| 2011 | BJ | ND | KPC-2, TEM-1 | 64 | 1 | 64 | >128 | 0.5 | >256 | 16 | 128 | 2 | 0.25 | 25 | ND | |
| 2011 | BJ | ND | KPC-2, TEM-1, CMY-2 | 128 | 0.5 | 256 | >128 | 1 | >128 | 16 | 128 | 2 | 0.25 | 256 | ND | |
| Klebsiella pneumoniae (7) | 2011 | ZJ | 11 | KPC-2, TEM-1b | 128 | 2 | 64 | >128 | 0.5 | >256 | 4 | 256 | 16 | 0.25 | 2 | 16 |
| 2011 | SH | 11 | KPC-2, TEM-1, SHV-12 | >128 | 1 | >128 | >128 | 0.25 | >512 | 8 | 256 | 16 | 0.25 | >256 | 128 | |
| 2012 | ZJ | 11 | KPC-2, TEM-1, SHV-11, CTX-M-14 | 64 | 1 | 64 | >128 | 0.25 | >512 | 128 | 256 | >32 | 0.25 | >256 | >64 | |
| 2012 | ZJ | 11 | KPC-2, TEM-1, SHV-11, CTX-M-14 | 64 | 0.5 | 128 | >128 | 0.25 | >512 | 128 | >256 | >32 | 0.25 | >256 | >64 | |
| 2011 | GZ | 11 | KPC-2, CMY-2, SHV-11, CTX-M-14 | >128 | 1 | >128 | >128 | 1 | >128 | 256 | >256 | 128 | 0.125 | 16 | 16 | |
| 2011 | GZ | 11 | KPC-2, CMY-2, SHV-11, CTX-M-14 | >128 | 2 | >64 | >128 | 2 | >64 | 256 | >256 | 128 | 0.125 | 32 | 16 | |
| 2011 | ZJ | 11 | KPC-2, SHV-11, CTX-M-14 | 128 | 2 | 64 | >128 | 0.5 | >256 | 128 | >256 | 256 | 0.25 | >256 | 32 | |
| Citrobacter freundii (1) | 2011 | BJ | ND | IMP-4, TEM-1 | >128 | >128 | 1 | 128 | 0.5 | 256 | 8 | 8 | 4 | 0.5 | 2 | ND |
| Escherichia coli (2) | 2011 | BJ | ND | IMP-4 | >128 | >128 | 1 | 2 | ≤0.064 | >32 | 8 | 4 | 0.5 | 0.25 | 8 | ND |
| 2011 | BJ | ND | IMP-4, TEM-1, CTX-M-14 | >128 | >128 | 1 | 64 | 0.125 | 512 | 32 | 4 | 1 | 0.125 | 8 | ND | |
| Enterobacter cloacae (2) | 2012 | SY | ND | IMP-4, ACT-14 | >128 | >128 | 1 | 1 | 0.25 | 4 | 64 | 8 | 4 | 0.125 | 1 | 2 |
| 2011 | GZ | ND | IMP-4, ACT-15, TEM-1, SHV-12, CTX-M-14 | >128 | >128 | 1 | >128 | 0.125 | >1,024 | 32 | 64 | 2 | 0.125 | 4 | 128 | |
| Klebsiella pneumoniae (5) | 2011 | BJ | 147 | IMP-4, TEM-1 | 64 | 1 | 64 | >128 | 0.5 | >256 | 64 | 16 | 1 | 0.25 | 8 | ND |
| 2011 | BJ | 876 | IMP-4, TEM-1 | >128 | >128 | 1 | 8 | 0.125 | 64 | 16 | 32 | 4 | 0.25 | 1 | ND | |
| 2011 | BJ | 1100 | IMP-4, SHV-11 | 0.25 | 0.25 | 1 | ≤0.064 | ≤0.064 | 1 | 16 | 16 | 2 | 0.25 | 2 | ND | |
| 2011 | BJ | 876 | IMP-4, DHA-1 | >128 | >128 | 1 | 16 | 0.125 | 128 | 64 | >256 | 32 | 0.25 | 4 | ND | |
| 2011 | SY | 876 | IMP-4, SHV-107 | >128 | >128 | 1 | 0.125 | 0.125 | 1 | 8 | 16 | 2 | 0.125 | 1 | 0.064 | |
| Enterobacter cloacae (1) | 2011 | FZ | ND | IMP-8 | 128 | 1 | 128 | 128 | 0.5 | 256 | 32 | 64 | 0.5 | 0.125 | 1 | ND |
| Klebsiella pneumoniae (3) | 2012 | SX | 147 | NDM-1, SHV-12 | >128 | >128 | 1 | >128 | 0.5 | >256 | >256 | >256 | >32 | 0.25 | 2 | 32 |
| 2012 | SX | 147 | NDM-1, SHV-12, TEM-1 | >128 | >128 | 1 | >128 | 0.5 | >256 | 128 | >256 | 16 | 0.25 | 1 | 32 | |
| 2012 | SX | 147 | NDM-1, SHV-12, TEM-1, CTX-M-3 | >128 | >128 | 1 | >128 | ≤0.064 | >2,000 | 128 | >256 | 8 | 0.125 | 2 | 16 | |
| Enterobacter aerogenes (1) | 2011 | BJ | ND | TEM-1, ACT-15, CTX-M-3 | 128 | 4 | 32 | >128 | 2 | >64 | 128 | 256 | 16 | 0.125 | 4 | 64 |
| Escherichia coli (1) | 2011 | BJ | ND | TEM-1, CTX-M-14 | >128 | 4 | >32 | >128 | 2 | >64 | 256 | >256 | 4 | 0.125 | 2 | 32 |
| Nonfermenters | ||||||||||||||||
| Acinetobacter baumannii (11) | 2012 | NAc | NA | OXA-23-like, TEM-1 | >128 | 32 | >4 | >128 | 64 | >2 | 256 | >256 | 64 | 0.25 | >256 | 16 |
| Pseudomonas aeruginosa (8) | 2012 | NA | NA | Only resident bla genes detected | 8 | 8 | 1 | >128 | >128 | 1 | 32 | >256 | 32 | 2 | >256 | 16 |
| Pseudomonas aeruginosa (3) | 2012 | SH | ND | TEM-1 | 8 | 2 | 4 | 32 | 32 | 1 | 32 | >256 | 32 | 0.25 | >256 | 0.25 |
| 2012 | SH | ND | TEM-1 | 8 | 4 | 2 | 128 | 64 | 2 | 64 | >256 | >32 | 0.5 | >256 | 16 | |
| 2012 | SH | ND | TEM-1 | 8 | 2 | 4 | 32 | 16 | 2 | 32 | 256 | 16 | 0.5 | >256 | 0.5 | |
MICs were determined in milligrams per liter. The 48 isolates consisted of 11 P. aeruginosa and 11 A. baumannii isolates listed in Table 2 and 26 additional Enterobacteriaceae isolates not included in the collection for which data are shown in Table 2. The Enterobacteriaceae are listed by acquired carbapenem resistance mechanism as follows: first blaKPC-containing isolates, then isolates that contained genes encoding metallo-β-lactamases, and finally isolates in which no acquired carbapenemase-encoding gene could be detected. CAZ, ceftazidime; ATM, aztreonam; AVI, avibactam; FEP, cefepime; TZP, piperacillin-tazobactam; MEM, meropenem; POL, polymyxin; AMK, amikacin; LVX, levofloxacin.
BJ, Beijing; ZJ, Zhejiang; SH, Shanghai; GZ, Guangzhou; SY, Shenyang; FZ, Fuzhou; SX, Shanxi.
NA, not applicable for more than one isolate.
MLST, multilocus sequence type. ND, not detected.
MIC measurements were performed by the reference broth microdilution method as described by the Clinical and Laboratory Standards Institute (CLSI) M7-A9 (2012) (12). MICs of ceftazidime and aztreonam alone and in combination with avibactam (AstraZeneca Pharmaceuticals) at a fixed concentration of 4 mg/liter (CLSI M100-S23, 2013) (13) were measured. Antibiotic solutions for susceptibility testing were prepared fresh; i.e., dried panels were not used in this study. MICs of ceftazidime, aztreonam, and comparator agents were interpreted according to CLSI criteria in M100-S23, 2013 (13).
For meropenem-nonsusceptible Gram-negative bacilli (Table 1), PCR was used to amplify carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM, blaOXA-48, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, and blaOXA-50-like) and other β-lactamase genes (blaTEM, blaSHV, blaCTX-M, blaAmpC, blaDHA, and blaCMY) according to procedures described in previous studies (14). All PCR products were sequenced with an ABI7570 sequencer (Applied Biosystems, Foster City, CA).
In this study, avibactam potentiated the activities of ceftazidime and aztreonam in vitro against most species of clinical isolates of Enterobacteriaceae tested. For isolates of Enterobacteriaceae that were nonsusceptible to ceftazidime and/or cefotaxime (33%; 97 of 291), the addition of 4 mg/liter avibactam greatly increased the activities of ceftazidime and aztreonam against most species (64- to 1,024-fold MIC90 reduction) (Table 2). For the ceftazidime- and/or cefotaxime-nonsusceptible isolates of Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Providencia rettgeri in the present study, both clavulanic acid (data not shown) and avibactam restored the activity of ceftazidime. For the isolates of Citrobacter freundii, Enterobacter aerogenes, Enterobacter cloacae, and Morganella morganii that were nonsusceptible to ceftazidime and/or cefotaxime, the activity of ceftazidime could not be protected by clavulanic acid (with no MIC90 reduction; data not shown) but avibactam retained its protection of ceftazidime and aztreonam, with clear MIC90 reductions (64- to 256-fold). It is possible that a large proportion of the ceftazidime/cefotaxime-nonsusceptible isolates of these species produced stably derepressed AmpC β-lactamases (15–17).
TABLE 2.
MIC50s, MIC90s, and ranges of MICs of ceftazidime-avibactam and aztreonam-avibactam for 346 Gram-negative bacilli isolated from clinical specimensa
| Organism(s) and drug resistance phenotype(s)b (no. of isolates) | CAZ |
CAZ-AVI |
CAZ-AVI MIC90 reduction (fold) | ATM |
ATM-AVI |
ATM-AVI MIC90 reduction (fold) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |||
| Escherichia coli (25) | 1 | 64 | 0.064–>128 | 0.125 | 0.5 | 0.064–0.5 | 128 | 8 | 128 | 0.064–>128 | 0.064 | 0.125 | 0.064–0.5 | 1,024 |
| CAZ-NS and/or CTX-NS (18) | 2 | 64 | 0.5–>128 | 0.25 | 0.5 | 0.064–0.5 | 128 | 32 | >128 | 1–>128 | 0.064 | 0.5 | 0.064–0.5 | >256 |
| Klebsiella oxytoca (25) | 0.25 | 2 | 0.064–4 | 0.25 | 0.5 | 0.064–0.5 | 4 | 0.5 | 8 | 0.064–>128 | 0.064 | 0.064 | 0.064–0.25 | 128 |
| Klebsiella pneumoniae (25) | 1 | 64 | 0.125–>128 | 0.125 | 1 | 0.064–2 | 64 | 1 | 64 | 0.064–>128 | 0.064 | 0.125 | 0.064–0.25 | 512 |
| CAZ-NS and/or CTX-NS (14) | 4 | 64 | 0.5–>128 | 0.125 | 1 | 0.064–2 | 64 | 8 | 128 | 1–>128 | 0.064 | 0.125 | 0.064–0.25 | 1,024 |
| Citrobacter diversus (27) | 0.125 | 0.5 | 0.064–64 | 0.125 | 0.25 | 0.064–0.5 | 2 | 0.064 | 0.25 | 0.064–128 | 0.064 | 0.064 | 0.064–0.125 | 4 |
| Citrobacter freundii (24) | 8 | 128 | 0.064–>128 | 0.25 | 1 | 0.064–2 | 128 | 2 | 64 | 0.064–128 | 0.064 | 0.5 | 0.064–1 | 128 |
| CAZ-NS and/or CTX-NS (13) | 32 | >128 | 1–256 | 0.5 | 1 | 0.125–2 | >128 | 32 | 64 | 2–128 | 0.125 | 0.5 | 0.064–1 | 128 |
| Enterobacter aerogenes (26) | 0.5 | 64 | 0.064–128 | 0.25 | 0.5 | 0.064–0.5 | 128 | 0.5 | 32 | 0.064–32 | 0.064 | 0.125 | 0.064–0.5 | 256 |
| CAZ-NS and/or CTX-NS (13) | 16 | 64 | 0.5–128 | 0.25 | 0.5 | 0.125–0.5 | 128 | 16 | 32 | 0.064–32 | 0.125 | 0.25 | 0.064–0.5 | 128 |
| Enterobacter cloacae (26) | 0.5 | 64 | 0.125–128 | 0.25 | 0.5 | 0.125–1 | 128 | 0.25 | 32 | 0.064–64 | 0.064 | 0.25 | 0.064–1 | 128 |
| CAZ-NS and/or CTX-NS (10) | 32 | 128 | 0.125–128 | 0.5 | 1 | 0.125–1 | 128 | 16 | 64 | 0.25–64 | 0.125 | 0.25 | 0.064–1 | 256 |
| Morganella morganii (25) | 1 | 64 | 0.064–128 | 0.125 | 1 | 0.064–1 | 64 | 0.5 | 16 | 0.064–64 | 0.064 | 0.25 | 0.064–1 | 64 |
| CAZ-NS and/or CTX-NS (10) | 8 | 128 | 0.5–128 | 0.125 | 1 | 0.064–1 | 128 | 2 | 32 | 0.5–64 | 0.064 | 0.5 | 0.064–1 | 64 |
| Proteus mirabilis (24) | 0.064 | 64 | 0.064–>128 | 0.064 | 0.25 | 0.064–1 | 256 | 0.064 | 4 | 0.064–>128 | 0.064 | 0.064 | 0.064–0.125 | 64 |
| CAZ-NS and/or CTX-NS (11) | 0.25 | >128 | 0.064–>128 | 0.064 | 0.25 | 0.064–1 | >512 | 0.5 | 64 | 0.064–>128 | 0.064 | 0.064 | 0.064–0.125 | 1,024 |
| Proteus vulgaris (27) | 0.064 | 0.5 | 0.064–1 | 0.064 | 0.125 | 0.064–0.125 | 4 | 0.064 | 0.125 | 0.064–32 | 0.064 | 0.064 | 0.064–0.064 | 2 |
| Providencia spp.c (12) | 0.5 | 256 | 0.064–>128 | 0.5 | 1 | 0.064–>128 | 256 | 0.125 | 64 | 0.064–>128 | 0.064 | 0.125 | 0.064–>128 | 512 |
| CAZ-NS and/or CTX-NS (4) | NAd | NA | >128–>128 | NA | NA | 0.5–>128 | NA | NA | NA | 16–>128 | NA | NA | 0.064–>128 | NA |
| Serratia marcescens (25) | 0.25 | 8 | 0.125–8 | 0.125 | 1 | 0.125–1 | 8 | 0.25 | 64 | 0.064–128 | 0.064 | 0.25 | 0.064–0.25 | 256 |
| CAZ-NS and/or CTX-NS (4) | NA | NA | 8–8 | NA | NA | 0.25–1 | NA | NA | NA | 64–128 | NA | NA | 0.25–0.25 | NA |
| Pseudomonas aeruginosa (25) | 4 | 16 | 0.5–64 | 4 | 8 | 1–16 | 2 | 16 | 128 | 4–>128 | 16 | 64 | 4–>128 | 2 |
| MEM-NS (11) | 8 | 8 | 2–16 | 4 | 8 | 2–8 | 1 | 32 | >128 | 4–>128 | 32 | >128 | 8–>128 | 1 |
| MDR (6) | 8 | 8 | 1–16 | 4 | 8 | 1–8 | 1 | 32 | >128 | 4–>128 | 32 | >128 | 4–>128 | 1 |
| CAZ-NS and/or ATM-NS (18) | 8 | 16 | 2–64 | 4 | 8 | 2–16 | 2 | 32 | >128 | 16–>128 | 32 | >128 | 8–>128 | 1 |
| Acinetobacter baumannii (28) | 8 | >128 | 2–>128 | 8 | 32 | 1–>128 | >4 | 32 | >128 | 8–>128 | 16 | 64 | 4–>128 | >2 |
| MEM-NS (11) | 128 | >128 | 32–>128 | 16 | 32 | 16–128 | >4 | 64 | >128 | 8–>128 | 32 | 64 | 32–64 | >2 |
| MDR (12) | 128 | >128 | 32–>128 | 16 | 32 | 16–>128 | >4 | 64 | >128 | 8–>128 | 32 | 64 | 32–>128 | >2 |
| CAZ-NS (13) | 128 | >128 | 32–>128 | 16 | 32 | 4–128 | >4 | 64 | >128 | 8–>128 | 32 | 64 | 16–64 | >2 |
| Acinetobacter calcoaceticus (2) | NA | NA | >128–>128 | NA | NA | >128–>128 | NA | NA | NA | >128–>128 | NA | NA | >128–>128 | NA |
MICs are expressed in milligrams per liter. Organisms were isolated in 2011 or 2012 and tested using freshly prepared panels.
CAZ, ceftazidime; ATM, aztreonam; AVI, avibactam; CTX, cefotaxime; MEM, meropenem; NS, nonsusceptible; MDR, multidrug resistant. Multidrug-resistant isolates were defined as isolates demonstrating resistance to at least one antimicrobial agent from three or more different classes.
Four Providencia rettgeri isolates were ceftazidime nonsusceptible and aztreonam nonsusceptible; avibactam restored the activities of ceftazidime and aztreonam against 3 of these isolates with 128- and 256- to 1,000-fold MIC reduction, respectively.
NA, not applicable. MIC50s and MIC90s are not presented for groups of fewer than 6 isolates.
Avibactam potentiated the activities of ceftazidime and aztreonam against carbapenem-nonsusceptible Enterobacteriaceae that coharbored genes encoding KPC-2 and other class A enzymes (CTX-M-14/3, SHV-11/12, and TEM-1/1b) or class C enzymes (ACT-15, and CMY-2) (Table 1). Against carbapenem-nonsusceptible Enterobacteriaceae that carried genes encoding MBLs, such as IMP-4/8 and NDM-1, avibactam did not restore the activity of ceftazidime; however, the aztreonam-avibactam combination remained active (8).
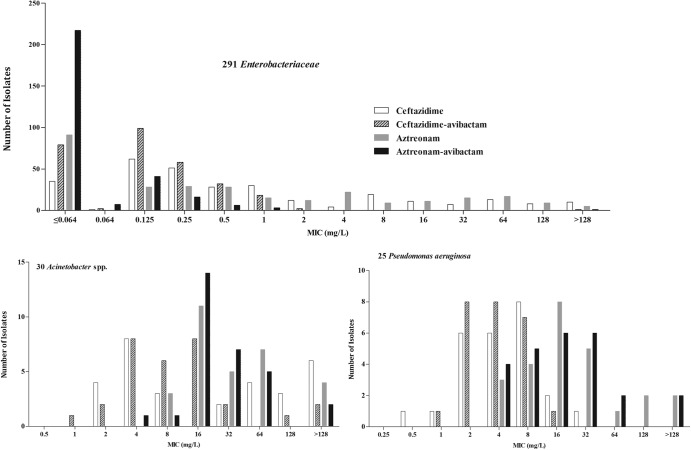
The MIC90 of ceftazidime-avibactam for 25 unselected clinical isolates of P. aeruginosa was 8 mg/liter (Table 2), as was found previously in studies from Canada (18) and France (19). The MIC of ceftazidime-avibactam was ≤8 mg/liter for 24 of 25 isolates (96%) (data not shown). For 18 P. aeruginosa isolates that were ceftazidime and/or aztreonam nonsusceptible, the addition of avibactam reduced the MIC90 of ceftazidime from 16 to 8 mg/liter (Table 2; Fig. 1). The MIC of aztreonam alone was ≤8 mg/liter for 7 of 25 P. aeruginosa isolates (28%), and the MIC of aztreonam-avibactam was ≤8 mg/liter for 9 of 25 isolates (36%) (data not shown). Eleven P. aeruginosa isolates were phenotypically identified as meropenem nonsusceptible; however, the mechanisms of nonsusceptibility were not clarified by bla gene analysis. The only nonresident β-lactamase gene identified was blaTEM-1, in 3 of the isolates (Table 1). Regardless of mechanism, more than 90% of ceftazidime-avibactam MICs for these 11 carbapenem-nonsusceptible P. aeruginosa isolates were ≤8 mg/liter (Table 1).
FIG 1.
Ceftazidime, ceftazidime-avibactam, aztreonam, and aztreonam-avibactam MIC distributions for 291 Enterobacteriaceae isolates (excluding meropenem-nonsusceptible isolates), 30 Acinetobacter isolates, and 25 P. aeruginosa isolates.
MIC90s of ceftazidime and aztreonam against Acinetobacter baumannii were lowered by >4-fold and >2-fold, respectively, upon combining with avibactam at 4 mg/liter (Table 2). For the 11 meropenem-nonsusceptible A. baumannii isolates that harbored nonresident genes encoding OXA-23-like and TEM-1 β-lactamases, the ranges of MICs of ceftazidime-avibactam and aztreonam-avibactam were relatively high, at 16 to 128 and 32 to 64 mg/liter, respectively, even though avibactam reduced the MIC90s somewhat (Tables 1 and 2).
In conclusion, the in vitro activity of ceftazidime-avibactam against bacteria isolated from patients in China supports further evaluation of ceftazidime-avibactam in clinical studies against ESBL-, AmpC-, and serine carbapenemase-producing Enterobacteriaceae isolates. In vitro, ceftazidime-avibactam showed more activity than a carbapenem against carbapenem-nonsusceptible and KPC-producing isolates. At the same time, aztreonam-avibactam could serve as a candidate for the treatment of infections with MBL-producing Enterobacteriaceae, especially NDM-producing organisms (Table 1) (19). In both cases, additional studies are needed to establish what the potential roles of ceftazidime-avibactam and aztreonam-avibactam might be as substitutes for carbapenems to reduce the dissemination of carbapenemases in the future.
ACKNOWLEDGMENTS
We express our appreciation to the staff members of the Department of Clinical Laboratory of Peking University People's Hospital for assistance in performing the study.
Footnotes
Published ahead of print 16 December 2013
REFERENCES
- 1.Ehmann DE, Jahić H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non–β-lactam β-lactamase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 109:11663–11668. 10.1073/pnas.1205073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman K. 2011. Diazabicyclooctanes (DBOs): a potent new class of non-β-lactam β-lactamase inhibitors. Curr. Opin. Microbiol. 14:550–555. 10.1016/j.mib.2011.07.026 [DOI] [PubMed] [Google Scholar]
- 3.Shlaes DM. 2013. New β-lactam-β-lactamase inhibitor combinations in clinical development. Ann. N. Y. Acad. Sci. 1277:105–114. 10.1111/nyas.12010 [DOI] [PubMed] [Google Scholar]
- 4.Zhanel GG, Lawson CD, Adam H, Schweizer F, Zelenitsky S, Lagacé-Wiens PR, Denisuik A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, III, Karlowsky JA. 2013. Ceftazidime-avibactam: a novel cephalosporin/β-lactamase inhibitor combination. Drugs 73:159–177. 10.1007/s40265-013-0013-7 [DOI] [PubMed] [Google Scholar]
- 5.Sader HS, Flamm RK, Jones RN. 2013. Antimicrobial activity of ceftaroline-avibactam tested against clinical isolates collected from U.S. medical centers in 2010–2011. Antimicrob. Agents Chemother. 57:1982–1988. 10.1128/AAC.02436-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stachyra T, Levasseur P, Péchereau MC, Girard AM, Claudon M, Miossec C, Black MT. 2009. In vitro activity of the β-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemases. J. Antimicrob. Chemother. 64:326–329. 10.1093/jac/dkp197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endimiani A, Choudhary Y, Bonomo RA. 2009. In vitro activity of NXL104 in combination with beta-lactams against Klebsiella pneumoniae isolates producing KPC carbapenemases. Antimicrob. Agents Chemother. 53:3599–3601. 10.1128/AAC.00641-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore DM, Mushtaq S, Warner M, Zhang J, Maharjan S, Doumith M, Woodford N. 2011. Activities of NXL104 combinations with ceftazidime and aztreonam against carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 55:390–394. 10.1128/AAC.00756-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagacé-Wiens PR, Tailor F, Simner P, DeCorby M, Karlowsky JA, Walkty A, Hoban DJ, Zhanel GG. 2011. Activity of NXL104 in combination with beta-lactams against genetically characterized Escherichia coli and Klebsiella pneumoniae isolates producing class A extended-spectrum beta-lactamases and class C beta-lactamases. Antimicrob. Agents Chemother. 55:2434–2437. 10.1128/AAC.01722-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktas Z, Kayacan C, Oncul O. 2012. In vitro activity of avibactam (NXL104) in combination with β-lactams against Gram-negative bacteria, including OXA-48 β-lactamase-producing Klebsiella pneumoniae. Int. J. Antimicrob. Agents. 39:86–89. 10.1016/j.ijantimicag.2011.09.012 [DOI] [PubMed] [Google Scholar]
- 11.Crandon JL, Nicolau DP. 2013. Human simulated studies of aztreonam and aztreonam-avibactam to evaluate activity against challenging Gram-negative organisms, including metallo-β-lactamase producers. Antimicrob. Agents Chemother. 57:3299–3306. 10.1128/AAC.01989-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A9, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 13.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement. M100-S23. Clinical and Laboratory Standards, Wayne, PA [Google Scholar]
- 14.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders CC. 1987. Chromosomal cephalosporinases responsible for multiple resistance to newer β-lactam antibiotics. Annu. Rev. Microbiol. 41:573–593. 10.1146/annurev.mi.41.100187.003041 [DOI] [PubMed] [Google Scholar]
- 16.Pfaller MA, Jones RN, Marshall SA, Coffman SL, Hollis RJ, Edmond MB, Wenzel RP. 1997. Inducible ampC β-lactamase producing Gram-negative bacilli from bloodstream infections: frequency, antimicrobial susceptibility, and molecular epidemiology in a national surveillance program (SCOPE). Diagn. Microbiol. Infect. Dis. 28:211–219 [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. 1999. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob. Agents Chemother. 43:769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walkty A, DeCorby M, Lagacé-Wiens PRS, Karlowsky JA, Hoban DJ, Zhanell GG. 2011. In vitro activity of ceftazidime combined with NXL104 versus Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals: CANWARD 2009. Antimicrob. Agents Chemother. 55:2992–2994. 10.1128/AAC.01696-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levasseur P, Girard A-M, Claudon M, Goossens H, Black MT, Coleman K, Miossec C. 2012. In vitro antibacterial activity of the ceftazidime-avibactam (NXL104) combination against Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 56:1606–1608. 10.1128/AAC.06064-11 [DOI] [PMC free article] [PubMed] [Google Scholar]