Abstract
Identifying the mechanism of action of new potential antibiotics is a necessary but time-consuming and costly process. Phenotypic profiling has been utilized effectively to facilitate the discovery of the mechanism of action and molecular targets of uncharacterized drugs. In this research, Raman spectroscopy was used to profile the phenotypic response of Escherichia coli to applied antibiotics. The use of Raman spectroscopy is advantageous because it is noninvasive, label free, and prone to automation, and its results can be obtained in real time. In this research, E. coli cultures were subjected to three times the MICs of 15 different antibiotics (representing five functional antibiotic classes) with known mechanisms of action for 30 min before being analyzed by Raman spectroscopy (using a 532-nm excitation wavelength). The resulting Raman spectra contained sufficient biochemical information to distinguish between profiles induced by individual antibiotics belonging to the same class. The collected spectral data were used to build a discriminant analysis model that identified the effects of unknown antibiotic compounds on the phenotype of E. coli cultures. Chemometric analysis showed the ability of Raman spectroscopy to predict the functional class of an unknown antibiotic and to identify individual antibiotics that elicit similar phenotypic responses. Results of this research demonstrate the power of Raman spectroscopy as a cellular phenotypic profiling methodology and its potential impact on antibiotic drug development research.
INTRODUCTION
The ability to quickly identify the mechanism of action and cellular target(s) of new antibiotic compounds is a critical development for antibiotic drug research. It is well established that antibiotic drug development is a time-consuming and costly process, and much of the difficulty originates in identifying the mechanism of action of putative antibiotic compounds (1, 2). Identifying the mechanism of action of an antibiotic is difficult due to (i) the complexity and interdependence of the cellular system (3), (ii) the possible occurrence of multiple cellular targets (4–6), and (iii) the possibility of pleiotropic effects (1, 3). Determining the mechanism of action of putative drugs relies on affinity-based (direct) and phenotypic profiling (indirect) approaches (2, 7). Direct approaches utilize affinity chromatography, expression cloning, protein microarray, and mass spectrometry to bind, isolate, and identify target protein(s). This approach is limited to compounds that can maintain sufficient binding affinity following immobilization, such that the target protein(s) can be isolated and identified (7). The direct approach also suffers from nonspecific binding (2). A more recent and increasingly popular direct approach for determining the mechanism of action includes selecting for resistant mutants and identifying the mutations by whole-genome sequencing (8, 9). However, creating resistant mutants (not associated with efflux pumps or other generic cellular stress responses) can be challenging, and whole-genome sequencing remains expensive for the time being. Indirect approaches to determining the mechanism of action of putative drug compounds are based on the phenotypic response of a cell to a compound. With the indirect approach, the search for the mechanism of action is facilitated by comparing the phenotypic profile of a culture exposed to a putative compound to a database of phenotypic profiles resulting from culture exposure to well-characterized compounds. This provides associative information regarding the mechanism of action of the putative drug, including candidate molecular targets and plausible pathways (2, 10). Phenotypes induced by various compounds can be profiled using the currently available profiling systems, including transcriptional-, proteomic-, metabolomic-, and cell imaging-based profiling. Detailed reviews of direct and indirect approaches to target identification are available in published literature (2, 5–7). For example, transcriptional profiling, which is based on examining the genome-wide expression level of mRNAs in a given cell population, was successfully used to identify acetyl coenzyme A carboxylase as a molecular target of the natural product moiramide B (10). This research elucidated the mechanism of action of moiramide B by examining the transcriptional profile of moiramide B-treated Bacillus subtilis against a database that included profiles resulting from treatment with 14 well-characterized antibiotics and profiles of mutants with downregulation of genes coding for known antibacterial targets (10). Transcriptional profiling was also utilized to identify the heat shock protein 90 as the molecular target of the triterpenoid natural product gedunin, which exhibits antimalarial, insecticidal, and anticancer activity (11). The researchers were able to generate testable hypotheses regarding the mechanism of action of gedunin by examining the transcriptional profile of gedunin-treated cells against a database containing profiles of cells exposed to 164 small molecules with known molecular targets (11). Similarly, proteomic profiling provides a global view of the protein composition of the cell. This can be used to determine the mechanism of action of a novel antibiotic by comparing the proteomic profile of a cell treated with the novel antibiotic to that induced by well-characterized antibiotics (12). For example, the proteomic profile of B. subtilis treated with the novel pyrimidinone compound Bay 50-2369 was found to be highly similar to the profiles induced by erythromycin, chloramphenicol, tetracycline, and fusidic acid. Because these antibiotics are known to directly or indirectly inhibit peptidyl-transferase activity, it was concluded that Bay 50-2369 inhibits peptidyl-transferase. In independent experiments, peptidyl-transferase was confirmed as the molecular target of TAN 1057 A/B, a natural compound structurally related to Bay 50-2369 (13). Metabolomic profiling is based on comprehensive analysis of global metabolism and has been enabled by recent technological advances in the quantification of cellular metabolites (2, 7, 14). Yu et al. (14) investigated the mechanism of action of Radix Tinosporae extract by comparing the metabolomic profile of treated Staphylococcus aureus to profiles induced by nine antibiotics with known mechanisms of action. Principal component analysis (PCA) of metabolomic profiles, obtained using high-performance liquid chromatography-mass spectrometry (HPLC-MS) data, revealed that the mechanism of action of Radix Tinosporae was similar to that of rifampin and norfloxacin (14). Techniques for multiparameter imaging-based phenotypic profiling rely on high-throughput fluorescence microscopy (15) or morphological analysis (16) and have shown great promise in drug discovery research (2, 7). For example, bright-field cell imaging coupled with nuclear staining was used to construct a library of morphological profiles for cancer cells exposed to 54 anticancer drugs with known molecular targets (16). The library was then used with multivariate statistical analysis to identify the mechanistically unknown drugs NPD6689, NPD8617, and NPD8969 as novel tubulin inhibitors. Despite significant progress, current phenotypic profiling methodologies (i) require substantial time and material recourses, (ii) cannot be applied universally, and (iii) will require significant development to allow automated high-throughput analysis of drug targets (2). In the present research, the use of Raman spectroscopy was investigated as an alternative phenotypic profiling technique. The Raman spectroscopy-based approach offers significant advantages over other phenotypic profiling methodologies, because it is (i) noninvasive, (ii) label free, (iii) prone to automation, and (iv) able to return results in real time during an experiment.
Analyzing cell phenotypes using Raman spectroscopy.
Raman spectroscopy has been identified as a powerful tool for analyzing bacterial phenotypes due to its (i) sensitivity, (ii) short analysis time, and (iii) nondestructive nature (17, 18). It has also been shown to be an effective tool for discriminating among several bacterial phenotypes (19–22) and for in vivo metabolic analysis (17, 23–25). Compared to the considerable volume of research published on applying Raman spectroscopy in biotechnology research, little has been reported on applying Raman spectroscopy in antibiotic drug research. Nonetheless, the relatively few articles published to date on the topic point to a significant potential for Raman spectroscopy to become a powerful tool aiding antibiotic drug research. First, it has been shown that Raman spectroscopy can distinguish between bacterial strains resistant and sensitive to a specific antibiotic (22, 26). For example, Maquelin et al. (22) used Raman spectroscopy with an 830-nm excitation wavelength to analyze Enterococcus faecalis strains sensitive and resistant to vancomycin. Using multivariate statistical analysis, it was found that Raman spectroscopy data could discriminate between resistant and sensitive strains in vancomycin-treated samples. Walter et al. (26) showed that Raman spectroscopy, conducted using 244- and 532-nm excitation wavelengths, can differentiate E. coli cultures according to the presence of a plasmid containing an ampicillin resistance gene, even in the absence of ampicillin. The authors found the resulting Raman spectra contained spectral features suggesting higher lipid and RNA content and lower cytochrome content in E. coli harboring the plasmid. In addition, Raman spectroscopy has been used to monitor the metabolic state under antibiotic stress (18, 27–29). For example, Neugebauer et al. (28) used Raman spectroscopy with a 244-nm excitation wavelength to track the ratio of protein to nucleic acids during the growth of Bacillus pumilus treated with ciprofloxacin. The use of the 244-nm excitation wavelength is known to enhance Raman bands specific to nucleic acids and protein building blocks (28). Raman spectroscopy has been used with a similar excitation wavelength to study the effects of the protein synthesis inhibitor amikacin on Pseudomonas aeruginosa. As the concentration of amikacin was increased, the intensity of protein-related Raman peaks decreased, whereas the intensity of nucleic acid bands increased, consistent with the mechanism of action of amikacin (29). Similar results were reported, as Raman spectroscopy was used to monitor the relative protein and DNA content of individual E. coli cells treated with cefazolin (18). However, in this case, Raman spectra were acquired using a 785-nm excitation wavelength from individual, optically trapped, E. coli cells. This technique is often referred to as “laser tweezers” Raman spectroscopy (30). Finally, it has been suggested that Raman spectroscopy could provide critical insight into the mechanisms of action of different antibiotics (27, 29, 30). For example, Moritz et al. (30) used laser tweezers Raman spectroscopy to study the response of E. coli to penicillin G-streptomycin and cefazolin. The researchers identified Raman bands specific to the response of E. coli to cefazolin and/or penicillin G-streptomycin treatment and extended those to aminoglycosides and β-lactams in general. Although significant to the development of Raman methodologies for studying cell phenotypes, most existing studies have involved either one or two antibiotics. The ability of Raman spectroscopy to discriminate between greater numbers of antibiotic treatments has not been investigated to our knowledge. Furthermore, the experimental setups of the previously discussed studies all differed in significant ways, including the (i) bacterial species and strains used, (ii) methods of sample preparation, and (iii) excitation wavelength(s) used in the Raman spectroscopy analysis. This means that the available published studies should be considered individually, and observed changes in Raman spectra cannot be compared across the multiple studies. For a rigorous assessment of the ability of Raman spectroscopy to discriminate among bacterial phenotypes resulting from antibiotic treatment (so it can be used as a phenotypic profiling tool), multiple antibiotics must be examined with the same bacterial strain under identical experimental conditions, including excitation wavelength(s).
Raman spectroscopy-based phenotypic profiling.
In this research, profiles of E. coli phenotypes resulting from treatment with 15 well-characterized antibiotics from four antibiotic classes were characterized using Raman spectroscopy analysis data obtained from dried culture samples. Using discriminate analysis of processed Raman spectral data, it was possible to distinguish among culture phenotypic profiles according to antibiotic function. In fact, the resulting Raman spectra contained sufficient biochemical information to distinguish between profiles induced by individual antibiotics belonging to the same class. In addition, a systematic study was conducted to evaluate the ability of Raman spectroscopy-based phenotypic profiling with discriminant analysis to predict the functional class of an unknown antibiotic compound with an uncharacterized mechanism of action. Results showed the ability of Raman spectroscopy to identify the functional class of the unknown antibiotic compound as well as the individual antibiotics that elicit a similar phenotypic response.
MATERIALS AND METHODS
Antibiotic treatments.
The class designations for antibiotics used in this research (Table 1) are based on the overall effects on phenotype produced by each antibiotic according to known mechanisms of action (1). For example, gentamicin, streptomycin, kanamycin, clindamycin, chloramphenicol, and tetracycline were grouped together as protein synthesis inhibitors (antibiotic class name “PROT” in Table 1). Gentamicin, streptomycin, and kanamycin are aminoglycosides that primarily inhibit protein synthesis by targeting the 16S rRNA component of the 30S ribosome subunit. Clindamycin (lincosamide) and chloramphenicol (amphenicol) also inhibit protein synthesis by targeting the 50S ribosome subunit. Tetracycline inhibits the 30S ribosome by blocking the access of aminoacyl tRNAs to the ribosome (1). Ciprofloxacin and nalidixic acid are quinolone antibiotics known to inhibit DNA synthesis (class name “DNA”) by targeting DNA gyrase. Trimethoprim inhibits synthesis of tetrahydrofolic acid, which is needed by bacteria to synthesize DNA. Ampicillin, amoxicillin, and cefotaxime are β-lactams that inhibit cell wall (class name “CW”) biosynthesis by targeting to penicillin-binding proteins involved in peptidoglycan synthesis. Rifampin, rifapentine, and rifaximin inhibit RNA synthesis (class name “RNA”) by binding to the β-subunit of a DNA-bound and actively transcribing RNA polymerase. The MIC of each antibiotic used in this study required to inhibit growth of E. coli cultures was determined experimentally, as described below. These results are also listed in Table 1.
TABLE 1.
Antibiotic class designations and MICs against E. coli
| Antibiotic class and name | MIC, in mg/liter (μM) |
|---|---|
| Protein synthesis inhibition (PROT) | |
| Gentamicin (GE) | 1.91 (4) |
| Streptomycin (ST) | 5.83 (4) |
| Clindamycin (CL) | 118.13 (256) |
| Chloramphenicol (CH) | 5.17 (16) |
| Kanamycin (KA) | 9.32 (16) |
| Tetracycline (TT) | 0.89 (2) |
| Cell wall synthesis inhibition (CW) | |
| Ampicillin (AM) | 2.97 (8) |
| Amoxicillin (AX) | 2.92 (8) |
| Cefotaxime (CF) | 0.06 (0.125) |
| DNA synthesis inhibition (DNA) | |
| Trimethoprim (TR) | 1.16 (4) |
| Nalidixic acid (NX) | 1.86 (8) |
| Ciprofloxacin (CI) | <0.005 (<0.015) |
| RNA synthesis inhibition (RNA) | |
| Rifampin (RI) | 13.17 (16) |
| Rifapentine (RP) | 22.95 (32) |
| Rifaximin (RX) | 12.57 (16) |
| No antibiotic treatment (control) | |
| None | None |
Determination of MICs.
E. coli Mach1 cells (Invitrogen Life Technologies, Grand Island, NY) were grown and maintained at 37°C in Trypticase soy broth (TSB) or on Trypticase soy agar (BD Biosciences San Jose, CA). Antibiotic stock solutions (10 mM) were prepared by dissolving ampicillin, amoxicillin, tetracycline, chloramphenicol, clindamycin, rifampin, rifaximin, rifapeutine, trimethoprim, and nalidixic acid powders in dimethylsulfoxide (DMSO). Cefotaxime, kanamycin, gentamicin, and streptomycin were dissolved in water. Ciprofloxacin was dissolved in 0.1 N HCl. MICs were determined in triplicate using the broth microdilution method. The MIC is defined as the lowest concentration of antibiotic leading to no visible bacterial growth after 16 h.
Preparation of cultures for Raman spectroscopy analysis.
E. coli Mach1 cells were grown in liquid Luria broth medium at 37°C, and cell growth was monitored by measuring the optical density at 600 nm (OD600). The culture was grown to an OD600 of 0.5 and then divided into subcultures. Antibiotics were added to the subcultures to a final concentration of three times the MIC (termed 3× MIC). After 30 min, cells were harvested by centrifugation at 5,000 rpm for 1 min, discarding the supernatant. The cells were then washed three times with cold phosphate-buffered saline (PBS) to remove residual growth media. For each wash, the pellet was resuspended in 1 ml PBS and centrifuged for 1 min at 5,000 rpm. Finally, 0.5 μl of the cell suspension was deposited on aluminum foil and dried at room temperature for subsequent Raman spectroscopy analysis. Raman spectra were collected from areas where E. coli cells formed a thin layer covering the aluminum foil surface.
Raman spectroscopy.
Dried E. coli Mach1 cells were analyzed using a Bruker Senterra dispersive Raman spectrometer equipped with a confocal microscope (Bruker Optics, Billerica, MA). Measurements were carried out using a 10-mW, 532-nm laser focused through a 100× objective. A spectral resolution of 9 to 15 cm−1 was used. The total exposure time was 15 s, which was found to be sufficient to obtain spectra with a good signal-to-noise ratio without causing visible sample damage. Lengthy exposure of dried bacterial sample to high laser power may cause graphitization, which dominates the Raman spectrum and obscures other contributions. If not directly visible on the sample, this kind of sample damage can be detected indirectly in the Raman spectrum by noting an unusual rise of a wide doublet peak around 1,500 cm−1 (31). An average of 64 spectra were collected for each treatment.
Data processing.
All spectra were normalized in MATLAB (v2012a; The MathWorks, Inc., Natick, MA) and baseline corrected using OPUS software (Bruker Optics, Billerica, MA). Normalization of Raman spectra is necessary to remove variation of the Raman signal caused by differences in the focal volumes among samples (32). Initially, statistical analyses were performed with Raman spectra normalized using vector normalization, because this method is relatively unbiased toward any single band or group of bands that could possibly be affected by changes in the bacterial phenotype (32). A separate study was then performed in which multiple procedures for spectral normalization were applied and compared. Details are given in Results. All spectra were baseline corrected using the Concave Rubberband correction method, available in OPUS software, with 20 iterations and 64 baseline points.
Chemometric analysis.
The chemometric analysis was performed using MATLAB (v2012a; The MathWorks, Inc., Natick, MA). Chemometric analysis was primarily conducted by linear discriminant analysis with processed Raman spectral data. Different methods of data normalization were investigated and are discussed in Results. Discriminant analysis was used to build a statistical model that (i) requires input of Raman spectra of an E. coli culture exposed to an antibiotic compound and (ii) returns the class of antibiotic that elicits the phenotypic response represented by the input spectra. Thus, discriminant analysis was used to predict a classification variable (X) (e.g., antibiotic class, such as protein synthesis inhibitor, etc.) based on measured responses (Y) (e.g., band intensities in Raman spectra of an exposed culture). Discriminant analysis determines the distance from each point of the data set to the multivariate mean of each class and then assigns the point to the closest class (33). Essentially, discriminant analysis is divided into two steps: (i) to build (i.e., “train”) a model (i.e., classifier) using Raman spectra of cultures exposed to antibiotics of known class assignments, and (ii) to classify (i.e., predict the class of) a new Raman spectrum of an antibiotic-exposed culture based on the distance to the multivariate mean of the closest class. Two class assignment schemes were used with discriminant analysis to achieve the following two distinct objectives.
(i) Identify the class of an antibiotic eliciting the observed phenotypic response.
Antibiotics were grouped into classes based on the overall effects on phenotype according to known mechanisms of action (1). The class designation for all antibiotics used in this research is given in Table 1. In this case, the developed discriminant analysis model classified Raman spectra according to antibiotic class.
(ii) Identify individual antibiotics that elicit a phenotypic response closely related to the observed.
In this case, antibiotics were not grouped according to class, and the discriminant analysis model was trained to classify Raman spectra according the individual antibiotic treatments. This approach proved to be particularly useful when the tested antibiotic exhibits multiple mechanisms of action.
RESULTS
Raman spectroscopy can detect changes in E. coli phenotypes caused by various antibiotics.
Raman spectra of E. coli cultures consist of bands that correspond to all biomolecules present in culture. Examples are shown in Fig. 1. These bands represent both chemical and physical signatures of molecular structures. The tentative band assignments are shown in Table 2. A lateral shift in the position of a peak reflects a biochemical or biophysical change in the phenotype caused by antibiotic treatment. In addition, a change in the intensity of a particular peak (i.e., peak height) reflects a change in the abundance of the biomolecule represented by that peak. However, variation in peak intensity may also result from differences in the focal volumes among samples. This variation is not related to phenotypic changes caused by antibiotic treatments and must be removed to ensure the analysis is independent of different sample densities and geometries (32). Removing, or at least significantly reducing, this undesired variation is achieved through normalization of Raman spectra to a specific peak or set of peaks. The effect of spectral normalization is also illustrated in Fig. 1. The averages and standard deviations for 131 independently collected spectra of the control E. coli culture (i.e., not treated with antibiotic) are shown before and after normalization (Fig. 1a and b). Before normalization (Fig. 1a), greater variation in spectral intensity is indicated by the prominent standard deviation spectrum with relatively high intensity peaks. This variation was largely removed upon normalization (Fig. 1b). Sample variation can also arise from various background signals originating from fluorescence of the sample or thermal fluctuations on the signal detector (32). To minimize the effect of this variation, background signal was removed through baseline correction (Fig. 1c).
FIG 1.
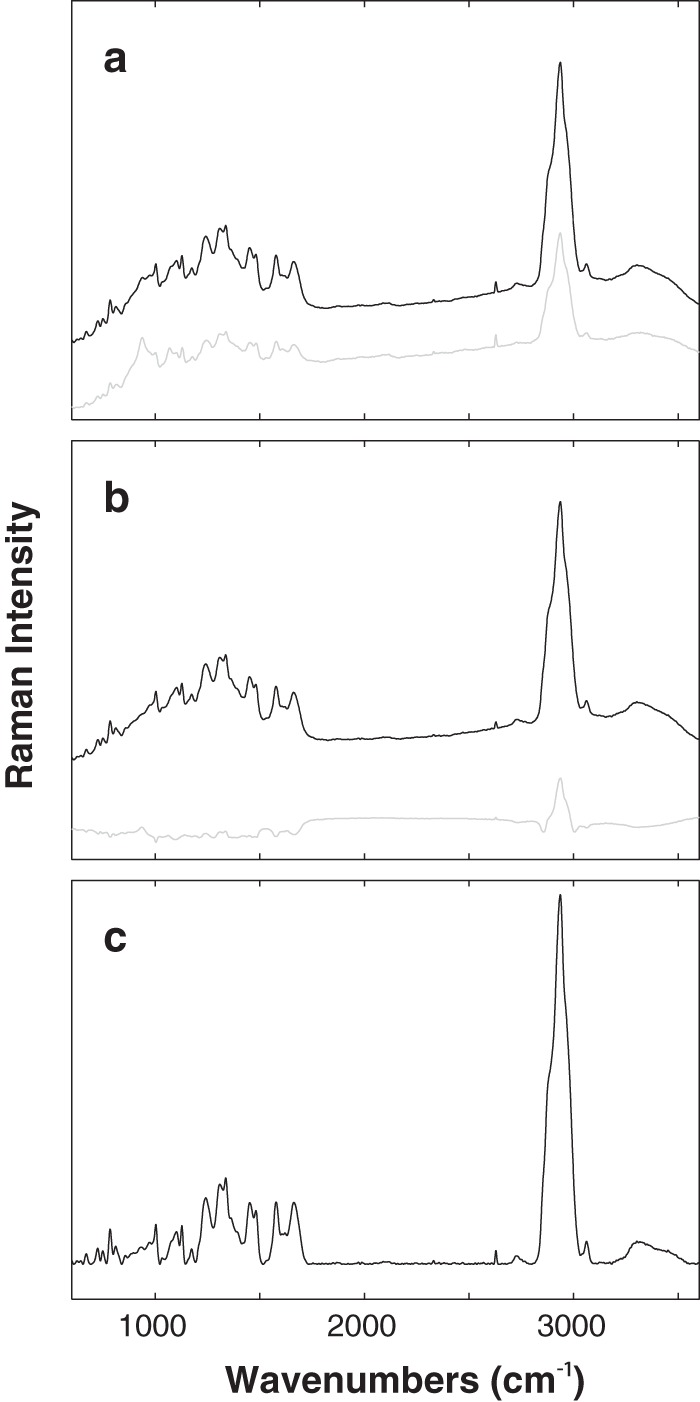
Preprocessing Raman spectroscopy data. The average spectrum (black line) of 131 independently obtained Raman spectra of the control E. coli culture (i.e., untreated with antibiotic) before normalization (raw data) (a), after normalization (b), and after baseline correction (c). Intensity variation between individual spectra is indicated by the standard deviation spectra (gray line) shown in panels a and b.
TABLE 2.
Reduced set of biologically relevant Raman peaks
| Wavenumber (cm−1) | Peak assignmenta |
|---|---|
| 3,061 | ν(C = C-H)Aromatic |
| 2,961 | ν(CH2/CH3); lipids |
| 2,936 | ν(CH2/CH3); proteins |
| 2,880 | ν(CH2/CH3); lipids |
| 2,850 | ν(CH2/CH3); lipids |
| 1,660 | Amid I; proteins |
| 1,621 | Try, Trp, Phe |
| 1,577 | ν(ring); guanine, adenine |
| 1,481 | ν(ring); guanine, adenine |
| 1,451 | γ(CH) |
| 1,336 | δ(CH) |
| 1,309 | δ(CH2) |
| 1,242 | Amid III; proteins |
| 1,173 | ν(C-C) |
| 1,127 | Cytochrome |
| 1,032 | ν(C-C) |
| 1,003 | Phe, Trp |
| 857 | ν(C-C); COC 1,4-glycosidic link |
| 811 | ν(PO3)-2; RNA |
| 784 | ν(ring); Cys, uracil |
| 749 | Cytochrome |
| 725 | Adenine; DNA |
| 670 | Guanine; DNA |
ν, stretching vibration; δ, twisting deformation; γ, bending deformation.
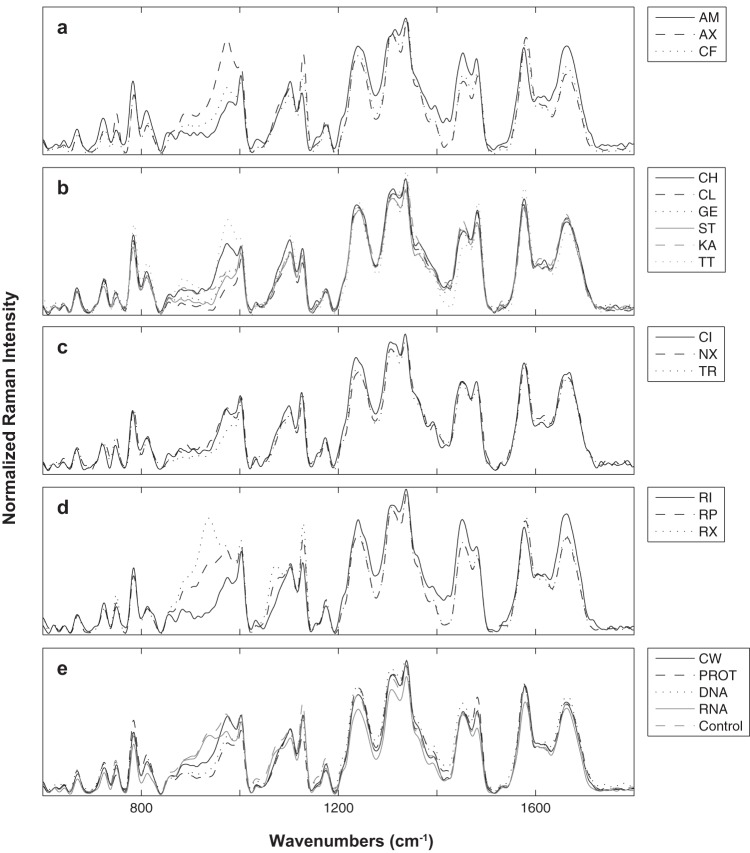
E. coli cultures treated with antibiotic at 3× MIC, prior to analysis by Raman spectroscopy, continued to grow at a lower rate than the wild type, as was evident by OD600 measurements following 30 min of antibiotic exposure (data not shown). This slowed-growth response was desired, since the goal was to examine the phenotypic effects of the antibiotic without killing the cultures or arresting growth completely. The phenotypic effects of each of the antibiotics used in this research were evident in the Raman spectra of treated E. coli cultures. Normalized and baseline-corrected spectra are shown in Fig. 2. Raman spectroscopy was sensitive enough to detect variations among and within antibiotic classes. For example, the average spectra of the three cultures treated with cell wall synthesis inhibitors were different from each other (Fig. 2a). At the same time, the average spectrum of all cell wall synthesis inhibitors was different from the average spectrum of other antibiotic classes (Fig. 2e). Due to the complexity of the Raman spectra of E. coli cultures, it is difficult to analyze the effects of antibiotics by comparing changes in individual Raman bands. Instead, chemometric analysis is more suitable because it provides a method to compare changes to the Raman spectra as a whole. Chemometric analysis was employed to leverage the sensitivity of Raman spectroscopy in order to develop a statistical model capable of predicting the phenotypic effects of an unknown antibiotic.
FIG 2.
Comparison of Raman spectra among and within antibiotic classes. Normalized and baseline-corrected Raman spectra of antibiotic-treated phenotypes averaged by antibiotic (n = ∼64) and grouped by class: CW (a), PROT (b), DNA (c), and RNA (d). Raman spectra of antibiotic-treated phenotypes averaged by antibiotic class are shown in panel e. All abbreviations are defined in Table 1.
Chemometric method development: can Raman spectroscopy data be used to discriminate among multiple E. coli phenotypes resulting from different antibiotics?
In this section, the development of the chemometric approach to utilizing Raman spectroscopic data for phenotypic profiling E. coli cultures according to antibiotic treatment is discussed. The developed chemometric approach is used in subsequent sections to build a predictive statistical model capable of identifying the phenotypic effects of unknown antibiotic agents. Before the predictive model was developed and utilized, it was first necessary to establish the ability of Raman spectroscopy with chemometrics to discriminate among the multiple E. coli phenotypes that result from treatment with different antibiotics. To achieve this goal, the ability to discriminate E. coli phenotypes based on (i) the class of antibiotics and (ii) individual antibiotics (even among those belonging to the same class) was investigated.
(i) Raman spectrum discrimination based on the class of antibiotic treatment.
E. coli cultures exposed to different antibiotics (Table 1) were analyzed by Raman spectroscopy. Discriminant analysis was applied to the normalized and baseline-corrected Raman spectra in an attempt to discriminate according to the class of antibiotic applied. Initially, discriminant analysis was performed on the Raman spectra in three ways: (i) using the band intensity values from the full biologically relevant spectral region from 600 to 1,800 cm−1 and from 2,800 to 3,100 cm−1 (this approach is referred to as FR-DA), (ii) using band intensity values for the reduced number of biologically relevant Raman bands listed in Table 2 (this approach is referred to as R-DA), and (iii) using a specified number of principal components (PCs) calculated from principal component analysis of the band intensity values from the entire Raman spectrum (600 to 3,200 cm−1) (this approach is referred to as PC-DA). Principal component analysis is a multivariate statistical technique that is often used to analyze spectroscopic data, because the number of variables of interest (i.e., peaks or intensity at each wavenumber) can be in the thousands (32). It reduces the dimensionality of the data (i.e., the number of variables) while preserving variance among treatment groups (34). Each of the three approaches was used to discriminate the Raman spectra of treated cultures according to antibiotic class. The quality of the discriminant analysis models (i.e., their ability to correctly classify spectra according to antibiotic class) was validated using the “leave-one-out” cross-validation method. This means that Raman spectra were removed, one spectrum at a time, from the training data set used to build a classifier model. The model was then used to classify the removed spectrum. Thus, leave-one-out cross-validation provides an unbiased estimate of the ability of the classifier model (35). The percentages of all analyzed samples classified correctly (i.e., classified into the same antibiotic class as that applied to the culture) are shown in Table 3. PC-DA proved the best at discriminating cultures according to antibiotic class, with an 83.6% correct classification rate. With five classes of antibiotics tested (including the control with no antibiotic treatment), the probability of correct assignment at random is 20%. Discrimination results for PC-DA are visualized in the three-dimensional canonical plot shown in Fig. 3a. The canonical plot shows the location of each point relative to the multivariate means of each group. The ability of Raman spectroscopy and discriminate analysis to distinguish among E. coli cultures based on antibiotic class is evident in this figure. As expected, increasing the number of PCs included in the analysis increased the percentage of spectra classified correctly. However, beyond ∼50 PCs, no additional improvement was observed (Fig. 3b).
TABLE 3.
Correct classification rates estimated using leave-one-out cross-validation of discriminant analysisa
| Data set | % Classified correctly by: |
|
|---|---|---|
| Class | Antibiotic | |
| FR-DA | 45.2 | 45.0 |
| R-DA | 68.8 | 47.8 |
| PC-DAb | 83.6 | 71.3 |
| Randomc | 20.0 | 6.3 |
The analysis was performed using the (i) full biologically relevant spectral region (FR-DA), (ii) Raman bands listed in Table 2 (R-DA), and (iii) principal components (PCs) of the full spectrum (PC-DA).
Performed using the first 50 PCs, which preserved 83.4% of variance in the original data.
The probability of correct classifications by random assignment.
FIG 3.
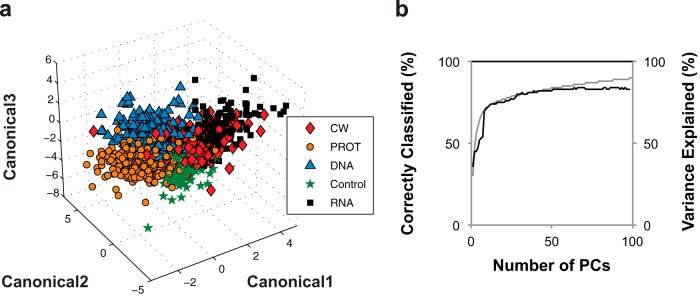
Discrimination of Raman spectra according to antibiotic class. (a) Canonical plot for discrimination of antibiotic-treated cultures based on antibiotic class using the first 50 PCs from PCA of the full Raman spectrum. (b) The relationship between the number of PCs included in PC-DA and the percent correct classification (black line). The secondary axis represents variance of the original data set preserved by PCs (gray line). All abbreviations are defined in Table 1.
(ii) Raman spectrum discrimination based on specific antibiotic treatment.
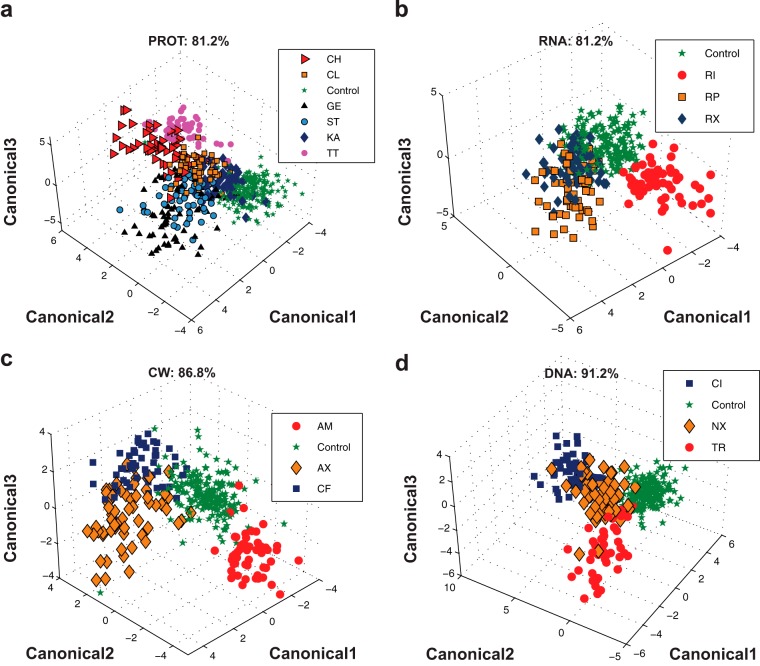
In this part of the analysis, discriminant analysis was performed without grouping Raman spectra according to antibiotic class. In this approach, discriminant analysis was used to classify Raman spectra according to individual antibiotic treatments. Again, PC-DA was found to be superior to FR-DA and R-DA for this task (Table 3). The percentage of samples classified correctly was 71.3% for PC-DA, 47.8% for FR-DA, and 45.0% for R-DA. With 15 antibiotics tested and the untreated control, the probability for correct assignment at random is 6.3%. Although PC-DA discriminated individual antibiotics with 71.3% accuracy, it was hard to visualize the discrimination on a three-dimensional plot due to the large number of groups (15 antibiotics) that need to be represented on one plot. To visually demonstrate the ability to discriminate based on individual antibiotics, PC-DA was performed on each antibiotic class individually; results are shown in Fig. 4. The percentage of correct classification for each antibiotic class is indicated on the corresponding figure panel. Note that the percentage of correct classification for each antibiotic class reported in Fig. 4 is much better than the overall correct classification reported in Table 3. This is because Fig. 4 shows the results of discriminant analyses performed separately on each antibiotic class. Table 3 reports the results of discriminant analysis performed on the 15 antibiotics all at once. Discriminant analysis results demonstrate the ability of Raman spectroscopy to profile antibiotic-induced E. coli phenotypes according to both the antibiotic class (Table 3 and Fig. 3) and the individual antibiotic treatment (Table 3 and Fig. 4). This result is consistent with previous studies; however, in this research, the number of antibiotics examined simultaneously was significantly larger. The library of Raman spectra from E. coli phenotypes induced by the 15 well-characterized antibiotics represents the beginning of a database of phenotypic profiles against which the profiles of putative drugs can be searched and compared. In the next section, the utility of this Raman spectroscopy-based phenotypic profiling approach is demonstrated through its use to predict the mechanism of action of well-characterized antibiotics that were not used to build the model.
FIG 4.
Discrimination by individual antibiotic. PC-DA discrimination of E. coli phenotypes based on individual antibiotic treatment for protein (a), RNA (b), cell wall (c), and DNA (d) synthesis inhibitors. PC-DA was performed using the first 50 PCs from PCA of the full Raman spectrum. Correct classification rates are indicated on each graph. All abbreviations are defined in Table 1.
Chemometric method validation by identifying the phenotypic response of E. coli to antibiotics unknown to the classification model.
The results presented thus far have demonstrated the ability of Raman spectroscopy and discriminant analysis to distinguish among E. coli phenotypes caused by treatment with different classes of antibiotics and even among antibiotics belonging to the same class. However, it is worth noting that the discriminant analysis models were built using spectra representing all antibiotic treatments (i.e., the model training data set included spectra representing all antibiotic treatments except a single spectrum used for model validation). This methodology is consistent with previously published studies and is the accepted methodology for training and validating discriminate analysis results (36). However, the question remains, can Raman spectroscopy with discriminant analysis identify the phenotypic effects caused by a new antibiotic compound completely unknown to the model (i.e., the model training data set does not include spectra representing the new antibiotic)? This question is addressed in the following sections by evaluating the ability of Raman spectroscopy with discriminant analysis to (i) identify the class of an unknown antibiotic compound and (ii) identify individual antibiotics that elicit a phenotypic response similar to that of the unknown antibiotic compound.
(i) Identifying the class of an unknown antibiotic.
Raman spectroscopy with discriminant analysis was used to identify the functional antibiotic class of several unknown antibiotics (i.e., they were excluded completely from the model training data set). In other words, the methodology described above was applied to identify the phenotypic effects of a new antibiotic compound given the Raman spectra of the treated cells. To achieve this goal, Raman spectra of cultures treated with each of the 15 antibiotics used in the study were individually considered unknown. Systematically, Raman spectra of cultures treated with these antibiotics were excluded, all spectra for one antibiotic at a time, from the training data set used to build the discriminant analysis classifier model. In this approach, a new discriminant analysis model was created for every case. Thus, the data set used for model training did not contain any spectra representing the excluded antibiotic. The new discriminant analysis models were then used to classify Raman spectra of E. coli cultures exposed to the unknown antibiotic. This approach should not be confused with the leave-one-out cross-validation often used to evaluate discriminant analysis. In the validation approach used here, the complete set of spectra from the antibiotic in question was removed from the training set. With the leave-one-out cross-validation method, only one spectrum was removed at a time, leaving other spectra belonging to the same antibiotic. The percentages of correct antibiotic class identification for Raman spectra of unknown antibiotics classified using R-DA and PC-DA are listed in Table 4. Chloramphenicol is used as an example to further explain the results given in this table. First, all Raman spectra from the E. coli culture treated with chloramphenicol were removed from the discriminant analysis model training data set. A new discriminant analysis model was built using the spectra from all other antibiotics (and the untreated control) as the training set. Separate models were constructed using R-DA, PC-DA with 50 PCs, and PC-DA with 9 PCs (the choice of 9 PCs is explained below). Second, following training, R-DA and PC-DA models were used to classify the Raman spectra obtained from the chloramphenicol-treated culture. All three models correctly classified these spectra as resulting from an antibiotic that inhibits protein synthesis with 89.8% (for R-DA), 93.9% (for PC-DA with 50 PCs), and 93.9% (for PC-DA with 9 PCs) accuracy. The 49 Raman spectra were taken from the chloramphenicol-treated culture and analyzed by the classification models. This procedure was repeated for the other antibiotics (and control) used in this research.
TABLE 4.
Percent correct prediction of the class of an antibiotic compound unknown to the PC-DA model with 9 and 50 PCs included in building the model
| Antibiotic | Class | % Correct prediction |
||
|---|---|---|---|---|
| R-DA | PC-DA (50 PCs) | PC-DA (9 PCs) | ||
| Chloramphenicol | PROT | 89.8 | 93.9 | 93.9 |
| Clindamycin | PROT | 40.0 | 70.9 | 83.6 |
| Gentamicin | PROT | 42.7 | 78.7 | 69.3 |
| Kanamycin | PROT | 27.8 | 50.0 | 63.0 |
| Streptomycin | PROT | 59.2 | 67.3 | 71.4 |
| Tetracycline | PROT | 5.7 | 60.4 | 75.5 |
| Ciprofloxacin | DNA | 5.8 | 10.1 | 47.8 |
| Nalidixic acid | DNA | 22.0 | 28.8 | 67.8 |
| Trimethoprim | DNA | 11.3 | 24.5 | 64.2 |
| Amoxicillin | CW | 32.8 | 45.9 | 41.0 |
| Ampicillin | CW | 9.6 | 0.0 | 0.0 |
| Cefotaxime | CW | 36.8 | 29.8 | 38.6 |
| Rifampin | RNA | 1.8 | 1.8 | 3.5 |
| Rifapentine | RNA | 49.2 | 78.0 | 66.1 |
| Rifaximin | RNA | 63.1 | 80.0 | 80.0 |
| Overall accuracy (%) | 33.2 | 48.0 | 57.7 | |
| Randoma | 20.0 | 20.0 | 20.0 | |
The probability of correct classifications by random assignment. These values were not used in calculating the overall accuracy.
The results reported in Table 4 suggest that the PC-DA method with 9 PCs is superior to RDA and PC-DA with 50 PCs in predicting the class of a new antibiotic (i.e., one not represented in model training). Initially, it was presumed that PC-DA with 50 PCs or higher would perform better, since this method was superior at discriminating samples with known class assignment (Table 3 and Fig. 3b). However, when PC-DA was used to predict the class of Raman spectra from cultures treated with new antibiotics completely unknown to the model, it was evident that using 50 or more PCs did not produce optimum results. The relationship between the accuracy of PC-DA and the number of PCs used to build the model (not shown) revealed that prediction accuracy peaked at 9 PCs and then declined as more PCs were included. It is possible that including higher numbers of PCs in the PC-DA resulted in overfitting, a common problem of multivariate statistical analysis (36). With a smaller number of antibiotics, the R-DA, which is based on using a selected number of biologically relevant Raman bands, produced the best prediction results. However, as the number of antibiotics and complexity of the data set increased, R-DA did not perform as well, as shown in Table 4. Overall, PC-DA with 9 PCs was 57.5% accurate at determining the antibiotic class of unknowns. Raman spectra of phenotypes resulting from treatment by chloramphenicol, clindamycin, gentamicin, streptomycin, tetracycline, and kanamycin were all classified correctly as protein synthesis inhibitors, with an average of 76.1% accuracy. The average prediction accuracies for other antibiotic classes were 59.9, 49.9, and 26.5% for DNA, RNA, and cell wall synthesis inhibitors, respectively.
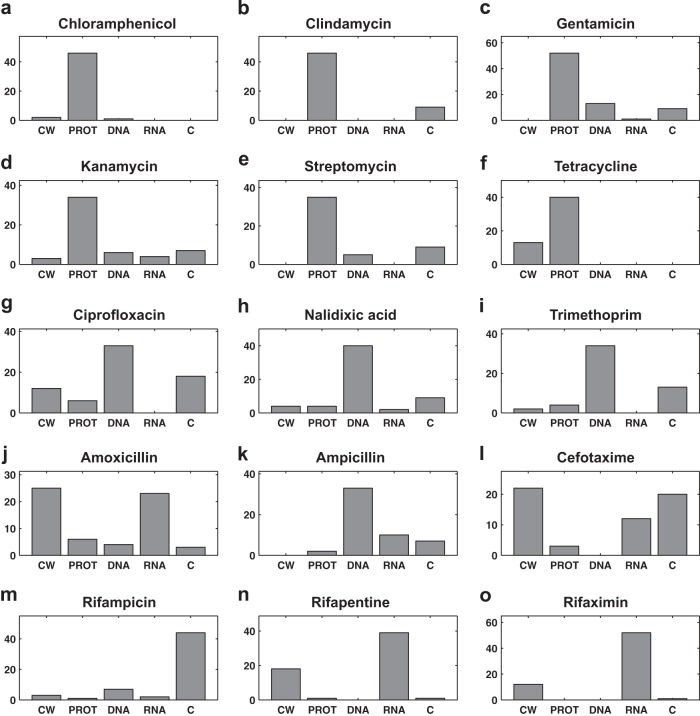
While the results given in Table 4 provide a convenient way to assess the overall performance of the discriminant analysis model, it does not provide access to all information that can be learned from the model classifications. For example, results in Table 4 show that only 47.8% of the Raman spectra of cultures treated with ciprofloxacin were classified correctly as DNA synthesis inhibitors when using PC-DA with 9 PCs. However, another representation of the data is shown in Fig. 5 and reveals additional details of this classification. When considering the detailed classification result shown in Fig. 5g, it was found that the dominant classification count for ciprofloxacin was for the correct class (i.e., DNA synthesis inhibitors). Similarly, the prediction accuracy for amoxicillin and cefotaxime were found to be 41.0 and 38.6% (Table 4), respectively. This seems low given that the random classifier accuracy (with no model) is 20%. However, the highest prediction counts for these two cell wall synthesis inhibitors were in the correct class (Fig. 5j and l). Prediction accuracy was not as good for the third cell wall synthesis inhibitor, ampicillin, which was consistently predicted as a DNA synthesis inhibitor. Poor prediction results also persisted for the rifampin-induced phenotype, which was consistently seen by the model as a phenotype with no antibiotic treatment. This result is clearly wrong but points to some interesting observations. First, the phenotypes induced by ampicillin and rifampin must be significantly different from those induced by other antibiotics of their respective classes. This suggests that the designation of antibiotic class needs to become more specific, or that there are critical members of each antibiotic class that must be included in the model to cover all potential resulting phenotypes. Furthermore, the model consistently recognized that the rifampin-induced phenotype was different from the phenotypes caused by other antibiotics. Results shown in Fig. 2, 3, and 4 suggest that Raman spectroscopy was sensitive enough to distinguish rifampin-induced phenotypes from the control phenotypes. Taking these results together, it is reasonable to conclude that the quality of prediction was limited by data processing and chemometric analysis and not the sensitivity of Raman spectroscopy. As the data set of antibiotics continues to grow with future implementations, these occurrences will become fewer.
FIG 5.
Prediction of antibiotic class for the unknown antibiotics. The prediction count by class for each antibiotic when treated as an unknown is shown in panels a to o. The name of the unknown is indicated above each panel. Here, the untreated control is abbreviated as C. All other abbreviations are defined in Table 1.
(ii) Identifying individual antibiotics that elicit a phenotypic response similar to that of the unknown antibiotic.
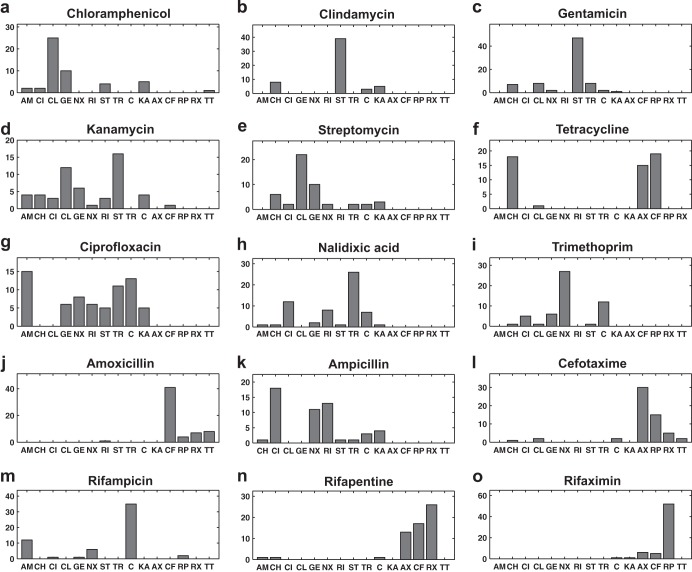
PC-DA was used to classify Raman spectra of cultures treated with antibiotics unknown to the model according to individual antibiotic treatment rather than antibiotic class. By doing so, PC-DA identifies the individual antibiotic that elicits a phenotypic response closest to that of the unknown antibiotic. Details of the PC-DA antibiotic-based classification results are shown in Fig. 6. Results presented in Fig. 6a suggest that the phenotypic effects of chloramphenicol on E. coli are predominantly similar to those caused by clindamycin, gentamicin, kanamycin, tetracycline, and streptomycin. Since all five identified antibiotics are protein synthesis inhibitors, it can be concluded from the modeling exercise that chloramphenicol also acts by inhibiting protein synthesis. Indeed, it is well known that chloramphenicol is a lincosamide antibiotic that inhibits protein synthesis (1). The discriminant analysis classification model used to produce these results had no prior knowledge from Raman spectra related to chloramphenicol treatment. In general, all protein synthesis inhibitors were predicted to have phenotypic effects similar to each other except for tetracycline, which was classified as producing a phenotype similar to that caused by chloramphenicol, amoxicillin, and cefotaxime (Fig. 6a to f). However, this result was consistent with the fact that tetracycline, although primarily a protein synthesis inhibitor, has damaging effects on the cytoplasmic membrane (37, 38). It is also shown in Fig. 6g that out of the 69 acquired Raman spectra of E. coli cells treated with ciprofloxacin, (i) 33.3% (23/69) were classified as similar to nalidixic acid, a DNA synthesis inhibitor; (ii) 26.0% (18/69) were classified as similar to trimethoprim, also a DNA synthesis inhibitor; (iii) 17.4 (12/69) were classified as similar to ampicillin, a cell wall synthesis inhibitor; and (iv) 14.5% (10/69) were classified as similar to rifampin, an RNA synthesis inhibitor. Previous research has shown that although ciprofloxacin primarily inhibits DNA synthesis, it also affects various macromolecules, including RNA, and cell wall integrity (39). Thus, these results suggest that the discriminant analysis classifier model also has the potential to recognize multiple mechanisms of action of putative antibiotic compounds. The discriminant analysis results also suggested that phenotypes induced by nalidixic acid and kanamycin treatment have characteristics similar to those caused by ampicillin. It has been reported that some quinolones and aminoglycosides disorganize the cell wall (40). Clindamycin was ineffective against E. coli compared to other protein synthesis inhibitors used in this research, as indicated by the relatively high MIC (Table 1). However, at 3× MIC, clindamycin slowed E. coli growth significantly compared to the control culture, as was evident in OD600 measurements (data not shown). The clindamycin data also support the central hypothesis of this research: Raman spectroscopy can detect global phenotypic changes induced by an antibiotic compound regardless of mechanism or strength. Raman spectroscopy was able to detect the limited changes induced by clindamycin. In addition, chemometric analysis showed that the clindamycin-induced profile was most similar to those caused by other protein synthesis inhibitors. The fact that Raman spectroscopy was able to detect the changes induced by a weak antibiotic speaks to the exceptional sensitivity of this methodology. Furthermore, removing clindamycin from the analysis did not change fundamental findings and conclusions presented in this research.
FIG 6.
Prediction of antibiotic similarity for the unknown antibiotics. The prediction count for each antibiotic when treated as an unknown is shown in panels a to o. The name of the unknown is indicated above each subfigure. C, untreated control. All other abbreviations are defined in Table 1.
Impact of Raman spectrum normalization methodology on discriminant analysis results.
The effects of Raman spectrum normalization methods on the classification accuracy of the PC-DA model were also evaluated. As discussed earlier, normalization removes intensity variation that is related to experimental setup and not actual sample variation. Four commonly used normalization methods were examined. These include (i) vector normalization and normalizing Raman signal intensities over an entire spectrum, (ii) the signal intensity of the phenylalanine band at 1,003 cm−1, (ii) the signal intensity of the band at 1,450 cm−1, which originates from CHx vibrations abundant in biomass, and (iv) the signal resulting from the C-H stretching vibration from 2,900 to 3,100 cm−1. Prediction results using raw and normalized data are given in Table 5. Without spectrum normalization, the lowest prediction accuracy (43.8%) was observed. The best results were obtained when Raman spectra were normalized using vector normalization. It is noted that the case for vector normalization can also be made since it is relatively unbiased toward any single band or group of bands that could be affected by changes in the bacterial phenotype. However, results using other normalization methods were not substantially different, which highlights the robustness of the Raman spectroscopy-based discriminant analysis model and its ability to compensate for experimental variation. It is noted that the experimental data analyzed in this research were obtained by the same instrument within a relatively short time frame. Comparisons made on data obtained from different instruments and time frames would likely show a larger influence of normalization method on classification accuracy.
TABLE 5.
Impact of Raman spectra normalization methods
| Antibiotic | Class | % Correct predictiona |
||||
|---|---|---|---|---|---|---|
| No normalization | Vector normalization | 1,003 cm−1 | 1,445–1,455 cm−1 | 2,900–3,100 cm−1 | ||
| Chloramphenicol | PROT | 46.9 | 93.9 | 81.6 | 89.8 | 89.8 |
| Clindamycin | PROT | 23.6 | 83.6 | 61.8 | 58.2 | 87.3 |
| Gentamicin | PROT | 16.0 | 69.3 | 60.0 | 36.0 | 73.3 |
| Kanamycin | PROT | 13.0 | 63.0 | 40.7 | 37.0 | 61.1 |
| Streptomycin | PROT | 30.6 | 71.4 | 71.4 | 71.4 | 71.4 |
| Tetracycline | PROT | 11.3 | 75.5 | 34.0 | 37.7 | 67.9 |
| Ciprofloxacin | DNA | 62.3 | 47.8 | 47.8 | 36.2 | 34.8 |
| Nalidixic acid | DNA | 88.1 | 67.8 | 52.5 | 71.2 | 67.8 |
| Trimethoprim | DNA | 81.1 | 64.2 | 64.2 | 67.9 | 62.3 |
| Amoxicillin | CW | 63.9 | 41.0 | 57.4 | 42.6 | 42.6 |
| Ampicillin | CW | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Cefotaxime | CW | 61.4 | 38.6 | 40.4 | 40.4 | 38.6 |
| Rifampin | RNA | 0.0 | 3.5 | 3.5 | 3.5 | 1.8 |
| Rifapentine | RNA | 78.0 | 66.1 | 59.3 | 72.9 | 66.1 |
| Rifaximin | RNA | 80.0 | 80.0 | 86.2 | 83.1 | 78.5 |
| Overall accuracy (%) | 43.8 | 57.7 | 50.7 | 49.9 | 56.2 | |
| Randomb | 20.0 | 20.0 | 20.0 | 20.0 | 20.0 | |
Percentage of correct PC-DA prediction of antibiotic class for the spectra representing each antibiotic excluded from the training data set. Raman signal intensities were normalized using vector normalization and with respect to the signal intensity of the phenylalanine band at 1,003 cm−1, the signal intensity of the C-H band at 1,450 cm−1, and the signal intensity of the C-H stretching vibration region at 2,900 to 3,100 cm−1.
The probability of correct classifications by random assignment.
DISCUSSION
The results of this research have shown that Raman spectroscopy with appropriate chemometrics is a powerful tool for profiling the phenotypic response of bacteria to antibiotics. This technique will have significant value to antibiotic drug development research. With the relatively diverse data set analyzed, the ability of Raman spectroscopy to profile the effects of an unknown antibiotic compound on the phenotype of an E. coli culture was demonstrated. First, it was shown that Raman spectroscopy data could be used to discriminate cultured samples based on antibiotic treatment. This was achieved by performing discriminate analysis on Raman spectroscopy data with results validated using the leave-one-out cross-validation method. Second, it was shown that Raman spectroscopy with discriminate analysis could be applied to predict the phenotypic effects of a mechanistically unknown antibiotic compound on an E. coli culture. To demonstrate this capability, individual antibiotics were excluded from the model training data set and were tested as unknowns. In this approach, none of the spectra representing the excluded antibiotic were included in model building (32). This more realistically mimics real-life situations where the discriminant analysis model will have no prior knowledge of the new antibiotic compound being examined. In several papers published on the topic of Raman spectroscopy-based classification of bacterial samples, discriminant analysis results are usually presented as evidence of the ability of Raman spectroscopy to classify individual samples into predetermined classes (19, 24, 32, 41, 42). These discriminant analysis results are often cross-validated using the K-fold or leave-one-out method (32). Cross-validation (i) removes a subset of Raman spectra from the data set, (ii) reconstructs a discriminant model using remaining spectra, and then (iii) uses the model to predict the class of the removed spectra. This method of validation often produces positive results, because spectra from the class to which the removed sample belongs are still included in the model training data set. The critical question remains whether classification accuracy is as good for samples from a completely new antibiotic treatment not included in model training. For example, PC-DA with 50 PCs distinguished among cultures based on the class of antibiotic treatments with 83.6% accuracy estimated using leave-one-out cross-validation (Table 3). However, PC-DA with 50 PCs did not perform as well in predicting the class of new antibiotic (new to the model), with 48.0% overall accuracy (Table 4). In contrast, PC-DA with 9 PCs, which performed relatively poorly in the leave-one-out cross-validation (70.6%), performed better than PC-DA with 50 PCs when used to predict the group of unknown antibiotics, with 57.7% overall accuracy. Using a smaller number of PCs preserved the variance in the original data and was sufficient to develop a discriminant analysis model with good predictive capabilities. Using a smaller number of PCs (9 versus 50) also substantially reduced computation time. This is significant because computation time becomes a more critical issue as more antibiotics are added to the data set and chemometric analysis becomes more sophisticated.
Identifying the target and the mechanism of action of a putative antibiotic is difficult and time-consuming. The Raman spectroscopy-based phenotypic profiling system can aid this process by revealing important information about the phenotypic effects of the mechanistically unknown antibiotic compound. This information can be used to generate testable hypotheses and will further inform the search for the mechanism of action and cellular target(s) of the putative antibiotic compound. Indeed, as more antibiotic response profiles are added to the database, the accuracy and usefulness of the Raman spectroscopy-based phenotypic profiling system will improve. In addition, this database not only can become more diversified by adding data from additional antibiotic treatments but also has the potential to be expanded by including other microbial strains.
ACKNOWLEDGMENTS
A.A. was supported by research grants from the Institute for Critical Technologies and Applied Science (ICTAS) at Virginia Tech and from the Biodesign and Bioprocessing Research Center at Virginia Tech.
Footnotes
Published ahead of print 2 December 2013
REFERENCES
- 1.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8:423–435. 10.1038/nrmicro2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Futamura Y, Muroi M, Osada H. 2013. Target identification of small molecules based on chemical biology approaches. Mol. Biosyst. 9:897–914. 10.1039/c2mb25468a [DOI] [PubMed] [Google Scholar]
- 3.Toledo-Arana A, Solano C. 2010. Deciphering the physiological blueprint of a bacterial cell. Bioessays 32:461–467. 10.1002/bies.201000020 [DOI] [PubMed] [Google Scholar]
- 4.Silver LL. 2011. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 24:71–109. 10.1128/CMR.00030-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazas MD, Hancock REW. 2005. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 10:1245–1252. 10.1016/S1359-6446(05)03566-X [DOI] [PubMed] [Google Scholar]
- 6.Rix U, Superti-Furga G. 2009. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 5:616–624. 10.1038/nchembio.216 [DOI] [PubMed] [Google Scholar]
- 7.Hart CP. 2005. Finding the target after screening the phenotype. Drug Discov. Today 10:513–519. 10.1016/S1359-6446(05)03415-X [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Charles Gill J, Sang Lee H, Mann P, Zuck P, Timothy Meredith C, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Christopher Tan M, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem. Biol. 20:272–284. 10.1016/j.chembiol.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amini S, Tavazoie S. 2011. Antibiotics and the post-genome revolution. Curr. Opin. Microbiol. 14:513–518. 10.1016/j.mib.2011.07.017 [DOI] [PubMed] [Google Scholar]
- 10.Freiberg C, Fischer HP, Brunner NA. 2005. Discovering the mechanism of action of novel antibacterial agents through transcriptional profiling of conditional mutants. Antimicrob. Agents Chemother. 49:749–759. 10.1128/AAC.49.2.749-759.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. 2006. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313:1929–1935. 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- 12.Bandow J, Hecker M. 2007. Proteomic profiling of cellular stresses in Bacillus subtilis reveals cellular networks and assists in elucidating antibiotic mechanisms of action. Progress Drug Res. 64:79–101. 10.1007/978-3-7643-7567-6_4 [DOI] [PubMed] [Google Scholar]
- 13.Böddeker N, Bahador G, Gibbs C, Mabery E, Wolf J, Xu L, Watson J. 2002. Characterization of a novel antibacterial agent that inhibits bacterial translation. RNA 8:1120–1128. 10.1017/S1355838202024020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Yi ZB, Liang YZ. 2007. Main antimicrobial components of Tinospora capillipes, and their mode of action against Staphylococcus aureus. FEBS Lett. 581:4179–4183. 10.1016/j.febslet.2007.07.056 [DOI] [PubMed] [Google Scholar]
- 15.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. 2004. Multidimensional drug profiling by automated microscopy. Science 306:1194–1198. 10.1126/science.1100709 [DOI] [PubMed] [Google Scholar]
- 16.Futamura Y, Kawatani M, Kazami S, Tanaka K, Muroi M, Shimizu T, Tomita K, Watanabe N, Osada H. 2012. Morphobase, an encyclopedic cell morphology database, and Its use for drug target identification. Chem. Biol. 19:1620–1630. 10.1016/j.chembiol.2012.10.014 [DOI] [PubMed] [Google Scholar]
- 17.Huang WE, Li M, Jarvis RM, Goodacre R, Banwart SA. 2010. Shining light on the microbial world: the application of Raman microspectroscopy. Adv. Appl. Microbiol. 70:153–186. 10.1016/S0065-2164(10)70005-8 [DOI] [PubMed] [Google Scholar]
- 18.Moritz TJ, Taylor DS, Polage CR, Krol DM, Lane SM, Chan JW. 2010. Effect of cefazolin treatment on the nonresonant Raman signatures of the metabolic state of individual Escherichia coli cells. Anal. Chem. 82:2703–2710. 10.1021/ac902351a [DOI] [PubMed] [Google Scholar]
- 19.Notingher I. 2007. Raman spectroscopy cell-based biosensors. Sensors 7:1343–1358. 10.3390/s7081343 [DOI] [Google Scholar]
- 20.López-Díez EC, Goodacre R. 2004. Characterization of microorganisms using UV resonance Raman spectroscopy and chemometrics. Anal. Chem. 76:585–591. 10.1021/ac035110d [DOI] [PubMed] [Google Scholar]
- 21.Goodacre R, Timmins EM, Burton R, Kaderbhai N, Woodward AM, Kell DB, Rooney PJ. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology 144:1157–1170. 10.1099/00221287-144-5-1157 [DOI] [PubMed] [Google Scholar]
- 22.Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, Puppels GJ. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255–271. 10.1016/S0167-7012(02)00127-6 [DOI] [PubMed] [Google Scholar]
- 23.Athamneh AIM, Senger RS. 2012. Peptide-guided surface-enhanced Raman scattering probes for localized cell composition analysis. Appl. Environ. Microbiol. 78:7805–7808. 10.1128/AEM.02000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall EK, Singer GA, Polzl M, Hammerle I, Schwarz C, Daims H, Maixner F, Battin TJ. 2011. Looking inside the box: using Raman microspectroscopy to deconstruct microbial biomass stoichiometry one cell at a time. ISME J. 5:196–208. 10.1038/ismej.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster KC, Urlaub E, Gapes JR. 2000. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J. Microbiol. Methods 42:29–38. 10.1016/S0167-7012(00)00169-X [DOI] [PubMed] [Google Scholar]
- 26.Walter A, Reinicke M, Bocklitz T, Schumacher W, Rösch P, Kothe E, Popp J. 2011. Raman spectroscopic detection of physiology changes in plasmid-bearing Escherichia coli with and without antibiotic treatment. Anal. Bioanal. Chem. 400:2763–2773. 10.1007/s00216-011-4819-4 [DOI] [PubMed] [Google Scholar]
- 27.Neugebauer U, Schmid U, Baumann K, Holzgrabe U, Schmitt M, Popp J. Towards an understanding of the mode of action of fluoroquinolone drugs, poster. Abstr. Eur. Conf. Biomed. Optics, poster 6633-68. 2007 [Google Scholar]
- 28.Neugebauer U, Schmid U, Baumann K, Holzgrabe U, Ziebuhr W, Kozitskaya S, Kiefer W, Schmitt M, Popp J. 2006. Characterization of bacterial growth and the influence of antibiotics by means of UV resonance Raman spectroscopy. Biopolymers 82:306–311. 10.1002/bip.20447 [DOI] [PubMed] [Google Scholar]
- 29.López-Díez EC, Winder CL, Ashton L, Currie F, Goodacre R. 2005. Monitoring the mode of action of antibiotics using Raman spectroscopy: investigating subinhibitory effects of amikacin on Pseudomonas aeruginosa. Anal. Chem. 77:2901–2906. 10.1021/ac048147m [DOI] [PubMed] [Google Scholar]
- 30.Moritz TJ, Polage CR, Taylor DS, Krol DM, Lane SM, Chan JW. 2010. Evaluation of Escherichia coli cell response to antibiotic treatment by use of Raman spectroscopy with laser tweezers. J. Clin. Microbiol. 48:4287–4290. 10.1128/JCM.01565-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efrima S, Zeiri L. 2009. Understanding SERS of bacteria. J. Raman Spectrosc. 40:277–288. 10.1002/jrs.2121 [DOI] [Google Scholar]
- 32.Bocklitz T, Walter A, Hartmann K, Rösch P, Popp J. 2011. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 704:47–56. 10.1016/j.aca.2011.06.043 [DOI] [PubMed] [Google Scholar]
- 33.Fisher RA. 1936. The use of multiple measurements in taxonomic problems. Ann. Hum. Genet. 7:179–188. 10.1111/j.1469-1809.1936.tb02137.x [DOI] [Google Scholar]
- 34.Ringner M. 2008. What is principal component analysis? Nat. Biotechnol. 26:303–304. 10.1038/nbt0308-303 [DOI] [PubMed] [Google Scholar]
- 35.Chapelle O, Vapnik V, Bousquet O, Mukherjee S. 2002. Choosing multiple parameters for support vector machines. Mach. Learn. 46:131–159. 10.1023/A:1012450327387 [DOI] [Google Scholar]
- 36.Sullivan R. 2012. Introduction to data mining for the life sciences. Humana Press, Totowa, NJ [Google Scholar]
- 37.Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44:827–834. 10.1128/AAC.44.4.827-834.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pato ML. 1977. Tetracycline inhibits propagation of deoxyribonucleic acid replication and alters membrane properties. Antimicrob. Agents Chemother. 11:318–323. 10.1128/AAC.11.2.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma I, Rohilla A, Khuller GK. 1999. Alterations in macromolecular composition and cell wall integrity by ciprofloxacin in Mycobacterium smegmatis. Lett. Appl. Microbiol. 29:113–117. 10.1046/j.1365-2672.1999.00597.x [DOI] [PubMed] [Google Scholar]
- 40.Bryskier A. 1993. Fluoroquinolones: mechanisms of action and resistance. Int. J. Antimicrob. Agents 2:151–183. 10.1016/0924-8579(93)90052-7 [DOI] [PubMed] [Google Scholar]
- 41.Schmid U, Rösch P, Krause M, Harz M, Popp J, Baumann K. 2009. Gaussian mixture discriminant analysis for the single-cell differentiation of bacteria using micro-Raman spectroscopy. Chemometr. Intell. Lab. Syst. 96:159–171. 10.1016/j.chemolab.2010.07.008 [DOI] [Google Scholar]
- 42.Notingher I, Jell G, Notingher PL, Bisson I, Tsigkou O, Polak JM, Stevens MM, Hench LL. 2005. Multivariate analysis of Raman spectra for in vitro non-invasive studies of living cells. J. Mol. Struct. 747:179–185. 10.1016/j.molstruc.2004.12.046 [DOI] [Google Scholar]