Abstract
Resistance of Enterococcus faecalis against antimicrobial peptides, both of host origin and produced by other bacteria of the gut microflora, is likely to be an important factor in the bacterium's success as an intestinal commensal. The aim of this study was to identify proteins with a role in resistance against the model antimicrobial peptide bacitracin. Proteome analysis of bacitracin-treated and untreated cells showed that bacitracin stress induced the expression of cell wall-biosynthetic proteins and caused metabolic rearrangements. Among the proteins with increased production, an ATP-binding cassette (ABC) transporter with similarity to known peptide antibiotic resistance systems was identified and shown to mediate resistance against bacitracin. Expression of the transporter was dependent on a two-component regulatory system and a second ABC transporter, which were identified by genome analysis. Both resistance and the regulatory pathway could be functionally transferred to Bacillus subtilis, proving the function and sufficiency of these components for bacitracin resistance. Our data therefore show that the two ABC transporters and the two-component system form a resistance network against antimicrobial peptides in E. faecalis, where one transporter acts as the sensor that activates the TCS to induce production of the second transporter, which mediates the actual resistance.
INTRODUCTION
Enterococcus faecalis is a member of the normal gut microflora in animals and humans. While it is mostly a harmless commensal, opportunistic infections, particularly of immunocompromised hospital patients, are a major health concern. Due to the rise of vancomycin-resistant strains (VREs), treatment of such infections is becoming increasingly difficult, and a better understanding of the resistance of E. faecalis against cell wall-targeting antibiotics is urgently needed. In addition to vancomycin resistance, enterococci display a high intrinsic resistance against other inhibitors of cell wall biosynthesis, including cephalosporins and bacitracin (1–3). Bacitracin is not used clinically for the treatment of enterococcal infections, yet resistance against this and other peptide antibiotics is nevertheless biologically relevant in the human intestinal environment. For example, gut bacteria are exposed to defensins that are part of the innate immune defense in the gastrointestinal tract (4). Additionally, other members of the microflora, especially lactic acid bacteria, produce a range of bacteriocins as a means of interspecies competition (5). Thus, resistance of E. faecalis against peptide antibiotics is likely to be important for the success of the bacterium in the human host, which in turn affects the risk of opportunistic infections.
In other Gram-positive bacteria, several resistance mechanisms that combat the action of cell wall-active peptides have been identified. One strategy can be alterations in the cell's surface charge, for example, by alanylation of teichoic acids catalyzed by the DltABCD system (6, 7). Accordingly, in a recent study, the dlt operon of E. faecalis was shown to be upregulated in response to bacitracin and vancomycin (8). Alternatively, upregulation of the enzyme inhibited by the antibiotic, e.g., in the case of bacitracin, undecaprenyl-pyrophosphate phosphatases, can confer resistance, as has been shown for Bacillus subtilis and E. faecalis (2, 9). A third resistance mechanism is the expression of specific ATP-binding cassette (ABC) transporters. One type of such transporters possesses permeases with six transmembrane helices and is exemplified by the bacitracin transporter BcrAB, which is used for self-immunity by bacitracin-producing strains of Bacillus licheniformis (10) but has also been shown to confer acquired high-level bacitracin resistance to E. faecalis (3). An unrelated group of transporters, characterized by permeases with 10 transmembrane helices and a large extracellular domain, are involved in resistance against a range of antimicrobial peptides. The best-understood example is BceAB of B. subtilis, which confers resistance against bacitracin, mersacidin, and actagardine (11, 12). These transporters form the Peptide-7-Exporter family in the Transport Classification Database and are collectively referred to as BceAB-type systems (13, 14). They are usually found in the genetic neighborhood of a two-component regulatory system (TCS; BceRS type) that controls expression of the transporter operon (15, 16). Importantly, these transporters not only mediate resistance but play an additional role as sensors, because the TCSs alone are unable to detect their substrate peptides (11, 17). Together, transporters and TCSs form peptide antibiotic resistance modules and are found widely distributed among low-G+C Gram-positive organisms (13, 16). A comparative genomics analysis identified two BceAB-type transporters in the genome of E. faecalis (16), but no functional data are available on these systems, nor has a TCS been identified as the corresponding regulator.
In accordance with the need for a better understanding of the response of E. faecalis to antibiotics that inhibit cell wall synthesis, a recent transcriptomic study identified a large number of genes that were upregulated after exposure to ampicillin, cephalothin, vancomycin, and bacitracin (8). Of these compounds, bacitracin elicited the broadest response, affecting genes with functions in cell wall maintenance, metabolism, and transport processes. Here we report on a proteomic analysis of the bacitracin response of E. faecalis. Among the differentially produced proteins, we again identified those involved in cell wall maintenance and energy metabolism as important factors. Additionally, one BceAB-type transporter was found at higher levels following bacitracin exposure, and subsequent investigations revealed the existence of a regulatory and resistance network comprised of two such transporters and one TCS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli MC1061 was used for cloning with vector pTCVlac; strains DH5α and XL1-Blue were used for all other cloning. E. coli and B. subtilis were grown routinely in Luria-Bertani (LB) medium at 37°C with agitation (200 rpm). Enterococcus faecalis was grown routinely in brain heart infusion (BHI) at 37°C without agitation. E. faecalis was transformed by electroporation as previously described (18). B. subtilis was transformed by natural competence as previously described (19). Selective media contained ampicillin (100 μg ml−1), chloramphenicol (10 μg ml−1 for E. coli; 15 μg ml−1 for E. faecalis; 5 μg ml−1 for B. subtilis), kanamycin (50 μg ml−1 for E. coli; 1,000 μg ml−1 for E. faecalis), 1 μg ml−1 erythromycin with 25 μg ml−1 lincomycin (for macrolide-lincosamide-streptogramin B [MLS] resistance), or spectinomycin (150 μg ml−1 for E. faecalis; 100 μg ml−1 for B. subtilis). Bacitracin was supplied as the Zn2+ salt. Solid media contained 1.5% (wt/vol) agar. Growth was measured as optical density at 600 nm (OD600).
Proteomic analysis.
For proteomic analyses, exponentially growing cultures (OD600 = 0.4) of E. faecalis V583 in BHI medium were exposed to 64 μg ml−1 bacitracin for 1 h. The cytoplasmic protein fraction was extracted and analyzed by isoelectric focusing, followed by second-dimension (2D) SDS-PAGE. Differentially produced proteins were identified by mass spectrometry. Detailed experimental and analytical procedures are described in the supplemental text.
Construction of plasmids and genetic techniques.
All primer sequences used for cloning or transcriptional start site mapping are listed in Table S2 in the supplemental material.
Transcriptional promoter fusions to lacZ in E. faecalis were constructed in the vector pTCV-lac (20). All fragments were cloned via the EcoRI and BamHI sites of the vector. The exact regions amplified as well as the primers used are given in Tables S1 and S2 in the supplemental material. For complementation of the strain of E. faecalis with a deletion of the transporter operon EF2050-2049, the entire operon, including its native promoter region, was cloned into the EcoRI and BamHI sites of vector pAT28 (21). Transcriptional promoter fusions in B. subtilis of EF2752 (PEF2752) or EF2050 (PEF2050) to the bacterial luciferase genes (luxABCDE) were cloned into the EcoRI and SpeI sites of the vector pAH328 (22), creating plasmids pCF135 and pCF134. The exact regions contained in the construct are given in Table S1 in the supplemental material.
Plasmids for heterologous, xylose-inducible expression of EF2752-2751 or EF2050-2049 in B. subtilis (pCF129 and pCF130) were constructed in the vector pXT using the BamHI and EcoRI restriction sites, placing the genes under the control of the vector's xylA promoter (23).
The construct for the heterologous expression of EF0926-0927 in B. subtilis was cloned according to the BioBrick standard (24). To facilitate constitutive expression in B. subtilis, a BioBrick of the bceRS operon promoter, PbceR, of B. subtilis was amplified and cloned into the EcoRI and SpeI sites of vector pSB1A3, creating pCF144. A BioBrick of EF0926-0927 containing an optimal Shine-Dalgarno sequence for B. subtilis was similarly cloned into pSB1A3 via EcoRI and SpeI, creating plasmid pCF143. Assembly of both BioBricks in vector pBS2E (25) created plasmid pCF145, where expression of EF0926-0927 is controlled by PbceR.
Constructs for unmarked gene deletions in E. faecalis were cloned in the vector pLT06 (26). For each gene or operon to be deleted, 700- to 1,000-bp fragments located immediately before the start codon of the first gene (“up” fragment) and after the stop codon of the last gene (“down” fragment) were amplified. The primers were designed to create a 17- to 20-bp overlap between the PCR products (see Table S2 in the supplemental material), facilitating fusion of the fragments by PCR overlap extension (27) and subsequent cloning into pLT06. Gene deletions were performed as previously described (26).
All constructs were checked for PCR fidelity by sequencing, and all created strains were verified by PCR using appropriate primers.
To determine the transcriptional start sites of the EF2050-2049 and EF2752-2751 operons, total RNA was isolated from E. faecalis JH2-2 using a RNeasy minikit and QIAshredder columns (Qiagen) according to the manufacturer's instructions. DNA was removed with a TURBO DNA-free kit (Ambion). Transcriptional start sites were determined by 5′ rapid amplification of cDNA ends (5′ RACE) as described previously (28). The sets of nested primers used are listed in Table S2 in the supplemental material.
Antimicrobial susceptibility assays.
For antibiotic susceptibility assays, all cultures were grown in Mueller-Hinton (MH) medium. MICs of bacitracin were determined by broth dilution assays. Freshly grown colonies of E. faecalis were suspended in sterile saline to 0.5 McFarland standard turbidity and diluted 1:1,000 into MH medium containing serial 2-fold dilutions of bacitracin. For B. subtilis, freshly grown overnight cultures in MH broth were used as inoculum at a dilution of 1:500. After 24 h of incubation, the MIC was scored as the lowest concentration where no growth was observed. MICs obtained from broth dilution assays were subsequently confirmed using bacitracin Etest strips (bioMérieux) according to the manufacturer's instructions.
Additionally, bacitracin susceptibility of B. subtilis was determined by growth inhibition of exponentially growing cultures in LB. Strains were inoculated 1:500 from overnight cultures and grown in 100-μl volumes in a 96-well plate. Cultures were incubated at 37°C with shaking (medium intensity) in a Synergy2 multimode microplate reader from BioTek controlled by the software Gen5, and OD600 was monitored every 5 min. At an OD600 of 0.02 (4 to 5 doublings since inoculation; corresponding to an OD600 of 0.1 in cuvettes with a 1-cm light path length), bacitracin was added to a final concentration of 2, 4, 8 or 16 μg ml−1 with one well left untreated, and growth was monitored for another 2.5 h.
β-Galactosidase assays.
Qualitative assays for induction of LacZ reporter constructs in E. faecalis were performed by the disk diffusion method. A suspension of cells from freshly grown colonies at 0.5 McFarland standard turbidity was spread onto MH agar containing appropriate antibiotics for selection and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (100 μg ml−1) using cotton swabs. Filter discs containing 5 μl antibiotic test solution (100 mg ml−1) were placed onto the plates. After overnight incubation, plates were scored for formation of a blue ring around the filter discs.
For quantitative assays, exponentially growing cells in BHI medium were exposed to different concentrations of bacitracin for 1 h as previously described (2). β-Galactosidase activities were assayed in permeabilized cells as described previously and were expressed in Miller units (MU) (29, 30).
Luciferase assays.
Luciferase activities of B. subtilis strains containing pCF135 and pCF134 were assayed using the microplate reader described above. LB medium was inoculated 1:500 from overnight cultures, and each strain was grown in 100-μl volumes in a 96-well plate. Cultures were incubated at 37°C with shaking (medium intensity), and the OD600 was monitored every 10 min. At an OD600 of 0.02 (see above), bacitracin was added to a final concentration of 1, 3, or 10 μg ml−1, with one well left untreated. Cultures were further incubated for 2 h, and the OD600 and luminescence (endpoint reads; 1-s integration time; sensitivity, 200) were monitored every 5 min. OD600 values were corrected using wells containing 100 μl LB medium as blanks. Raw luminescence output (relative luminescence units [RLU]) was normalized to cell density by dividing each data point by its corresponding corrected OD600 value (RLU/OD).
RESULTS
Proteome analysis of the bacitracin stress response of E. faecalis.
To investigate the response of E. faecalis to bacitracin, we analyzed the proteome of strain V583, whose genome has been fully sequenced, after 1 h exposure to 64 μg ml−1 bacitracin, which corresponded to the strain's MIC, compared to an untreated control. A detailed description of the findings is presented in the supplemental text. The 2D gels and detailed analyses of protein spots are available in Fig. S1 and in Tables S3 and S4, respectively, in the supplemental material. In brief, as expected, a number of proteins involved in cell wall metabolism were found in increased quantities after bacitracin stress. Additionally, several enzymes for energy metabolism or fatty acid synthesis were differentially expressed, indicating metabolic rearrangements in response to bacitracin. Further proteins with increased production after bacitracin exposure likely indicated a general response to stress. The two most strongly upregulated proteins are homologous to a protein of unknown function, YvlB of B. subtilis, and to cobyric acid synthase. Their roles in the response to bacitracin are not clear. The third strongest effect was observed with EF2050. This protein is the ATPase component of a BceAB-like ABC transporter (EF2050-2049), a group of proteins that to date have been identified only in the context of resistance against peptide antibiotics (13, 16). As mentioned above, operons for two such transporters were previously identified in the genome of E. faecalis by a comparative genomics study, namely, EF2050-2049 and EF2752-2751 (16). Although our proteomic study identified only the former transporter, both loci were found to be upregulated in response to bacitracin by transcriptome analysis (8). We therefore decided to investigate these two transporters in more detail, regarding both their role in bacitracin resistance and their regulation.
Identification of orthologous genes in E. faecalis JH2-2.
Because strain V583 is a vancomycin-resistant clinical isolate, we chose the laboratory strain JH2-2 for all further studies. For this, we first needed to identify the genes corresponding to EF2050-2049 and EF2752-2751 from strain V583. Using PCR primers designed according to the V583 genome sequence, we readily obtained amplicons of the correct size from strain JH2-2. DNA sequencing confirmed that all four genes possessed the same sequence in both strains. For simplicity and consistency with the existing literature, we maintained the strain V583 nomenclature for all genes from strain JH2-2 throughout this study.
As described in the introduction, BceAB-type transporters are usually regulated by a BceRS-like TCS encoded in the genomic neighborhood of the transporter. However, neither of the two transporters investigated here possessed such a regulatory system. Because the sensor kinases of these TCSs have a characteristic domain architecture of two transmembrane helices with a very short intervening extracellular linker (ca. 3 to 10 amino acids) and no additional cytoplasmic domains besides the catalytic domains for autophosphorylation, candidate TCSs can be identified via genome analyses (16). Indeed, the genome of E. faecalis V583 encodes a single BceRS-like TCS, EF0926-0927, and identical genes were identified by PCR and sequencing in strain JH2-2. Schematics of all three loci are shown in Fig. 1.
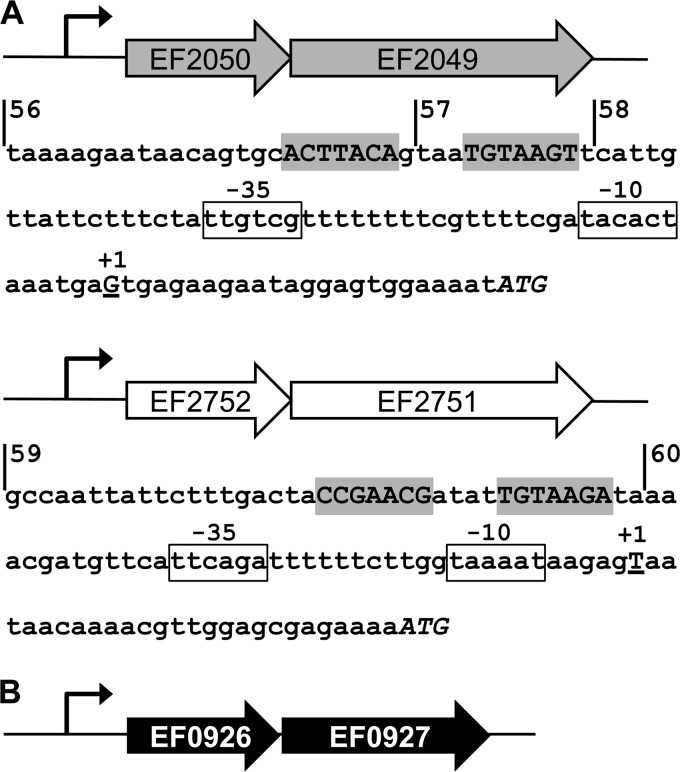
FIG 1.
Schematic of operon structures and promoter regions in the resistance circuit. (A) Operons for the ABC transporters. In each, the first gene encodes the ATPase and the second gene the permease; the bent arrow indicates the promoter. The nucleotide sequences of the promoter regions are given below the respective schematics. The proposed response regulator binding site is highlighted in gray, the −10 and −35 elements are boxed, the transcriptional start site (+1) determined by 5′ RACE is shown as an underlined capital letter, and the translational start is shown in italicized capitals. Vertical lines show the beginning of fragments used to construct transcriptional reporter fusions, and the numbers of the derived constructs are given. (B) Operon for the two-component system. The first gene encodes the response regulator, and the second gene encodes the sensor kinase; the promoter region is indicated by a bent arrow. No putative regulator binding site was identified in the promoter, and therefore no sequence is shown.
The transporter EF2050-2049, which was identified as bacitracin induced in our proteome analysis, is a member of phylogenetic group VII of BceAB-type transporters, which also includes the YxdLM system of B. subtilis (16). The second transporter, EF2752-2751, could not be assigned to any group. However, its branch of the phylogenetic tree includes four other transporters, one from E. faecium and three from Lactobacillus species (16), showing that the occurrence of close homologues is not restricted to E. faecalis. Interestingly, one of these transporters was recently shown to be involved in nisin resistance of Lactobacillus casei (31).
Both transporters are induced by bacitracin.
To confirm bacitracin-induced expression of the transporter operon EF2050-2049, and to test if EF2752-2751 was also inducible under these conditions, we first constructed transcriptional fusions of the promoter regions PEF2050 and PEF2752 to a lacZ reporter gene. The transcriptional start sites of both operons were determined by 5′ RACE, and putative −10 and −35 promoter elements could be identified within a suitable distance upstream of the +1 position (Fig. 1A). Additionally, sequence analysis of the regions upstream of the −35 element revealed the presence of inverted repeats with similarity to the binding consensus of BceR-like regulators (16) (Fig. 1A). The transcriptional fusions were therefore designed to contain all of these motifs (fragments 56 for PEF2050 and 59 for PEF2752 [Fig. 1A]).
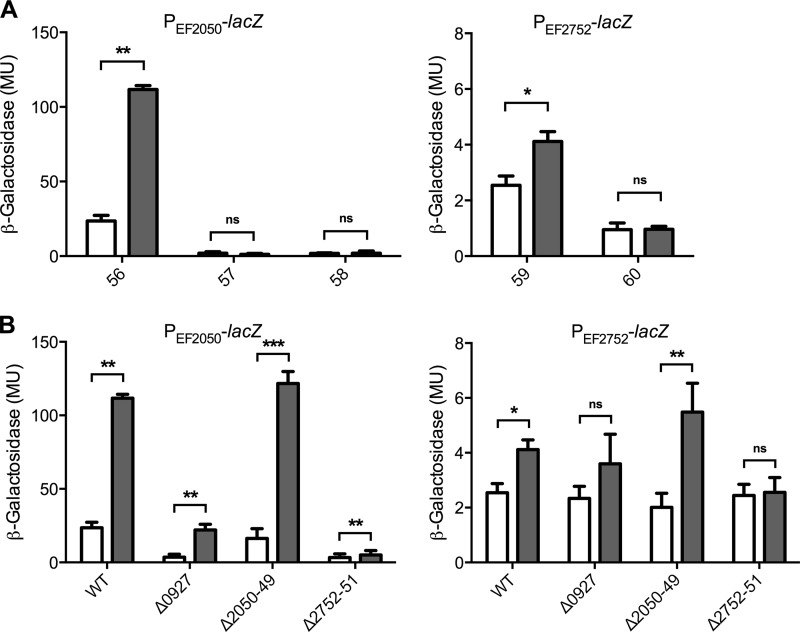
Initial assays were performed by disc diffusion on agar plates inoculated with a lawn of the reporter strains. Consistent with the proteome analysis, blue circles indicating promoter induction were observed around filter discs containing bacitracin (data not shown). Because BceAB-like transporters normally recognize several different substrates (12, 13, 17), we also tested several other cell wall-active antibiotics. Nisin, gallidermin, vancomycin, teicoplanin, and penicillin G did not elicit a response, but the lantibiotic mersacidin was able to induce both promoters (data not shown). These results were confirmed by quantitative β-galactosidase assays in liquid cultures and showed that maximum induction of both promoters was obtained at 20 to 50 μg ml−1 mersacidin or 4 μg ml−1 bacitracin but not with any of the other antibiotics tested (Fig. 2A and data not shown). Because mersacidin is not commercially available and bacitracin elicited the more sensitive response, all subsequent assays were performed using bacitracin as an inducer. Following 1 h exposure of exponentially growing cultures to 4 μg ml−1 bacitracin, PEF2050-lacZ was induced approximately 5-fold from 23 Miller units (MU) to 112 MU (Fig. 2A, left). PEF2752-lacZ was also induced by bacitracin, but the overall activities were low (2 to 4 MU) and induction was only 2-fold (Fig. 2B, right), explaining why this transporter had not been identified in the proteome analysis.
FIG 2.
Induction of the transporter operons by bacitracin. Promoter regions of the transporter operons EF2050-2049 (left) and EF2752-2751 (right) were fused to lacZ, and resulting strains of E. faecalis JH2-2 were assayed for β-galactosidase activity, expressed in Miller units (MU), after 1 h exposure of exponentially growing cultures to 0 μg ml−1 (white bars) or 4 μg ml−1 (gray bars) bacitracin. (A) Successive truncations from the 5′ end of the promoter region. Bars are labeled by the number of each construct, as shown in Fig. 1. Constructs 56 and 59 contain the full promoter region. (B) Full-length constructs of the promoter fusions from panel A assayed in the wild-type (WT; same data as in panel A) and mutant backgrounds. The genes deleted in each strain are indicated by the locus tags below the bars. Results are means plus standard deviations for three to four biological replicates. The significance of induction was calculated for each strain by one-tailed, pairwise t test analysis and is indicated by one (P < 0.05), two (P < 0.005), or three (P < 0.001) asterisks. ns, not significant.
Next, we constructed a series of truncated promoter fusions lacking part of (fragment 57) or the entire (fragments 58 and 60) proposed regulator binding sites (Fig. 1A). All strains carrying the derived promoter-lacZ fusions displayed activities near the detection limit (ca. 1 MU) and no induction by bacitracin (Fig. 2A), showing that these sequences were required for expression and thus indeed presented likely binding sites for a BceR-like regulator.
Both transporters and the TCS are required for full bacitracin resistance of E. faecalis.
To investigate the role of the two transporters in bacitracin resistance, we created unmarked deletions of the entire coding region of each transporter. Additionally, the gene for the sensor kinase, EF0927, was deleted. Despite repeated attempts, no deletion of the regulator gene EF0926 could be achieved. All three deletion strains displayed reduced bacitracin resistance compared to the wild-type strain JH2-2, albeit to different extents (Table 1). The strongest effect with a 2- to 4-fold-increased sensitivity was observed for EF2050-2049, while deletion of EF2752-2751 and of EF0927 resulted in changes of only up to 2-fold in the MIC. The very minor effect of the EF0927 deletion strain can possibly be explained by deletion of the sensor kinase alone with the regulator still present, as discussed in more detail below. Interestingly, deletion of both transporters simultaneously did not further reduce the MIC compared to deletion of EF2050-2049 alone. The lack of an additive effect might suggest that both transporters participate in the same pathway, rather than acting independently of each other. Importantly, none of the deletion strains displayed a growth defect compared to the wild-type strain (Table 1), showing that the differences in MIC were specifically due to loss of resistance determinants and not to altered growth kinetics. Complementation of the EF2050-2049 deletion mutant by supplying the transporter operon in trans (strain DLEf16) restored bacitracin resistance (Table 1). The higher MIC compared to the wild-type strain is most likely due to the increase in copy number by the plasmid-based complementation strategy and supports the role of EF2050-2049 in bacitracin resistance of E. faecalis.
TABLE 1.
Bacitracin sensitivity of E. faecalis strains
| Strain or genotype | MIC (μg ml−1) |
Growth rate (h−1)b | |
|---|---|---|---|
| Broth dilutiona | Etest | ||
| JH2-2 | 32 | 32 | 1.06 ± 0.067 |
| ΔEF0927 | 16–32 | 24 | 1.2 ± 0.377 |
| ΔEF2050-2049 | 8–16 | 8 | 1.11 ± 0.135 |
| ΔEF2752-2751 | 16 | 16 | 1.14 ± 0.146 |
| ΔEF2050-2049 ΔEF2752-2751 | 8 | 8 | 1.08 ± 0.033 |
| DLEf16 | 64 | 64 | ND |
Results are from three independent broth dilution experiments; where a range of concentrations is given, results varied between replicates.
Results are means ± standard errors from three to six independent experiments. ND, not determined.
Transporters and TCS form a regulatory network.
BceAB-type transporters are often required for their own regulation by acting as the actual sensors of antimicrobial peptides that somehow communicate with the TCS to trigger activation of the signaling cascade (11, 17). In such cases, expression of the transporter operon is abolished in strains carrying transporter deletions (11, 12, 32). In some bacteria, exemplified by Staphylococcus aureus, two separate transporters exist, where one acts as the sensor, while the second is responsible for resistance (17, 32). We therefore wanted to investigate the role of the two enterococcal transporters in regulation of their own promoters. Additionally, the TCS EF0926-0927 had so far only been implicated in regulation of the transporters based on sequence predictions, which had to be validated experimentally.
Deletion of the sensor kinase EF0927 drastically reduced the expression levels of PEF2050-lacZ to less than 20% of wild-type activities (Fig. 2B, left). Interestingly, a significant induction by bacitracin could still be observed, which was again likely due to the presence of the response regulator as described below. Expression of PEF2752-lacZ was also reduced by deletion of the sensor kinase, but due to the overall low activities of this reporter, the differences were less pronounced. Nevertheless, the significant induction by bacitracin observed in the wild type was lost in the EF0927 deletion strain (Fig. 2B, right). These data show that the TCS EF0926-0927 indeed acts as the regulator for both transporter operons.
Deletion of the transporter EF2050-2049 had no effect on expression of either transporter (Fig. 2B), indicating that this transporter's role was restricted to mediating resistance. In contrast, deletion of EF2752-2751 severely reduced the activities of both lacZ fusions, and bacitracin-dependent induction was lost (Fig. 2B). Thus, the second transporter represents the antibiotic sensor of the resistance network.
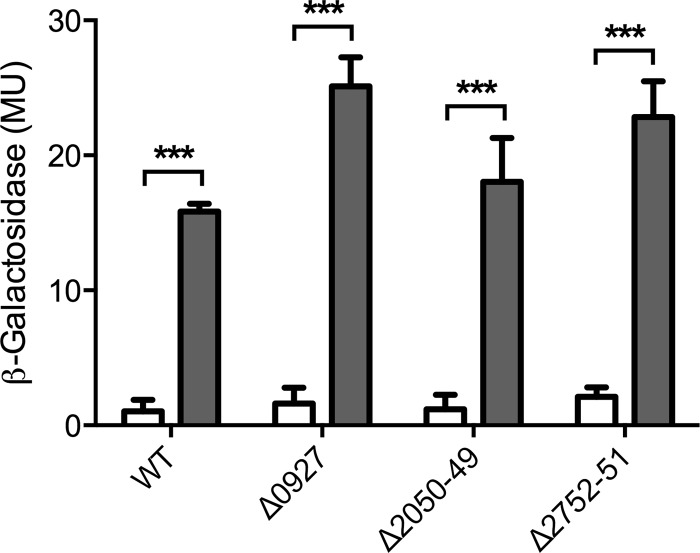
In B. subtilis, expression of BceRS, the TCS regulating bceAB expression, is not induced by bacitracin (33). However, the homologous system BraRS from S. aureus (referred to as NsaRS in reference 34) was shown to be upregulated in response to nisin, one of its substrate peptides (34). To test if EF0926-0927 was inducible by bacitracin, we constructed a transcriptional fusion of the promoter PEF0926 to lacZ and introduced it into E. faecalis JH2-2 and derived deletion strains. Exposure of exponentially growing cells to bacitracin resulted in a strong upregulation of promoter activities from 1 to 16 MU (Fig. 3). Interestingly, deletion of neither the TCS nor the two transporters affected promoter activities or bacitracin-dependent induction (Fig. 3), showing that regulation of the TCS is mediated by an as-yet-unidentified additional regulator and not due to autoregulation. Importantly, this regulation by a factor external to the resistance network may offer an explanation for the mild phenotypes of the EF0927 deletion strain compared to the transporter mutants: in the ΔEF0927 background, expression of the response regulator gene EF0926 is still upregulated in the presence of bacitracin. Because in the absence of their cognate sensor kinase many response regulators can be efficiently phosphorylated and thus activated by small-molecule phospho-donors such as acetyl-phosphate (35, 36), this increased production of EF0926 may indirectly lead to an induction of its target promoters by bacitracin.
FIG 3.
Induction of the two-component system operon by bacitracin. The promoter region of the two-component system operon EF0926-0927 was fused to lacZ and introduced into wild-type E. faecalis JH2-2 (WT) and mutant backgrounds. The resulting strains were assayed for β-galactosidase activity, expressed in Miller units (MU), after 1 h exposure of exponentially growing cultures to 0 μg ml−1 (white bars) or 4 μg ml−1 (gray bars) bacitracin. The genes deleted in each strain are indicated by locus tags below the bars. Results are means plus standard deviations for three biological replicates. The significance of induction and strain differences was calculated across the entire data set by two-way analysis of variance (ANOVA). Significant effects of bacitracin compared to uninduced cells are indicated by three asterisks (P < 0.001); the different mutant backgrounds caused significant differences between strains (P = 0.018; not depicted in the graph).
EF2050-2049 can mediate bacitracin resistance in B. subtilis.
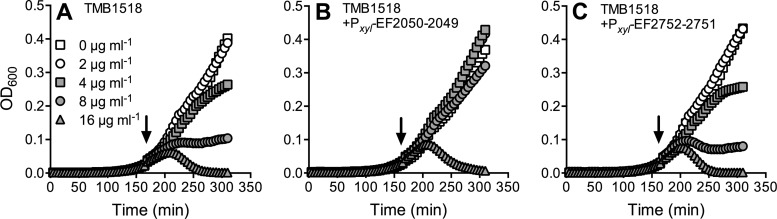
Because of the mild effects of the gene deletions generated in E. faecalis, we next attempted to transfer parts of the identified resistance network to B. subtilis to confirm the individual roles of the components in bacitracin resistance. As a chassis, we employed a strain of B. subtilis W168 carrying unmarked deletions of all three endogenous Bce-like modules (bceRS-bceAB, psdRS-psdAB, and yxdJK-yxdLM-yxeA). This strain, TMB1518, has been developed and validated as an appropriate platform to investigate resistance mechanisms against inhibitors of cell wall synthesis and the associated regulatory pathways from E. faecalis in a genetically highly accessible set-up (a detailed description will be published elsewhere). Expression of the EF2050-2049 operon under the control of a xylose-inducible promoter increased the MIC for bacitracin of TMB1518 from 2 μg ml−1 to 4 to 8 μg ml−1 (from 1 to 1.5 μg ml−1 to 3 μg ml−1 when determined by Etest assays), while expression of EF2752-2751 was unable to confer any resistance. As observed before for E. faecalis, the differences in MIC were again small. We therefore chose a different approach, assaying for growth inhibition of exponentially growing cultures that were challenged with increasing antibiotic concentrations, which can provide a more sensitive assay for cell wall-active antibiotics. B. subtilis strain TMB1518 as well as its derivative carrying the EF2752-2751 expression construct were unaffected by 2 μg ml−1 bacitracin, while 4 μg ml−1 or 8 μg ml−1 increasingly inhibited growth (Fig. 4A and C). In contrast, the strain harboring the expression construct for EF2050-2049 was not affected by concentrations up to 4 μg ml−1 and showed only slight growth inhibition at 8 μg ml−1 (Fig. 4B). Exposure to 16 μg ml−1 caused cell lysis in all strains tested (Fig. 4). These data confirm that EF2050-2049 is indeed directly capable of mediating bacitracin resistance, not only in E. faecalis but also in the heterologous host B. subtilis. EF2752-2751, on the other hand, is not directly responsible for bacitracin resistance.
FIG 4.
Transfer of bacitracin resistance to B. subtilis. Strain TMB1518 (A) and derived strains carrying expression constructs of the transporter operon EF2050-2049 (B) or EF2752-2751 (C) were grown to exponential phase and challenged with bacitracin, and growth was monitored as optical density (OD600). The time point of bacitracin addition is indicated by the arrow; concentrations are given in panel A. Representative results of two or three independent experiments are shown. Experiments were carried out in a 100-μl culture volume in 96-well plates; thus, OD values cannot be directly compared to measurements made in cuvettes with a 1-cm light path length.
EF2752-2751 and the TCS are sufficient for promoter induction in B. subtilis.
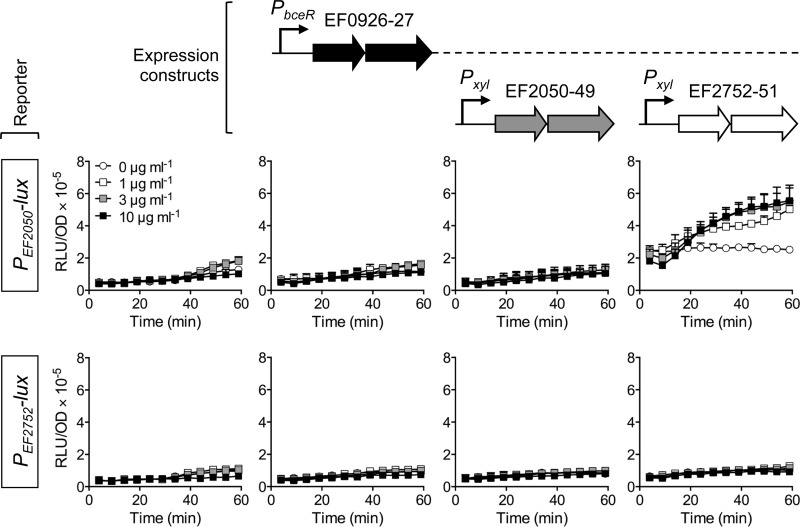
Following the successful transfer of bacitracin resistance to B. subtilis, we next wanted to test if the regulatory pathway could also be reconstituted in the heterologous host. To this end, both target promoters, PEF2050 and PEF2752, were fused to the bacterial luciferase operon luxABCDE as a reporter (22) and introduced into TMB1518. Both constructs resulted in basal luciferase activities that were not affected by addition of bacitracin (Fig. 5, leftmost panels). Thus, no endogenous B. subtilis system was able to induce either of the promoters. Next, the TCS operon EF0926-0927 was introduced into the reporter strains, controlled by the promoter of the homologous bceRS operon of B. subtilis to ensure appropriate expression levels. The presence of the TCS alone did not alter the activities of the transporter promoters, and bacitracin-dependent induction was still not observed (Fig. 5, left center panels), confirming that the TCS alone is unable to respond to the peptide, as has been shown for other BceRS-like systems (11, 12, 32). Additional introduction of the expression construct for EF2050-2049 also did not change the promoter activities (Fig. 5, right center panels), showing that the encoded transporter has no sensory function. In contrast, simultaneous presence of the TCS and the transporter EF2752-2751 increased the basal activities of the PEF2050-lux reporter, and addition of bacitracin resulted in a further 2-fold induction (Fig. 5, top rightmost panel). Expression of PEF2752-lux was not altered in the same genetic background (Fig. 5, bottom rightmost panel), consistent with the minor induction observed for this promoter in E. faecalis. These results clearly show that together, the transporter EF2752-2751 and the TCS EF0926-0927 constitute the sensory and regulatory component of the resistance network and that the second transporter operon, EF2050-2049, is their main target.
FIG 5.
Functional reconstitution of the regulatory circuit in B. subtilis. Promoter regions of the transporter operons EF2050-2049 (top graphs) and EF2752-2751 (bottom graphs) were fused to luxABCDE and introduced into B. subtilis TMB1518. Additionally, the two-component system and transporter operons were introduced under the control of a constitutive (PbceR) or xylose-inducible (Pxyl) promoter. The expression constructs present in each strain are illustrated above the graphs, using the same shading as in Fig. 1. Exponentially growing cultures were exposed to different concentrations of bacitracin, given in the top leftmost graph, and luminescence normalized to optical density (RLU/OD) was monitored over 60 min. Results are means plus standard deviations for two or three biological replicates.
DISCUSSION
Our proteomic analysis of bacitracin-exposed E. faecalis showed that the bacterium responds to the antibiotic with a specific reaction to the bacitracin-induced cell wall damage, as well as with a more general response to stress or growth inhibition. Overall, our data are largely consistent with the findings of a recent transcriptome analysis of E. faecalis treated with different inhibitors of cell wall synthesis, including bacitracin and vancomycin (8). A detailed comparison of our study and the previous one is presented in the supplemental text. Together, these two data sets provide a useful overview of the response of E. faecalis to inhibitors of cell wall synthesis such as bacitracin.
Among the differentially expressed genes identified from both studies was a putative ABC transporter, EF2050-2049, which is a homologue of the bacitracin resistance transporter BceAB of B. subtilis (11, 33). A second such transporter, EF2752-2751, had been previously identified in E. faecalis by a comparative genomics analysis of BceAB-type transporters in Firmicutes bacteria (16). While this second transporter was not found in our proteomic analysis, its ATP-binding cassette domain-encoding gene, EF2752, was slightly induced by bacitracin during the transcriptome study (8). Additionally, we could identify a TCS of E. faecalis with similarity to BceRS of B. subtilis, which regulates expression of BceAB (33). Again, the encoding genes EF0926-0927 had been reported as bacitracin inducible and were also found to be induced by the cell wall-active antibiotics cephalothin and vancomycin (8). Our subsequent characterization of these three gene loci in E. faecalis as well as heterologously in B. subtilis showed that they act together and form a resistance network against bacitracin. A schematic of the derived model is shown in Fig. 6.
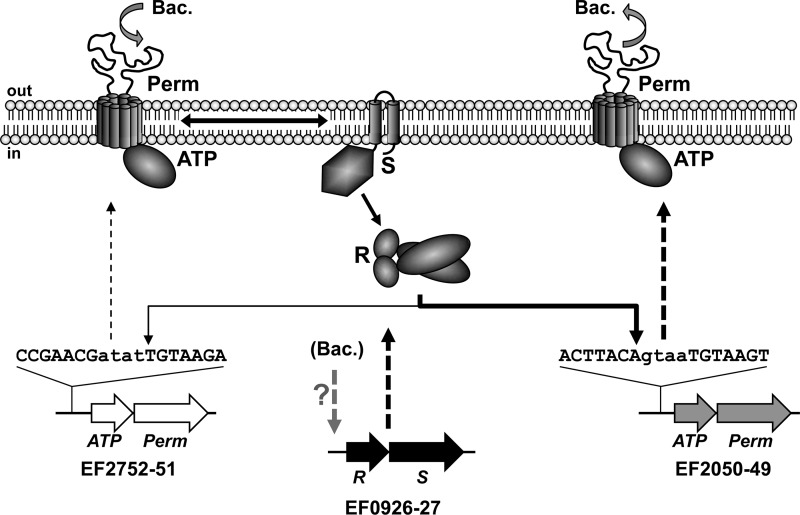
FIG 6.
Model of the bacitracin resistance network in E. faecalis. Schematic illustrations of the involved genes and proteins are shown and the sequences of regulator binding sites are given. Bacitracin is detected by EF2752-2751, indicated by the curved downward arrow. Communication between the transporter and two-component system is shown by a double arrow, and phospho-transfer as well as target promoter activation is shown by single arrows. Increased expression of operons is shown by dashed arrows. The differences in the strengths of induction are reflected by thickness of lines. Bacitracin resistance mediated by EF2050-2049 is indicated by the curved upward arrow. Bacitracin-dependent induction of the two-component system operon by an unknown mechanism is shown as a dashed arrow with a question mark. ATP, ATPase; Perm, permease; R, response regulator; S, sensor kinase; Bac., bacitracin.
The primary sensor of the network is the transporter EF2752-2751, which communicates the presence of bacitracin to the sensor kinase EF0927. Activation of the sensor kinase and phospho-transfer to the response regulator EF0926 then leads to activation of the main target promoter, PEF2050, and increased production of the transporter EF2050-2049. This transporter then removes the bacitracin from its site of action, thus ensuring resistance. Simultaneously, expression of the sensory transporter is slightly induced by the TCS, while expression of the TCS operon is induced by an as-yet-unidentified regulator that is not directly part of the resistance network. Upregulation of a BceRS-like TCS has so far been reported only for the BraRS (= NsaRS) system of S. aureus (34) and may lead to an increased sensitivity or stronger induction of the resistance transporter. Future studies will be directed at identification of the regulator for the TCS. Because the transcriptome study showed the TCS to be inducible by three of four tested inhibitors of cell wall biosynthesis (8), it appears likely that this regulation is part of the cell envelope stress response of E. faecalis. A number of candidate regulatory systems have been identified by comparative genomics and provide a good starting point for subsequent investigations (37).
Several BceRS-BceAB-type resistance modules have been characterized in detail and were always shown to be involved in peptide antibiotic resistance (13, 17). Importantly, the TCSs always rely on one of the transporters for stimulus perception and are unable to induce their target genes in the absence of their transporter (11, 12, 31, 32, 38). In most cases the transporter and TCS are encoded in adjacent operons (15, 16), but in S. aureus and Lactobacillus casei, some TCSs were shown to regulate the expression of a second transporter encoded elsewhere on the chromosome (31, 32). The situation in E. faecalis as identified in the present study is even more complex, with not only the target transporter but also the sensory transporter being encoded in a different locus from the TCS. To our knowledge, this is the first report where a regulatory interaction between a BceAB-like transporter and BceRS-like TCS was shown for two systems not encoded together. Our findings further emphasize the widespread occurrence of these resistance modules and show that the regulatory paradigm is conserved even if genomic arrangement is not.
As mentioned in the introduction, E. faecalis is likely exposed to a range of antimicrobial peptides in the gastrointestinal tract of humans and animals, which can be of host origin or produced by other bacteria of the gut microflora. This raises a question regarding the physiological substrate of the resistance network described here. Most Bce-like modules analyzed so far are not specific for a single substrate but instead recognize a range of often structurally diverse peptides (17). In S. aureus, the human beta-defensin hBD3 and cathelicidin LL-37 have been identified as substrates of the ApsRS-VraFG module (39), showing that the function of Bce-like modules is not restricted to bacterially derived antimicrobial peptides. Our initial screening experiments identified the lantibiotic mersacidin as a second inducer of both enterococcal transporters, and it is possible or even likely that other substrates exist. Particularly for a gut bacterium like E. faecalis, it will be interesting to test if any human antimicrobial peptides can induce expression of the transporters identified here and if the resistance network imparts a selective advantage to the bacterium in the intestinal environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lynn Hancock for supplying vector pLT06 and Richard Losick for vector pAH328. We are also grateful to Tanja Schneider for her generous gift of purified mersacidin and gallidermin. Furthermore, we thank the Centre for Protein Research, Department of Biochemistry, University of Otago, for assistance with mass spectrometry.
Work in the laboratory of S.G. was supported by grants of the Deutsche Forschungsgemeinschaft (GE2164/3-1) and the Fonds der Chemischen Industrie. Research in the laboratory of G.M.C. was supported by the Health Research Council New Zealand and Marsden Fund, Royal Society of New Zealand. C.F. was supported by a Ph.D. scholarship from the China Scholarship Council. A.S. was supported by a Maldivian National Ph.D. scholarship.
D.J.L., M.R.W., and F.K. characterized the resistance network in E. faecalis; C.F. performed all work with B. subtilis; A.S. and A.C. performed the proteomic analysis; S.G. and G.M.C. designed the study and coordinated experimental work; S.G. wrote the manuscript.
Footnotes
Published ahead of print 16 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02111-13.
REFERENCES
- 1.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaaly A, Kalamorz F, Gebhard S, Cook GM. 2013. Undecaprenyl pyrophosphate phosphatase confers low-level resistance to bacitracin in Enterococcus faecalis. J. Antimicrob. Chemother. 68:1583–1593. 10.1093/jac/dkt048 [DOI] [PubMed] [Google Scholar]
- 3.Manson JM, Keis S, Smith JMB, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob. Agents Chemother. 48:3743–3748. 10.1128/AAC.48.10.3743-3748.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevins CL, Martin-Porter E, Ganz T. 1999. Defensins and innate host defence of the gastrointestinal tract. Gut 45:911–915. 10.1136/gut.45.6.911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788. 10.1038/nrmicro1273 [DOI] [PubMed] [Google Scholar]
- 6.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410. 10.1074/jbc.274.13.8405 [DOI] [PubMed] [Google Scholar]
- 7.McBride SM, Sonenshein AL. 2011. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157:1457–1465. 10.1099/mic.0.045997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abranches J, Tijerina P, Avilés-Reyes A, Gaca AO, Kajfasz JK, Lemos JA. 2013. The cell wall-targeting antibiotic stimulon of Enterococcus faecalis. PLoS One 8:e64875. 10.1371/journal.pone.0064875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao M, Helmann JD. 2002. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J. Bacteriol. 184:6123–6129. 10.1128/JB.184.22.6123-6129.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podlesek Z, Herzog B, Comino A. 1997. Amplification of bacitracin transporter genes in the bacitracin producing Bacillus licheniformis. FEMS Microbiol. Lett. 157:201–205. 10.1111/j.1574-6968.1997.tb12774.x [DOI] [PubMed] [Google Scholar]
- 11.Rietkötter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68:768–785. 10.1111/j.1365-2958.2008.06194.x [DOI] [PubMed] [Google Scholar]
- 12.Staroń A, Finkeisen DE, Mascher T. 2011. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55:515–525. 10.1128/AAC.00352-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhard S. 2012. ABC transporters of antimicrobial peptides in Firmicutes bacteria—phylogeny, function and regulation. Mol. Microbiol. 86:1295–1317. 10.1111/mmi.12078 [DOI] [PubMed] [Google Scholar]
- 14.Saier MH, Yen MR, Noto K, Tamang DG, Elkan C. 2009. The Transporter Classification Database: recent advances. Nucleic Acids Res. 37:D274–8. 10.1093/nar/gkn862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph P, Fichant G, Quentin Y, Denizot F. 2002. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 4:503–513 http://www.horizonpress.com/backlist/jmmb/v/v4/58.pdf [PubMed] [Google Scholar]
- 16.Dintner S, Staroń A, Berchtold E, Petri T, Mascher T, Gebhard S. 2011. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 193:3851–3862. 10.1128/JB.05175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebhard S, Mascher T. 2011. Antimicrobial peptide sensing and detoxification modules: unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81:581–587. 10.1111/j.1365-2958.2011.07747.x [DOI] [PubMed] [Google Scholar]
- 18.Shepard BD, Gilmore MS. 1995. Electroporation and efficient transformation of Enterococcus faecalis grown in high concentrations of glycine. Methods Mol. Biol. 47:217–226 [DOI] [PubMed] [Google Scholar]
- 19.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 20.Poyart C, Trieu-Cuot P. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193–198. 10.1016/S0378-1097(97)00423-0 [DOI] [PubMed] [Google Scholar]
- 21.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. 10.1093/nar/18.14.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, Muffler A, Rodriguez S, Perkins J, Losick R. 2010. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 192:5402–5412. 10.1128/JB.00534-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derré I, Rapoport G, Msadek T. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38:335–347. 10.1046/j.1365-2958.2000.02124.x [DOI] [PubMed] [Google Scholar]
- 24.Knight T. Idempotent vector design for standard assembly of BioBricks. MIT Artificial Intelligence Laboratory, MIT Synthetic Biology Working Group, Massachusetts Institute of Technology, Cambridge, MA [Google Scholar]
- 25.Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, Mascher T. 2013. The Bacillus BioBrick box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J. Biol. Eng. 7:29. 10.1186/1754-1611-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191:6203–6210. 10.1128/JB.00592-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 28.Gauntlett JC, Gebhard S, Keis S, Manson JM, Pos KM, Cook GM. 2008. Molecular analysis of BcrR, a membrane-bound bacitracin sensor and DNA-binding protein from Enterococcus faecalis. J. Biol. Chem. 283:8591–8600. 10.1074/jbc.M709503200 [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Gebhard S, Tran SL, Cook GM. 2006. The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate. Microbiology 152:3453–3465. 10.1099/mic.0.29201-0 [DOI] [PubMed] [Google Scholar]
- 31.Revilla-Guarinos A, Gebhard S, Alcántara C, Staroń A, Mascher T, Zúñiga M. 2013. Characterization of a regulatory network of peptide antibiotic detoxification modules in Lactobacillus casei BL23. Appl. Environ. Microbiol. 79:3160–3170. 10.1128/AEM.00178-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiron A, Falord M, Valle J, Débarbouillé M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81:602–622. 10.1111/j.1365-2958.2011.07735.x [DOI] [PubMed] [Google Scholar]
- 33.Ohki R, Giyanto, Tateno K, Masuyama W, Moriya S, Kobayashi K, Ogasawara N. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135–1144. 10.1046/j.1365-2958.2003.03653.x [DOI] [PubMed] [Google Scholar]
- 34.Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J, Elasri MO, Shaw LN. 2011. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 157:2206–2219. 10.1099/mic.0.049692-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamnongpol S, Groisman EA. 2000. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J. Mol. Biol. 300:291–305. 10.1006/jmbi.2000.3848 [DOI] [PubMed] [Google Scholar]
- 36.Schrecke K, Jordan S, Mascher T. 2013. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol. Microbiol. 87:769–788. 10.1111/mmi.12130 [DOI] [PubMed] [Google Scholar]
- 37.Jordan S, Hutchings MI, Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146. 10.1111/j.1574-6976.2007.00091.x [DOI] [PubMed] [Google Scholar]
- 38.Falord M, Karimova G, Hiron A, Msadek T. 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56:1047–1058. 10.1128/AAC.05054-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66:1136–1147. 10.1111/j.1365-2958.2007.05986.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.