Abstract
The methylenecyclopropane nucleoside (MCPN) analogs synguanol and its 6-alkoxy (MBX2168) and 6-alkylthio (MBX1616) derivatives retained good in vitro activities against several common ganciclovir-resistant UL97 kinase variants of human cytomegalovirus. Foscarnet-MCPN cross-resistance was observed among UL54 polymerase variants. UL54 exonuclease domain ganciclovir-cidofovir dual-resistant variants were remarkably more hypersensitive to these MCPNs than to cyclopropavir, with some 50% effective concentration ratios that were <0.1× the wild type. Different categories of MCPNs may have therapeutically exploitable mechanistic differences in viral DNA polymerase inhibition.
TEXT
As long-standing antiviral therapies for human cytomegalovirus (CMV) infection, ganciclovir and its oral prodrug, valganciclovir, have been extensively studied for treatment outcomes, including ganciclovir resistance, which develops in a minority of treated individuals (1). In clinical practice, UL97 kinase mutations that impair the initial phosphorylation of ganciclovir are by far the most frequent mechanism of resistance (2). UL54 DNA polymerase mutations typically add to preexisting UL97 mutations after prolonged ganciclovir therapy and increase the overall level of drug resistance. Strategies for management of drug-resistant CMV are limited, and new antiviral compounds without limiting toxicity or cross-resistance are sought (1).
The CMV DNA polymerase continues to be targeted for antiviral drug development. Recent developments include brincidofovir (CMX001), an orally bioavailable hexadecyloxypropyl conjugate of cidofovir with improved in vitro potency and evidence of efficacy in a phase II prophylaxis trial (3). The methylenecyclopropane nucleoside (MCPN) analog cyclopropavir (Fig. 1), which showed good anti-CMV activity in preclinical studies (4), has recently entered early-stage clinical trials. A related MCPN compound, synguanol, with one less hydroxymethyl group attached to the cyclopropane ring (Fig. 1), also has anti-CMV activity in vitro and in an animal model, but it was deferred for clinical development in favor of cyclopropavir because of its lower antiviral potency (5). However, synguanol was reported in an early study to retain activity against ganciclovir-resistant mutants (6), while more recent data have indicated cyclopropavir and ganciclovir cross-resistance of some CMV UL97 variants, such as M460I and H520Q (7). Additionally, 6-alkoxy and 6-alkylthio derivatives of synguanol have been synthesized that showed improved in vitro antiviral activities against CMV and other herpesviruses (8). As with ganciclovir, the CMV UL97 kinase is involved in the initial phosphorylation of these compounds, and drug-resistant UL97 mutants have been described (9). The aim of the present study was to compare the activities of synguanol, a 6-ether derivative (MBX2168), a 6-thioether derivative (MBX1616), and cyclopropavir (Fig. 1) against well-known CMV UL97 kinase and UL54 DNA polymerase mutants that confer resistance to one or more of the currently licensed anti-CMV drugs. We also propagated a CMV laboratory strain under selection pressure from the synguanol derivatives to determine if other mutations were preferentially selected in vitro.
FIG 1.
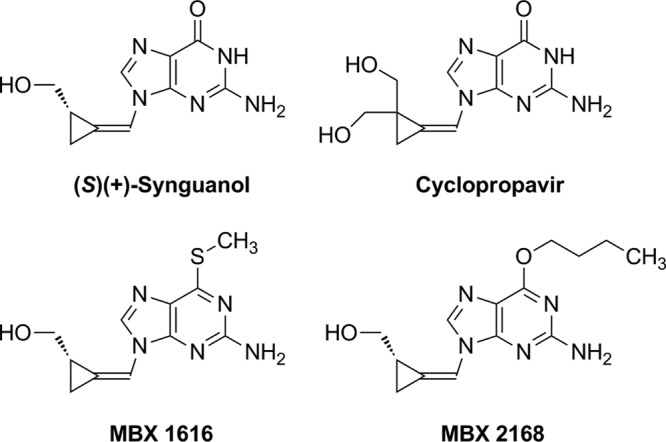
Structures of methylenecyclopropane nucleoside analogs.
Ganciclovir, cidofovir, and foscarnet were obtained as the licensed pharmaceuticals. The MCPN analogs cyclopropavir and synguanol, and derivatives, were synthesized at Microbiotix (Worcester, MA) as described previously (8, 10) and prepared as 10 mM stock solutions in dimethyl sulfoxide. CMV strain variants were derived from bacterial artificial chromosome (BAC) clones BA1, BA9, and BA33 (11, 12), modified from laboratory strain AD169 by introduction of a secreted alkaline phosphatase (SEAP) reporter gene and BAC vector BeloBAC11 at gene locus US6. Most existing CMV UL97 and UL54 mutants were constructed by recombineering of BAC clones BA9 and BA33, respectively, in Escherichia coli strain SW105 as described previously (7, 11–13). BA9 and BA33 contain deletions of their respective wild-type gene sequences from BA1 that prevent inadvertent incorporation into mutant recombinant progeny. Cell-free live virus stock was obtained after transfection of BAC DNA into human foreskin fibroblast (HFF) cell cultures (11). Additional UL54 variants were constructed for this study by using the same methods as for BA33: T4066 (D301N), T4053 (T700A), T4059 (A719V), and T4063 (K853R). Other variants contained UL97 amino acid substitutions M460V and L595S, derived from the strain AD169 derivative T2211 before it was cloned as BAC BA1 (14).
To facilitate the selection of drug resistance mutations, error-prone exonuclease variant T2294 (UL54 D413A) (15) or the newly constructed T3720 (in-frame deletion of UL54 codon 413) was propagated in HFF cultures under ascending concentrations of MBX1616 or MBX2168, starting at less than the 50% effective concentration (EC50) for wild-type virus, and passed weekly at least 15 times from infected cells to fresh subconfluent HFF cultures under gradually increasing drug concentrations that allowed for a similar extent of viral cytopathic effect 1 week later. Every 5 passages, DNA extracts of infected cell cultures were PCR amplified and sequenced to screen for mutations in the UL97 kinase (codons 335 to 615) and UL54 (codons 300 to 600 and 698 to 1000) polymerase genes.
Standardized reporter-based yield reduction drug susceptibility assays were performed as described previously (11, 13). Briefly, 24-well cluster plates of confluent HFF monolayers were inoculated with calibrated low-multiplicity viral inocula and incubated under various drug concentrations, and the SEAP activities in culture supernatants collected at 5 to 7 days, as assayed using a chemiluminescent substrate, were used to determine the EC50 for SEAP activity reduction. EC50 changes of 2-fold or greater are statistically significant in this phenotyping system (11).
Table 1 shows the susceptibilities of CMV variants to synguanol, MBX1616, and MBX2168. The UL97 mutants included the 7 most common ones encountered among ganciclovir-resistant clinical isolates (2) and the uncommon M460T. Variants M460V/I/T and H520Q showed moderate (5× to 7× EC50) resistance to these compounds but less so to MBX2168. Typical foscarnet-resistant UL54 pol variants T700A, E756D, and A809V (2) showed a 3- to 5-fold EC50 increase to the MCPNs, while M844V, which was selected in vitro under cyclopropavir pressure (13), was more resistant. In contrast, remarkable hypersensitivity of ganciclovir-cidofovir-resistant UL54 exonuclease domain (exo) mutants was observed, especially for N408K, with EC50 ratios of <0.1×. Variants T4053 (pol T700A) and T4066 (exo D301N) were also tested against cyclopropavir, with EC50s of 1.3 ± 0.4 μM (mean ± standard deviation; 5.6× wild type) and 0.11 ± 0.04 μM (0.5× wild type), respectively.
TABLE 1.
Genotypes and drug susceptibility phenotypes of recombinant CMV strains evaluated in HFF cells
| Strain type and numbera | Variantb | Synguanol |
MBX1616 |
MBX2168 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EC50c (μM; mean ± SD) | nd | Ratioe | EC50 (μM; mean ± SD) | n | Ratio | EC50 (μM; mean ± SD) | n | Ratio | ||
| Baseline AD169-derived strains | ||||||||||
| T3261 | Wild type | 0.95 ± 0.20 | 12 | 0.81 ± 0.22 | 58 | 0.46 ± 0.13 | 61 | |||
| T3265 | Wild type | 1.05 ± 0.31 | 32 | 1.01 ± 0.26 | 40 | 0.46 ± 0.15 | 46 | |||
| UL97 mutants | ||||||||||
| T3362 | M460I | 4.92 ± 1.25 | 23 | 5.2 | 5.28 ± 1.45 | 39 | 6.5 | 1.78 ± 0.44 | 39 | 3.9 |
| T3346 | M460T | 4.76 ± 1.60 | 11 | 5.0 | 4.91 ± 1.42 | 8 | 6.1 | 1.84 ± 0.43 | 8 | 4.0 |
| T2259 | M460V | 4.76 ± 1.27 | 12 | 5.0 | 4.45 ± 0.69 | 7 | 5.5 | 1.08 ± 0.23 | 8 | 2.3 |
| T3324 | H520Q | 5.30 ± 1.30 | 11 | 5.6 | 5.57 ± 1.19 | 10 | 6.9 | 1.69 ± 0.32 | 12 | 3.7 |
| T3259 | C592G | 1.84 ± 0.56 | 11 | 1.9 | 2.84 ± 0.87 | 12 | 3.5 | 0.93 ± 0.18 | 10 | 2.0 |
| T3252 | A594V | 1.64 ± 0.45 | 7 | 1.7 | 2.19 ± 0.40 | 9 | 2.7 | 0.67 ± 0.19 | 10 | 1.5 |
| T2260 | L595S | 1.04 ± 0.24 | 12 | 1.1 | 2.41 ± 0.55 | 8 | 3.0 | 0.90 ± 0.29 | 10 | 2.0 |
| T3329 | C603W | 2.00 ± 0.48 | 9 | 2.1 | 2.57 ± 0.37 | 11 | 3.2 | 1.01 ± 0.14 | 11 | 2.2 |
| UL54 pol mutants | ||||||||||
| T4053 | T700A | 3.83 ± 0.80 | 8 | 3.6 | 3.87 ± 1.36 | 12 | 3.8 | 2.10 ± 0.69 | 9 | 4.6 |
| T4059 | A719V | 5.66 ± 1.78 | 13 | 5.4 | 4.29 ± 1.93 | 8 | 4.2 | 2.45 ± 0.72 | 6 | 5.3 |
| T3658 | E756D | 2.59 ± 0.48 | 7 | 2.5 | 3.48 ± 0.70 | 16 | 3.4 | 1.74 ± 0.37 | 12 | 3.8 |
| T3271 | A809V | 2.56 ± 0.69 | 7 | 2.4 | 3.23 ± 0.77 | 10 | 3.2 | 1.86 ± 0.41 | 10 | 4.0 |
| T3652 | M844V | 6.50 ± 1.65 | 6 | 6.2 | 6.31 ± 0.45 | 12 | 6.2 | 3.19 ± 0.70 | 14 | 6.9 |
| T4063 | K853R | 1.10 ± 0.20 | 12 | 1.0 | 1.09 ± 0.24 | 7 | 1.1 | 0.47 ± 0.13 | 8 | 1.0 |
| T3429 | A987G | 0.55 ± 0.17 | 10 | 0.5 | 0.52 ± 0.10 | 8 | 0.5 | 0.18 ± 0.06 | 8 | 0.4 |
| UL54 exo mutants | ||||||||||
| T4066 | D301N | 0.12 ± 0.03 | 11 | 0.12 | 0.14 ± 0.04 | 7 | 0.13 | 0.06 ± 0.02 | 7 | 0.12 |
| T3313 | N408K | 0.04 ± 0.01 | 7 | 0.04 | 0.04 ± 0.01 | 7 | 0.04 | 0.03 ± 0.01 | 9 | 0.06 |
| T3267 | F412L | 0.10 ± 0.03 | 9 | 0.09 | 0.18 ± 0.07 | 8 | 0.18 | 0.05 ± 0.01 | 10 | 0.10 |
| T3005 | P522A | 0.26 ± 0.08 | 8 | 0.24 | 0.29 ± 0.05 | 8 | 0.29 | 0.09 ± 0.03 | 16 | 0.20 |
Recombinant virus serial numbers used as identifiers.
Amino acid substitution in the indicated gene.
The mean drug concentration required to reduce supernatant SEAP reporter activity by 50% at 1 week.
Number of assay replicates.
Ratio of the mean EC50 to that of the matching baseline strain (T3261 for UL97 mutants and T3265 for UL54 mutants). Values in bold indicate a >5-fold change from the baseline strain.
In vitro propagation of exo mutant T2294 under exposure to MBX1616 (2 experiments) or MBX2168 (2 experiments) and T4072 under exposure to MBX2168 (4 experiments) all eventually resulted in the selection of UL97 mutants: M460I in all except one instance, where M460T (Table 1) was selected instead. These variants were detected by passage 10 in 5 experiments and by passage 15 in the remainder, at drug concentrations ranging from 3 to 6 μM for MBX1616 and 0.8 to 2 μM for MBX2168. In one experiment (T2294/MBX1616), pol substitutions A719V and K853R, adding to UL97 M460I, were detected at passage 20 under 20 μM, and in another experiment, (T4072/MBX2168) pol substitution V812L added to UL97 M460T by passage 15 under 6 μM. UL54 V812L is known to confer low-grade foscarnet, ganciclovir, and cidofovir resistance (16). Variants A719V and K853R have not been reported in connection with CMV drug resistance, although A719V was described in early literature regarding drug-resistant herpes simplex virus (17). Therefore, CMV recombinants representing A719V (T4059) and K853R (T4063) were constructed from BA33, and the resulting phenotypes were compared with that of the wild-type baseline T3265, as summarized in Table 1. Mutant T4059 also showed increased EC50s for cyclopropavir (1.37 ± 0.34 μM; 6× wild type) and foscarnet (138 ± 34 μM; 3.5× wild type). K853R conferred no drug resistance and was presumably coselected with A719V in an error-prone strain.
This study shows similarities and important differences in cross-resistance of CMV mutants to synguanol MCPNs, cyclopropavir, and currently licensed antivirals. The 7 canonical UL97 resistance substitutions and M460T (Table 1) conferred 7- to 10-fold ganciclovir EC50 increases, except for C592G, which showed a 3-fold increase (11). As with cyclopropavir, UL97 M460I is preferentially selected in vitro by synguanol derivative MCPNs, but the relative levels of resistance conferred by M460I and H520Q are 4- to 7-fold above the baseline EC50 instead of 12- to 20-fold as reported for cyclopropavir (7). Several other ganciclovir-associated mutations confer little MCPN resistance, adding to an early report of no cross-resistance between synguanol and ganciclovir among a different selection of UL97 variants (6). Unlike cyclopropavir, MBX2168 is active against herpes simplex virus (8). Related synguanol 6-ether or -thioether derivatives retain activity against thymidine kinase-deficient strains highly resistant to acyclovir (8, 18), suggesting that cellular kinases contribute to significant initial phosphorylation of these compounds. This may also explain the relatively lesser resistance of UL97 knockout mutants (9) and ganciclovir-resistant CMV UL97 mutants (Table 1) to MBX2168.
The distinct cross-resistance phenotypes of UL54 exo and pol mutants are compared in Table 2, which summarizes current and published data (12, 13, 19–21). Mutations conferring foscarnet resistance that map to the Palm and Finger structural domains, which are involved in pyrophosphate exchange and recognition of the incoming base, conferred cross-resistance to MCPNs, including cyclopropavir (13). The selection of E756D under cyclopropavir pressure (13), and of additional substitutions in this study that conferred foscarnet cross-resistance (A719V and V812L) reinforced this association. The striking hypersensitivity of exo mutants such as N408K to the synguanol MCPNs, as opposed to their significant ganciclovir and cidofovir resistance, implies a major difference in drug resistance mechanisms that remains poorly understood. While it has been hypothesized that exo mutations facilitate the excision of misincorporated nucleoside analogs (19), recent data suggest that exo mutations may instead permit continued DNA chain elongation despite the misincorporation of ganciclovir (22), a mechanism that could not apply to obligate chain terminators such as synguanol. Cyclopropavir, while not an obligate chain terminator, may have structural features that prevent ganciclovir-resistant exo mutants from effectively exercising the same resistance mechanism, resulting in a lesser degree of hypersensitivity than that seen with synguanol (Table 2). Mechanistic differences in ganciclovir and MCPN resistance may support a therapeutic role for synguanol MCPNs against exo mutants, particularly compounds such as MBX2168, which has a low baseline EC50 for CMV, low cellular cytotoxicity (8), and likely a lesser dependence on a viral kinase for initial phosphorylation, thus mitigating the drug resistance of UL97 mutants.
TABLE 2.
Comparative drug resistance levels of selected UL54 variants
| Antiviral agent | Fold changea in the EC50 for the indicated domain/UL54 variant combination |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ExoI/D301N | ExoII/N408K | ExoII/F412L | ExoIII/P522A | Palm-1/T700A | Palm-1/E756D | Finger/A809V | Palm-2/M844V | Thumb/A987G | |
| Ganciclovir | 3 | 4 | 5 | 3 | 1 | 1 | 2 | 2 | 6 |
| Cidofovir | 3 | 21 | 9 | 4 | 1 | 1 | 2 | 2 | 5 |
| Foscarnet | 1 | 1 | 1 | 1 | 6 | 3 | 4 | 2 | 1 |
| Cyclopropavir | 0.5 | 0.6 | 0.5 | 0.5 | 6 | 3 | 3 | 4 | 0.5 |
| Synguanol | 0.1 | 0.04 | 0.09 | 0.2 | 4 | 2 | 2 | 6 | 0.5 |
| MBX1616 | 0.1 | 0.04 | 0.2 | 0.3 | 4 | 3 | 3 | 6 | 0.5 |
| MBX2168 | 0.1 | 0.06 | 0.1 | 0.2 | 5 | 4 | 4 | 7 | 0.4 |
Values were rounded to one significant digit. Data are from Table 1, the text, or previously published susceptibility phenotypes for cyclopropavir (13) and for ganciclovir, foscarnet, and cidofovir (12, 13, 19–21). ExoI to ExoIII, exonuclease subdomains I to III; Palm, Finger, and Thumb, polymerase catalytic subdomains. Characteristic phenotypes (i.e., increased EC50 values) for currently licensed antivirals are shown in bold.
ACKNOWLEDGMENTS
We thank Ronald Ercolani and Gail Marousek for technical assistance.
This work was supported by NIH grants (R44-AI82799 and R01-AI39938) and Department of Veterans Affairs research funds (I01-BX00925).
Footnotes
Published ahead of print 30 December 2013
REFERENCES
- 1.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Danziger-Isakov L, Humar A. 2013. Updated international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation 96:333–360. 10.1097/TP.0b013e31829df29d [DOI] [PubMed] [Google Scholar]
- 2.Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712. 10.1128/CMR.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marty FM, Winston DJ, Rowley SD, Vance E, Papanicolaou GA, Mullane KM, Brundage TM, Robertson AT, Godkin S, Mommeja-Marin H, Boeckh M. 2013. CMX001 to prevent cytomegalovirus disease in hematopoietic-cell transplantation. N. Engl. J. Med. 369:1227–1236. 10.1056/NEJMoa1303688 [DOI] [PubMed] [Google Scholar]
- 4.Kern ER, Bidanset DJ, Hartline CB, Yan Z, Zemlicka J, Quenelle DC. 2004. Oral activity of a methylenecyclopropane analog, cyclopropavir, in animal models for cytomegalovirus infections. Antimicrob. Agents Chemother. 48:4745–4753. 10.1128/AAC.48.12.4745-4753.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rybak RJ, Zemlicka J, Qiu YL, Hartline CB, Kern ER. 1999. Effective treatment of murine cytomegalovirus infections with methylenecyclopropane analogues of nucleosides. Antiviral Res. 43:175–188 [DOI] [PubMed] [Google Scholar]
- 6.Baldanti F, Sarasini A, Drach JC, Zemlicka J, Gerna G. 2002. Z-isomers of 2-hydroxymethylcyclopropylidenemethyl adenine (synadenol) and guanine (synguanol) are active against ganciclovir- and foscarnet-resistant human cytomegalovirus UL97 mutants. Antiviral Res. 56:273–278 http://dx.doi.org/10.1016/S0166-3542(02)00129-8 [DOI] [PubMed] [Google Scholar]
- 7.Chou S, Bowlin TL. 2011. Cytomegalovirus UL97 mutations affecting cyclopropavir and ganciclovir susceptibility. Antimicrob. Agents Chemother. 55:382–384. 10.1128/AAC.01259-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prichard MN, Williams JD, Komazin-Meredith G, Khan AR, Price NB, Jefferson GM, Harden EA, Hartline CB, Peet NP, Bowlin TL. 2013. Synthesis and antiviral activities of methylenecyclopropane analogs with 6-alkoxy and 6-alkylthio substitutions that exhibit broad-spectrum antiviral activity against human herpesviruses. Antimicrob. Agents Chemother. 57:3518–3527. 10.1128/AAC.00429-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komazin-Meredith G, Chou S, Prichard MN, Hartline CB, Cardinale SC, Comeau K, Williams JD, Khan AR, Peet NP, Bowlin TL. 21 October 2013, posting date. Human cytomegalovirus UL97 kinase is involved in the mechanism of action of methylenecyclopropane analogs with 6-ether and -thioether substitutions. Antimicrob. Agents Chemother. 58:274–278. 10.1128/AAC.01726-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou S, Breitenbach JM, Borysko KZ, Drach JC, Kern ER, Gullen E, Cheng YC, Zemlicka J. 2004. Synthesis and antiviral activity of (Z)- and (E)-2,2-[bis(hydroxymethyl)cyclopropylidene]methylpurines and -pyrimidines: second-generation methylenecyclopropane analogues of nucleosides. J. Med. Chem. 47:566–575. 10.1021/jm030316s [DOI] [PubMed] [Google Scholar]
- 11.Chou S. 2010. Recombinant phenotyping of cytomegalovirus UL97 kinase sequence variants for ganciclovir resistance. Antimicrob. Agents Chemother. 54:2371–2378. 10.1128/AAC.00186-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou S. 2011. Phenotypic diversity of cytomegalovirus DNA polymerase gene variants observed after antiviral therapy. J. Clin. Virol. 50:287–291. 10.1016/j.jcv.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou S, Marousek G, Bowlin TL. 2012. Cyclopropavir susceptibility of cytomegalovirus DNA polymerase mutants selected after antiviral drug exposure. Antimicrob. Agents Chemother. 56:197–201. 10.1128/AAC.05559-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou S, Van Wechel LC, Lichy HM, Marousek GI. 2005. Phenotyping of cytomegalovirus drug resistance mutations by using recombinant viruses incorporating a reporter gene. Antimicrob. Agents Chemother. 49:2710–2715. 10.1128/AAC.49.7.2710-2715.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou S, Marousek GI. 2008. Accelerated evolution of maribavir resistance in a cytomegalovirus exonuclease domain II mutant. J. Virol. 82:246–253. 10.1128/JVI.01787-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cihlar T, Fuller MD, Mulato AS, Cherrington JM. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382–393 [DOI] [PubMed] [Google Scholar]
- 17.Knopf CW. 1987. The herpes simplex virus type 1 DNA polymerase gene: site of phosphonoacetic acid resistance mutation in strain Angelotti is highly conserved. J. Gen. Virol. 68:1429–1433 [DOI] [PubMed] [Google Scholar]
- 18.Qiu YL, Geiser F, Kira T, Gullen E, Cheng YC, Ptak RG, Breitenbach JM, Drach JC, Hartline CB, Kern ER, Zemlicka J. 2000. Synthesis and enantioselectivity of the antiviral effects of (R,Z)-,(S,Z)-methylenecyclopropane analogues of purine nucleosides and phosphoralaninate prodrugs: influence of heterocyclic base, type of virus and host cells. Antivir. Chem. Chemother. 11:191–202 [DOI] [PubMed] [Google Scholar]
- 19.Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32–39. 10.1086/375743 [DOI] [PubMed] [Google Scholar]
- 20.Chou S, Marousek G, Li S, Weinberg A. 2008. Contrasting drug resistance phenotypes resulting from cytomegalovirus DNA polymerase mutations at the same exonuclease locus. J. Clin. Virol. 43:107–109. 10.1016/j.jcv.2008.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cihlar T, Fuller MD, Cherrington JM. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Coen DM. 2013. How do human cytomegalovirus DNA polymerase mutations affecting exonuclease activity cause resistance to ganciclovir, abstr 9.26. 38th Annu. Int. Herpesvirus Workshop, Grand Rapids, MI, . [Google Scholar]


