Abstract
The role of Acinetobacter baumannii ATCC 17978 UmuDC homologs A1S_0636-A1S_0637, A1S_1174-A1S_1173, and A1S_1389 (UmuDAb) in antibiotic resistance acquired through UV-induced mutagenesis was evaluated. Neither the growth rate nor the UV-related survival of any of the three mutants was significantly different from that of the wild-type parental strain. However, all mutants, and especially the umuDAb mutant, were less able to acquire resistance to rifampin and streptomycin through the activities of their error-prone DNA polymerases. Furthermore, in the A. baumannii mutant defective in the umuDAb gene, the spectrum of mutations included a dramatic reduction in the frequency of transition mutations, the mutagenic signature of the DNA polymerase V encoded by umuDC.
TEXT
The exposure of some strains of the nosocomial pathogen Acinetobacter baumannii (e.g., strain ATCC 17978) to DNA-damaging agents results in a much higher frequency of DNA damage-induced rifampin resistance (Rifr) mutations than spontaneous Rifr mutations (1). Rifampin resistance can be acquired through the recA-dependent DNA damage response, most commonly following DNA base-pair substitutions in the rpoB gene, which encodes the β subunit of RNA polymerase (1, 2). In Escherichia coli, Y-family error-prone DNA polymerases Pol IV and Pol V (encoded by dinB and umuDC, respectively) are induced as part of the SOS system, a coordinated cellular response to environmental stress (3).
The DNA damage-induced mutation frequency in a recA mutant of A. baumannii 17978 is significantly lower than in its isogenic parental strain (1). However, in a dinB mutant, a previous study showed the same frequency of DNA damage-induced Rifr mutants as in the parental ATCC 17978 strain (1). It is worth noting that in A. baumannii ATCC 17978, two complete umuDC operons (A1S_0636-A1S_0637 and A1S_1174-A1S_1173) and one unlinked umuD homolog (A1S_1389, referred to here as umuDAb) have been identified, and all of them, rather than only the dinB gene, are induced by DNA damage (1). The mutation signatures of DNA Pol V and DinB are transition mutations and −1 frameshifts, respectively (4–6). Interestingly, the most frequent mutations determined in Rifr mutants of the ATCC 17978 strain after UV-induced mutagenesis involve a C→T transition (T. Witkowski, A. Grice, and J. Hare, presented at the Ninth International Symposium on Biology of Acinetobacter, Cologne, Germany, 19 to 21 June 2013). Together, these findings suggest that the multiple DNA Pol V components encoded by the ATCC 17978 genome have an important function in DNA damage-induced mutagenesis. To investigate the role of the two putative umuDC operons, A1S_0636-A1S_0637 and A1S_1174-A1S_1173, and the unlinked umuDAb gene in the DNA damage-induced mutagenesis phenotype of strain ATCC 17978, we inactivated both putative operons and the umuDAb gene and then evaluated their abilities to generate DNA damage-induced mutations. The umuDAb mutant was described elsewhere (7). The A1S_0636-A1S_0637 and A1S_1174-A1S_1173 mutants were obtained as reported previously (8), using the oligonucleotides listed in Table 1. The growth rate of the three isogenic mutants (A1S_0636-A1S_0637, A1S_1174-A1S_1173, and umuDAb) was not significantly different from that of the wild-type (WT) ATCC 17978 parental strain (data not shown), as reported for E. coli and other bacteria (9). However, although the difference was not statistically significant, the UV survival rate of strains 0636-7 and 1174-3 was slightly lower than that of the WT parental strain whereas this was not the case for the umuDAb mutant (Fig. 1).
TABLE 1.
Oligonucleotides used in this work
| Name | Sequencea | Application |
|---|---|---|
| MUT0636Y7F | 5′-CTGGAATCGATATCAACG | Mutant construction |
| MUT0636Y7R | 5′-AATGTCTTGAGCGCATTG | Mutant construction |
| 0636Y7EXTF | 5′-TGATGGATATGAATGAGC | Mutant verification |
| 0636Y7EXTR | 5′-CGACCTATACCAACACAG | Mutant verification |
| MUT1174Y3F | 5′-ACAGGTAAGCCTAACCAC | Mutant construction |
| MUT1174Y3 | 5′-ACTATCAATGCTCAATGC | Mutant construction |
| EXT1174Y3F | 5′-CGTCGATAAGCGAGAAGT | Mutant verification |
| EXT1174Y3R | 5′-GATAAAGCTCTCGACATG | Mutant verification |
| M13FpU | 5′-GTTTTCCCAGTCACGAC | Mutant verification |
| M13RpUC | 5′-CAGGAAACAGCTATGAC | Mutant verification |
| UmuDAb-XbaI | 5′-ATCGTCTAGAATGCCAAAGAAGAAAGAA | umuDAb complementation |
| UmuDAb-NcoI | 5′-ATCGCCATGGTTATCTCATTCGTTTGAG | umuDAb complementation |
Restriction endonuclease recognition sites are underlined.
FIG 1.
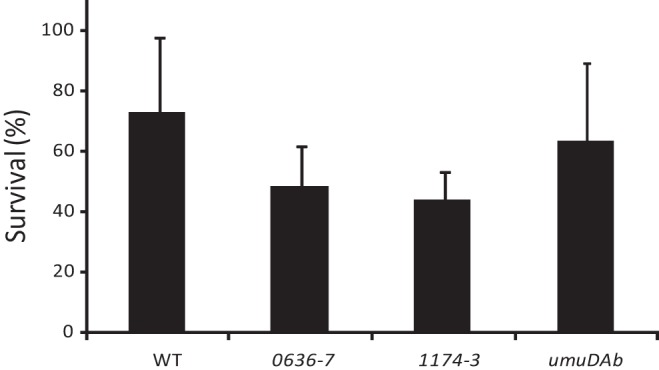
Survival of strain ATCC 17978 (WT) and mutants A1S_0636-A1S_0637 (0636-7), A1S_1174-A1S_1173 (1174-3), and A1S_1389 (umuDAb) after exposure to UV light (100 J/m2). The percentage of surviving cells in UV-irradiated cultures was determined by comparison with nonirradiated cells. Error bars represent the standard errors of the means of the results of three independent experiments. Data were analyzed by a two-tailed, one-way analysis of variance (ANOVA). In all cases, the difference from the WT parental strain (ATCC 17978) was not significant (P > 0.05).
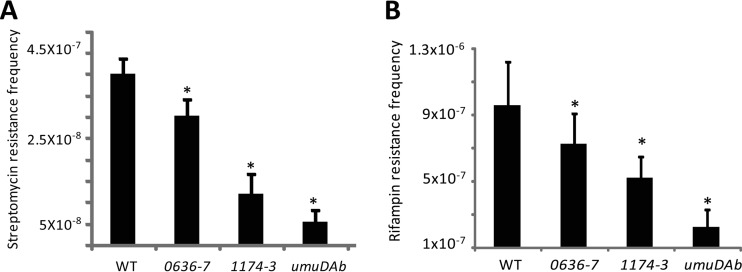
UV-induced mutagenesis was carried out using a modification of the protocol described by Norton et al. (1). Similarly to rifampin, chromosomally acquired streptomycin resistance (Strr) is frequently due to DNA base-pair substitutions but in genes encoding either ribosomal proteins or rRNAs (10). Overnight cultures of the corresponding strain (1 ml) were diluted in 19 ml of NaCl (0.9% [wt/vol]) and then thinly spread on a sterile petri plate. The samples were irradiated with UV-C light (254 nm) at a dose of 100 J/m2. After treatment, 1 ml of each sample was diluted in 9 ml of Luria Bertani (LB) broth and grown for 24 h at 37°C. The appropriate cell dilutions were then deposited on LB plates with and without streptomycin (100 μg/ml) to assess, respectively, the number of Strr mutants and the total number of CFU. The plates were incubated at 37°C for 24 h. Mutation frequency was calculated by dividing the number of Strr mutants by the total number of CFU. All experiments with UV-irradiated cells were performed in the dark to prevent the photoreactivation of pyrimidine dimers. Spontaneous Strr mutants from untreated cultures were determined as described above but without UV irradiation. The spontaneous Strr mutation frequency was approximately 10−10 in both WT and mutant strains. In contrast, the UV-induced Strr mutation frequency was at least 100-fold higher in the irradiated WT strain but significantly lower (P < 0.01) in all of the irradiated mutant strains (Fig. 2A). The most important UmuD homolog contributing to the UV-induced Strr mutation frequency was UmuDAb, followed by a DNA polymerase V encoded by the A1S_1174-A1S_1173 operon (Fig. 2A) and by the A1S_0636-A1S_0637 DNA polymerase V, which showed modest (but statistically significant) UV-induced Strr mutation acquisition compared to the WT parental strain (Fig. 2A). The spontaneous and UV-induced Rifr mutation frequencies were also studied as described above but using LB plates with and without rifampin (50 μg/ml). Under these conditions, the spontaneous Rifr mutation frequency was approximately 10−8 in both the WT and the mutant strains whereas the UV-induced Rifr mutation frequency was about 100-fold higher in the WT strain and significantly lower (P < 0.01) in all the mutant strains (Fig. 2B). Thus, the contributions of A1S_0636-A1S_0637, A1S_1174-A1S_1173, and UmuDAb to the acquisition of Rifr after UV treatment were similar to their contributions to Strr acquisition under the same conditions.
FIG 2.
Frequency of streptomycin (A) and rifampin (B) resistance after UV treatment of strain ATCC 17978 (WT) and mutants A1S_0636-A1S_0637 (0636-7), A1S_1174-A1S_1173 (1174-3), and A1S_1389 (umuDAb). Error bars represent the standard deviations of the means of the results for at least three independently tested cultures. Data were analyzed by a two-tailed, one-way analysis of variance (ANOVA), followed by the Tukey test for post hoc multiple group comparisons. *, P < 0.01 compared to the WT parental strain (ATCC 17978).
To analyze the spectra of mutations produced by each of the umu mutants after UV treatment, we used an established Rifr assay for A. baumannii (1). Colony PCR was performed on 10 individual A. baumannii Rifr mutants obtained from 10 independent experiments for each strain (Table 2). Oligonucleotides rpoB-1441F and rpoB-2095R amplify a 654-bp region of the gene rpoB (A1S_0287 in A. baumannii ATCC 17978), encoding the β subunit of the RNA polymerase, the site of frequent Rifr-inducing base-pair substitutions in this species (1). Sequencing (Macrogen) was carried out using the same oligonucleotide set, and the data were analyzed with SeqManII (DNAstar). Interestingly, most of the mutations detected in the WT and in the mutant strains A1S_0636-A1S_0637 and A1S_1174-A1S_1173 were C→T transitions (Table 2). For the WT strain, these results agree with those reported very recently (T. Witkowski, A. Grice, and J. Hare, presented at the Ninth International Symposium on Biology of Acinetobacter, Cologne, Germany, 19 to 21 June 2013). However, in the umuDAb mutant, only 1 of 10 (10%) mutations was a transition (Table 2). It is worth noting that the mutagenic signature for DNA polymerase V is its incorporation of guanine opposite the 3′ thymine of a thymine-thymine pyrimidine (6-4) pyrimidone photoproduct, i.e., a transition mutation (6). These data agree with the finding that in A. baumannii ATCC 17978 the most important UmuD homolog contributing to the UV-induced mutation frequency is UmuDAb (Fig. 2).
TABLE 2.
Spectrum of mutations produced by the indicated strain after UV treatment
| Strain and mutation category | Mutation type and rpoB nucleotide changea | RpoB amino acid substitution | Mutation frequency (%) |
|---|---|---|---|
| ATCC 17978 WT transversions | 1564 CAG→AAG | 522 Gln→Leu | 30 |
| 1619 TCT→TAT | 540 Ser→Tyr | 10 | |
| ATCC 17978 WT transitions | 1562 TCT→TTT | 521 Ser→Phe | 10 |
| 1613 CGT→CAT | 537 Arg→His | 10 | |
| 1619 TCT→TTT | 540 Ser→Phe | 30 | |
| 1718 CCT→CTT | 573 Pro→Leu | 10 | |
| 0636_7 mutant transversions | 1565 CAG→CTG | 522 Gln→Leu | 10 |
| 1741 ATC→TTC | 581 Ile→Phe | 10 | |
| 0636_7 mutant transitions | 1562 TCT→TTT | 521 Ser→Phe | 30 |
| 1603 CAT→TAT | 535 His →Tyr | 20 | |
| 1619 TCT→TTT | 540 Ser→Phe | 30 | |
| 1174_3 mutant transversions | 1604 CAT→CTT | 535 His→Leu | 10 |
| 1174_3 mutant transitions | 1562 TCT→TTT | 521 Ser→Phe | 40 |
| 1619 TCT→TTT | 540 Ser→Phe | 40 | |
| 1718 CCT→CTT | 573 Pro→Leu | 10 | |
| 1389 (umuDAb) mutant transversions | 1565 CAG→CTG | 522 Gln→Leu | 20 |
| 1574 GAC→GTC | 525 Asp→Val | 10 | |
| 1619 TCT→TAT | 540 Ser→Tyr | 40 | |
| 1741 ATC→TTC | 581 Ile→Phe | 20 | |
| 1389 (umuDAb) mutant transitions | 1718 CCT→CTT | 573 Pro→Leu | 10 |
Boldface and underlining indicates nucleotide changes.
To complement the umuDAb mutant, this gene was PCR amplified using the oligonucleotides UmuDAb-XbaI and UmuDAb-NcoI (Table 1). The resulting product was cloned into the XbaI-NcoI restriction sites of the pET-RA plasmid (11), yielding pETRA-UmuDAb, which was introduced into the A. baumannii umuDAb mutant by electroporation, as reported previously (11). Since this plasmid contains a gene encoding Rifr, we compared the abilities of the complemented umuDAb mutant and the same strain carrying the empty pET-RA plasmid to generate DNA damage-induced Strr mutants, as described above. The acquisition of Strr was approximately 100-fold higher in the UV-treated complemented umuDAb mutant than in the UV-treated umuDAb mutant carrying the empty pET-RA vector (P < 0.01). This result indicates that UmuDAb is an important DNA polymerase V component that probably binds to either of the products of the two unlinked UmuC homologs identified in the ATCC 17978 genome (A1S_2008 and/or A1S_2015) and/or to any of the umuC homologs belonging to the operons mentioned above, as all these genes are induced after DNA damage (1, 7).
To our knowledge, this is the first experimental demonstration that in A. baumannii ATCC 17978 a umuD homolog that acts as a LexA regulator analog (7) is also the most active error-prone DNA polymerase V component conferring resistance to both rifampin and streptomycin. Thus, in conclusion, the data presented in this work suggest that the two complete umuDC operons and a umuDAb gene, together acting as DNA polymerase V components, provide an adaptation mechanism for this nosocomial pathogen.
ACKNOWLEDGMENTS
This study was funded by grants from the Ministerio de Ciencia e Innovación (BFU2011-23478); the European Community, FP 7, ID: 278232 (Magic Bullet); the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III—cofinanced by the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) and FIS PI12/00552. J.A. is the recipient of a Sara Borrell postdoctoral grant from the Instituto de Salud Carlos III (Madrid, Spain).
We are deeply grateful for the helpful discussions with Pilar Cortés (UAB), Susana Campoy (UAB), and Alejandro Beceiro (CHUAC). We acknowledge the efforts of Joan Ruiz (UAB), Susana Escribano (UAB), Joan Colom (UAB), Jorge López (UAB), and Carmen Fernández (CHUAC) for excellent technical assistance.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 16 December 2013
REFERENCES
- 1.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J. Bacteriol. 195:1335–1345. 10.1128/JB.02176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didier JP, Villet R, Huggler E, Lew DP, Hooper DC, Kelley WL, Vaudaux P. 2011. Impact of ciprofloxacin exposure on Staphylococcus aureus genomic alterations linked with emergence of rifampin resistance. Antimicrob. Agents Chemother. 55:1946–1952. 10.1128/AAC.01407-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. 2007. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 15:70–77. 10.1016/j.tim.2006.12.004 [DOI] [PubMed] [Google Scholar]
- 4.Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058–1070. 10.1016/j.molcel.2007.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reuven NB, Tomer G, Livneh Z. 1998. The mutagenesis proteins UmuD' and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol. Cell 2:191–199. 10.1016/S1097-2765(00)80129-X [DOI] [PubMed] [Google Scholar]
- 6.Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. 2000. Roles of Escherichia coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014–1018. 10.1038/35010020 [DOI] [PubMed] [Google Scholar]
- 7.Aranda J, Poza M, Shingu-Vázquez M, Cortés P, Boyce JD, Adler B, Barbé J, Bou G. 2013. Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii. J. Bacteriol. 195:5577–5582. 10.1128/JB.00853-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Héritier C, Poirel L, Lambert T, Nordmann P. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198–3202. 10.1128/AAC.49.8.3198-3202.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodgate R, Sedgwick SG. 1992. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol. Microbiol. 6:2213–2218. 10.1111/j.1365-2958.1992.tb01397.x [DOI] [PubMed] [Google Scholar]
- 10.Springer B, Kidan YG, Prammananan T, Ellrott K, Bottger EC, Sander P. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45:2877–2884. 10.1128/AAC.45.10.2877-2884.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda J, Poza M, Pardo BG, Rumbo S, Rumbo C, Parreira JR, Rodríguez-Velo P, Bou G. 2010. A rapid and simple method for constructing stable mutants of Acinetobacter baumannii. BMC Microbiol. 10:279. 10.1186/1471-2180-10-279 [DOI] [PMC free article] [PubMed] [Google Scholar]