Abstract
Mycobacteria contain genes for several DNA-dependent RNA primases, including dnaG, which encodes an essential replication enzyme that has been proposed as a target for antituberculosis compounds. An in silico analysis revealed that mycobacteria also possess archaeo-eukaryotic superfamily primases (AEPs) of unknown function. Using a homologous recombination system, we obtained direct evidence that wild-type dnaG cannot be deleted from the chromosome of Mycobacterium smegmatis without disrupting viability, even in backgrounds in which mycobacterial AEPs are overexpressed. In contrast, single-deletion AEP mutants or mutants defective for all four identified M. smegmatis AEP genes did not exhibit growth defects under standard laboratory conditions. Deletion of native dnaG in M. smegmatis was tolerated only after the integration of an extra intact copy of the M. smegmatis or Mycobacterium tuberculosis dnaG gene, under the control of chemically inducible promoters, into the attB site of the chromosome. M. tuberculosis and M. smegmatis DnaG proteins were overproduced and purified, and their primase activities were confirmed using radioactive RNA synthesis assays. The enzymes appeared to be sensitive to known inhibitors (suramin and doxorubicin) of DnaG. Notably, M. smegmatis bacilli appeared to be sensitive to doxorubicin and resistant to suramin. The growth and survival of conditional mutant mycobacterial strains in which DnaG was significantly depleted were only slightly affected under standard laboratory conditions. Thus, although DnaG is essential for mycobacterial viability, only low levels of protein are required for growth. This suggests that very efficient inhibition of enzyme activity would be required for mycobacterial DnaG to be useful as an antibiotic target.
INTRODUCTION
Mycobacterium tuberculosis is a deadly pathogen that claims nearly 2 million lives annually and infects an estimated 2 billion people, who serve as a reservoir of latently infected individuals (1). Most tuberculosis (TB) cases are not the result of new infections but are caused by the reactivation of dormant M. tuberculosis (2). TB caused by drug-sensitive strains is fully treatable, but patients must take three or four drugs for approximately ≥6 months. Premature termination of drug therapy results in the emergence of resistant strains. The World Health Organization estimates that 50 million individuals harbor multidrug-resistant (MDR) M. tuberculosis, which is resistant to at least rifampin and isoniazid. Treating these MDR strains requires second-line drugs, which are expensive, have side effects, and take longer to work (up to 2 years). More disturbing is that strains of untreatable extensively drug-resistant (XDR) TB, which are additionally resistant to any fluoroquinolone and at least one of three injectable second-line drugs (capreomycin, kanamycin, or amikacin), have already been identified in 58 countries. This XDR form, together with totally drug-resistant (TDR) TB, seems to represent the greatest health threat (3). The options for treating MDR/XDR/TDR TB infections are becoming seriously limited, threatening to return TB control to the preantibiotic era (4, 5). The first-line drugs for treating TB are restricted to a few sensitive targets, including inhA (NADH-dependent enoyl-[acyl carrier protein] reductase) and kasA (3-oxoacyl-[acyl carrier protein] synthase 1) for isoniazid, rpoB (DNA-directed RNA polymerase subunit beta) for rifampin, and the embCAB operon for ethambutol. Also in this category are enzymes required for the intracellular activation of currently used drugs, such as katG (catalase peroxidase peroxynitritase T) for isoniazid, pncA (pyrazinamidase/nicotinamidase) for pyrazinamide, and etaA (monooxygenase) for ethionamide (6). The identification of new drugs and sensitive targets would appear to be indispensable for the control of drug-resistant forms of TB. One requirement for a promising antibacterial enzyme target is that it be essential for the organism and that it not be present in the host. Such candidates might be found among basic essential metabolism pathways, including DNA replication processes.
Bacterial DNA replication is performed by PolIII, which is unable to synthesize DNA de novo and therefore requires a primer to allow the initiation of DNA synthesis. The replication of leading strands requires at least a single primer to initiate the process, but replication of the lagging strand requires an individual starter for each Okazaki fragment. In Escherichia coli, the enzyme that synthesizes such primers is the RNA polymerase, DnaG. Eukaryotes also possess a distinct primase responsible for the synthesis of RNA primers. DNA primase is a single-strand DNA (ssDNA)-dependent RNA polymerase that plays a key role in DNA synthesis (7). The DNA primases of bacteria and bacteriophages are classified into one group, and the primases of eukaryotes and archaea belong to a second group. All primases share many catalytic properties, but the proteins in the two classes differ both in structure and in their relationship with other proteins in the replication complex (8, 9). The prokaryotic primase associates with the replicative DNA helicase. DnaG primase contains three distinct domains, an N-terminal zinc-binding region, a middle RNA polymerase domain, and a C-terminal domain containing either a DNA helicase (phage) or a region for interaction with the DNA helicase (bacteria) (10). In contrast to DnaG primase, which is monomeric, eukaryotic primase is a heterodimeric complex of DNA polymerase α and an accessory β subunit. The small primase subunit (PriS) belongs to the archaeo-eukaryotic primase (AEP) superfamily (11). The PriS complex contains an active site for RNA primer synthesis and the large primase accessory subunit (PriL), which may coordinate primase and polymerase action and is required for the initiation of primer synthesis (12). Previous studies have demonstrated that AEPs are also present in diverse bacteria (13, 14).
An AEP domain constitutes one of three domains in ATP-dependent ligase (LigD), which is a key protein in the nonhomologous end-joining (NHEJ) DNA repair system (11, 15, 16, 17, 18). The primase domain has terminal transferase, DNA-dependent RNA primase, and DNA-dependent DNA/RNA gap-filling polymerase activities (15, 16, 18, 19, 20). In mycobacteria, both DnaG and AEPs have been reported. The replicative DnaG primase is encoded by the dnaG gene, which is located in the dnaG operon (21).
The viability of DnaG primases as antibiotic targets rests on the presumption that these enzymes are essential for all bacteria because they are required for initiating DNA replication. However, it is difficult to definitively establish this indispensability, which is a fundamental prerequisite if these enzymes are to be considered potential antibiotic targets. In this report, we undertook a series of experiments that unequivocally demonstrate that dnaG is essential in Mycobacterium smegmatis, even in AEP-overexpressing backgrounds. We also characterized the enzymatic activities of M. smegmatis and M. tuberculosis DnaG proteins. A detailed analysis of the amount of DnaG in various strains revealed that the level of protein can vary by ∼6-fold without producing a major effect on growth under standard laboratory conditions. Strains engineered during this study will be useful in any future detailed evaluation of antibiotics targeting DnaG.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study were derived from M. smegmatis mc2155 (22) and were cultured in Middlebrook 7H9 broth supplemented with albumin-dextrose-sodium chloride or NB broth (8.0 g/liter nutrient broth [Difco], 10.0 g/liter glucose). Where required, further additions included 0.2% Tween 80 (pH 6.0 to 6.2), 50 μg/ml hygromycin (Hyg), 7.5 μg/ml gentamicin (Gen), and 25 μg/ml kanamycin (Kan). Mycobacterial transformants were selected on Middlebrook 7H10 agar plates enriched with albumin-dextrose-sodium chloride containing Kan (25 μg/ml), Gen (7.5 μg/ml), or Hyg (50 μg/ml). E. coli strains were cultured in LB medium (10 g/liter tryptone, 5 g/liter yeast extract, 10 g/liter NaCl, pH 7.0). Where required, further additions included 100 μg/ml ampicillin (Amp), 200 μg/ml Hyg, and 50 μg/ml Kan.
Gene-cloning strategies.
Standard molecular biology methods were used for all cloning protocols (23). All PCR products were obtained using thermostable AccuPrime Pfx DNA polymerase (Invitrogen), cloned initially into a blunt vector (pJet1.2; Fermentas), sequenced, and then released by digestion with the appropriate restriction enzymes before cloning into the final vectors. Subcloning into expression vectors was facilitated by incorporating restriction enzyme recognition sites into the sequence of the primers. The plasmids used in this work are listed in Table S1 in the supplemental material.
Construction of gene-replacement vectors.
To perform unmarked deletions of dnaG, prim2, prim3, and prim4 genes in M. smegmatis, we used a suicidal recombination delivery vector based on p2NIL (24). The recombination vector carried the 5′ end of dnaG with the upstream region connected to the 3′ end of the gene with the downstream region, amplified with the primers shown in Table S2 in the supplemental material. The aacC1 gene was then introduced between the upstream and downstream regions of dnaG. Finally, the PacI screening cassette from pGOAL17 was inserted into the constructs, resulting in the suicide delivery vectors pMK165, pAB148, pMK189, and pMK190 carrying ΔdnaG::aacC1R, Δprim2, Δprim3, and Δprim4, respectively; these were used to engineer the directed M. smegmatis mutants as described previously (25, 26). The resultant mutant strains were verified by PCR and Southern blot hybridization (see Fig. S1 in the supplemental material).
Construction of complementation plasmids.
M. smegmatis genes (dnaG, ligD, prim2, prim3, prim4), M. tuberculosis dnaG, and E. coli dnaG were amplified by PCR using the primers listed in Table S2 in the supplemental material and cloned into the BamHI-XbaI sites of the pJam2 vector downstream from the Pami promoter (see Table S2). Next, all genes with their Pami promoters were excised from these vectors with HindIII and XbaI and cloned into the integration vector pMV306, generating pMK172, pMK207, pAB145, pMK206, pMK193, pMK173, and pMK174 for dnaG, ligD, prim2, prim3, prim4 (M. smegmatis), dnaG (M. tuberculosis), and dnaG (E. coli), respectively. The M. smegmatis dnaG gene was amplified by PCR using the primers listed in Table S2 and cloned into the BamHI-HindIII sites of the pKW08Lx vector downstream from the Ptet promoter (see Table S2). Next, genes with their Ptet promoters were excised from this vector (pMK214) with HindIII and XbaI and cloned into the integration vector pKW08Lx-Int, generating the pMK215 vector.
Disruption of M. smegmatis genes encoding primases.
The protocol of Parish and Stoker (24) was used to disrupt the M. smegmatis dnaG prim2, prim3, prim4, and ligD genes at their native loci on the chromosome. The mutants were generated by subsequent disruption of individual genes. The suicidal recombination plasmid DNA (pMK165, pAB148, pMK189, pMK190, and pMK111) was treated with NaOH (0.2 mM) and integrated into the M. smegmatis mc2155 chromosome by homologous recombination. The resulting single-crossover recombinant (SCO) mutant colonies were blue, resistant to Kan and Gen, and sensitive to sucrose. The site of recombination was confirmed by PCR and Southern hybridization. The SCO strains were further processed to select for double-crossover (DCO) mutants that were white, sensitive to Kan, and resistant to sucrose (2%). PCR and Southern hybridization analyses were used to distinguish between the wild type and each mutant DCO. Probes that hybridized to each gene, labeled using a nonradioactive primer extension system (DIG-labeling system; Amersham, GE Healthcare, Sweden), were generated by PCR. The primers used for PCR amplification are listed in Table S2 in the supplemental material.
Protein purification.
For protein expression, all E. coli cultures were grown at 37°C in LB medium containing Amp. The expression constructs were made by PCR amplification of M. smegmatis dnaG (dnaGMs) and M. tuberculosis dnaG (dnaGMtb), cloning into pJet1.2, excising with BamHI/HindIII (for dnaGMs) or BamHI/EcoRI (for dnaGMtb), and cloning into the final vectors, pHIS and pGEX, respectively. The pHIS and pGEX derivatives were transformed into E. coli BL21(DE3), and cells were plated on LB agar containing antibiotics and grown overnight. Single colonies were inoculated into 5 ml liquid medium, grown overnight, and diluted 100-fold into fresh medium (500 ml). After the colonies were grown to midexponential phase (optical density at 600 nm [OD600], 0.8 to 1.0), protein expression was induced by the addition of IPTG (isopropyl β-d-1-thiogalactopyranoside) to 0.4 mM. After overnight incubation, cells were harvested, sonicated, and centrifuged to separate the soluble and insoluble fractions. DnaG DNA primase was purified from the soluble fraction by affinity column chromatography using Ni2+-charged His Bind resin (Novagen) for the His-tagged protein and glutathione agarose (Pierce) for the glutathione S-transferase (GST)-tagged protein. After concentration of protein using Amicon Ultra 4-ml concentrators with a 30,000 molecular-weight cut-off polyethersulfone (PES) membrane, protein sample concentrations were determined using the bicinchoninic acid (BCA) method (Bio-Rad protein assay). The purified DnaG and LigD (15) were used for rabbit immunization as described previously (26).
DnaG primase activity assay.
In vitro assays of primase activity were performed essentially as described previously (27). The reaction mixture (total volume, 25 μl) for the RNA primer synthesis assays contained 50 mM HEPES (pH 7.5), 100 mM potassium glutamate, 10 mM dithiothreitol, 10 mM magnesium acetate (or 10 mM manganese chloride), and 2 μM ssDNA template (5′-tactctcatcgtggaatcctgaca). DnaG primase was added to a final concentration of 3 μM, and the sample was preincubated for 10 min at 30°C. After the addition of ATP, CTP, GTP (each to a final concentration of 200 μM), UTP (to a final concentration of 20 μM), and 0.6 μCi of [α-32P]UTP (3,000 Ci/nmol), the sample was incubated for an additional 4 h. The reaction was stopped by adding 30 μl of 3 M sodium acetate. The RNA products were then precipitated overnight at −70°C with 96% cold ethanol in the presence of 40 μg glycogen. The precipitates were washed with 75% cold ethanol and dissolved in 8 μl loading buffer (95% formaldehyde, 0.05% bromophenol blue, 0.05% xylene cyanol, 20 mM EDTA). Samples were heated at 98°C for 10 min, and the reaction products were separated by electrophoresis in 18% urea-polyacrylamide gel. After electrophoresis, the results were visualized by autoradiography using X-ray film with intensifying screens overnight at −70°C. The sensitivity of DnaG to inhibitors (suramin and doxorubicin) relative to the controls was determined by adding an inhibitor to the reaction buffer at concentrations ranging from 1 to 100 μM.
AlamarBlue and CFU susceptibility tests.
The microplate alamarBlue assay (28) was used to test the sensitivity of mycobacteria to DnaG inhibitors. Suramin and doxorubicin were dissolved in NB medium at final concentrations of 0.039 to 5 mM (suramin) or 0.98 to 125 μM (doxorubicin), filtered (0.22 μm), and distributed in the wells of microtiter plates (100 μl/well). Next, 100 μl of wild-type M. smegmatis and complemented ΔdnaG mutants (OD600, 0.1) was added to the control wells and to the wells containing inhibitors. The plates were incubated at 37°C for 72 h in a humidified atmosphere. AlamarBlue reagent (25 μl; Invitrogen) was added, and the plates were incubated overnight at 37°C. A color change from blue to pink indicated bacterial growth. The MIC was defined as the lowest drug concentration that prevented a color change. The sensitivity of mycobacteria to suramin and doxorubicin was also determined by monitoring the numbers of CFU of the wild-type and mutant strains growing in the presence of different concentration of inhibitors as described previously (29).
RNA extraction and reverse transcription.
For quantitative real-time PCR (qRT-PCR) experiments with prim2, prim3, and prim4 transcripts, RNA was extracted from wild-type M. smegmatis strains and from the SCO strains (ΔdnaG::aacC1-dnaG) carrying an extra copy of an AEP gene under the control of a chemically inducible promoter (attB::Pamiprim2/prim3/prim4) using the TRIzol LS reagent (Invitrogen) as described previously (26). For reverse transcription, we used a SuperScript III first-strand synthesis system (Invitrogen) and performed the reactions in total volumes of 20 μl containing 1 μg of total RNA. Subsequently, 2 μl of cDNA (equivalent to 50 ng of RNA) was used in the qRT-PCR experiments (see below).
qRT-PCR.
qRT-PCR for the analysis of prim2, prim3, and prim4 gene expressions was performed using the Maxima SYBR green qPCR master mix (Fermentas) and a 7900HT real-time PCR system (Applied Biosystems). Each reaction mixture (final volume, 25 μl) was mixed on ice and contained 1× Maxima SYBR green qPCR master mix, 50 ng of cDNA, and 0.3 μM each primer (see Table S2 in the supplemental material for primer sequences). For expression analysis of the M. smegmatis prim2, prim3, and prim4 genes, we used a three-step cycling protocol in which the reaction mixtures were first heated to 95°C for 10 min and were then subjected to 45 cycles at 95°C for 15 s (denaturation), at 63°C for 30 s (annealing), and at 72°C for 30 s (extension). The data were acquired during the extension step. To verify the specificities and identities of the PCR products generated, melting curve analysis was performed at the end of each PCR. Each experiment was performed in triplicate, and the results are presented as the means and standard errors. The results were normalized with respect to sigA gene expression as the internal control and reflect the fold change in the expression of a given gene in the mutant strain versus the wild-type strain, as calculated using the double-delta method 2-ΔΔCT (30).
RESULTS
Rv2343c and Msmeg4482 genes encode active primases that are sensitive to doxorubicin and suramin.
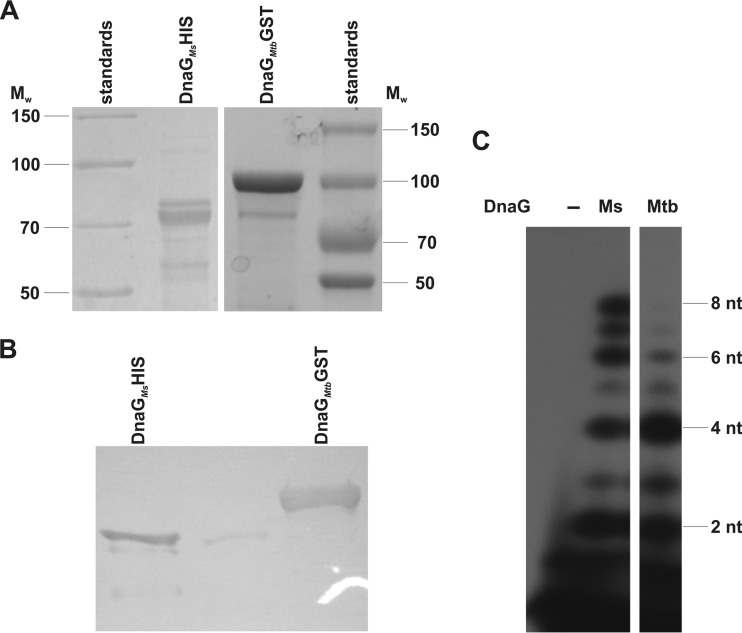
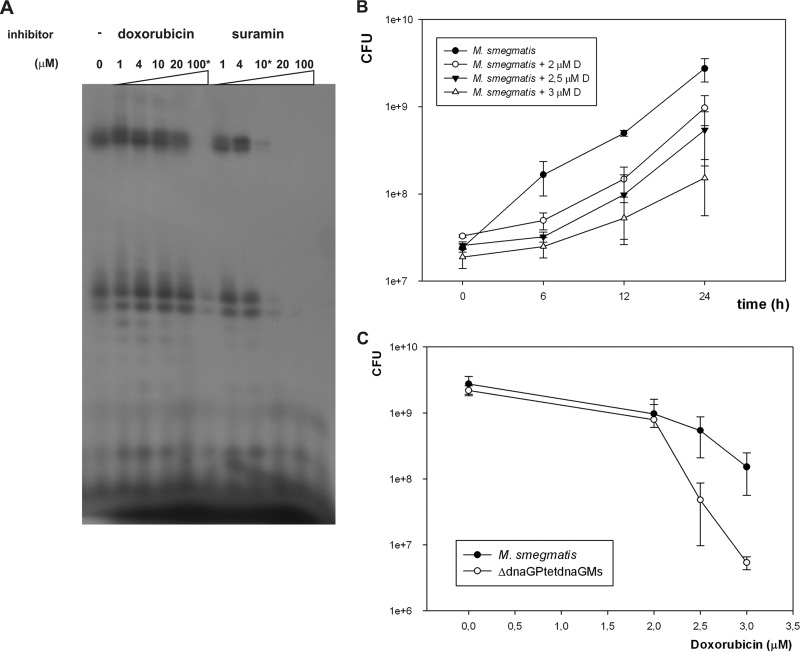
Bacterial primases (DnaG) are highly conserved across all bacterial genomes, allowing ready identification of homologous genes by bioinformatics analysis. The DnaG of M. tuberculosis displays 48% similarity and 32% identity with its E. coli counterpart and as much as 89% similarity and 82% identity with the DnaG of M. smegmatis (see http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). In contrast, bacterial primases are distinct from their eukaryotic and archaeal counterparts, which makes them an attractive antibacterial drug target (31, 32). The genomes of fast- and slow-growing mycobacteria each carry a single gene that is homologous to bacterial dnaG (Msmeg4482 and Rv2343c for fast and slow growers, respectively). The activity of M. tuberculosis DnaG was recently confirmed in an in vitro study (32). We used a pT7 Pol-based E. coli expression system to overproduce and purify DnaG from M. smegmatis and M. tuberculosis, and we subsequently used the purified DnaG of M. smegmatis for the vaccination of a rabbit to obtain polyclonal antibodies. The primase activities of DnaGs from these two strains were monitored by visualizing and quantifying radiolabeled RNA primer products on denaturing gels. We tested a number of templates and reaction parameters to identify the optimal conditions for DnaG activity. A reaction buffer supplemented with Mn2+ or Mg2+ and a 24-mer (5′-tactctcatcgtggaatcctgaca) ssDNA template allowed us to confirm the activity of both enzymes (Fig. 1). DnaG activity was suppressed in the presence of doxorubicin (100 μM) and suramin (10 μM), known inhibitors of DnaG (32), which inhibited the activities of M. smegmatis and M. tuberculosis DnaG in vitro by about 85% and 70%, respectively (Fig. 2A). Notably, doxorubicin, but not suramin, displayed antibacterial activity against M. smegmatis grown in liquid culture. The alamarBlue assay, which allowed us to monitor cell viability, showed that 8 μM doxorubicin inhibits the growth of mycobacteria. On the other hand, suramin was tolerated even at concentrations of 5 mM (see Fig. S2 in the supplemental material). The CFU analysis of M. smegmatis growing in the presence of different concentrations of doxorubicin revealed dose-dependent inhibition of growth (Fig. 2B). Moreover, the M. smegmatis mutant expressing as little as about 20% of the physiological level of DnaG appeared to be more sensitive to doxorubicin than the wild-type strain (Fig. 2C). In the presence of 2.5 μM and 3 μM doxorubicin, the numbers of viable cells were at 50% and 20%, respectively, of those viable in the wild-type culture.
FIG 1.
The purification and activity of M. smegmatis (Ms) and M. tuberculosis (Mtb) DnaG proteins. (A) SDS-PAGE analysis. Purified proteins were resolved on a 12% polyacrylamide gel followed by Instant Blue staining. Mw, molecular weight; nt, nucleotides. (B) Western blot analysis with antibodies raised against DnaG of M. smegmatis. (C) Protein activity assays with Mg2+, as described in Materials and Methods.
FIG 2.
(A) Synthesis of radiolabeled RNA primers by M. smegmatis DnaG and its inhibition by doxorubicin and suramin. Phosphorimager analysis of an 18% urea–polyacrylamide gel showing RNA products of the priming reaction. The priming reaction was performed with Mn2+ on a 24-mer ssDNA template (5′-tactctcatcgtggaatcctgaca) in the presence of a mixture of the four nucleotides containing [α-32P]UTP. (B) Time-dependent colony formation by wild-type M. smegmatis (M. smegmatis mc2155) growing in the presence of various concentrations of doxorubicin (0, 2, 2.5, and 3 μM). Colony formation values are means ± standard errors from three independent experiments. (C) The dose-dependent colony formation by wild-type M. smegmatis (M. smegmatis mc2155) and its mutant with downregulated DnaG expression (ΔdnaGPtetdnaG) growing in the presence of doxorubicin. The number of CFU of 24-h-old culture is shown. Colony formation values are means ± standard errors from three independent experiments.
The activity of DnaG, but not that of primases in the AEP family, is essential for M. smegmatis viability.
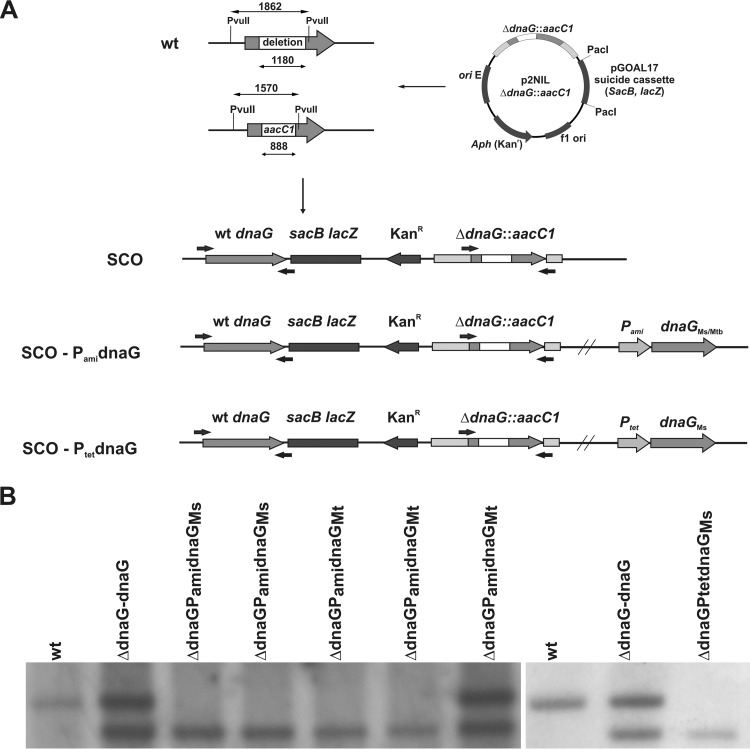
Bioinformatic analyses of the genomes of M. tuberculosis and M. smegmatis showed that, in addition to dnaG, mycobacteria also contain putative AEP-like primase genes. The best characterized of these is PolDom, an AEP that is part of a multidomain enzyme called ligase D (LigD), which is required for NHEJ DNA repair during the stationary phase (15, 19). The function of other genes displaying homology to AEPs remains to be elucidated. Similar to the case in other bacteria, the dnaG gene of M. smegmatis was previously reported to be essential for viability (21). Here, we used a gene-replacement strategy to verify that dnaG is essential in mycobacteria and tested whether AEPs are also essential. In contrast to dnaG, which could not be replaced by a nonfunctional copy without disrupting viability, individual or collective removal of intact AEP genes in M. smegmatis to generate ΔMSMEG_5570, ΔMSMEG_6301, ΔMSMEG_0597, ΔMSMEG_2105 or Δ(MSMEG_5570; MSMEG_6301; MSMEG_0597; MSMEG_2105) was well tolerated, establishing that the AEPs are not essential for the viability of M. smegmatis (see Fig. S1 in the supplemental material). To further confirm that dnaG is essential and to engineer a dnaG conditional mutant, we cloned the intact gene into a plasmid that placed it under the control of a tetracycline- or acetamide-inducible promoter (Ptet/Pami) and introduced this construct into the attB locus of M. smegmatis chromosomal DNA. The additional dnaG copy enabled us to replace the native dnaG gene with its mutated copy. The genotype of each mutant was confirmed by Southern blotting hybridization (Fig. 3). Additionally, the attB integrated, intact copy of dnaG provided with pMV306Hygr vector was subjected for replacement with an “empty” pMV306Kanr vector in six independent experiments. The lack of Kanr recombinants without an intact dnaG confirmed additionally the essentiality of DnaG for viability of M. smegmatis (33).
FIG 3.
Complementation of the M. smegmatis ΔdnaG SCO strain. (A) Schematic showing the restriction-digested DNA fragment (1,862 bp) and the size of the internal deletion in the mutated gene (1,180 bp). The dnaG gene is represented by gray arrows and the internal deletion by white rectangles. The aacC1 gene (gentamicin resistance cassette) was cloned within the dnaG gene to facilitate screening of DCO mutants. The dnaG is essential for the viability of M. smegmatis. SCO strains were enriched with intact dnaG from M. smegmatis or M. tuberculosis under the control of an inducible promoter (PamidnaGMs/dnaGMt or PtetdnaGMs). (B) The genotype of selected strains was confirmed by PCR and Southern hybridization analysis.
DnaG is not complemented by AEPs.
Since the only essential primase in mycobacteria is DnaG, it can be considered essential for replication of chromosomal DNA. To evaluate DnaG as a potential drug target, we tested whether it is still essential in an AEP-overexpressing background. Genes encoding AEPs were cloned to place them under the control of a chemically inducible promoter (Pami) and integrated into the attB locus of an M. smegmatis SCO mutant carrying both a functional dnaG gene and a dnaG gene disrupted by the aacC1R gene (ΔdnaG::aacC1R). The overexpression of AEPs was confirmed by Western blot analysis (LigD) or quantitative RT-PCR (prim2, prim3, and prim4) (see Fig. S3 in the supplemental material). Next, we selected for mutants lacking an intact copy of dnaG (i.e., those carrying ΔdnaG::aacC1R exclusively). In no case did the overexpression of AEPs rescue the viability defect of M. smegmatis lacking an intact chromosomal dnaG gene, confirming that dnaG is essential and demonstrating that these primases serve nonredundant functions. Next, we tested whether M. smegmatis dnaG could be replaced by its counterpart from M. tuberculosis or E. coli. The introduction of PamidnaGMtb into the attB locus allowed us to remove the native dnaG gene without disrupting viability (Fig. 3). Removal of the native copy of dnaG from M. smegmatis chromosomal DNA was not tolerated following introduction of E. coli dnaG (PamidnaGEc) into the attB site of M. smegmatis SCO strain, suggesting that the mycobacterial DnaG have distinct activities or interactions that are essential for replication in these organisms.
Depletion of DnaG only modestly affects the viability of M. smegmatis.
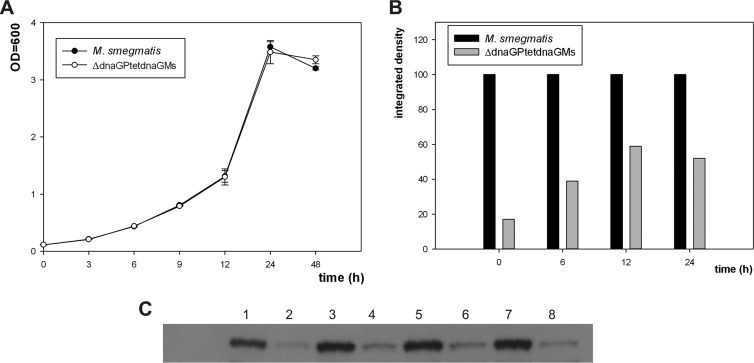
The depletion of an ideal drug target should result in bacterial cell death or at least in the inhibition of growth. Having confirmed that DnaG is essential for the viability of mycobacteria, even in AEP overproduction backgrounds, we sought to test the effect of controlled DnaG depletion on the viability of M. smegmatis. To this end, we grew a conditional DnaG expression mutant carrying only intact dnaGMs (or dnaGMtb) under the control of a chemically inducible Ptet promoter (or Pami), with and without anhydrotetracycline (or acetamide) as an inducer. The growth of the wild-type control strain and conditional mutants was monitored by measuring the optical density at 600 nm (OD600) and determining colony formation; the DnaG protein level was monitored by Western blotting (Fig. 4). Surprisingly, an approximately 83% reduction of DnaG protein levels in M. smegmatis did not (according to the OD600) or only modestly (0.5 to 0.8 log in CFU) affected the growth of bacterial cultures, suggesting that mycobacteria can tolerate substantial depletion of this protein, an observation that has important implications for evaluations of DnaG as a future drug target. The growth inhibition effect was not observed when mineral medium was used to culture wild-type M. smegmatis or conditional mutants (data not shown).
FIG 4.
Phenotypic analysis of M. smegmatis and the conditionally complemented mutant ΔdnaG. (A) Growth rate analysis of wild-type M. smegmatis and a strain complemented with an intact copy of dnaGMs under the control of a tetracycline promoter. Growth rate analyses were performed on rich medium (7H9/oleic acid-albumin-dextrose-catalase [OADC]). OD values are means ± standard errors from three independent experiments. (B) Densitometric analysis of DnaG protein levels in M. smegmatis and ΔdnaGPtetdnaGMs. (C) Western blot analysis with antibodies raised against DnaG of M. smegmatis proteins isolated from cells growing in rich medium in indicated time intervals. Lane 1, M. smegmatis, 0 h; lane 2, ΔdnaGPtetdnaGMs, 0 h; lane 3, M. smegmatis, 6 h; lane 4, ΔdnaGPtetdnaGMs, 6 h; lane 5, M. smegmatis, 12 h; lane 6, ΔdnaGPtetdnaGMs, 12 h; lane 7, M. smegmatis, 24 h; lane 8, ΔdnaGPtetdnaGMs, 24 h.
DISCUSSION
The ability to replicate DNA is essential for the viability of every living organism. Consequently, the proteins involved in replication should be essential for viability, at least as long as other proteins cannot complement their functions. Some antibiotics (e.g., doxorubicin, suramin) are known to interfere with the replication machinery of bacteria; however, neither first-line anti-TB drugs nor second-line anti-TB drugs target the mycobacterial replisome (6). We previously evaluated NAD+-dependent DNA ligase (LigA), which appeared to be essential for mycobacterial viability, as a putative drug target (34). LigA inhibitors that do not affect ATP-dependent ligases were also identified (35, 36). Unexpectedly, depletion of LigA did not significantly affect the growth of mycobacteria (34), largely precluding LigA as a target for new anti-TB drugs. Recently, Biswas et al. developed a colorimetric primase-phosphatase assay as a tool for screening for efficient DnaG inhibitors (32). These researchers used this assay to screen 2,556 small molecules and identified suramin, doxorubicin, and ellagic acid as potential DnaG inhibitors. Here, we evaluated DnaG from mycobacteria as a drug target and engineered an M. smegmatis strain carrying only intact M. tuberculosis dnaG under the control of a chemically inducible promoter. As previously reported (21), dnaG appeared to be essential for the viability of M. smegmatis. The genomes of both M. tuberculosis and M. smegmatis contain multiple copies of a second class of DNA primases belonging to the AEP family. However, we found that DnaG is still essential, even in strains that overexpress these AEP enzymes, establishing that they cannot complement the priming activity of DnaG. The overexpressions of AEPs were confirmed at the protein (LigD) or RNA (Prim2, Prim3, and Prim4) level, and we are not able to exclude the possibility that, in the latter case, some troubles at the translation step occurred. Conversely, the overexpressing genes were originally from the same strain, and common problems (e.g., different GC content, codon usage) for heterogeneous protein expression should not have a place. In contrast to DnaG, individual or even all AEPs were inactivated without affecting the growth of M. smegmatis mutants. This observation would suggest that AEPs identified in mycobacteria do not participate in DNA replication and more likely have other roles, including DNA repair.
DnaGs of M. smegmatis and M. tuberculosis were expressed in E. coli and purified to near homogeneity for biochemical studies. The two enzymes were shown to possess primase activity in the presence of Mn2+ and ssDNA (24-mer) that was significantly inhibited in the presence of suramin or doxorubicin. However, only doxorubicin appeared to inhibit the growth of wild-type M. smegmatis and a ΔdnaG M. smegmatis mutant carrying an intact dnaG from M. smegmatis. The observed resistance to a high concentration of suramin might reflect the limited ability of this compound to permeate mycobacterial cell walls, which are well known for presenting a permeability barrier for hydrophobic and hydrophilic compounds (37, 38). The efficiency of a given inhibitor as an antibacterial drug depends on the intracellular ratio of inhibitor to its target. Thus, the concentration of target protein and the minimal concentration required to reduce cell viability are determinants of the success of treatment. In cases where bacilli are highly sensitive to reductions in the concentration of a given molecule, the molecule in question is a promising drug target. Mycobacterial DnaG may not satisfy this criterion because as little as 17% of wild-type DnaG levels appeared to be sufficient to support the growth of M. smegmatis; thus, DnaG may not be a good target for potential anti-TB drugs. As noted above, we observed a similar effect for mycobacterial LigA, another essential replication protein (34). This might suggest that replication proteins are overexpressed in mycobacteria under normal growth conditions, possibly owing to their relatively slow doubling times (39). It has also been reported that 1 to 3% of LigA is sufficient to support E. coli growth under laboratory conditions (40, 41). It is not clear why bacteria express “extra” replication proteins, but it may be related to additional functions performed by these proteins in the cell. The extensive use of LigA or DnaG in DNA damage and repair/recombination pathways might dictate that cells produce a much larger amount of these proteins. The overcapacity in terms of the amount of DnaG available in M. smegmatis suggests that an irreversible inhibitor would be required to eliminate DnaG activity. This likely inhibitory requirement should be taken into account during the screening of new chemicals to target this and related essential replication-associated proteins. The high level of identity between M. smegmatis and M. tuberculosis DnaG and the complementation of M. smegmatis ΔdnaG by intact dnaGMtb without a detectable effect on viability or the growth rate suggest that our findings are not limited to nonpathogenic mycobacteria. Thus, a fast-growing nonpathogenic strain expressing DnaG from M. tuberculosis would appear to be very useful for the initial testing of DnaG inhibitors identified by random in vitro screening or through rational drug design. Unlike LigA, mycobacterial DnaG was not replaced with its E. coli counterpart. This is consistent with the limited identity between M. smegmatis and E. coli DnaG (32%), which might preclude the interaction of E. coli DnaG with the mycobacterial replisome. The evaluation of DnaG as a putative drug target and construction of M. smegmatis conditional mutants should help in future studies to identify chemicals that efficiently target this essential replication protein in mycobacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the State Committee for Scientific Research (N303 3582 33) and the European Regional Development Fund, under Operational Programme Innovative Economy (POIG.01.01.02-10-107/09). A.D. was supported by a grant from the Biotechnology and Biological Sciences Research Council.
Footnotes
Published ahead of print 30 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01721-13.
REFERENCES
- 1.Dye C, Williams BG. 2010. The population dynamics and control of tuberculosis. Science 328:856–861. 10.1126/science.1185449 [DOI] [PubMed] [Google Scholar]
- 2.Gengenbacher M, Kaufmann SH. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36:514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136:420–425. 10.1378/chest.08-2427 [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention 2006. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs worldwide, 2000-2004. MMWR Morb. Mortal. Wkly. Rep. 55:301–305 [PubMed] [Google Scholar]
- 5.Raviglione M. 2006. XDR-TB: entering the post-antibiotic era? Int. J. Tuberc. Lung Dis. 10:1185–1187 [PubMed] [Google Scholar]
- 6.Zhang Y, Vilcheze C, Jacobs WR., Jr 2005. Mechanisms of drug resistance in Mycobacterium tuberculosis, p 115–142 In Cole ST, Eisenach KD, McMurray DN, Jacobs WR., Jr (ed), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC [Google Scholar]
- 7.Griep MA. 1995. Primase structure and function. Indian J. Biochem. Biophys. 32:171–178 [PubMed] [Google Scholar]
- 8.Lao-Sirieix SH, Pellegrini L, Bell SD. 2005. The promiscuous primase. Trends Genet. 21:568–572. 10.1016/j.tig.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 9.Lao-Sirieix SH, Nookala RK, Roversi P, Bell SD, Pellegrini L. 2005. Structure of the heterodimeric core primase. Nat. Struct. Mol. Biol. 12:1137–1144. 10.1038/nsmb1013 [DOI] [PubMed] [Google Scholar]
- 10.Corn JE, Berger JM. 2006. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 34:4082–4088. 10.1093/nar/gkl363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875–3896. 10.1093/nar/gki702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frick DN, Richardson CC. 2001. DNA primases. Annu. Rev. Biochem. 70:39–80. 10.1146/annurev.biochem.70.1.39 [DOI] [PubMed] [Google Scholar]
- 13.Weller GR, Doherty AJ. 2001. A family of DNA repair ligases in bacteria? FEBS Lett. 505:340–342. 10.1016/S0014-5793(01)02831-9 [DOI] [PubMed] [Google Scholar]
- 14.Aravind L, Koonin EV. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11:1365–1374. 10.1101/gr.181001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. 2005. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 351:531–544. 10.1016/j.jmb.2005.06.038 [DOI] [PubMed] [Google Scholar]
- 16.Pitcher RS, Wilson TE, Doherty AJ. 2005. New insights into NHEJ repair processes in prokaryotes. Cell Cycle 4:675–678. 10.4161/cc.4.5.1676 [DOI] [PubMed] [Google Scholar]
- 17.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d'Adda di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686–1689. 10.1126/science.1074584 [DOI] [PubMed] [Google Scholar]
- 18.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306:683–685. 10.1126/science.1099824 [DOI] [PubMed] [Google Scholar]
- 19.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. 2007. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 366:391–405. 10.1016/j.jmb.2006.10.046 [DOI] [PubMed] [Google Scholar]
- 20.Bartlett EJ, Brissett NC, Doherty AJ. 2013. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc. Natl. Acad. Sci. U. S. A. 110:E1984–E1991. 10.1073/pnas.1302616110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klann AG, Belanger AE, Abanes-De Mello A, Lee JY, Hatfull GF. 1998. Characterization of the dnaG locus in Mycobacterium smegmatis reveals linkage of DNA replication and cell division. J. Bacteriol. 180:65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911–1919. 10.1111/j.1365-2958.1990.tb02040.x [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24.Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 25.Brzostek A, Pawelczyk J, Rumijowska-Galewicz A, Dziadek B, Dziadek J. 2009. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 191:6584–6591. 10.1128/JB.00488-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawelczyk J, Brzostek A, Kremer L, Dziadek B, Rumijowska-Galewicz A, Fiolka M, Dziadek J. 2011. AccD6, a key carboxyltransferase essential for mycolic acid synthesis in Mycobacterium tuberculosis, is dispensable in a nonpathogenic strain. J. Bacteriol. 193:6960–6972. 10.1128/JB.05638-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swart JR, Griep MA. 1995. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry 34:16097–16106. 10.1021/bi00049a025 [DOI] [PubMed] [Google Scholar]
- 28.Collins LA, Franzblau SG. 1997. Microplate Alamar Blue assay versus Bactec 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 41:1004–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciarrocchi G, MacPhee DG, Deady LW, Tilley L. 1999. Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob. Agents Chemother. 41:2766–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 31.Kuchta RD, Stengel G. 2010. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804:1180–1189. 10.1016/j.bbapap.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswas T, Resto-Roldán E, Sawyer SK, Artsimovitch I, Tsodikov OV. 2013. A novel non-radioactive primase-pyrophosphatase activity assay and its application to the discovery of inhibitors of Mycobacterium tuberculosis primase DnaG. Nucleic Acids Res. 41:e56. 10.1093/nar/gks1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. 2006. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62:132–147. 10.1111/j.1365-2958.2006.05333.x [DOI] [PubMed] [Google Scholar]
- 34.Korycka-Machala M, Rychta E, Brzostek A, Sayer HR, Rumijowska-Galewicz A, Bowater RP, Dziadek J. 2007. Evaluation of NAD(+)-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 51:2888–2897. 10.1128/AAC.00254-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava SK, Tripathi RP, Ramachandran R. 2005. NAD+-dependent DNA Ligase (Rv3014c) from Mycobacterium tuberculosis. Crystal structure of the adenylation domain and identification of novel inhibitors. J. Biol. Chem. 280:30273–30281. 10.1074/jbc.M503780200 [DOI] [PubMed] [Google Scholar]
- 36.Dwivedi N, Dube D, Pandey J, Singh B, Kukshal V, Ramachandran R, Tripathi RP. 2008. NAD(+)-dependent DNA ligase: a novel target waiting for the right inhibitor. Med. Res. Rev. 28:545–568. 10.1002/med.20114 [DOI] [PubMed] [Google Scholar]
- 37.Trias J, Benz R. 1994. Permeability of the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 14:283–290. 10.1111/j.1365-2958.1994.tb01289.x [DOI] [PubMed] [Google Scholar]
- 38.Draper P. 1998. The outer parts of the mycobacterial envelope as permeability barriers. Front. Biosci. 3:1253–1261 [DOI] [PubMed] [Google Scholar]
- 39.Dziadek J, Rutherford SA, Madiraju MV, Atkinson MA, Rajagopalan M. 2003. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology 149:1593–1603. 10.1099/mic.0.26023-0 [DOI] [PubMed] [Google Scholar]
- 40.Lehman IR. 1974. DNA ligase: structure, mechanism, and function. Science 186:790–797. 10.1126/science.186.4166.790 [DOI] [PubMed] [Google Scholar]
- 41.Modrich P, Lehman IR. 1973. Deoxyribonucleic acid ligase. A steady state kinetic analysis of the reaction catalyzed by the enzyme from Escherichia coli. J. Biol. Chem. 248:7502–7511 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.