Abstract
The in vitro antibacterial activity of solithromycin (CEM-101) against macrolide-resistant isolates (n = 62) of Streptococcus agalactiae (group B streptococcus [GBS]) was determined. Phenotypic characterization of macrolide-resistant strains was performed by double-disc diffusion testing. A multiplex PCR was used to identify the erm(B), erm(TR), and mef(A/E) genes, capsular genotypes, and alpha-like (Alp) protein genes from the GBS strains. Determination of MIC was carried out using the microdilution broth method. The Etest method was used for penicillin, azithromycin, clarithromycin, and erythromycin. Solithromycin had a MIC50 of ≤0.008 μg/ml and a MIC90 of 0.015 μg/ml against macrolide-susceptible S. agalactiae. These MICs were lower than those displayed by penicillin (MIC50 of 0.032 μg/ml and MIC90 of 0.047 μg/ml), the antibiotic agent of choice for prophylaxis and treatment of GBS infections. Against macrolide-resistant S. agalactiae, solithromycin had a MIC50 of 0.03 μg/ml and a MIC90 of 0.125 μg/ml. Against erm(B) strains, solithromycin had a MIC50 of 0.03 μg/ml and a MIC90 of 0.06 μg/ml, while against mef(A) strains, it had a MIC50 of 0.03 μg/ml and a MIC90 of 0.125 μg/ml. Most erythromycin-resistant GBS strains were of serotype V (64.5%) and associated significantly with alp2-3. Moreover, a statistically significant association was observed between the constitutive macrolide-lincosamide-streptogramin B resistance (cMLSB) phenotype and the erm(B) gene-carrying strains, the alp2-3 gene and the M phenotype, and the mef(A/E) gene and epsilon. Overall, our results show that solithromycin had lower or similar MICs than penicillin and potent activity against macrolide-resistant strains independent of their genotype or phenotype, representing a valid therapeutic alternative where β-lactams cannot be used.
INTRODUCTION
Streptococcus agalactiae (group B streptococcus [GBS]) is a common cause of severe infections in neonates, such as sepsis and meningitis. It is also an important pathogen causing bacteremia and endocarditis in elderly patients, patients with diabetes, and immunocompromised subjects (1, 2). The highest GBS mortality and morbidity result from invasive infections in neonates, particularly in those with very low birth weight (3, 4). Due to the severity of disease resulting from S. agalactiae infections in neonates, the elderly, diabetics, and immunocompromised patients, the U.S. Food and Drug Administration (FDA) has recently proposed S. agalactiae as a qualified infectious diseases pathogen (5).
Penicillin is the first-line antibiotic for treatment of GBS infection, as well as for intrapartum antibiotic prophylaxis to prevent early-onset infection, because resistance to this agent has not been reported so far among GBS clinical isolates. Macrolides are the recommended second-line drugs and the first alternative in cases of β-lactam allergy.
However, in 2008, GBS clinical isolates were identified with reduced penicillin susceptibility, in which an increase was observed in the MICs of β-lactam antibiotics, including penicillin (MICs of 0.25 to 1 mg/liter) (6, 7). In addition, the rates of erythromycin resistance have increased at different levels in various regions in the world (8, 9). There are two mechanisms of resistance to macrolides: one is a modification of the ribosomal target site by a dimethylation of an adenine residue in the 23S rRNA, encoded by erm genes, and the other involving increased efflux of the drug outside the organism by macrolide efflux pumps, encoded by mef genes. Target site modification confers inducible (iMLSB) or constitutive (cMLSB) resistance to all antibiotics in the macrolide-lincosamide-streptogramin B group, while the presence of the efflux pump confers resistance only to 14- and 15-membered macrolides (M phenotype).
To overcome the macrolide resistance of Gram-positive cocci, the ketolides, which are macrolide analogs, were developed to treat respiratory infections due to microorganisms (Streptococcus pneumoniae and Streptococcus pyogenes) that are macrolide resistant.
Telithromycin was the first ketolide introduced as the drug able to address the macrolide resistance problem and received FDA approval in 2004. However, because of severe adverse events (10, 11), it is approved for use only in community-acquired bacterial pneumonia (CABP).
Solithromycin (CEM-101) is a novel fluoroketolide that shows activity comparable or superior to those of telithromycin, azithromycin, erythromycin, and clarithromycin, with high potency against Gram-positive and Gram-negative bacteria, as well as activity against most macrolide-resistant bacteria (12–14). It is currently being evaluated in a phase 3 trial as monotherapy for CABP.
The aim of this study was to evaluate the in vitro activity of solithromycin against a spectrum of S. agalactiae strains with different macrolide resistance genotypes and phenotypes compared to those of penicillin G, erythromycin, azithromycin, and clarithromycin. This collection of strains was further characterized for surface proteins and capsular type, which represent important virulence factors of GBS.
MATERIALS AND METHODS
Strain collection.
A total of 72 clinical isolates of S. agalactiae, which had been collected from Brescia's main hospital (Spedali Civili) between 2005 and 2012, were used in the MIC determination study. The isolates were recovered from different specimens (23 urine samples, 43 vaginal samples, 3 urethral swabs, and 3 rectal swabs). GBS strains were isolated by streak plating 1 to 10 μl of transport medium on ChromID streptoB agar plate (bioMérieux, St. Louis, MO). The plates were incubated at 37°C for 18 to 24 h under aerobic conditions. GBS was selected by the production of a pink pigment when grown aerobically on ChromID streptoB agar. GBS identification was performed by means of the Vitek system (bioMérieux).
Capsular gene typing.
The capsular genotype (Ia, Ib, and II to IX) of S. agalactiae was identified by a multiplex PCR assay as previously described (15). DNA was extracted from each strain using a DNeasy kit (Qiagen). Approximately 1 ng of DNA was used in the PCRs with primers and conditions as described elsewhere (15). Serotypes of strains were identified by analyzing the unique banding pattern following 1.5% (wt/vol) agarose gel electrophoresis.
Alp genes.
The alpha-like protein (Alp) genes bca, alp1 (Epsilon), alp2/3, Rib, and alp4 in the strains were detected by using a multiplex PCR as previously described (16). In brief, the PCR mixture (total volume, 25 μl) contained 1 ng of DNA template, 1× PCR buffer, 2 mmol/liter of MgCl2, 200 μmol/liter of deoxynucleoside triphosphates (dNTPs), 400 nmol/l of each of the five pairs of primers, and 0.3 U of AmpliTaq Gold (Roche). Amplification conditions were as previously described (16). Amplification of the alpha-like protein genes was evaluated by agarose gel (2%, wt/vol) electrophoresis of the PCR products.
Antimicrobial resistance phenotype and genotype.
Phenotypic characterization of macrolide-resistant strains was performed by double-disc diffusion testing as described previously (17). Erythromycin (15 μg) and clindamycin (2 μg) discs were placed 20 mm apart. Isolates resistant to erythromycin with blunting of the clindamycin inhibition were of the iMLSB phenotype, isolates that demonstrated resistance to both erythromycin and clindamycin were of the cMLSB phenotype, isolates showing resistance to erythromycin without blunting of the clindamycin inhibition zone were of the M phenotype, and isolates resistant to clindamycin yet susceptible or intermediate to erythromycin belonged to the L phenotype. Interpretive criteria were according to CLSI guidelines (18). A multiplex PCR was used to identify the erm(B), erm(TR), and mef(A/E) genes from the GBS strains, using primers and conditions previously reported (17–19), and a separate PCR was used to amplify the lin(B) gene (20, 21).
Antimicrobial agents and MIC determination.
Solithromycin (CEM-101) was obtained from Cempra, Inc., Chapel Hill, NC. Determination of MIC was carried out using the microdilution broth method according to CLSI guidelines (22). In brief, an inoculum of approximately 5 × 105 to 5 × 106 CFU/ml was incubated with a concentration of solithromycin ranging from 0.008 to 4 μg/ml. S. pneumoniae ATCC 49619 was used as a quality control. Results were observed after 18 h of incubation at 37°C. For comparison to solithromycin, penicillin, azithromycin, clarithromycin, and erythromycin were used. The Etest method (Liofilchem, Italy) was used for all of the reference antibiotics. The test was performed according to the manufacturer's instructions. Antibiotic concentrations ranged from 0.002 to 32 μg/ml for penicillin and from 0.016 to 256 μg/ml for azithromycin, clarithromycin, and erythromycin. Erythromycin was also tested by an automated microdilution broth method (Vitek2; bioMérieux). The concentrations ranged from 0.25 to 8 μg/ml. Breakpoint interpretation was done according to EUCAST guidelines (23), and breakpoints were as follows: penicillin, ≤0.25 and >0.25 μg/ml, susceptible and resistant, respectively; erythromycin, azithromycin, and clarithromycin, ≤0.25 and >0.5 μg/ml, susceptible and resistant, respectively.
Statistical analysis.
The χ2 test was used to evaluate the differences in distributions of surface proteins, serotypes, genotypes and phenotypes. A P value of <0.05 was considered significant, and a P value of <0.01 was considered highly significant.
RESULTS
MICs of antimicrobial agents for clinical strains.
The activities of solithromycin and the comparator antimicrobial agents against clinical strains are shown in Table 1. The MIC50 and the MIC90 of solithromycin were ≤0.008 and 0.015 μg/ml against erythromycin-susceptible strains, which were respectfully at least 4-fold and 3-fold lower than that of penicillin, the first-line agent both for intrapartum antibiotic prophylaxis and for the treatment of GBS infections in adults. On the other hand, erythromycin and clarithromycin had a MIC50 and a MIC90 comparable to that of penicillin, while azithromycin had both a MIC50 and MIC90 of ≤0.125 μg/ml. Against erythromycin-resistant strains, solithromycin had a MIC50 of 0.03 μg/ml and a MIC90 of 0.125 μg/ml. The MIC50 of penicillin was 0.032 and comparable to that of solithromycin, whereas the MIC90 of penicillin was 2.7-fold lower than that of solithromycin against erythromycin-resistant strains.
TABLE 1.
Activities of solithromycin and comparator antimicrobial agents against Streptococcus agalactiae
| Organism | Antimicrobial drug | MIC (mg/liter) |
|||
|---|---|---|---|---|---|
| 50% | 90% | Range observed | Range tested | ||
| Erythromycin-resistant GBS (n = 62) | Solithromycin | 0.03 | 0.125 | ≤0.008–1 | 0.008–4 |
| Penicillin | 0.032 | 0.047 | 0.012–0.06 | 0.002–32 | |
| Erythromycina | >8 | >8 | 0.5->8 | 0.25–8 | |
| Erythromycinb | >256 | >256 | 0.5->256 | 0.016–256 | |
| Azithromycin | >256 | >256 | 0.19->256 | 0.016–256 | |
| Clarithromycin | >256 | >256 | 0.25->256 | 0.016–256 | |
| Erythromycin-susceptible GBS (n = 10) | Solithromycin | ≤0.008 | 0.015 | ≤0.008–0.03 | 0.008–4 |
| Penicillin | 0.032 | 0.047 | 0.012–0.047 | 0.002–32 | |
| Erythromycina | ≤0.25 | ≤0.25 | ≤0.25 | 0.25–8 | |
| Erythromycinb | 0.047 | 0.047 | 0.012–0.064 | 0.016–256 | |
| Azithromycin | ≤0.125 | ≤0.125 | 0.019–0.19 | 0.016–256 | |
| Clarithromycin | 0.047 | 0.047 | 0.023–0.047 | 0.016–256 | |
Broth microdilution test.
Etest.
Evaluation of macrolide-resistant genotypes and phenotypes of GBS.
The determination of macrolide-resistant genotypes in GBS was performed to evaluate the differences in the activities between solithromycin and the other antimicrobial agents tested. Among the 62 macrolide-resistant clinical strains, 30 displayed the cMLSB phenotype, 21 the M phenotype, 7 the iMLSB phenotype, and 4 the L phenotype. Regarding L phenotypes, three were erythromycin-intermediate and clindamycin-resistant strains and one was erythromycin susceptible and clindamycin resistant by the disc diffusion test. To identify the cause of macrolide resistance, we screened for the presence of several genes. Most of the screened strains possessed a single resistance gene. Among these strains, the erm(B) gene was present in 26 strains and was mostly associated with the cMLSB phenotype, with a MIC of >256 μg/ml for almost all of the reference macrolides. The mef(A/E) gene was present in 22 strains, while erm(A) [subclass erm(TR)] was identified in 3 strains. The lin(B) gene was not detected in any GBS strains, and the L phenotypes observed were associated with the erm(B) gene (3 strains) and the mef(A/E) gene (1 strain). Eleven strains possessed more than one resistance gene. There were five isolates with a susceptible phenotype in which the presence of a resistance gene was detected, including the erm(B) gene (2 isolates), the erm(A) [subclass erm(TR)] (2 isolates), and one isolate that had both erm(B) and erm(A) [subclass erm(TR)].
Activities of the different antimicrobial agents against the various macrolide-resistant genotypes and phenotypes of GBS.
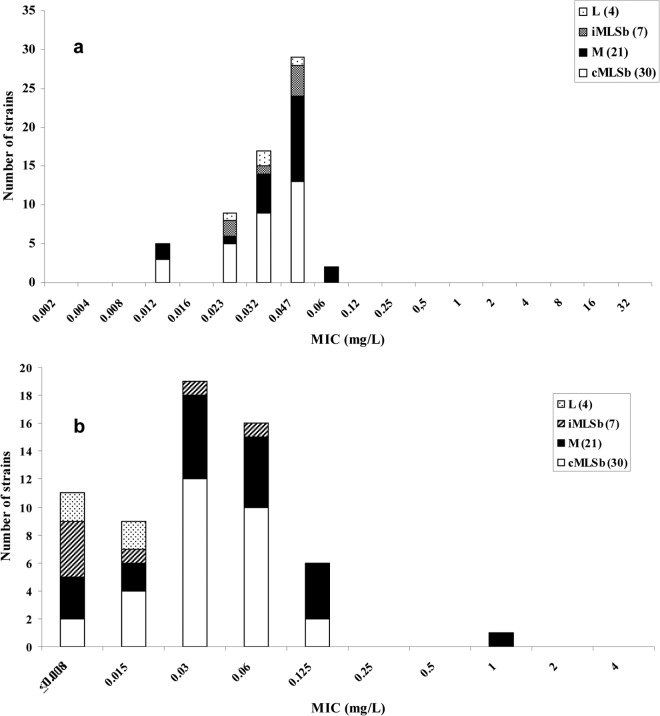
MIC distributions of solithromycin and penicillin for the different phenotypes of GBS are shown in Fig. 1. For solithromycin, most of the strains that displayed the cMLSB phenotype had a MIC between 0.03 and 0.06 μg/ml, while for penicillin the MIC range was between 0.03 and 0.047 μg/ml. Similar MIC distributions were observed for strains with the M phenotype. In contrast, most of the strains that had the iMLSB phenotype had a MIC of 0.047 μg/ml for penicillin and a MIC of ≤0.008 μg/ml for solithromycin.
FIG 1.
MIC distribution of penicillin (a) and solithromycin (b) for the different phenotypes of macrolide-resistant Streptococcus agalactiae strains.
Strains with the L phenotype had a MIC distribution between ≤0.008 and 0.015 μg/ml for solithromycin and between 0.023 and 0.047 μg/ml for penicillin (Fig. 1).
The majority of the erm(B) gene-carrying strains of S. agalactiae showed high resistance (MIC > 256 μg/ml) to clarithromycin and azithromycin. In contrast, solithromycin showed a MIC90 of 0.06 μg/ml against the same strains (Table 2). The MIC90 of solithromycin against mef(A/E) strains was 0.125 μg/ml, whereas the MIC90 for erm(B) gene-carrying strains was 2-fold lower (0.06 μg/ml).
TABLE 2.
MIC50s and MIC90s of solithromycin and comparator drugs against S. agalactiae strains with defined macrolide-resistant genotypes
| Drug | MIC (mg/liter) for strains with different macrolide resistance mechanisms (no. of strains) |
|||
|---|---|---|---|---|
|
erm(B) (26) |
mef(A/E) (22) |
|||
| MIC50 | MIC90 | MIC50 | MIC90 | |
| Solithromycin | 0.03 | 0.06 | 0.03 | 0.125 |
| Penicillin | 0.032 | 0.047 | 0.047 | 0.047 |
| Erythromycina | >8 | >8 | >8 | >8 |
| Erythromycinb | >256 | >256 | 6 | >256 |
| Azithromycin | >256 | >256 | 12 | >256 |
| Clarithromycin | >256 | >256 | 6 | >256 |
Broth microdilution test.
Etest.
Capsule serotyping and the alp family genes.
The detection of genes encoding particular capsular serotypes was performed by multiplex PCR. Overall, the most represented serotypes isolated were types V (n = 40 [55.5%]), III (n = 12 [16.6%]), and Ia (n = 10 [13.8%]), followed by serotypes Ib, II, and IV, which were represented by three isolates each (4%). One strain belonged to serotype VI. No strains of serotypes VII and VIII were found in this pool of isolates.
Surface proteins of GBS are likely to play an important role in the pathogenesis of S. agalactiae infection; therefore, they were evaluated by PCR. The presence of a particular alp gene in relation to the serotype was noted (Table 3).
TABLE 3.
Distribution of the alp genes among the observed GBS serotypes
| Surface alpha-like protein gene (no. of isolates) | No. of isolates per serotypea |
||||||
|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | |
| alp2-3 (30) | 2 | 3 | 25** | ||||
| rib (33) | 4 | 3 | 3 | 9* | 2 | 12 | |
| epsilon (22) | 6* | 2 | 14 | ||||
| alpha c (10) | 2 | 2 | 6 | ||||
| alp 4 (14) | 1 | 4 | 8 | 1 | |||
*, P < 0.05; **, P < 0.01.
Of the 30 alp2-3-positive strains isolated, 25 were of serotype V; 14 of 22 epsilon-positive strains corresponded to serotype V, and 6 corresponded to serotype Ia. rib-positive strains were present in almost all serotypes isolated. Conversely, a certain serotype commonly corresponded to a particular Alp gene: serotype Ib and II presented rib, serotype IV carried either rib or epsilon, many serotype V strains (62.5%) possessed alp2-3, and serotype Ia predominantly carried epsilon (6/15 isolates), while rib was the most common surface protein associated with the serotype III (9/18) (P < 0.05). Different associations of alp genes were present in a single strain. Regarding the phenotypes and genotypes, we found a statistically significant association between the cMLSB phenotype and the erm(B) gene-carrying strains (P < 0.05), between the cMLSB phenotype and alp2-3 (P < 0.05), between the M phenotype and epsilon (P < 0.05), and between the mef(A/E) gene-carrying strains and epsilon (P < 0.001) (data not shown).
DISCUSSION
The recent emergence of S. agalactiae strains with reduced penicillin susceptibility in Japan and the United States constitutes a problem for the use of this drug in prophylaxis (6, 24). The increasing importance of S. agalactiae has been noted by its inclusion in the list of proposed qualified pathogens by the FDA. The molecular analysis of these particular strains showed a mutagenic pathway comparable to that observed when the first β-lactam-resistant S. pneumoniae strains were isolated. The emergence of a physiologically GBS pbp2x (Q557E) mutant is worrying, because the accumulation of additional mutations might lead to complete penicillin resistance. This suggests a potential risk of therapeutic failure of intrapartum prophylaxis in the near future.
Traditionally the macrolides, and in particular erythromycin, have been considered the second-line choice of antibiotic in patients allergic to β-lactams. However, resistance to macrolides and lincosamides has risen during the last decades, with 19% of the S. agalactiae isolates resistant to erythromycin and 53% of these showing resistance to clindamycin (25). Regarding the erythromycin resistance among strains of S. agalactiae, we have previously found a resistance rate of 15% (26), a result similar to what has been observed in Spain, Portugal, Germany, France, and Canada (27–30), but resistance rates differ considerably between regions, with a rate of only 3.8% reported in the Czech Republic (31) and 38% to 41.9% in the United States (8).
To address the resistance problem, new macrolide antibiotics called ketolides have been developed that have potent activity against erythromycin-resistant streptococci.
In the collection of S. agalactiae isolates used in this study, there was a predominance of cMLSB and M phenotypes, indicating that erythromycin resistance was mediated by the two principal mechanisms: methylation of 23S rRNA, determined by erm genes, and active drug efflux by pumps encoded by mef genes. These strains showed cross-resistance to clarithromycin and azithromycin, with MIC90s of >256 μg/ml.
The novel fluoroketolide solithromycin tested in this study demonstrated superior potency over older macrolides against all macrolide-resistant strains, with a MIC90 of 0.125 μg/ml. The enhanced activity of solithromycin over other ketolide compounds is likely due to a higher binding affinity to bacterial ribosomes based on an 11,12-carbamate-butyl-[1,2,3]-triazolyl-amino-phenyl side chain as well as a 2-fluoro modification (32). Solithromycin demonstrated potent activity against macrolide-susceptible GBS, with a MIC90 of 0.015 μg/ml, which was 3-fold lower than that of penicillin. Although strains with either mef(A/E) or erm(B) have slightly higher solithromycin MICs than susceptible strains, the solithromycin MIC for macrolide-resistant GBS rarely exceeds 0.125 μg/ml. This lower MIC suggests that this drug may be useful in the treatment of infections caused by these pathogens. There were five isolates of the cMLSB phenotype that had both erm(A) subclass erm(TR) and erm(B) genes; the coexistence of both genes has been documented previously (33). Furthermore, three isolates that displayed the cMLSB phenotype harbored both mef(A/E) genes and erm(B), and one isolate that had the iMLSB phenotype had both erm(A) subclass erm(TR) and mef(A/E). This finding implies differential gene expression, as only the erm(B) gene and erm(TR) gene were expressed in the different isolates, respectively. Exceptionally and for the first time, to our knowledge, we found one strain that harbored all three macrolide resistance genes and displayed the cMLSB phenotype.
We observed that all of the GBS strains that had the iMLSB phenotype and harbored the erm(B) or the erm(A) (subclass ermTR) gene expressed low-level resistance to erythromycin (MICs, 1 to 12 μg/ml) but high azithromycin MICs in absolute terms (2 to >256 μg/ml). This unusual resistance pattern has been previously identified in macrolide-resistant S. pyogenes strains harboring the erm(A) gene with point mutations in the erm(A) regulatory region leading to constitutive methylase expression (34). Whether or not this was the case for the strains isolated in this study requires further evaluation.
Further, we identified five macrolide-susceptible strains that contained the erm(B) or erm(TR) gene or both, as has been reported previously (17). Whether it is possible for these susceptible strains carrying macrolide resistance genes to become resistant upon environmental stimulus or over time is unknown.
It has been hypothesized previously that the spread of strains of particular surface protein profiles and serotypes reflects the selection of the best evolutionary lineages by the immune system (35). In this study, we found that our isolates presented serotype-surface protein gene combinations (serotype V-alp2-3 and serotype III-rib) already reported (35, 36) and a different combination (serotype Ia-epsilon) that we observed in a previous study (26), suggesting that new successfully selected clones may be emerging. Moreover, statistically significant associations were observed between the cMLSB phenotype and the erm(B) gene-carrying strains, alp2-3 and the M phenotype, and the mef (A/E) gene-carrying strains and epsilon.
Among strains resistant to macrolides, the V serotype dominated (40/62 [64.5%]), an association previously reported (37). Our results are consistent with the literature and underline the spread of a phenomenon during the past years, which is an increasing number of GBS isolates being resistant to erythromycin, representing serotype V. Given this trend, the excellent activity of solithromycin against macrolide-susceptible and macrolide-resistant GBS observed in this study becomes more relevant, as this compound may represent a valid alternative in the treatment of infections caused by this pathogen, in particular if there is a limitation of therapeutic options.
ACKNOWLEDGMENTS
This study was supported by Cempra Pharmaceuticals, Inc., Chapel Hill, NC.
We thank Kara Keedy for her assistance in the preparation of the manuscript.
Footnotes
Published ahead of print 30 December 2013
REFERENCES
- 1.Baker CJ. 2000. Group B streptococcal infections, p 222–237 In Stevens DL, Kaplan EL. (ed), Streptococcal infections: clinical aspects, microbiology, and molecular pathogenesis. Oxford University Press, New York, NY [Google Scholar]
- 2.Jackson LA, Hilsdon R, Farley MM, Harrison LH, Reingold AL, Plikaytis BD, Wenger JD, Schuchat A. 1995. Risk factors for group B streptococcal disease in adults. Ann. Intern. Med. 123:415–420. 10.7326/0003-4819-123-6-199509150-00003 [DOI] [PubMed] [Google Scholar]
- 3.Heath PT, Balfour G, Weisner AM, Efstratiou A, Lamagni Tl, Tighe H, O'Connell LA, Cafferkey M, Verlander NQ, Nicoll A, McCartney AC, PHLS GBS Working Group. 2004Group B streptococcal disease in UK and Irish infants younger than 90 days. Lancet 363:292–294. 10.1016/S0140-6736(03)15389-5 [DOI] [PubMed] [Google Scholar]
- 4.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK. 2002. Changes in pathogens causing early-onset sepsis in very low-birth weight infants. N. Engl. J. Med. 347:240–247. 10.1056/NEJMoa012657 [DOI] [PubMed] [Google Scholar]
- 5.Federal Register 2013. Establishing a list of qualifying pathogens under the Food And Drug Administration Safety and Innovation Act. Proposed rule. Fed. Regist. 78:35155–35173 [PubMed] [Google Scholar]
- 6.Kimura K, Suzuki S, Wachino J, Kurokawa H, Yamane K, Shibata N, Nagano N, Kato H, Shibayama K, Arakawa Y. 2008. First molecular characterization of group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:2890–2897. 10.1128/AAC.00185-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagano N, Nagano Y, Kimura K, Tamai K, Yanagisawa H, Arakawa Y. 2008. Genetic heterogeneity in pbp genes among clinically isolates group B streptococci with reduced penicillin susceptibility. Antimicrob. Agents Chemother. 52:4258–4267. 10.1128/AAC.00596-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borchardt S, DeBusscher JH, Tallman PA, Manning SD, Marrs CF, Kurzynski TA, Foxman B. 2006. Frequency of antimicrobial resistance among invasive and colonising group B streptococcal isolates. BMC Infect. Dis. 6:57–64. 10.1186/1471-2334-6-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinheiro S, Radhouani H, Coelho C, Gonçalves A, Carvalho JA, Ruiz-Larrea F, Torres C, Igrejas G, Poeta P. 2009. Prevalence and mechanisms of erythromycin resistance in Streptococcus agalactiae from healthy pregnant women. Microb. Drug Resist. 15:121–124. 10.1089/mdr.2009.0895 [DOI] [PubMed] [Google Scholar]
- 10.Brinker AD, Wassel RT, Lyndly I, Serrano J, Avigan M, Lee WM, Seef LB. 2009. Telithromycin-associated hepatotoxicity: clinical spectrum and causality assessment of 42 cases. Hepatology 49:250–257. 10.1002/hep.22620 [DOI] [PubMed] [Google Scholar]
- 11.Onur O, Guneysel O, Denizbasi A, Celikel C. 2007. Acute hepatitis attack after exposure to telithromycin. Clin. Ther. 29:1725–1729. 10.1016/j.clinthera.2007.08.004 [DOI] [PubMed] [Google Scholar]
- 12.McGhee P, Clark C, Kosowska-Shick K, Nagai K, Dewasse B, Beachel L, Appelbaum PC. 2010. In vitro activity of CEM-101 against Streptococcus pneumoniae and Streptococcus pyogenes with defined macrolide resistance mechanisms. Antimicrob. Agents Chemother. 54:230–238. 10.1128/AAC.01123-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woosley LN, Castanheira M, Jones RN. 2010. CEM-101 activity against Gram-positive organisms. Antimicrob. Agents Chemother. 54:2182–2187. 10.1128/AAC.01662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putnam SD, Castanheira M, Moet GJ, Farrell DJ, Jones RN. 2010. CEM-101, a novel fluoroketolide: antimicrobial activity against a diverse collection of Gram-positive and Gram-negative bacteria. Diagn. Microbiol. Infect. Dis. 66:393–401. 10.1016/j.diagmicrobio.2009.10.013 [DOI] [PubMed] [Google Scholar]
- 15.Imperi M, Pataracchia M, Alfarone G, Baldassari L, Orefici G, Creta R. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 80:212–214. 10.1016/j.mimet.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 16.Creti R, Fabretti F, Orefici G, von Hunolstein C. 2004. Multiplex PCR assay for direct identification of group B streptococcal α-protein-like protein genes. J. Clin. Microbiol. 42:1326–1329. 10.1128/JCM.42.3.1326-1329.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gygax SE, Schuyler JA, Kimmel LE, Trama JP, Mordechai E, Adelson ME. 2006. Erythromycin and clindamycin resistance in group B streptococcal clinical isolates. Antimicrob. Agents Chemother. 50:1875–1877. 10.1128/AAC.50.5.1875-1877.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute 2010. Performance standard for antimicrobial susceptibility testing, M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 19.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Azavedo JCS, Yeung RH, Bast DJ, Duncan CL, Borgia SB, Low DE. 1999. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob. Agents Chemother. 43:2144–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desjardins M, Delgaty KL, Ramotar K, Seetaram C, Toye B. 2004. Prevalence and mechanisms of erythromycin resistance in group A and group B streptococcus: implications for reporting susceptibility results. J. Clin. Microbiol. 42:5620–5623. 10.1128/JCM.42.12.5620-5623.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, M7-A9. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 23.EUCAST 2012. Breakpoint tables for interpretation of MICs and zone diameters, version 2.0. European Committee on Antimicrobial Susceptibility Testing. 2012. http://www.eucast.org/clinical_breakpoints/.
- 24.Dahesh S, Hensler ME, Van Sorge NM, Gertz RE, Jr, Schrag S, Nizet V, Beall BW. 2008. Point mutation in the group B streptococcal pbp2x gene conferring decreased susceptibility to beta-lactam antibiotics. Antimicrob. Agents Chemother. 52:2915–2918. 10.1128/AAC.00461-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florindo C, Viegas S, Paulino A, Rodrigues E, Gomes JP, Borrego MJ. 2010. Molecular characterization and antimicrobial susceptibility profiles in Streptococcus agalactiae colonizing strains: association of erythromycin resistance with subtype III-1 genetic clone family. Clin. Microbiol. Infect. 16:1458–1463. 10.1111/j.1469-0691.2010.03106.x [DOI] [PubMed] [Google Scholar]
- 26.De Francesco MA, Caracciolo S, Gargiulo F, Manca N. 2012. Phenotypes, genotypes, serotypes and molecular epidemiology of erythromycin resistant Streptococcus agalactiae in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 31:1741–1747. 10.1007/s10096-011-1495-4 [DOI] [PubMed] [Google Scholar]
- 27.Fluegge K, Supper S, Siedler A, Berner R. 2004. Antibiotic susceptibility in neonatal invasive isolates of Streptococcus agalactiae in a 2-year nationwide surveillance study in Germany. Antimicrob. Agents Chemother. 48:4444–4446. 10.1128/AAC.48.11.4444-4446.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez JJ, Andreu A, the Spanish Group for the Study of Perinatal infection from the Spanish Society for Microbiology and Infectious Diseases 2005. Multicenter study of the mechanisms of resistance and clonal relationships of Streptococcus agalactiae isolates resistant to macrolides, lincosamides, and ketolides in Spain. Antimicrob. Agents Chemother. 49:2525–2527. 10.1128/AAC.49.6.2525-2527.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Azavedo JC, McGavin M, Duncan C, Low DE, McGeer A. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504–3508. 10.1128/AAC.45.12.3504-3508.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitoussi F, Loukil C, Gros I, Clermont O, Mariani P, Bonacorsi S, Le Thomas I, Deforche D, Bingen E. 2001. Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45:1889–1891. 10.1128/AAC.45.6.1889-1891.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motlová J, Strakova L, Urbaskova P, Sak P, Sever T. 2004. Vaginal and rectal carriage of Streptococcus agalactiae in the Czech Republic: incidence, serotypes distribution and susceptibility to antibiotics. Indian J. Med. Res. 119:84–87 [PubMed] [Google Scholar]
- 32.Llano-Soleto B, Klepacki D, Mankinb AS. 2008. Binding and action of CEM-101, a new macrolide/ketolide in development for treating infections with macrolide-resistant and macrolide-susceptible bacteria, abstr. F-3983. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., 25 to 28 October 2008, Washington, DC [Google Scholar]
- 33.Bingen E, Fitoussi F, Doit C, Cohen R, Tanna A, George R, Loukil C, Brahimi N, Le Thomas I, Deforche D. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. 10.1128/AAC.44.6.1453-1457.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malhotra-Kumar S, Mazzariol A, Van Heirstraeten L, Lammens C, de Rijk P, Cornaglia G, Goossens H. 2009. Unusual resistance patterns in macrolide-resistant Streptococcus pyogenes harbouring ermA. J. Antimicrob. Chemother. 63:42–46. 10.1093/jac/dkn432 [DOI] [PubMed] [Google Scholar]
- 35.Lindahl G, Stalhammar-Carlemalm M, Areschoug T. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102–127. 10.1128/CMR.18.1.102-127.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quentin R, Huet H, Wang FS, Geslin P, Goudeau A, Selander RK. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 33:2576–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gherardi G, Imperi M, Baldassarri L, Pataracchia M, Alfarone G, Recchia S, Orefici G, Dicuonzo G, Creti R. 2007. Molecular epidemiology and distribution of serotypes, surface proteins and antibiotic resistance in Italy. J. Clin. Microbiol. 45:2909–2916. 10.1128/JCM.00999-07 [DOI] [PMC free article] [PubMed] [Google Scholar]