Abstract
Omadacycline is a novel first-in-class aminomethylcycline with potent activity against important skin and pneumonia pathogens, including community-acquired methicillin-resistant Staphylococcus aureus (MRSA), β-hemolytic streptococci, penicillin-resistant Streptococcus pneumoniae, Haemophilus influenzae, and Legionella. In this work, the mechanism of action for omadacycline was further elucidated using a variety of models. Functional assays demonstrated that omadacycline is active against strains expressing the two main forms of tetracycline resistance (efflux and ribosomal protection). Macromolecular synthesis experiments confirmed that the primary effect of omadacycline is on bacterial protein synthesis, inhibiting protein synthesis with a potency greater than that of tetracycline. Biophysical studies with isolated ribosomes confirmed that the binding site for omadacycline is similar to that for tetracycline. In addition, unlike tetracycline, omadacycline is active in vitro in the presence of the ribosomal protection protein Tet(O).
INTRODUCTION
Omadacycline is the first of the novel aminomethylcyclines, which are semisynthetic compounds related to the tetracyclines, to undergo clinical development (Fig. 1). The tetracycline family of antimicrobials has been in clinical use for over 60 years and includes tetracycline, doxycycline, and minocycline. As a class, they are well tolerated; have a broad spectrum of antimicrobial activity, including against Gram-positive bacteria, Gram-negative bacteria, anaerobes, and atypical bacteria; and have proven effective in the treatment of a variety of bacterial infections involving respiratory tract, skin and skin structure, urinary tract, and intra-abdominal sites (1, 2).
FIG 1.
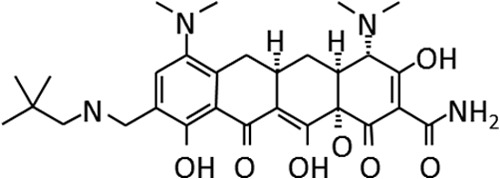
Chemical structure of omadacycline.
When first released in the 1950s to 1960s, the tetracyclines were an important component of the antibiotic armamentarium. Their clinical use declined in subsequent years, primarily due to the increasing prevalence of tetracycline resistance and the availability of effective alternative therapies. There are two major mechanisms of tetracycline resistance: efflux and ribosome protection. The two mechanisms have been described in Gram-positive and Gram-negative bacteria either separately or together, with ribosome protection generally more common in Gram-positive bacteria and efflux in Gram-negative bacteria (3). The most common genotypes of ribosome protection are tet(M) and tet(O). Efflux is determined by a family of related genotypes, in particular, tet(K) and tet(B) (2).
Omadacycline has potent activity against important skin and lung pathogens, including community-acquired methicillin-resistant Staphylococcus aureus (MRSA), β-hemolytic streptococci, penicillin-resistant Streptococcus pneumoniae, Haemophilus influenzae, and Legionella. The compound specifically overcomes tetracycline resistance mechanisms and is not affected by mechanisms of resistance to other classes of antibiotics. Omadacycline is entering phase 3 development for treatment of acute bacterial skin and skin structure infections (ABSSSI), community-acquired bacterial pneumonia (CABP), and complicated urinary tract infection (cUTI). We present here data examining the mechanism of action (MOA) for omadacycline.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains were purchased from the American Type Culture Collection or came from the laboratory of Stuart Levy. Characterization of strains for the mechanism of tetracycline resistance was done as previously described (4).
Source of omadacycline and tetracycline standards.
Chemistries for the synthesis of novel tetracyclines have been previously described (5). Briefly, reagents and catalysts were used without further purification. Reactions were monitored by high-performance liquid chromatography (HPLC) with UV at 280 nm, using C18 reverse-phase 4-μm-pore-size analytical columns (50 mm in length by 4.5 mm in diameter). Omadacycline was purified by preparative HPLC separations. 1H nuclear magnetic resonance (NMR) spectra were recorded at 300 or 400 MHz, and the chemical shift values are expressed in δ values. The tetracycline standards (tetracycline, doxycycline, and minocycline) and other antibiotics were purchased from Sigma-Aldrich.
MIC determinations.
Susceptibility testing was performed according to the M7-A5 CLSI-recommended microdilution method. Cation-adjusted Mueller-Hinton broth (MHB) was used. To prepare the inoculum, organisms were grown to a 0.5 McFarland standard, which was measured with a Microscan turbidity meter. Microplates were incubated at 35°C for 18 to 24 h as specified by CLSI M07-08.
Macromolecular synthesis.
Macromolecular synthesis analysis was performed as previously described (6, 7) with the following modifications. Isogenic strains of Staphyloccocus aureus, RN450 (tetracycline sensitive) and RN4250 [tetracycline resistant, carrying the tet(K) resistance element], were used. The tests were performed using 2-fold dilutions of test compounds (ranging from 0.03 to 32 μg/ml) in a 96-well format. Overnight Mueller-Hinton broth cultures of bacterial strains were diluted to an optical density at 530 nm (OD530) of 0.4 and incubated with shaking for 1 h. These cells were used to inoculate a test plate containing diluted test and control compounds. Radiolabeled precursors were added, and the plates were incubated with shaking at 37°C. The reactions were quenched with 50% trichloroacetic acid (TCA) and refrigerated for 1 h before harvesting to a Filtermat was performed. The Filtermat with scintillant was counted for 1 min per sample in a PerkinElmer Wallac (Boston, MA) Microbeta 1450 instrument.
In vitro translation.
Ribosomes were collected from Escherichia coli MRE600 and further purified on a sucrose gradient (8). Poly(U)-dependent Poly(Phe) synthesis (in vitro translation) was carried out using binding buffer conditions (20 mM HEPES-KOH [pH 7.6], 6 mM Mg acetate, 150 mM NH4Cl, 4 mM 2-mercaptoethanol, 0.05 mM spermine, 2 mM spermidine) at 30°C for 1 h as detailed in reference 9. Tet(O) protein was purified by nickel-affinity chromatography (10) and added in a 1:1 molar ratio with ribosomes as appropriate. Poly(Phe) chains were precipitated by adding 100 ml of 1% bovine serum albumin and 2 ml of 5% trichloroacetic acid (TCA) and incubated at 90°C for 15 min. The mixture was filtered through glass fiber filters, washed twice with 2 ml of 5% trichloroacetic acid, and washed once with 2 ml of a 1:1 mixture of ether and ethanol. Filters were dried and counted after the addition of 300 ml of Soluene 350 (Packard, Meriden, CT) and 7 ml of scintillation fluid.
Ribosomal binding.
A series of tubes were prepared with increasing concentrations (in μM) of the competitor test compound and a fixed concentration (3 μM) of radiolabeled tetracycline ([3H]tetracycline). A total of 24 pmol of ribosomes in binding buffer was incubated at 37°C for 5 min. The labeled tetracycline/test competitor solutions were added, and the competition assay was incubated at 37°C for 40 min. The samples were collected on 45-μM-pore-size nitrocellulose filters and washed 3 times. The filters were allowed to dry and added to a scintillation vial with scintillation fluid, and counts per minute were measured (11). The percentage of [3H]tetracycline binding in the absence of competitor was taken as 100%. Data were fitted to a model for one-site competitive binding to determine the IC50 value (the concentration of competitor at which half-maximal binding of [3H]tetracycline occurs) using SigmaPlot (SSPC Inc., Chicago, IL).
RESULTS
The activity of omadacycline against tetracycline-sensitive and -resistant strains.
Unlike tetracycline and doxycycline, omadacycline is active against strains expressing either efflux or ribosomal mechanisms of tetracycline resistance. MIC values for omadacycline are comparable among tetracycline-susceptible or -resistant strains. In addition, omadacycline is active against strains expressing various forms of tetracycline efflux or ribosomal protection (Table 1).
TABLE 1.
Activity of omadacycline against tetracycline-susceptible and -resistant strainsa
| Strain | Tetr determinant | Mechanism type | MIC (μg/ml) |
||
|---|---|---|---|---|---|
| Omadacycline | Tetracycline | Doxycycline | |||
| S. aureus RN450 | None | None | 0.125 | ≤0.06 | ≤0.06 |
| S. aureus ATCC 29213 | None | None | 0.25 | 0.125 | 0.125 |
| S. aureus MRSA5 | Tet(M) | Ribosomal protection | 0.125 | >64 | 4 |
| S. aureus RN4250 | Tet(K) | Efflux | 0.25 | 32 | 4 |
| S. pneumoniae PBS382 | Tet(O) | Ribosomal protection | <0.06 | 32 | 4 |
Antibacterial MICs were determined using CLSI methodology for antibacterial susceptibility testing. Tetracyline resistance determinants were confirmed by PCR as previously described (4).
Omadacycline is an inhibitor of protein synthesis.
Tetracyclines are known inhibitors of protein synthesis. To confirm the activity of omadacycline as a protein synthesis inhibitor and to demonstrate that a new mechanism of action was not responsible for activity against tetracycline-resistant bacteria, we used a whole-cell assay to assess inhibition of macromolecular synthesis of protein, DNA, RNA, or cell wall precursors into macromolecules (Table 2). Omadacycline and other tetracyclines exhibit preferential inhibition of protein synthesis. While tetracycline displays higher IC50s in strains carrying tetracycline resistance mechanisms, omadacycline is unaffected by the presence of either efflux [tet(K)] or ribosome protection [tet(M)], consistent with MIC determinations. In addition to their effects on protein synthesis, tetracyclines also exhibit moderate inhibition of peptidoglycan synthesis, which may be secondary to the effect on protein synthesis.
TABLE 2.
Effect of omadacycline on macromolecular synthesis in susceptible and resistant S. aureus strainsa
| Compound | Strain | Tetr | MIC (μg/ml) | IC50 protein synthesis (μg/ml) | IC50 RNA synthesis (μg/ml) | IC50 DNA synthesis (μg/ml) | IC50 PG synthesis (μg/ml) |
|---|---|---|---|---|---|---|---|
| Omadacycline | RN450 | None | 0.125 | <0.03 | >32 | >32 | 11.6 |
| ATCC 29213 | None | 0.25 | 0.19 | >32 | >32 | 15.7 | |
| RN4250 | tet(K) | 0.25 | 0.08 | >32 | >32 | >32 | |
| MRSA5 | tet(M) | 0.125 | 0.11 | >32 | >32 | 15.6 | |
| Tetracycline | RN450 | None | <0.06 | 0.04 | 31.4 | 25.7 | 8.8 |
| ATCC 29213 | None | 0.125 | 0.09 | 23.7 | >32 | 7.6 | |
| RN4250 | tet(K) | 32 | 13.8 | 32 | >32 | 22.7 | |
| MRSA5 | tet(M) | >64 | 1.8 | >64 | >32 | >32 | |
| Doxycycline | RN450 | None | <0.06 | 0.02 | >32 | >32 | 3.3 |
| ATCC 29213 | None | 0.125 | 0.08 | >32 | >32 | 2.9 | |
| Rifampin | RN450 | None | NT | 0.01 | 0.01 | >32 | >32 |
| ATCC 29213 | None | NT | <0.01 | 0.01 | >32 | >32 | |
| Ciprofloxacin | RN450 | None | 0.5 | 14.0 | >32 | 0.4 | >32 |
| ATCC 29213 | None | 0.5 | >32 | >32 | 0.3 | >32 | |
| Fosfomycin | RN450 | None | NT | >32 | >32 | >32 | 7.8 |
| ATCC 29213 | None | NT | >32 | >32 | >32 | 11.9 |
Antibacterial MICs were determined using CLSI methodology for antibacterial susceptibility testing. Tetracycline resistance determinants were confirmed by PCR as described in Materials and Methods. Macromolecular synthesis was performed as described previously (6, 7) and as described in Materials and Methods. Tetracycline and doxycycline are known protein synthesis inhibitors of the tetracycline class. Rifampin inhibits RNA polymerase, ciprofloxacin DNA synthesis, and fosfomycin peptidoglycan (PG) synthesis. The effects of rifampin on protein synthesis are secondary to the effects on RNA production (19). “Tetr” refers to the tetracycline-resistant element carried by the strain. IC50 data represent the calculated concentration of drug that reduces activity in the assay by 50% relative to the no-drug control. NT, not tested.
Omadacycline inhibits protein synthesis in the presence of Tet(O).
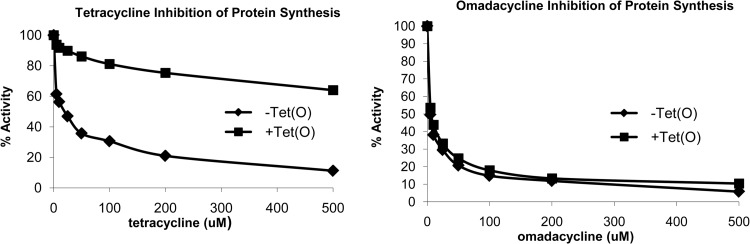
The ribosomal protection protein Tet(O) interacts with the 70S ribosome and promotes the release of bound tetracycline. Using a cell-free in vitro protein synthesis reaction, the ability of omadacycline to inhibit protein synthesis was investigated in both the presence and absence of the Tet(O). As shown in Fig. 2, omadacycline effectively inhibited protein synthesis in a cell-free system regardless of whether Tet(O) was present or not. This result is in marked contrast to those determined for tetracycline, whose activity was inhibited by the presence of the ribosomal protection protein Tet(O) (Fig. 2).
FIG 2.
Effect of Tet(O) protein on the protein synthesis activity of omadacycline in vitro. Poly(U)-dependent Poly(Phe) synthesis (in vitro translation) was carried out in the presence or absence of purified Tet(O) added in a 1:1 molar ratio with ribosomes and various concentrations of either omadacycline or tetracycline. Percent activity relative to the control reaction (no omadacycline or tetracycline) is plotted.
Omadacycline has a ribosomal binding site similar to that of tetracycline.
To address the binding site for omadacycline, competition experiments were performed where [3H]tetracycline and either omadacycline or minocycline competed for binding to purified ribosomes. In these studies, labeled tetracycline and unlabeled omadacycline or minocycline were given equal opportunities to bind to the ribosome. As shown in Fig. 3, both omadacycline and minocycline efficiently competed with labeled tetracycline for binding to 70S ribosomes. The IC50 values for omadacycline and minocycline were comparable. Previous studies have shown minocycline to have a greater affinity for binding to the ribosome than tetracycline (12).
FIG 3.

Ribosomal binding competition studies with [3H]tetracycline and unlabeled minocycline (A) or omadacycline (B). A series of tubes were prepared containing purified 70S ribosomes and increasing concentrations of the competitor test compound and a fixed concentration (3 μM) of [3H]tetracycline. Data are plotted as the log of the unlabeled competitor concentration in nM versus the percent binding relative to the reaction mixture containing no competitor. IC50 values were calculated by nonlinear curve fitting to a one-site competitive binding model and were determined to be 1.63 ± 0.01 μM (standard error) for minocycline and 1.96 ± 0.01 μM for omadacycline.
DISCUSSION
The tetracyclines are a family of structurally related compounds that have been in clinical use for over 50 years. It is known that the antibacterial action of the tetracyclines can be classified into two categories on the basis of mechanism of action (13, 14). Clinically used bacteriostatic tetracyclines such as tetracycline, doxycycline, minocycline, or tigecycline are considered the “typical” tetracyclines with inhibition of protein synthesis as a known mechanism of action. However, other tetracyclines such as chelocardin or anhydrotetracycline are known as atypical tetracyclines. These atypical compounds are poor inhibitors of protein synthesis and instead have cytotoxic perturbation of the cytoplasmic membrane as their primary mechanism of action (13, 14). This work demonstrates that omadacycline specifically inhibits protein synthesis consistent with the known mechanism of action for the typical tetracyclines. Biophysical experiments using purified ribosomes demonstrated that omadacycline is bound to 70S ribosomes with an affinity comparable to that of minocycline, which is known to have an affinity greater than that of tetracycline (12). For tetracycline, it has been shown that there is a single high-affinity binding site and that there are additional low-affinity sites (9, 15). While our studies suggest that omadacycline competes with tetracycline for binding to the ribosome, we cannot distinguish whether the competition is at the primary and/or at other secondary sites.
In contrast to other tetracyclines such as tetracycline or doxycycline, but in similarity to tigecycline, omadacycline is unaffected by the two major mechanisms of tetracycline resistance efflux or ribosomal protection. This ability of omadacycline to function in the presence of these resistance mechanisms was demonstrated in vivo with favorable MIC values in bacterial strains carrying known resistance mechanisms, through the use of macromolecular synthesis studies, and finally using in vitro protein translation. The mechanism by which omadacycline avoids efflux is not known. Tetracyclines are known to differ in their abilities to be substrates for the different tetracycline efflux pumps (16). Studies with tigecycline suggest that, despite being able to induce the expression of efflux pumps, the compound is no longer a substrate for the efflux transporters (17). It is likely that omadacycline is also not a substrate, but confirmation of this would require studies using inverted vesicles containing tetracycline efflux proteins. The precise mechanism of ribosomal protection is not entirely understood, although studies suggest that the binding of ribosomal protection proteins such as tet(M) and tet(O) alters ribosomal conformation and causes release of compounds such as tetracycline and minocycline (11). With this mechanism in mind, there are two ways in which omadacycline could overcome ribosomal protection-mediated resistance. It could be the result of increased affinity of binding to the ribosome, or, alternatively, as has been suggested for tigecycline, the compound might bind to the ribosome in a unique way that would circumvent ribosomal protection by tet(M) or tet(O) (12, 18). The data in this study support the latter possibility, as omadacycline seems to bind the ribosome with affinity similar to that of minocycline, which, unlike omadacycline, is susceptible to tet(M)-mediated ribosomal protection. Like tigecycline, omadacycline structurally contains a modification at the 9 position of the tetracycline core. Data suggest that with tigecycline, the additional sites of interaction between tigecycline and the ribosome contribute substantially to the ability of tigecycline to overcome ribosomal protection and to maintain activity against tetracycline-resistant bacteria (17). It is likely that similar interactions between omadacycline and the ribosome contribute to its ability to resist protection by Tet(M). Additional studies are under way to better characterize the interactions of omadacycline with the ribosome and understand further the mechanism by which it evades both efflux and ribosomal protection.
ACKNOWLEDGMENT
C.A.T. received financial support from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print 16 September 2013
REFERENCES
- 1.Speer BS, Shoemaker NB, Salyers AA. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaker M, Spanogiannopoulos P, Wright GD. 2010. The tetracycline resistome. Cell. Mol. Life Sci. 67:419–431. 10.1007/s00018-009-0172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts MC. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1–24. 10.1111/j.1574-6976.1996.tb00251.x [DOI] [PubMed] [Google Scholar]
- 4.Ng LK, Martin I, Alfa M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209–215. 10.1006/mcpr.2001.0363 [DOI] [PubMed] [Google Scholar]
- 5.Nelson ML, Ismail MY, McIntyre L, Bhatia B, Viski P, Hawkins P, Rennie G, Andorsky D, Messersmith D, Stapleton K, Dumornay J, Sheahan P, Verma AK, Warchol T, Levy SB. 2003. Versatile and facile synthesis of diverse semisynthetic tetracycline derivatives via Pd-catalyzed reactions. J. Org. Chem. 68:5838–5851. 10.1021/jo030047d [DOI] [PubMed] [Google Scholar]
- 6.Hughes J, Mellows G. 1978. On the mode of action of pseudomonic acid: inhibition of protein synthesis in Staphylococcus aureus. J. Antibiot. (Tokyo) 31:330–335. 10.7164/antibiotics.31.330 [DOI] [PubMed] [Google Scholar]
- 7.Wilson JM, Oliva B, Cassels R, O'Hanlon PJ, Chopra I. 1995. SB 205952, a novel semisynthetic monic acid analog with at least two modes of action. Antimicrob. Agents Chemother. 39:1925–1933. 10.1128/AAC.39.9.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bommer U, Burkhardt N, Jünemann R, Spahn C, Triana F, Nierhaus K. 1996. Ribosomes and polysomes, p 271–301 In Graham J, Rickwoods D. (ed), Subcellular fractionation. A practical approach. IRL Press, Washington, DC [Google Scholar]
- 9.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143–1154 [DOI] [PubMed] [Google Scholar]
- 10.Trieber CA, Burkhardt N, Nierhaus KH, Taylor DE. 1998. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 379:847–855 [DOI] [PubMed] [Google Scholar]
- 11.Connell SR, Trieber CA, Dinos GP, Einfeldt E, Taylor DE, Nierhaus KH. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945–953. 10.1093/emboj/cdg093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson MW, Ruzin A, Feyfant E, Rush TS, III, O'Connell J, Bradford PA. 2006. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob. Agents Chemother. 50:2156–2166. 10.1128/AAC.01499-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliva B, Chopra I. 1992. Tet determinants provide poor protection against some tetracyclines: further evidence for division of tetracyclines into two classes. Antimicrob. Agents Chemother. 36:876–878. 10.1128/AAC.36.4.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen B, Noller HF, Daubresse G, Oliva B, Misulovin Z, Rothstein DM, Ellestad GA, Gluzman Y, Tally FP, Chopra I. 1991. Molecular basis of tetracycline action: identification of analogs whose primary target is not the bacterial ribosome. Antimicrob. Agents Chemother. 35:2306–2311. 10.1128/AAC.35.11.2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829–1839. 10.1093/emboj/20.8.1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMurry LM, Cullinane JC, Levy SB. 1982. Transport of the lipophilic analog minocycline differs from that of tetracycline in susceptible and resistant Escherichia coli strains. Antimicrob. Agents Chemother. 22:791–799. 10.1128/AAC.22.5.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Someya Y, Yamaguchi A, Sawai T. 1995. A novel glycylcycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, is neither transported nor recognized by the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Antimicrob. Agents Chemother. 39:247–249. 10.1128/AAC.39.1.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 110:3812–3816. 10.1073/pnas.1216691110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid P, Speyer J. 1970. Rifampicin inhibition of ribonucleic acid and protein synthesis in normal and ethylenediaminetetraacetic acid-treated Escherichia coli. J. Bacteriol. 104:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]