Abstract
Acinetobacter baumannii has emerged as a nosocomial pathogen with an increased prevalence of multidrug-resistant strains. The role of the outer membrane protein A (OmpA) in antimicrobial resistance remains poorly understood. In this report, disruption of the ompA gene led to decreased MICs of chloramphenicol, aztreonam, and nalidixic acid. We have characterized, for the first time, the contribution of OmpA in the antimicrobial resistance phenotype of A. baumannii.
TEXT
Acinetobacter baumannii is one of the most important species associated with nosocomial infections, such as ventilator-associated pneumonia, bacteremia, urinary tract infections, skin and wound infections, and meningitis (1). During the last decade, this pathogen has become increasingly resistant to most antimicrobials, including broad-spectrum cephalosporins, penicillins, carbapenems, fluoroquinolones, and aminoglycosides. Several resistance mechanisms contribute to the multidrug resistance (MDR) phenotype in A. baumannii: decreased outer membrane protein (OMP) permeability, overexpression of efflux pumps, and acquisition of genetic elements carrying resistance determinants, such as plasmids, integrons, transposons, and resistance islands.
Gram-negative bacteria typically display diverse porins in their outer membrane that participate in the modulation of cellular permeability, outer membrane protein A (OmpA) being one of the most abundant. OmpA is a β-barrel porin highly conserved among bacterial species and in A. baumannii has been associated with a variety of interesting biological properties in in vitro model systems (2). OmpA has been shown to bind host epithelia, target mitochondria, translocate to the nucleus, and induce cell death and can also bind factor H, which may allow A. baumannii to develop serum resistance (3–6). Furthermore, OmpA has also been associated with antimicrobial resistance in related pathogens (7), although only two studies have shown the involvement of OmpA as the slow porin for β-lactams (8, 9). A clear role for OmpA in antimicrobial resistance, however, has not yet been fully demonstrated.
To evaluate the involvement of OmpA in antimicrobial resistance, an internal fragment of the ompA gene (578 bp) from A. baumannii strain ATCC 17978 was amplified by PCR using the primers given in Table 1 and cloned into the pGEM-T Easy (Promega) vector by A/T cloning (10). The resulting construct was transformed in Escherichia coli strain DH5α, and plasmid was then extracted and electroporated into A. baumannii strain ATCC 17978 in order to knock out its ompA gene by insertional mutagenesis. Transformants were selected on LB agar plates containing 80 μg/ml ticarcillin. ompA gene disruption within the resulting strain, designated JPAB01, was verified by PCR using a combination of primers matching the upstream region of ompA and the pGEM-T Easy vector as well as by analyzing the outer membrane protein (OMP) profile by SDS-PAGE and Western blotting with OmpA-specific antibodies (Fig. 1).
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| OmpAint up | GTTAAAGGCGACGTAGACG |
| OmpAint lw | CCAGTGTTATCTGTGTGACC |
| OmpAext up | GGAATGGCTATAACTGACATAATC |
| OmpAext lw | GAATCAGGAGATTTACAAATGACC |
| OmpA-EcoRI F | GACTAGGAATTCGGAATGGCTATAACTGACATAATC |
| OmpA-EcoRI R | GACTAGGAATTCGAATCAGGAGATTTACAAATGACC |
| M13-Fa | GTAAAACGACGGCCAGT |
| M13-Ra | CAGGAAACAGCTATGAC |
Primer designed in pGEM-T.
FIG 1.
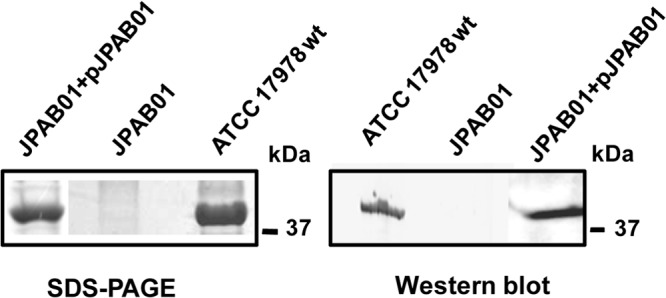
OMP profile and immunodetection of OmpA of A. baumannii strains. OMPs were extracted from the outer membrane of ATCC 17978 wt, JPAB01, and JPAB01+pJPAB01 strains and stained with Simply Blue SafeStain (SDS-PAGE) or electrotransferred onto a nitrocellulose membrane (Western blot). OMPs were probed with mouse anti-OmpA and horseradish peroxidase-conjugated goat anti-mouse IgG. Molecular mass standards (kDa) are shown on the right.
PCR, SDS-PAGE, and Western blot analysis confirmed the disruption of the ompA gene and the absence of OmpA expression in JPAB01 (Fig. 1 and data not shown). Etest testing of the susceptibilities of the wild-type (wt) and mutant strains to quinolones (ciprofloxacin and nalidixic acid), chloramphenicol, tetracycline, tigecycline, aztreonam, imipenem, erythromycin, trimethoprim, kanamycin, and ceftazidime was performed in duplicate. The JPAB01 strain was more susceptible than the ATCC 17978 strain to chloramphenicol (>8-fold), aztreonam (8-fold), and nalidixic acid (3-fold) (Table 2), all of which are substrates of efflux pumps.
TABLE 2.
Determination of MICs of different antimicrobial agents for A. baumannii strains used in this study (mean values of duplicate assays)
| Antimicrobial agent | MIC (μg/ml) |
Difference in susceptibility (fold) |
|||
|---|---|---|---|---|---|
| ATCC 17978 wt | JPAB01 | JPAB01+pJPAB01 | ATCC 17978 wt vs JPAB01b | ATCC 17978 wt vs JPAB01+pJPAB01 | |
| Ceftazidime | 2 | 3 | 4 | 0.67 | 0.5 |
| Imipenem | 0.5 | 0.38 | 0.38 | 1.31 | 1.31 |
| Ciprofloxacin | 0.25 | 0.38 | 0.25 | 0.66 | 1 |
| Kanamycin | 3 | 1.5 | 2 | 2 | 1.5 |
| Tetracycline | 6 | 4 | 32 | 1.5 | 0.19 |
| Erythromycin | 12 | 6 | NDa | 2 | ND |
| Trimethoprim | 32 | 32 | ND | 1 | ND |
| Tigecycline | 1 | 0.5 | 0.5 | 2 | 2 |
| Colistin | 0.5 | 0.25 | 1 | 2 | 0.5 |
| Aztreonam | 16 | 2 | 16 | 8 | 1 |
| Aztreonam + PAβN | 4 | 0.5 | 2 | 8 | 2 |
| Nalidixic acid | 16 | 6 | 16 | 2.67 | 1 |
| Nalidixic acid + PAβN | 2 | 2 | 2 | 1 | 1 |
| Chloramphenicol | >256 | 32 | >256 | >8 | ND |
| Chloramphenicol + PAβN | 32 | 32 | 32 | 1 | 1 |
ND, not determined.
Values in bold indicate a difference in susceptibility of greater than twofold.
To demonstrate that the increased chloramphenicol, aztreonam, and nalidixic acid susceptibilities observed for the JPAB01 strain were due to the lack of a functional OmpA protein, the complementation of JPAB01 has been performed. The ompA gene was amplified with the OmpA-EcoRI F and OmpA-EcoRI R primers (Table 1) from the ATCC 17978 wild-type (wt) genome and cloned into the EcoRI restriction site of the pWH1266 vector (11), yielding the pWH1266-OmpA plasmid called pJPAB01. pJPAB01 was transformed into DH5α and electroporated into JPAB01. Transformants were selected on LB agar plates containing 25 μg/ml tetracycline. The complementation of OmpA in the JPAB01 strain restored the MICs of chloramphenicol, aztreonam, and nalidixic up to >256, 16, and 16 μg/ml, respectively, demonstrating that OmpA was indeed involved in the chloramphenicol, aztreonam, and nalidixic acid resistance phenotypes. Moreover, the presence of the efflux pump inhibitor phenyl-arginine-β-naphthylamide (PAβN) reduced the MICs of chloramphenicol, aztreonam, and nalidixic acid at least >8-, 4-, and 8-fold in the wild-type strain and the complemented strain.
A similar reduction in the MICs of aztreonam and nalidixic acid, but not of chloramphenicol, was observed for the mutant JPAB01 strain in the presence of the inhibitor (Table 2). These results suggest the presence of additional mechanisms contributing to the reduced susceptibility to these antibiotics, likely related to the expression of efflux pumps.
This is the first description of the involvement of OmpA in antimicrobial resistance in A. baumannii. The exact mechanism of action is not clear, but it is possible that OmpA participates in the extrusion of compounds from the periplasmic space through the outer membrane and couples with inner membrane efflux systems, such as major facilitator superfamily (MFS) efflux pumps or RND systems lacking the OMP component. A similar situation has been described for the MexXY system in Pseudomonas aeruginosa, which, despite lacking a gene coding for an outer membrane porin, can associate with OprM and likely other OMPs, such as OpmB, OpmG, or OpmH, to form a functional tripartite efflux system (12, 13). A. baumannii contains several RND tripartite systems, such as AdeABC, AdeIJK, and AdeFGH. In AdeABC, the presence of AdeC is not required for antimicrobial resistance, suggesting that AdeAB can utilize another outer membrane constituent such as AdeK (14). Analysis of the genome of the ATCC 17978 strain suggests that the adeC gene is missing and that, thus, other OMPs have to compensate for the lack of adeC. Interestingly, a knockout mutant of the CraA MFS pump in A. baumannii displays a decrease in the MIC of chloramphenicol similar to that observed for JPAB01 (15), and no further effect is observed in the presence of efflux inhibitors.
In E. coli and Citrobacter freundii, increased expression of OmpA homologues has been associated with decreased susceptibility to tetracycline (7) and carbapenems (16), respectively. Taking into account that OmpA is the major outer membrane protein in A. baumannii, we cannot rule out the possibility that its absence in the knockout mutant disturbs membrane processes, including the transport of antimicrobial agents. To our knowledge, there are no studies regarding the overall modification of gene expression upon OmpA loss in A. baumannii, and this matter should be further investigated. In our study, only three nonrelated antimicrobials have shown a reduced MIC in the knockout strain, and thus, we do not believe such disturbance to be a major issue.
It is clear that additional studies are needed to evaluate the clinical relevance of OmpA expression in MDR, but we have shown the participation of OmpA in the antimicrobial resistance phenotype of A. baumannii.
ACKNOWLEDGMENTS
Y. Smani is funded by the Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, cofinanced by the European Development Regional Fund “A way to achieve Europe” ERDF, Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008).This study was supported by the Consejería de Innovación, Ciencia y Empresa (CTS 6317/10).
Footnotes
Published ahead of print 30 December 2013
REFERENCES
- 1.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 2.Smith SG, Mahon V, Lambert MA, Fagan RP. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273:1–11. 10.1111/j.1574-6968.2007.00778.x [DOI] [PubMed] [Google Scholar]
- 3.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 10:309–319. 10.1111/j.1462-5822.2007.01041.x [DOI] [PubMed] [Google Scholar]
- 4.Choi CH, Lee EY, Lee Y, Park T, Kim H, Hyun S, Kim S, Lee S, Lee J. 2005. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 7:1127–1138. 10.1111/j.1462-5822.2005.00538.x [DOI] [PubMed] [Google Scholar]
- 5.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150–3160. 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SW, Moon DC, Jin JS, Lee JH, Shin JH, Kim JM, Lee YC, Seol SY, Cho DT, Lee JC. 2009. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 301:224–231. 10.1111/j.1574-6968.2009.01820.x [DOI] [PubMed] [Google Scholar]
- 7.Viveiros M, Dupont M, Rodrigues L, Couto I, Davin-Regli A, Martins M, Pagès JM, Amaral L. 2007. Antibiotic stress, genetic response and altered permeability of Escherichia coli. PLoS One 2:e365. 10.1371/journal.pone.0000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nitzan Y, Deutsch EB, Pechatnikov I. 2002. Diffusion of beta-lactam antibiotics through oligomeric or monomeric porin channels of some gram-negative bacteria. Curr. Microbiol. 45:446–455. 10.1007/s00284-002-3778-6 [DOI] [PubMed] [Google Scholar]
- 9.Sugawara E, Nikaido H. 2012. OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. J. Bacteriol. 194:4089–4096. 10.1128/JB.00435-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roca I, Espinal P, Martí S, Vila J. 2011. First identification and characterization of an AdeABC-like efflux pump in Acinetobacter genomospecies 13TU. Antimicrob. Agents Chemother. 55:1285–1286. 10.1128/AAC.01142-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. 10.1016/0378-1119(90)90494-C [DOI] [PubMed] [Google Scholar]
- 12.Chuanchuen R, Murata T, Gotoh N, Schweizer HP. 2005. Substrate-dependent utilization of OprM or OpmH by the Pseudomonas aeruginosa MexJK efflux pump. Antimicrob. Agents Chemother. 49:2133–2136. 10.1128/AAC.49.5.2133-2136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murata T, Gotoh N, Nishino T. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol. Lett. 217:57–83. 10.1111/j.1574-6968.2002.tb11456.x [DOI] [PubMed] [Google Scholar]
- 14.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48:3298–3304. 10.1128/AAC.48.9.3298-3304.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roca I, Marti S, Espinal P, Martínez P, Gibert I, Vila J. 2009. CraA, a major facilitator superfamily efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:4013–4014. 10.1128/AAC.00584-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Yang L, Cai JC, Zhou HW, Chen GX. 2008. High-level carbapenem resistance in a Citrobacter freundii clinical isolate is due to a combination of KPC-2 production and decreased porin expression. J. Med. Microbiol. 57:332–337. 10.1099/jmm.0.47576-0 [DOI] [PubMed] [Google Scholar]


