Abstract
The attenuated Lassa vaccine candidate ML29 is a laboratory-produced reassortant between Lassa and Mopeia viruses, two Old World arenaviruses that differ by 40% in nucleic acid sequence. In our previous studies, ML29 elicited sterilizing immunity against Lassa virus challenge in guinea pigs and marmosets and virus-specific cell-mediated immunity in both simian immunodeficiency virus (SIV)-infected and uninfected rhesus macaques. Here, we show that ML29 is stable after 12 passages in vitro without losing its plaque morphology or its attenuated phenotype in suckling mice. Additionally, we used deep sequencing to characterize the viral population comprising the original stock of ML29, the stock of ML29 after 12 passages in Vero cells, and the ML29 isolates obtained from vaccinated animals. Twenty-seven isolates bore approximately 77 mutations that exceeded 20% of the single-nucleotide polymorphism (SNP) changes at any single locus. Of these 77 mutations, 5 appeared to be host specific, for example, appearing in mice but not in primates. None of these mutations were reversions of ML29 to the sequences of the parental Lassa and Mopeia viruses. The host-specific mutations indicate viral adaptations to virus-host interactions, and such interactions make reasonable targets for antiviral approaches. Variants capable of chronic infection did not emerge from any of the primate infections, even in immune-deficient animals, indicating that the ML29 reassortant is reasonably stable in vivo. In conclusion, the preclinical studies of ML29 as a Lassa virus vaccine candidate have been advanced, showing high levels of protection in nonhuman primates and acceptable stability both in vitro and in vivo.
INTRODUCTION
Arenaviruses are zoonotic pathogens that infect rodents and occasionally human beings, sometimes with fatal consequences. The prototype virus is lymphocytic choriomeningitis virus (LCMV), which can cause disease in humans, from minor febrile episodes to severe aseptic meningitis (1). Arenavirus particles contain single-stranded RNA (ssRNA) in which the viral genome is divided into two ambisense segments (2); the large (L) segment encodes an RNA-dependent RNA polymerase (RdRp) and a small zinc-binding “Z” protein, and the short (S) segment encodes the viral nucleocapsid protein (NP) and the glycoprotein precursor (GPC).
Lassa virus (LASV) is a highly pathogenic member of the family that can cause Lassa fever (LF) in people who inhabit rodent-infested dwellings in West Africa. Some studies report around 300,000 cases per year, with a mortality rate between 1 and 16% (3, 4). Lassa fever is prevalent in all age groups, and it is not gender specific. The highest-risk populations are (i) those who live in crowded rural areas infested by the rodent Mastomys natalensis (5), (ii) those working in a slaughterhouse, (iii) those bitten or scratched by animals, (iv) those hospitalized in areas of endemicity with poor medical practices, and (v) health care workers in contact with LF cases (6, 7).
The LF incubation period ranges from 6 to 21 days. Around 80% of the infections are asymptomatic, but the remaining 20% can evolve to flu-like disease, followed by the development of severe multisystemic damage to liver, spleen, and kidneys. Some severe cases develop hemorrhagic symptoms characterized by mouth, nose, gastrointestinal, or vaginal tract bleeding; low blood pressure; and, finally, death (8).
The annual number of infections and deaths, the fear of imported infections, and the lack of effective specific treatment make a strong case for LF vaccine development. We reported the development of an attenuated vaccine candidate called Mopeia-Lassa clone 29 (ML29), which is a reassortant virus between the nonpathogenic Mopeia virus (MOPV) and the pathogenic LASV (9, 10). ML29 conferred sterilizing protection in rodents and nonhuman primates after lethal challenge with homologous and distantly related LASV strains (11). Additionally, ML29 conferred postexposure protection in guinea pigs (11).
Like most RNA viruses, arenaviruses have an RNA-dependent RNA polymerase lacking proofreading activity. This feature can potentially affect the safety profile of live attenuated vaccines containing RNA genomes. Here, we evaluate the phenotype and genotype of ML29 after several passages in Vero cells. Additionally, ML29 was evaluated for variations after passage in vivo. ML29 clones were isolated from vaccinated rhesus macaques, marmosets, and mice, and these isolates were subjected to pyrosequencing. There were no major changes at any nucleotide position after the in vitro passages. However, we were able to detect some host-specific changes after passage in vivo, suggesting the emergence of host-specific adaptive mutations. The few detected changes suggest that ML29 is very stable after passages in vitro and in vivo. Notably, none of the major changes observed were reversions to wild-type parental virus (MOPV or LASV) sequences, and none of the ML29 isolates succeeded in escaping the host immune response to establish persistent infections in primates.
MATERIALS AND METHODS
Cells and virus.
Vero E6 (CRL-1586; ATCC) cells were grown in Dulbeccos's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL), 1% penicillin-streptomycin, and 2 mM l-glutamine. MOPV and ML29 were grown in Vero E6 cells and collected 72 h postinfection, as described previously (10). Briefly, monolayers of Vero E6 cells were infected with MOPV or ML29 at a multiplicity of infection (MOI) of 0.01, incubated for 1 h at 37°C in 5% CO2, and then washed with phosphate-buffered saline (PBS) and covered in DMEM-2% FBS. Medium was collected every 24 h for 3 days and stored at −80°C for titration on Vero cells.
ML29 virus stock was passaged 12 times by infecting Vero E6 cells at an MOI of 0.01 and by collecting media 72 h after each passage. Samples were stored at −80°C prior to virus titration, plaque morphology comparison, infection studies, viral RNA extraction, quantitation, and sequencing.
Virus passage in vivo. (i) Murine infection.
Five groups of suckling outbred white mice were intracerebrally (i.c.) inoculated with 50 μl of minimal essential medium (MEM) (18 mice); 103 PFU of MOPV (12 mice); or 103 PFU of ML29 passage 2 (28 mice), passage 4 (36 mice), or passage 12 (44 mice) and then followed for 30 days, and the mortality rates were recorded. Tissues from brain, liver, and spleen were frozen for virus isolation and reverse transcription (RT)-PCR amplification. In addition, five C57BL/6 mice and five CBA/J mice (4 weeks old) were inoculated i.c. Sera and brain samples were collected at day 8 postinfection for viral isolation and subsequent pyrosequencing analysis. To detect trace amounts of virus in plasma that was negative by direct titration, samples were put through a second biological amplification with Vero cells, as previously described (12, 13). The RNA samples from the C57BL/6 mice were designated IGS-17, IGS-18, IGS-19, IGS-20, and IGS-21, and the samples from the CBA/J mice were designated IGS-22, IGS-23, IGS-24, IGS-25, and IGS-26.
(ii) Primate infection.
Eleven serum samples from monkeys inoculated with simian immunodeficiency virus (SIV) SIVmac251 (as previously described [14]) and vaccinated 1 year later with ML29 (15) were also subjected to virus isolation. Five samples (IGS-1, IGS-2, IGS-3, IGS-4, and IGS-7) were positive for ML29, together with two samples (IGS-9 and IGS-10) from SIV-negative control monkeys. Four additional serum samples (IGS-11, IGS-12, IGS-13, and IGS-14) from vaccinated marmosets were ML29 positive. All samples were stored for further pyrosequencing analysis. The pyrosequencing control samples were ML29 initial inoculum and virus passed 12 times in Vero cells. Subsequently, the sequences obtained from samples and controls were compared with the ML29 published sequence (16). ML29 was detected in monkeys up to 3 weeks after vaccination, but no virus could be isolated after that. Due to low levels of virus, all ML29 isolates were passed once in Vero cells before sequencing. All the experimental protocols were approved by the Institutional Animal Care and Use Committee at the University of Maryland School of Medicine.
RNA extraction, cDNA preparation, and sequence analysis.
ML29 RNA was extracted from 1 ml of serum or infected cell supernatants (passages 2, 6, and 12) containing approximately 107 PFU of virus in 1 ml. TRIzol extraction was performed as described by the manufacturer (Invitrogen, Carlsbad, CA). To obtain full-length sequences for the L and S segments, each segment was separately RT-PCR amplified with specific primers using the Superscript III first-strand synthesis protocol, as previously described (16). Briefly, cDNA was synthesized using 10 μl (1 μg) of purified RNA, specific primers, deoxynucleoside triphosphates (dNTPs), and Superscript III (Invitrogen) in 20-μl reaction mixtures. Amplification reactions were done using 5 μl (1 μg) of cDNA, specific primers (Reverse, 3′-S segment-CGCACAGTGGATCCTAAGG, 3′-L segment-ACAAGTTGGAGGGAACAGGGACT, 3′-L segment-ACAAGTGTGTGGTGATGGAGGAGT, or 3′-L segment-CGCACCGGGGATCCTAGGC; Forward, 5′-S segment-CGCACCGGGGATCCTAGG, 5′-L segment-ACCGGGGATCCTAGGCAATTT, 5′-L segment-CTTGTGAATGTGCCACGACAGCAA, or 5′-L segment-ACAAGTTGGAGGGAACAGGGACT), dNTPs, and Platinum Taq DNA polymerase High Fidelity (Invitrogen) in 50-μl reaction mixtures. The amplified products (L segment, approximately 7.4 kb, and S segment, 3.4 kb) were separated in Tris-borate-EDTA (TBE)-agarose gels and purified using a QIAquick Gel Extraction Kit (Qiagen), and each DNA strand was sequenced using a primer-walking method (1, 16, 17). All sequencing reactions were performed in the Applied Biosystems 3130xl automated sequencer using BigDye terminators (Applied Biosystems), and the sequences were edited and assembled using Sequencher v4.6 (Gene Codes Corporation, Ann Arbor, MI). The results were aligned with L (GenBank accession number NC_006572) and S (GenBank accession number NC_006573) segments from ML29 using MacVector.
Single-nucleotide polymorphism (SNP) analysis.
For a more detailed comparison, total RNA was isolated from concentrated ML29-infected cell supernatants by the TRIzol method and then diluted 1:50 and quantified using an Eppendorf biophotometer. One hundred nanograms of RNA was amplified with random hexamers using the Ovation RNA-Seq System (San Carlos, CA), and then the cDNA product was quantified using Quant-It HS reagents (Invitrogen, Life Technologies, Carlsbad, CA) and used for the generation of sequencing libraries. Fragmentation was performed according to the manufacturer's protocol. Libraries representing each sample were subjected to emulsion PCR, and enriched DNA beads were loaded onto a picotiter plate and pyrosequenced with a Roche/454 GS Junior sequencer using titanium chemistry (454 Life Sciences, Branford, CT).
Pyrosequencing data were analyzed using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark). Initially, reads were trimmed to remove short and low-quality reads and then assembled using the CLC de novo assembler. As a cutoff for the variance analysis, it was established that variations of >10% from the genome consensus sequence would be included in the multiple alignment. Therefore, any sites with <10% variation were treated as below the error threshold. The resulting consensus sequences from FASTA were used in a reference-guided alignment with published ML29 S and L segment sequences using Vector NTI software.
Nucleotide sequence accession numbers.
Sequencing data have been released to GenBank's Sequence Read Archive (SRA) as IGS_1, SRA046428; IGS_2, SRA046422; IGS_3, SRA046430; IGS_4, SRA046423; IGS_7, SRA046431; IGS_9, SRA046473; IGS_10, SRA046474; IGS_11, SRA046475; IGS_12, SRA046485; IGS_13, SRA046493; IGS_14, SRA046495; IGS_15, SRA046488; IGS_16, SRA046484; IGS_17, SRA046481; IGS_18, SRA046482; IGS_19, SRA046486; IGS_20, SRA046487; IGS_21, SRA046483; IGS_22, SRA046489; IGS_23, SRA046490; IGS_24, SRA046491; IGS_25, SRA046492; IGS_26, SRA046494; IGS_27, SRA051861; IGS_28, SRA051889.
RESULTS
The ML29 phenotype and sequence remained stable after multiple in vitro passages.
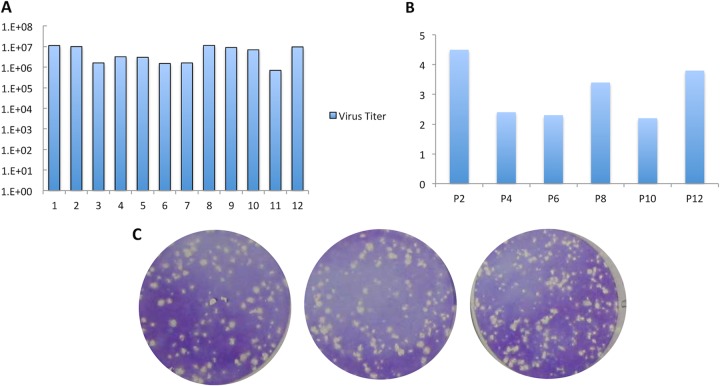
To ensure that mutations occurring in vitro, during the vaccine production process, would not affect the attenuated phenotype, ML29 was passaged 12 consecutive times in Vero cells, following the standard protocols for vaccine production. There were no significant differences in viral titers (less than 1 log unit), RNA concentrations, or plaque morphology between passages (Fig. 1A, B, and C). The cytopathic effects (CPE) produced by ML29 were similar to those described for other arenaviruses: healthy Vero cells became spindle shaped and refractile, and infected cells detached from the monolayer, forming progressively larger holes (or plaques) in the monolayer. LASV plaques are around 2 mm, while for MOPV, 1 mm is the average diameter. ML29 has the small-plaque morphology of MOPV, a phenotype controlled by its MOPV-derived L RNA segment (9). After 12 passages, the ML29 plaque size remained stable, ranging from 0.8 mm to 2.0 mm, with a mean of 1.1 mm, in agreement with previous results (Fig. 1C) (9).
FIG 1.
Comparison of different ML29 isolates from 12 passages in vitro. Vero cells were infected at an MOI of 0.01 with ML29. The supernatant was collected 72 h later for the subsequent infection. After 12 passages, all isolated viruses were compared for ML29 virus titer (A), RNA concentrations (μg/ml) (B), and plaque morphology (C), as well as for the phenotype in suckling mice (Fig. 2).
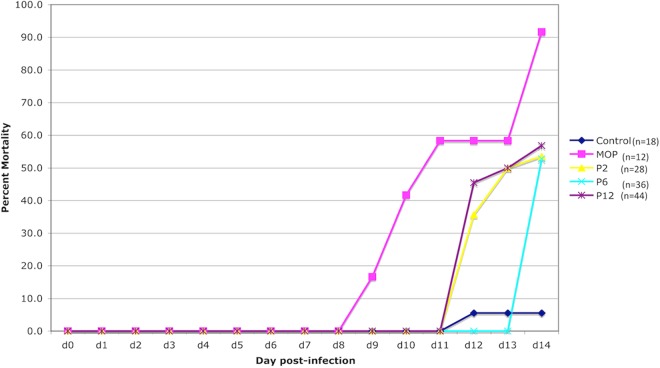
ML29 virus stocks were tested for phenotypic stability by i.c. inoculation of suckling mice in comparison to MOPV and LASV. In this model, MOPV kills between 50 and 100% of mice after 2 weeks of i.c. infection. LASV is rarely lethal, and ML29 has intermediate pathogenicity in suckling mice (9). Here, mice were inoculated i.c. with ML29 passages 2, 6, and 12; the mortality rates were approximately 50%, compared to 90% in MOPV-inoculated mice and 5% in the noninfected control group (Fig. 2). These data agree with previous studies and suggest that the ML-29 attenuated phenotype is not compromised after 12 passages in vitro (9).
FIG 2.
Accumulated mortality rates of suckling mice after MOPV or ML29 i.c. inoculation. The mortality rates were as expected for MOPV (around 90%) and ML29 (around 50%) after 14 days of infection.
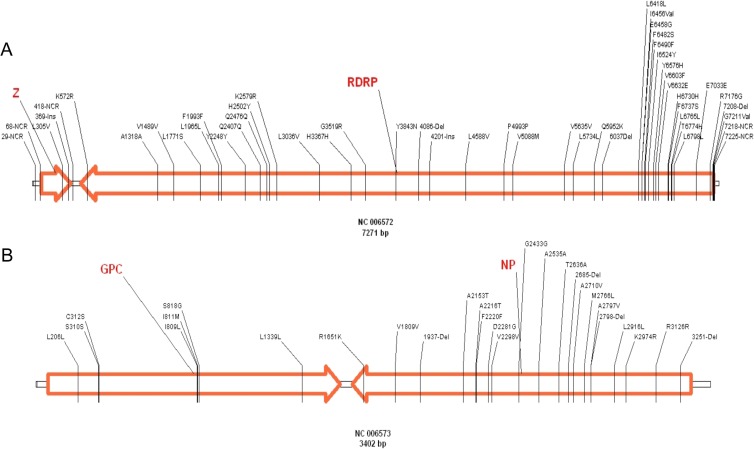
In order to detect changes in the virus genome after several passages in vitro, ML29 isolates were sequenced by the primer-walking method. The sequence analysis of both ML29 segments (L and S) from passages 2, 6, and 12 did not deviate from the consensus sequence of the ML29 viral stock, confirming 16 out of 18 mutations previously published in GenBank (NC_006572 and NC_006573) (Fig. 3A and B). By using this method, it was not possible to confirm the two mutations at the 5′ end of the L segment. These results showed that the ML29 consensus sequence is stable even after 12 passages in Vero cells.
FIG 3.
Distribution of mutations detected in all ML29 isolates compared with parental viruses MOPV and LASV (the genome GenBank identification number and size are shown below). The small boxes between the arrows represent the ML29 noncoding sequence, and the orange arrows correspond to open reading frames. The numbers represent the position of each change. The first letter is the previous amino acid, and the last letter is the new amino acid. NCR, noncoding region; Ins, insertion; Del, deletion.
The ML29 sequence undergoes some variation after inoculation of mice, marmosets, and rhesus macaques.
ML29 was isolated from serum samples collected from vaccinated animals: five SIV-positive (ISG-1, ISG-2, ISG-3, ISG-4, and ISG-7) and two SIV-negative (ISG-9 and ISG-10) monkeys, four marmosets (ISG-11, ISG-12, ISG-13, and ISG-14), five C57BL/6 i.c.-inoculated mice, and five CBA/J i.c.-inoculated mice.
After viral RNA extraction and cDNA preparation, ML29 isolates were tested by pyrosequencing. This method allowed the detection and characterization of single-nucleotide variants and the evaluation of their frequency in each viral preparation. The sequences obtained from animal samples, viral stock (IGS-15), ML29-purified viral stock (IGS-27), and ML29 isolated after 12 passages in vitro (IGS-15) were compared with the published ML29 sequences (16). Each passage represents 48 h of infection at an MOI of 0.01. During that time, the titer went from 1 × 103 PFU/ml to 1 × 107 PFU/ml. This 4-log-unit increase was maintained in IGS-16 during 12 passages in Vero cells.
Comparison of all samples with the reference ML29 sequences showed 27 different mutations in the S segment (all in coding areas) and 50 in the L segment (46 located in coding regions and 4 in noncoding areas) (Tables 1 and 2). SNPs were considered only if they represented >20% of the viral subspecies at that locus and were described as synonymous (not affecting codon coding) or as nonsynonymous (changing the encoded amino acid). Not all SNPs were present in all samples, except for a synonymous change in nucleotide 6490 (F/F) with a frequency higher than 95%. The viral stock sequence revealed 2 SNP differences compared with the previously published data: the F6490F synonymous change and a K2974R change with low frequency (36%). Wider sequence diversity was observed in the rest of the samples, including the passages in vitro, all in agreement with the viral species present in a viral population (Tables 1 and 2).
TABLE 1.
SNPs detected in the ML29 S segmenta
All samples were pyrosequenced, and the results were assembled using the published ML29 consensus sequence. The first row shows the nucleotide site and the corresponding amino acid change. Del represents deletion, and Ins represents insertion. The second row shows the site changed in the protein. The first column contains the sample identification as follows: IGS-1 to IGS-7 are ML29 serum isolates from SIV-positive monkeys, IGS-9 and IGS-10 are ML29 serum isolates from healthy monkeys, IGS-11 to IGS-14 are ML29 serum isolates from marmosets, IGS-15 corresponds to the ML29 viral stock, IGS-16 is ML29 after 12 passages in Vero cells, IGS-17 to IGS-21 are ML29 serum isolates from C57/BL6 mice, IGS-22 to IGS-26 are ML29 serum isolates from CBA/J mice, and IGS-27 is ML29 viral stock purified by ultracentrifugation. IGS-28 is the GenBank sequence of Mopeia virus. The color of each box represents the frequency of that SNP in the population.
TABLE 2.
SNPs detected in ML29 -L segment

All samples were pyrosequenced, and the results were assembled using the published ML29 consensus sequence. The first row shows the nucleotide site and the corresponding amino acid change. Del represents deletion, and Ins represents insertion. The second row shows the site changed in the protein. The first column contains the sample identification as follows: IGS-1 to IGS-7 are ML29 serum isolates from SIV-positive monkeys, IGS-9 and IGS-10 are ML29 serum isolates from healthy monkeys, IGS-11 to IGS-14 are ML29 serum isolates from marmosets, IGS-15 corresponds to the ML29 viral stock, IGS-16 is ML29 after 12 passages in Vero cells, IGS-17 to IGS-21 are ML29 serum isolates from C57/BL6 mice, IGS-22 to IGS-26 are ML29 serum isolates from CBA/J mice, and IGS-27 is ML29 viral stock purified by ultracentrifugation. IGS-28 is the GenBank sequence of Mopeia virus. The color of each box represents the frequency of that SNP in the population.
The highest variation was seen in the isolates from nonhuman primates, with a total of 35 SNPs in monkeys (GP, 4; NP, 9; Z, 1; L, 20), followed by 27 changes in marmosets (GP, 2; NP, 6; Z, 1; L, 18) and 17 in Vero cells (GP, 0; NP, 6; Z, 1; L, 10). The rodent system showed similar numbers of changes. In the isolates from CBA/J mice, there were 17 nucleotide substitutions (GP, 3; NP, 5; Z, 1; L, 8), and the C57BL/6 mice had the lowest number of changes, 13 (GP, 1; NP, 4; Z, 1; L, 7) (Table 3).
TABLE 3.
Types of mutations in the mutant spectrum of ML29 in vivo and in vitroa
| Mutation | No. |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vero cells |
Rhesus monkeys |
Marmosets |
C57 mice |
CBA/J mice |
||||||||||||||||
| GP | NP | Z | L | GP | NP | Z | L | GP | NP | Z | L | GP | NP | Z | L | GP | NP | Z | L | |
| A→C | 1 | 1 | ||||||||||||||||||
| A→G | 7 | 1 | 4 | 4 | 1 | 1 | 1 | |||||||||||||
| A→U | 1 | 1 | 2 | 1 | 1 | |||||||||||||||
| C→A | 1 | 1 | ||||||||||||||||||
| C→G | 1 | 1 | ||||||||||||||||||
| C→U | 1 | 1 | 3 | 4 | 1 | 3 | 1 | 2 | 1 | |||||||||||
| G→A | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | ||||||||||||
| G→C | ||||||||||||||||||||
| G→U | 1 | 1 | ||||||||||||||||||
| U→A | 1 | 1 | ||||||||||||||||||
| U→C | 2 | 4 | 3 | 1 | 2 | 2 | 1 | 3 | 1 | |||||||||||
| U→G | 1 | 1 | 1 | 1 | ||||||||||||||||
| Deletions | 1 | 1 | 1 | 2 | 1 | |||||||||||||||
| Insertions | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||
| Noncoding | 1 | 2 | 3 | 1 | 1 | 1 | ||||||||||||||
| Total no. of mutations | 6 | 1 | 10 | 4 | 9 | 1 | 20 | 2 | 6 | 1 | 18 | 1 | 4 | 1 | 7 | 3 | 5 | 1 | 8 | |
| Transitions | 6 | 9 | 2 | 8 | 13 | 4 | 10 | 1 | 2 | 0 | 4 | 3 | 5 | 3 | ||||||
| Transversions | 2 | 1 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | |||||||||
| Nonsynonymous | 4 | 3 | 2 | 5 | 1 | 8 | 2 | 2 | 8 | 2 | 1 | 2 | 1 | 3 | 2 | |||||
| Synonymous | 1 | 6 | 2 | 3 | 8 | 3 | 1 | 5 | 1 | 1 | 3 | 1 | 2 | 4 | ||||||
| Selection rate | 4 | 0.5 | 1 | 1.6 | 1 | 0.7 | 1.6 | 2 | 0.7 | 1 | 1.5 | 0.5 | ||||||||
The table shows the types of changes observed in each host within each viral protein. The selection rate was obtained from the dN/dS, ratio in which dN represents nonsynonymous and dS synonymous changes at the protein level.
There were a total of 67 transitions divided as follows. In nonhuman primates, there were 32 transitions in the L segment, all located in the L protein-coding sequence. The S segment had 20 transitions, with 2 in GP and 18 in NP. In the isolates from mice, the L segment had 7 transitions, also located in the L protein-coding region, and the S segment had 11 transitions, 4 in GP and 7 in NP. The total number of transversions was 18. In nonhuman primates, the L segment had 2 changes in Z and 5 in L protein-coding sequences; the S segment had 4 in GP and 1 in NP. In mice, the L segment had 1 transversion in Z and 4 in L, while the S segment had only 1 in NP (Table 3).
The most common mutations were A/G, with 19 substitutions. Most of them (17 substitutions) were in the L protein sequence. There were 19 U/C changes (12 in NP and 7 in L). C/U changes were also significant, with 17 substitutions (9 in L, 7 in NP, and 1 GP) (Table 3).
After including any variation of >10% in the individual genome consensus sequences and then performing multiple alignments, the estimated mutation frequencies per nucleotide were 2.17 × 10−3 for IGS-15, 2.37 × 10−3 for IGS-16, and from 4.73 × 10−4 to 1.98 × 10−3 for all of the isolates.
Some ML29 variation is host specific.
There were a total of 41 synonymous changes: 14 in the S segment (4 in GP and 10 in NP) and 27 in the L segment (1 in Z and 26 in L). The nonsynonymous mutations were also 46:21 in the S segment (5 in GP and 16 in NP) and 25 in the L segment (2 in Z and 23 in L). The selection rates in vivo were compared with those seen in vitro using Vero cells (Table 3). For Vero cell ML29 isolates, the nonsynonymous/synonymous ratio (selection rate dN/dS) was not affected for GP and Z proteins. For NP, the dN/dS ratio was 4, showing positive selection, while for the L protein, the ratio of 0.5 suggests negative selection. Selection rates for GP were similar in both in vitro and in vivo models. dN/dS ratios for NP were lower than that seen in vitro but still higher than 1 for monkeys and C57BL/6 and CBA/J mice (1.6, 2, and 1.5, respectively), while for marmosets it was less than 1 (0.7). Selection rates for the L segment were 1 or more than 1 in monkeys (1) and marmosets (1.6), while for Vero cells and C57BL/6 and CBA/J mice, the ratios were comparable and less than 1 (0.5, 0.7, and 0.5, respectively). The selection of changes varies with each host, suggesting that the virus sequence adapts to a specific host molecule (Table 4). Changes in GP were very close to the cleavage site (residue 260) (18), changes in NP were either close to the ssRNA binding site (residue 171) in the N domain or close to the immune evasion and Z protein binding sites (residues 59, 223, 341, and 551) in the C domain (18), and changes in L were close to the endonuclease domain (residue 266) or to the globular domain (1572). Protein prediction analysis showed that those mutations, at the amino acid level, induced structural changes in GP and NP (18). The L protein predicted structure was not obtained due to its large size.
TABLE 4.
Host-specific SNPs found after ML29 vaccination in different animalsa
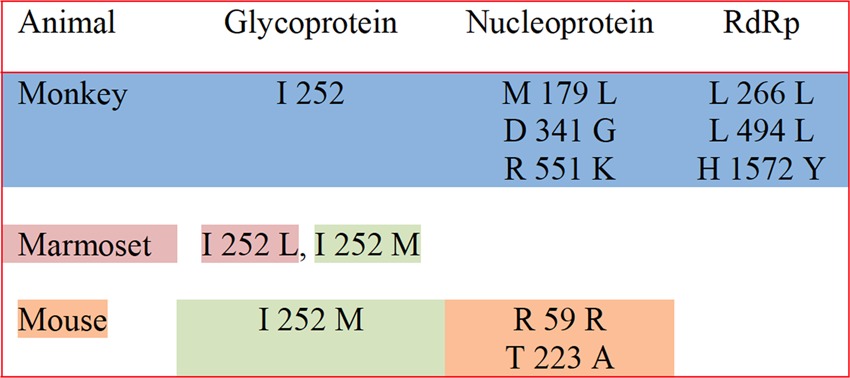
Specific mutations were found in monkeys (blue shading), marmosets (pink shading), or mice (orange shading) or were common to marmosets and mice (green shading). The data were previously published in a review paper (18); however, the table summarizes the original data presented in this article.
DISCUSSION
ML29 vaccination elicits specific immune responses and completely protects experimental rodents (mice and guinea pigs) and nonhuman primates against fatal LASV disease by inducing sterilizing cell-mediated immunity. A single injection of ML29 reassortant vaccine for LF induces low, transient viremia and low or moderate levels of ML29 replication in tissues of common marmosets, depending on the dose of the vaccination (13).
Due to the high mutation rates associated with RNA viruses, it is important to make sure that successive passages during the production process do not affect the ML29 attenuated phenotype as a result of the accumulation of genetic changes. Following the FDA guidelines (19), ML29 was tested in vitro and in vivo. The plaque morphology and the studies in suckling mice indicated that after 12 passages in Vero cells, ML29 did not lose its attenuated phenotype. Those observations were in agreement with the initial characterization of ML29 (9).
In healthy ML29-vaccinated monkeys, evidence of virus infection is difficult to obtain. Although it was easier to isolate ML29 virus from SIV-infected macaques, the viremia and levels of ML29 remained transient and low (15). Given the attenuated nature of ML29, those animals did not show increased signs of pathology or shortened life spans due to the SIV infection; however, they did show both humoral and cell-mediated immune responses to LASV GP and NP similar to those of non-SIV-infected animals (15).
Previous studies have shown tissue-specific variations in different viral models, including arenavirus sequences isolated from the same animal (20–30). To avoid this bias, ML29 was isolated only from sera of vaccinated animals. Additionally, sequences from cloned DNA virus genes were compared with S viral RNA linearly amplified and sequenced directly by pyrosequencing. These methodologies minimized polymerase misincorporations and reduced the artificial inflation of intrahost diversity (29).
The early-passage, highly plaque-purified ML29 viral stock contains very low numbers of viral subspecies (only 2 SNPs were detected), indicating some degree of stability in vitro after a low number of passages. This can be attributed to the fact that Vero cells do not exert much pressure on ML29; however, after 12 passages, there were 10 changes in the L segment and 6 in the S segment, albeit at low frequency for each altered locus (it is important to emphasize that the attenuated phenotype is preserved). Of those, only 1 in the L segment (L266L in the L protein) and 3 in the S segment (D341G, M179L, and K110R, all in NP) increased their percentages in the viral population after vaccination. These results suggest that low-frequency mutations under very low selective pressure in vitro can be positively selected in vivo, as demonstrated by their increased frequency in the viral population and in the dN/dS ratios. The selected mutations were more frequently seen in NP than in the L protein. This could be due to NP's variety of functions and interactions during the viral replication cycle: it encapsidates the genome segments, interacts with L protein to form the RNP core for RNA transcription, associates with Z protein for viral assembly, plays an important role in suppressing the innate immune response, and has exonuclease and nucleotide binding activities (31–38). In contrast to NP, the L protein is less immunogenic, and its RdRp activity has to be more conservative.
In order to include the maximum number of changes in the comparison of the mutation frequencies, we included any variation of >10% in the individual genome consensus sequences. The estimated frequencies did not changed significantly after 12 passages in vitro (2.17 × 10−3 for IGS-15 versus 2.37 × 10−3 for IGS-16), and they were reduced in vivo after vaccination (4.73 × 10−4 to 1.98 × 10−3 for all of the isolates), indicating high stability of ML29 after in vitro and in vivo replication. The high mutation frequencies seen in these isolates (10−3) in comparison with those reported for other arenaviruses (10−4) (39) are due to the inclusion criteria (>10%).
The nonsynonymous-to-synonymous mutation ratio (dN/dS) indicates the tolerance of an organism for genetic changes (40). Since the nonsynonymous changes are reflected in amino acid changes, the ratio can be considered a measure of selective pressure at the protein level (41). As seen in Table 3, the dN/dS ratio for NP is significantly reduced when comparing in vitro virus with virus recovered after vaccination, while there is a smaller variation in the dN/dS ratio for the L protein. This suggests that NP changes faster than the L protein but that those changes are not enough to overcome the host immune system. This observation could have a therapeutic application for LASV infections, since viruses with low dN/dS ratios may have increased sensitivity to lethal mutagenesis and can become targets for mutagenesis-based antiviral approaches (41). However, synonymous changes cannot be completely ignored, since some codons can be translated more efficiently than others or affect the RNA secondary structure (42). There is evidence that viruses can evolve toward the use of host-preferred codons (43, 44, 45).
Single-nucleotide polymorphisms (>20%) have been described here but not functionally characterized. Several ML29 isolates were derived from rhesus macaques 1 or 2 weeks after vaccination but were cleared from plasma by 3 weeks after vaccination. For these isolates, we speculate that they were unable to adapt to host restrictions and became extinct, as described for other RNA viruses (46–49).
In conclusion, this study provides in vitro and in vivo evidence of the stability of the reassortant virus ML29, as well as information regarding the compositions of viral subpopulations. There was no evidence of ML29 reversion to the parental sequences of MOPV or LASV, and there was no evidence of escape mutants that persisted in vaccinated primates, indicating that ML29 vaccine remained susceptible to host immune surveillance. We did not perform functional experiments to see the effect of each SNP in viral replication or pathogenesis. Further analyses using plaque-purified isolates and reverse genetics are required to clarify the role of each of the mutations in the attenuated phenotype displayed by this vaccine candidate.
ACKNOWLEDGMENTS
This project was funded in whole or in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, to C. M. Fraser under contract number HHSN272200900009C. Funding for the experiments with primates was provided by National Institutes of Health grants to M. S. Salvato (AI074790) and to I. S. Lukashevich (AI052367).
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Barton LL, Hyndman NJ. 2000. Lymphocytic choriomeningitis virus: reemerging central nervous system pathogen. Pediatrics 105:E35. 10.1542/peds.105.3.e35 [DOI] [PubMed] [Google Scholar]
- 2.Salvato MS, Clegg JCS, Buchmeier MJ, Charrel RN, Gonzalez JP, Lukashevich IS, Peters CJ, Romanowski V. 2012. Arenaviridae, p 715–723 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 3.McCormick JB. 1988. Lassa fever: epidemiology, therapy and vaccine development. Kansenshogaku Zasshi 62(Suppl):353–366 [PubMed] [Google Scholar]
- 4.McCormick JB, Fisher-Hoch SP. 2002. Lassa fever. Curr. Top. Microbiol. Immunol. 262:75–109. 10.1007/978-3-642-56029-3_4 [DOI] [PubMed] [Google Scholar]
- 5.Ogbu O, Ajuluchukwu E, Uneke CJ. 2007. Lassa fever in West African sub-region: an overview. J. Vector Borne Dis. 44:1–11 [PubMed] [Google Scholar]
- 6.Kerneis S, Koivogui L, Magassouba N, Koulemou K, Lewis R, Aplogan A, Grais RF, Guerin PJ, Fichet-Calvet E. 2009. Prevalence and risk factors of Lassa seropositivity in inhabitants of the forest region of Guinea: a cross-sectional study. PLoS Negl. Trop. Dis. 3:e548. 10.1371/journal.pntd.0000548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher-Hoch SP, Tomori O, Nasidi A, Perez-Oronoz GI, Fakile Y, Hutwagner L, McCormick JB. 1995. Review of cases of nosocomial Lassa fever in Nigeria: the high price of poor medical practice. BMJ 311:857–859. 10.1136/bmj.311.7009.857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCormick JB. 1987. Epidemiology and control of Lassa fever. Curr. Top. Microbiol. Immunol. 134:69–78 [DOI] [PubMed] [Google Scholar]
- 9.Lukashevich IS. 1992. Generation of reassortants between African arenaviruses. Virology 188:600–605. 10.1016/0042-6822(92)90514-P [DOI] [PubMed] [Google Scholar]
- 10.Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, Brasky K, Geiger R, Hubbard GB, Bryant J, Salvato MS. 2005. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J. Virol. 79:13934–13942. 10.1128/JVI.79.22.13934-13942.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrion R, Jr, Bredenbeek PJ, Jiang X, Tretyakova I, Pushko P, ISL 2012. Vaccine platforms to control arenaviral hemorrhagic fevers. J. Vaccines Vaccin. 3:1000160. 10.4172/2157-7560.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrion R, Jr, Brasky K, Mansfield K, Johnson C, Gonzales M, Ticer A, Lukashevich I, Tardif S, Patterson J. 2007. Lassa virus infection in experimentally infected marmosets: liver pathology and immunophenotypic alterations in target tissues. J. Virol. 81:6482–6490. 10.1128/JVI.02876-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukashevich IS, Carrion R, Jr, Salvato MS, Mansfield K, Brasky K, Zapata J, Cairo C, Goicochea M, Hoosien GE, Ticer A, Bryant J, Davis H, Hammamieh R, Mayda M, Jett M, Patterson J. 2008. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine 26:5246–5254. 10.1016/j.vaccine.2008.07.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poonia B, Salvato MS, Yagita H, Maeda T, Okumura K, Pauza CD. 2009. Treatment with anti-FasL antibody preserves memory lymphocytes and virus-specific cellular immunity in macaques challenged with simian immunodeficiency virus. Blood 114:1196–1204. 10.1182/blood-2009-02-202655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zapata JC, Poonia B, Bryant J, Davis H, Ateh E, George L, Crasta O, Zhang Y, Slezak T, Jaing C, Pauza CD, Goicochea M, Moshkoff D, Lukashevich IS, Salvato MS. 2013. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol. J. 10:52. 10.1186/1743-422X-10-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moshkoff DA, Salvato MS, Lukashevich IS. 2007. Molecular characterization of a reassortant virus derived from Lassa and Mopeia viruses. Virus Genes 34:169–176. 10.1007/s11262-006-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvato M, Shimomaye E, Oldstone MB. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169:377–384. 10.1016/0042-6822(89)90163-3 [DOI] [PubMed] [Google Scholar]
- 18.Zapata JC, Salvato MS. 2013. Arenavirus variations due to host-specific adaptation. Viruses 5:241–278. 10.3390/v5010241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FDA. 2010. Guidance for industry. Characterization and qualification of cell substrates and other biological materials used in the production of viral vaccines for infectious disease indications. http://www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/UCM202439.pdf
- 20.Cheng-Mayer C, Weiss C, Seto D, Levy JA. 1989. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc. Natl. Acad. Sci. U. S. A. 86:8575–8579. 10.1073/pnas.86.21.8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed R, Oldstone MB. 1988. Organ-specific selection of viral variants during chronic infection. J. Exp. Med. 167:1719–1724. 10.1084/jem.167.5.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trivedi P, Meyer KK, Streblow DN, Preuninger BL, Schultz KT, Pauza CD. 1994. Selective amplification of simian immunodeficiency virus genotypes after intrarectal inoculation of rhesus monkeys. J. Virol. 68:7649–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall JS, French R, Hein GL, Morris TJ, Stenger DC. 2001. Three distinct mechanisms facilitate genetic isolation of sympatric wheat streak mosaic virus lineages. Virology 282:230–236. 10.1006/viro.2001.0841 [DOI] [PubMed] [Google Scholar]
- 24.Cabot B, Martell M, Esteban JI, Sauleda S, Otero T, Esteban R, Guardia J, Gomez J. 2000. Nucleotide and amino acid complexity of hepatitis C virus quasispecies in serum and liver. J. Virol. 74:805–811. 10.1128/JVI.74.2.805-811.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanjuan R, Codoner FM, Moya A, Elena SF. 2004. Natural selection and the organ-specific differentiation of HIV-1 V3 hypervariable region. Evolution 58:1185–1194. 10.1111/j.0014-3820.2004.tb01699.x [DOI] [PubMed] [Google Scholar]
- 26.Deforges S, Evlashev A, Perret M, Sodoyer M, Pouzol S, Scoazec JY, Bonnaud B, Diaz O, Paranhos-Baccala G, Lotteau V, Andre P. 2004. Expression of hepatitis C virus proteins in epithelial intestinal cells in vivo. J. Gen. Virol. 85:2515–2523. 10.1099/vir.0.80071-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jelcic I, Hotz-Wagenblatt A, Hunziker A, Zur Hausen H, de Villiers EM. 2004. Isolation of multiple TT virus genotypes from spleen biopsy tissue from a Hodgkin's disease patient: genome reorganization and diversity in the hypervariable region. J. Virol. 78:7498–7507. 10.1128/JVI.78.14.7498-7507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jridi C, Martin JF, Marie-Jeanne V, Labonne G, Blanc S. 2006. Distinct viral populations differentiate and evolve independently in a single perennial host plant. J. Virol. 80:2349–2357. 10.1128/JVI.80.5.2349-2357.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown RJ, Peters PJ, Caron C, Gonzalez-Perez MP, Stones L, Ankghuambom C, Pondei K, McClure CP, Alemnji G, Taylor S, Sharp PM, Clapham PR, Ball JK. 2011. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J. Virol. 85:6024–6037. 10.1128/JVI.00131-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright CF, Morelli MJ, Thebaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. 2011. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J. Virol. 85:2266–2275. 10.1128/JVI.01396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinschewer DD, Perez M, de la Torre JC. 2003. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 77:3882–3887. 10.1128/JVI.77.6.3882-3887.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eichler R, Strecker T, Kolesnikova L, ter Meulen J, Weissenhorn W, Becker S, Klenk HD, Garten W, Lenz O. 2004. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res. 100:249–255. 10.1016/j.virusres.2003.11.017 [DOI] [PubMed] [Google Scholar]
- 33.Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J. Virol. 83:7029–7039. 10.1128/JVI.00329-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shtanko O, Imai M, Goto H, Lukashevich IS, Neumann G, Watanabe T, Kawaoka Y. 2010. A role for the C terminus of Mopeia virus nucleoprotein in its incorporation into Z protein-induced virus-like particles. J. Virol. 84:5415–5422. 10.1128/JVI.02417-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80:9192–9199. 10.1128/JVI.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. 2009. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 83:11330–11340. 10.1128/JVI.00763-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. U. S. A. 108:2396–2401. 10.1073/pnas.1016404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. 10.1038/nature09605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grande-Perez A, Sierra S, Castro MG, Domingo E, Lowenstein PR. 2002. Molecular indetermination in the transition to error catastrophe: systematic elimination of lymphocytic choriomeningitis virus through mutagenesis does not correlate linearly with large increases in mutant spectrum complexity. Proc. Natl. Acad. Sci. U. S. A. 99:12938–12943. 10.1073/pnas.182426999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes EC. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77:11296–11298. 10.1128/JVI.77.20.11296-11298.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graci JD, Cameron CE. 2008. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 3:553–566. 10.2217/17460794.3.6.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plotkin JB, Robins H, Levine AJ. 2004. Tissue-specific codon usage and the expression of human genes. Proc. Natl. Acad. Sci. U. S. A. 101:12588–12591. 10.1073/pnas.0404957101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhi N, Wan Z, Liu X, Wong S, Kim DJ, Young NS, Kajigaya S. 2010. Codon optimization of human parvovirus B19 capsid genes greatly increases their expression in nonpermissive cells. J. Virol. 84:13059–13062. 10.1128/JVI.00912-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL. 2010. Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 10:253. 10.1186/1471-2148-10-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandit A, Sinha S. 2011. Differential trends in the codon usage patterns in HIV-1 genes. PLoS One 6:e28889. 10.1371/journal.pone.0028889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeiffer JK, Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1:e11. 10.1371/journal.ppat.0010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pariente N, Sierra S, Lowenstein PR, Domingo E. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723–9730. 10.1128/JVI.75.20.9723-9730.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crotty S, Cameron CE, Andino R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U. S. A. 98:6895–6900. 10.1073/pnas.111085598 [DOI] [PMC free article] [PubMed] [Google Scholar]