ABSTRACT
The NSvc2 glycoproteins encoded by Rice stripe tenuivirus (RSV) share many characteristics common to the glycoproteins found among Bunyaviridae. Within this viral family, glycoproteins targeting to the Golgi apparatus play a pivotal role in the maturation of the enveloped spherical particles. RSV particles, however, adopt a long filamentous morphology. Recently, RSV NSvc2 glycoproteins were shown to localize exclusively to the ER in Sf9 insect cells. Here, we demonstrate that the amino-terminal NSvc2 (NSvc2-N) targets to the Golgi apparatus in Nicotiana benthamiana cells, whereas the carboxyl-terminal NSvc2 (NSvc2-C) accumulates in the endoplasmic reticulum (ER). Upon coexpression, NSvc2-N redirects NSvc2-C from the ER to the Golgi bodies. The NSvc2 glycoproteins move together with the Golgi stacks along the ER/actin network. The targeting of the NSvc2 glycoproteins to the Golgi bodies was strictly dependent on functional anterograde traffic out of the ER to the Golgi bodies or on a retrograde transport route from the Golgi apparatus. The analysis of truncated and chimeric NSvc2 proteins demonstrates that the Golgi targeting signal comprises amino acids 269 to 315 of NSvc2-N, encompassing the transmembrane domain and 24 adjacent amino acids in the cytosolic tail. Our findings demonstrate for the first time that the glycoproteins from an unenveloped Tenuivirus could target Golgi bodies in plant cells.
IMPORTANCE NSvc2 glycoprotein encoded by unenveloped Rice stripe tenuivirus (RSV) share many characteristics in common with glycoprotein found among Bunyaviridae in which all members have membrane-enveloped sphere particle. Recently, RSV NSvc2 glycoproteins were shown to localize exclusively to the ER in Sf9 insect cells. In this study, we demonstrated that the RSV glycoproteins could target Golgi bodies in plant cells. The targeting of NSvc2 glycoproteins to the Golgi bodies was dependent on active COP II or COP I. The Golgi targeting signal was mapped to the 23-amino-acid transmembrane domain and the adjacent 24 amino acids of the cytosolic tail of the NSvc2-N. In light of the evidence from viruses in Bunyaviridae that targeting Golgi bodies is important for the viral particle assembly and vector transmission, we propose that targeting of RSV glycoproteins into Golgi bodies in plant cells represents a physiologically relevant mechanism in the maturation of RSV particle complex for insect vector transmission.
INTRODUCTION
Rice stripe virus (RSV) is the type member of the genus Tenuivirus (1). RSV has caused severe damage to rice crops in China and is known to be transmitted by Laodelphax striatellus in a persistent, circulative-propagative manner (2). The RSV genome consists of four negative-sense single-stranded RNA segments, designated RNA1, 2, 3 and 4, which encode seven ORFs using a negative or ambisense coding strategy (3). RNA1 is negative sense and encodes an RNA-dependent RNA polymerase (RdRp) (4). The other three segments adopt an ambisense coding strategy. RNA2 encodes a 22.8-kDa protein (NSs2) from the viral RNA (vRNA) and a 94-kDa protein (NSvc2) from the viral cRNA (vcRNA) (5). RNA3 encodes a viral suppressor (NSs3, 23.9 kDa) from the vRNA (6) and a nucleocapsid protein (NSvc3, 35 kDa) from the vcRNA (7, 8). RNA4 encodes a 20.5-kDa protein (NSs4) from the vRNA and a movement protein (NSvc4, 32 kDa) from the vcRNA (9).
Based on the phylogenetic relationship and their genome organization and gene expression strategies, tenuiviruses are more closely related to the animal-infecting viruses in the genus Phlebovirus of the family Bunyaviridae than they are to plant tospoviruses (10). The NSvc2 protein encoded by RSV (hereinafter referred to as the NSvc2 glycoprotein) shares many characteristics in common with the glycoproteins found in the Bunyaviridae family of viruses in which all members adopt an enveloped spherical virion form (10). The glycoprotein encoded by the Bunyaviridae viruses is processed into two proteins, Gn (the amino-terminal glycoprotein) and Gc (the carboxyl-terminal glycoprotein), which together form the surface spikes of the mature enveloped virion (11–14). The Gn protein of several viruses, including Uukuniemi virus (UUKV) (15), Punta torovirus (16), and Rift valley fever virus (RVFV) (17) in the genus Phlebovirus, as well as Tomato spotted wilt tospovirus (TSWV) (18), has been shown to accumulate in the Golgi apparatus, while the Gc protein localizes to the endoplasmic reticulum (ER). Upon coexpression, both glycoproteins localize to the Golgi apparatus (16–19), suggesting that Gn can retarget Gc from the ER to the Golgi apparatus. The targeting of the viral glycoproteins to the Golgi apparatus plays a pivotal role in the maturation of the viral particles. The NSvc2 glycoprotein encoded by RSV was predicted to be functionally similar to the glycoproteins found on other Bunyaviridae viruses. RSV particles, however, adopt a long filamentous morphology unenveloped (19, 20). The enveloped nature of Bunyaviridae versus the unenveloped nature of Tenuivirus raises the question of what common or unique strategies have evolved for them to form different morphology of viral particle. Zhao et al. (21) recently reported that the NSvc2 protein, or its two processing products, the amino terminus of NSvc2 (NSvc2-N) and the carboxyl terminus of NSvc2 (NSvc2-C), exclusively localized to the ER membrane in Spodoptera frugiperda (Sf9) insect cells. It remains poorly understood whether the ER localization (the inability to target to the Golgi apparatus) of the NSvc2 glycoproteins is the key step determining the adoption of a long filamentous particle in RSV. It is also unknown why does a nonenveloped tenuivirus encode glycoproteins.
RSV systemically infects Nicotiana benthamiana by mechanical inoculation (9, 22). In the present study, the subcellular targeting of the NSvc2 glycoproteins and the requirements for their targeting were extensively characterized in N. benthamiana. We demonstrated that the NSvc2-N glycoprotein alone is able to target to the Golgi apparatus in N. benthamiana, whereas NSvc2-C localizes to the ER membrane in the absence of NSvc2-N. Upon coexpression, NSvc2-N redirects NSvc2-C to the Golgi apparatus. The NSvc2 glycoproteins were found to move together with the Golgi stacks along the ER/actin network in N. benthamiana epidermal cells. Using dominant-negative mutants, we demonstrated that the targeting of the NSvc2 proteins from the ER to the Golgi bodies was strictly dependent on COP I and COP II early secretion pathways. The analysis of truncated and chimeric NSvc2 proteins demonstrated that the Golgi targeting signal localized to amino acids 269 to 315, encompassing the 23-amino-acid transmembrane domain and the 24 adjacent amino acids of the cytosolic tail. Our findings provide novel insights into the cellular properties of RSV glycoproteins in plant cells.
MATERIALS AND METHODS
Plasmid constructs and organelle markers. (i) p1300S-NSvc2-N-YFP and p1300S-NSvc2-C-YFP.
NSvc2-N and NSvc2-C were amplified from total RNA isolated from rice infected by RSV using reverse transcription-PCR (RT-PCR) and the primers XT746/XT747 and XT800/XT388 (see Table S1 in the supplemental material). The NSvc2-N and NSvc2-C PCR fragments were digested with KpnI and BamHI and inserted into p1300S-YFP using the same restriction sites to obtain p1300S-NSvc2-N-YFP and p1300S-NSvc2-C-YFP, respectively.
(ii) p1300S-NSvc2-Intron-YFP.
A potato ST-LS1 intron (23) was inserted into the AG/GT site at nucleotide (nt) position 1182 of NSvc2. The ST-LS1 intron, N-terminal fragment (1,182 nt), and C-terminal fragment (1,423 nt) of NSvc2 were amplified using the primers XT957/XT958, XT746/XT959, and XT960/XT388, respectively. The three PCR fragments were mixed and amplified using XT746/XT388 to obtain NSvc2-Intron, which was then digested with KpnI and BamHI and inserted into p1300S-YFP using the same restriction sites.
(iii) p1300S-NSvc2-N-46del-YFP and p1300S-NSvc2-N-63del-YFP.
NSvc2-N containing either a 46- or 63-amino-acid deletion at the C terminus was amplified using the primer pairs XT746/XT807 or XT746/XT835, and the PCR products were inserted into the KpnI and BamHI sites of p1300S-YFP, respectively.
(iv) p1300S-SSNTMDNCTN-YFP, p1300S-SSNTMDNCTNdel46-YFP, and p1300S-SSNTMDNCTNdel63-YFP.
The signal peptide (SSN), transmembrane domain (TMDN) containing the full-length cytosolic domain (CTN), TMDN containing the CTN with a 46-amino-acid deletion, and the TMDN with the CTN containing a 63-amino-acid deletion at the C terminus of NSvc2-N were amplified using the corresponding primer pairs (XT746/XT837, XT836/XT747, XT836/XT807, and XT836/XT835). The SSNTMDNCTN, SSNTMDNCTNdel46, and SSNTMDNCTNdel63 fragments were fused using overlap PCR and the primers XT746/XT747, XT746/XT807, and XT746/XT835, and were inserted into the KpnI and BamHI sites of p1300S-YFP, respectively.
(v) p1300S-NSvc-C(TMDNCTN)-YFP and p1300S-NSvc-C(TMD-CT-del46)-YFP.
A fragment of NSvc2-C lacking the TMDC and the CTC was amplified using the primers XT800 and XT869. The TMDN fragment with the full-length CTN and the TMDN fragment with the CTN containing a 46-amino-acid deletion at the C terminus of NSvc2-N were amplified with the primer pairs XT747/XT868 and XT807/XT868. They were then fused using overlap PCR and the primers XT800/XT747 and XT800/XT807, respectively. The products of overlap PCR were digested with KpnI and BamHI and cloned into p1300S-YFP.
(vi) p1300S-CFP-Sec24 and p1300S-Arf1-CFP.
The full-length Sec24 (AT3G07100) and Arf1 genes were amplified using RT-PCR, and the total RNA was extracted from the Col ecotype of Arabidopsis thaliana using the primer pairs XT743/XT754 and XT784/XT785, respectively. The Sec24 PCR fragments were digested with BamHI and cloned into the BglII site of p1300S-CFP, while Arf1 was digested with BamHI and cloned into the BamHI site of p1300S-CFP.
(vii) p1300S-Arf1 (T31N).
To construct p1300S-ArfI (T31N), site-directed mutagenesis was used to introduce the mutation into Arf1 using the primers XT784/XT795 and XT794/XT785 and overlap PCR. The PCR product was digested with BamHI and cloned into p1300S.
The ER marker mCherry-HDEL (24) and the Golgi marker Man49-mCherry (24) were obtained from the Arabidopsis Biological Resource Center. The Sar1 dominant-negative mutant construct Sar1 (H74L) was kindly provided by Taiyun Wei (25).
Plant material, transient expression, and treatment.
RSV (Jiangsu isolate) was collected from infected rice in a field in Nanjing and frozen at −80°C until use. All transient expression experiments were performed using 6- to 8-week-old N. benthamiana plants. Agrobacterium tumefaciens cells (C58C1 containing various RSV constructs and organelle markers) were grown using kanamycin selection. The Agrobacterium cells were treated with infiltration buffer (10 mM MgCl2, 10 mM MES [pH 5.9], and 150 μM acetosyringone) for 3 h at room temperature before being infiltrated (optical density at 600 nm = 0.5) into the abaxial surface of N. benthamiana leaves. All agroinfiltrated plants were grown in growth chambers (model GXZ500D; Jiangnan Motor Factory, Ningbo, People's Republic of China) under a 16-h light/8-h dark cycle and a constant temperature of 25°C. The agroinfiltrated leaves were examined for fluorescence expression between 24 to 72 h postinfiltration (hpi). When applicable, LatB (Sigma) was infiltrated at a final concentration of 10 μM into N. benthamiana leaves before fluorescence observation.
Confocal laser scanning microscopy.
Leaf discs were dissected from the agroinfiltrated leaf area of N. benthamiana leaves and mounted in water between two coverslips. Images and movies were captured using a Carl Zeiss LSM 710 confocal laser scanning microscope and ×20, ×63 oil or ×63 water immersion objective lenses. Cyan fluorescent protein (CFP) fluorescence was excited at 405 nm and emission captured at 440 to 470 nm, yellow fluorescent protein (YFP) fluorescence was excited at 488 nm and emission captured at 497 to 520 nm, and mCherry was excited at 561 nm and emission captured at 585 to 615 nm. Images were processed using the Zeiss 710 CLSM and Adobe Photoshop programs (San Jose, CA). Movies were edited using the Corel Video Studio Pro X4 software (Ottawa, Ontario, Canada).
Western blot analysis.
Plant leaves from N. benthamiana agroinfiltrated with NSvc2-N-YFP, NSvc2-C-YFP, and NSvc2-YFP constructs were ground in a 1:3 (wt/vol; 0.1 g/300 μl) ratio of extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, and 1× plant protease inhibitor). After centrifugation for 10 min at 3,000 × g, the supernatant of the total protein preparation was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis for immunoblot analysis. The blots were probed with anti-YFP (polyclonal antibody, 1:1,000 dilution; Biyuntian, Shanghai, China) and visualized with AP-conjugated goat anti-rabbit secondary antibodies (1:1,000 dilution; Biyuntian) followed by nitroblue tetrazolium (NBT)–5-bromo-4-chloro-3-indolylphosphate (BCIP) (ready-made solutions; Shenggong, Shanghai, China) staining.
For subcellular fractionations, the soluble and microsomal fractions were isolated from N. benthamiana leaves agroinfiltrated with NSvc2-N-YFP, NSvc2-C-YFP, and NSvc2-YFP constructs as described by Peremyslov et al. (26). The antigens on the membranes were blotted with anti-YFP (rabbit). It was detected by DyLight 680-coupled goat anti-rabbit antibodies (1:10,000 dilution; Pierce, Rockford, IL) and then visualized by using a Licor Odyssey scanner.
RESULTS
The NSvc2-N protein is targeted to the Golgi apparatus.
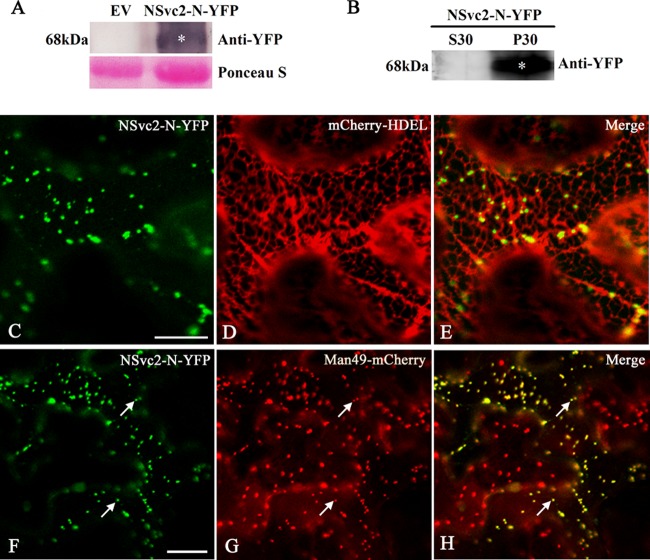
N. benthamiana is an ideal plant species in which to assess the subcellular localization of viral proteins. To characterize the subcellular target of the NSvc2 glycoproteins in plant cells, we first fused the YFP to the C terminus of NSvc2-N (Fig. 1) and then agroinfiltrated the construct into N. benthamiana epidermal cells. Western blot analysis showed that NSvc2-N-YFP fusion protein was expressed as a size of 68-kDa protein (Fig. 2A), indicating a proper expression of the NSvc2-N-YFP construct. To investigate the intracellular localization of the NSvc2-N-YFP protein, we isolated soluble (S30) and microsomal (P30) protein fractions from N. benthamiana leaves agroinfiltrated with NSvc2-N-YFP. We found that NSvc2-N-YFP was localized exclusively in microsomal fractions that are known to contain ER membrane structures and Golgi bodies (Fig. 2B).
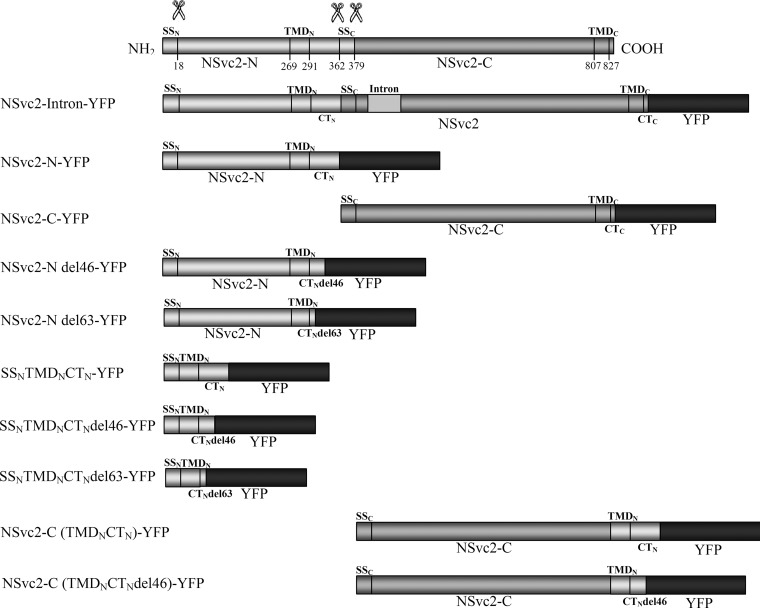
FIG 1.
Schematic diagrams of the viral constructs used for expression analysis (the glycoprotein constructs are aligned below the precursor). Predicted cleavage sites (scissor symbols) and amino acid positions are indicated. SS, TMD, and CT refer to the signal sequence, the transmembrane domain, and the cytosolic tail, respectively. SSN and SSC refer to the SSs of NSvc2-N and NSvc2-C, respectively. TMDN and TMDC refer to the TMD of NSvc2-N and NSvc2-C, respectively. CTN and CTC refer to the CTs of NSvc2-N and NSvc2-C, respectively. An intron of the potato ST-LS1 was inserted at the nucleotide position of 1182 on NSvc2. In all constructs, the YFP fluorophore was fused in frame at the site of the stop codon.
FIG 2.
Subcellular localization of the NSvc2-N protein in N. benthamiana leaf epidermal cells. (A) Immunoblot analysis of NSvc2-N-YFP fusion proteins expressed by agroinfiltration in N. benthamiana leaves. The blots were probed by using anti-YFP. Empty vector (EV) was used as a negative control. Ponceau-S was used as a loading control. (B) Subcellular fractionation analysis of NSvc2-N-YFP fusion protein. The soluble (S30) and microsomal (P30) fractions were isolated from agroinfiltrated leaves of N. benthamiana. The membrane blots were probed using anti-YFP. (C to E) Colocalization of NSvc2-N-YFP (C) with ER labeled by mCherry-HDEL at 36 hpi (D). (E) A merged image of panels C and D is also shown. (F to H) Colocalization of the NSvc2-N-YFP (F) with the Golgi apparatus labeled by Man49-mCherry at 36 hpi (G). A merged image (H) illustrates the NSvc2-N protein targeted to the Golgi apparatus. Scale bars, 20 μm.
To further characterize the subcellular localization of NSvc2-N-YFP, the infiltrated leaves were examined using Zeiss 710 confocal laser scanning microscopy. At 36 h postinfiltration (hpi), NSvc2-N-YFP was observed as numerous small bodies in the cortical cytoplasm of the cells (Fig. 2C). To determine whether NSvc2-N accumulated in the ER membrane, we coexpressed the NSvc2-N-YFP protein with the HDEL signal fused to the N terminus of mCherry (mCherry-HDEL) in N. benthamiana (24). The merge of NSvc2-N-YFP with mCherry-HDEL images revealed that the NSvc2-N-YFP signal did not colocalize with the ER marker, while those NSvc2-N-YFP punctate bodies were still associated with the ER membrane (Fig. 2C to E).
To determine whether the NSvc2-N-YFP bodies colocalized with the Golgi stacks, we coinfiltrated the Golgi marker construct Man49-mCherry (24) with NSvc2-N-YFP in N. benthamiana epidermal cells. At 36 hpi, we found that the NSvc2-N-YFP bodies colocalized with the Golgi stacks (Fig. 2F to H), suggesting that the NSvc2-N-YFP protein targets to the Golgi apparatus. We then examined the NSvc2-N-YFP protein signal at three time points—24, 48, and 72 hpi—and found that NSvc2-N-YFP was targeted to the Golgi body as early as 24 hpi (data not shown).
The NSvc2-C protein accumulates in the ER membrane.
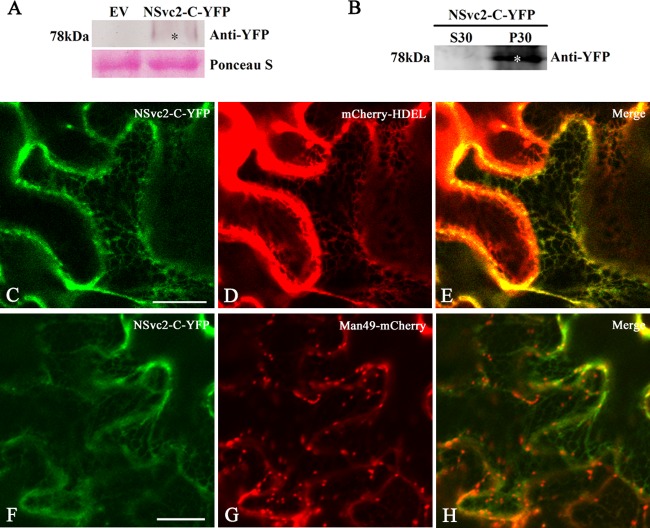
We also fused NSvc2-C protein with YFP at its C terminus (Fig. 1) and infiltrated the construct into N. benthamiana epidermal cells. Immunoblot analysis showed that NSvc2-C-YFP protein expressed as 78-kDa protein which is same as the predicted size of NSvc2-C-YFP fusion protein (Fig. 3A). Fractionation analysis revealed that NSvc2-C-YFP protein was localized only in the microsomal membrane fractions (Fig. 3B). To precisely define the intracellular distribution of NSvc2-C, the infiltrated leaves were characterized using confocal laser scanning microscopy. The green fluorescent signal of the NSvc2-C-YFP fusion protein appeared to be very weak, but was still detectable in an ER-like network structure observed at 36 hpi (Fig. 3C). To determine whether these fluorescent signals colocalized with the ER structure, the cortical ER marker mCherry-HDEL was coinfiltrated with NSvc2-C-YFP. As shown in Fig. 3C to E, the NSvc2-C-YFP protein colocalized with the ER membrane network.
FIG 3.
Subcellular localization of the NSvc2-C protein in N. benthamiana leaf epidermal cells. (A) Western blot analysis of NSvc2-C-YFP fusion proteins expressed by agroinfiltration in N. benthamiana leaves. The blots were probed using anti-YFP. Ponceau-S was used as a loading control. Empty vector (EV) was used as a negative control. (B) Subcellular distribution of NSvc2-C-YFP protein by fractionation analysis. The soluble (S30) and microsomal (P30) fractions were isolated from agroinfiltrated leaves of N. benthamiana. The membrane blots were probed using anti-YFP. (C to E) Colocalization of the NSvc2-C-YFP (C) with the ER labeled by mCherry-HDEL at 36 hpi (D). A merged image (E) shows that NSvc2-C-YFP aligns well with the ER membrane. (F to H) Colocalization of the NSvc2-C-YFP (F) with the Golgi apparatus labeled by Man49-mCherry at 36 hpi (G). (H) Merged image of panels F and G. Scale bars, 20 μm.
To examine whether NSvc2-C-YFP accumulated in the Golgi stacks, we coinfiltrated N. benthamiana cells with NSvc2-C-YFP and the Golgi marker Man49-mCherry. As shown in Fig. 3F to H, no fluorescent signal associated with NSvc2-C-YFP was found to accumulate in the Golgi apparatus. To confirm whether NSvc2-C-YFP exhibits any accumulation in the Golgi stacks, we checked the fluorescent signal of NSvc2-C-YFP at 24, 48, and 72 hpi. The NSvc2-C-YFP protein did not form any small bodies that could target to the Golgi body at the three time points examined (data not shown). These results suggest that NSvc2-C-YFP was arrested in the ER in N. benthamiana.
The NSvc2-N protein recruits NSvc2-C from the ER to the Golgi apparatus.
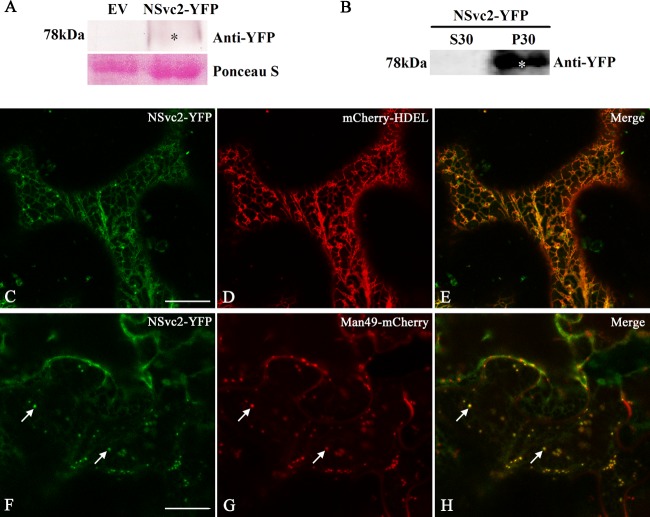
To determine the localization and trafficking of the NSvc2 glycoproteins when expressed from their precursor, we fused YFP to the C terminus of the NSvc2 precursor protein. However, the construct containing the full-length NSvc2 gene cannot grow in E. coli cells, suggesting that the full-length NSvc2 gene is toxic to E. coli. We therefore inserted a potato ST-LS1 intron (23) into the AG/GT site at nucleotide position 1182 of NSvc2. The intron-containing construct, NSvc2-Intron-YFP (Fig. 1), can successfully generate a green fluorescence signal in N. benthamiana epidermal cells after agroinfiltration. Total RNA was then isolated from infiltrated leaves and the NSvc2-Intron-YFP RT-PCR products were sequenced to confirm that the intron had been precisely processed from the inserted site of NSvc2 (NSvc2-Intron-YFP is hereinafter referred to as NSvc2-YFP). Immunoblot analysis showed that NSvc2-C-YFP has been efficiently processed from precursor protein NSvc2-YFP and expressed as 78-kDa protein (Fig. 4A). The processed protein was distributed exclusively in the microsomal fractions which are known to contain ER membranes and Golgi bodies (Fig. 4B).
FIG 4.
Subcellular localization of NSvc2-YFP in N. benthamiana leaf epidermal cells. (A) Immunoblot analysis of NSvc2-YFP fusion proteins (NSvc2-N and NSvc2-C-YFP glycoproteins were processed from its common glycoprotein precursor NSvc2-YFP) expressed by agroinfiltration in N. benthamiana leaves. The membrane blots were probed using anti-YFP. Ponceau S was used as a loading control. Empty vector (EV) was used as a negative control. (B) Subcellular distribution analysis of NSvc2-YFP protein by fractionation. The soluble (S30) and microsomal (P30) fractions were isolated from agroinfiltrated leaves of N. benthamiana. The membrane blots were probed using anti-YFP. (C to E) Coexpression of the NSvc2-YFP (NSvc2-C-YFP was processed from this glycoprotein precursor) (C) with mCherry-HDEL (D) at 48 hpi. (E) Merged image of panels C and D. (F to H) Colocalization of the NSvc2-YFP (NSvc2-C-YFP was processed from the glycoprotein precursor) (F) with the Golgi apparatus labeled by Man49-mCherry (G) at 48 hpi. (H) The merged image shows the NSvc2-C protein targeted to the Golgi apparatus in the presence of NSvc2-N. Scale bars, 20 μm.
We then coexpressed NSvc2-YFP with the ER marker mCherry-HDEL in N. benthamiana and the infiltrated leaves were examined using Zeiss confocal laser scanning microscopy. Monitoring of NSvc2-C-YFP (NSvc2-C-YFP processed from the NSvc2 precursor) showed that the fluorescent signal highlighted by NSvc2-C-YFP colocalized in the ER network at 24 to 48 hpi. At 48 to 72 hpi, NSvc2-C-YFP began to induce punctate structures along the ER membrane in the presence of NSvc2-N (Fig. 4C to E). To identify whether the newly formed bodies targeted to the Golgi apparatus, we coinfiltrated N. benthamiana with NSvc2-YFP and the Golgi marker Man49-mCherry. As shown in Fig. 4F to H, NSvc2-C-YFP bodies were indeed found to be targeted to the Golgi apparatus. These results strongly suggest that NSvc2-N is able to recruit NSvc2-C from the ER to the Golgi apparatus.
Targeted NSvc2 glycoproteins move together with the Golgi stacks in N. benthamiana.
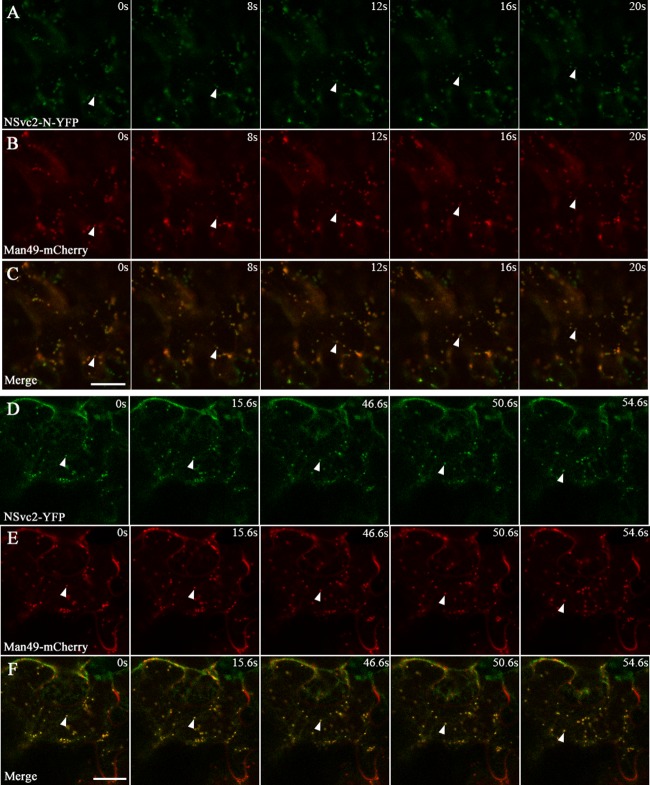
In tobacco leaf cells, Golgi bodies traffic on an underlying ER track in an actin-dependent manner (27, 28). To examine whether the targeted RSV NSvc2 glycoproteins move with the Golgi bodies, we utilized time-lapse confocal microscopy to monitor the movement of NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP (processed from the NSvc2-YFP precursor) in the presence of the Golgi marker. Figure 5A to C and D to F show examples of the movement of the NSvc2-N-YFP and NSvc2-N/NSvc2-C-YFP bodies with the Golgi stacks, and the arrows mark the progressive movement of these bodies in each sequence. We found that both NSvc2-N-YFP and NSvc2-N/NSvc2-C-YFP moved together with the Golgi bodies (Fig. 5A to C and D to F; see Videos S1 and S2 in the supplemental material).
FIG 5.
NSvc2 glycoproteins trafficking together with the Golgi stacks along the ER track in N. benthamiana leaf epidermal cells. (A to C) Time-lapse confocal images showing the movement of NSvc2-N-YFP (A) and the Golgi apparatus (B) labeled by Man49-mCherry at the times indicated. The position of the tracked signal is marked with an arrow. (C) Merged image of panels A and B. (D to F) Time-lapse confocal images showing the movement of NSvc2-YFP (NSvc2-N and NSvc2-C-YFP were processed from the glycoprotein precursor NSvc2-YFP) (D) and the Golgi apparatus (E) at the times indicated. The position of the tracked signal is marked with an arrow. The merged images (F) demonstrate that the NSvc2 proteins move together with the Golgi apparatus along the ER track. Scale bars, 20 μm.
To determine whether the movement of bodies labeled with NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP is dependent on similar forces driving the movement of the Golgi bodies, we treated agroinfiltrated leaves at 48 hpi with 10 μM latrunculin B, an actin depolymerizing agent (29). After 3 h of chemical treatment, we found that movement of the NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP, as well as Golgi bodies, was completely inhibited. However, NSvc2-N-YFP, NSvc2-N/NSvc2-C-YFP, and the Golgi bodies remained colocalized (see Videos S3 and S4 in the supplemental material). These data suggest that the NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP bodies move together with the Golgi stacks along the ER/actin network.
ER-to-Golgi targeting of NSvc2 glycoproteins is dependent on a functional COP II complex.
Given that the RSV NSvc2-N-YFP and NSvc2-YFP fusion proteins targeted to the Golgi, we ask whether the Golgi targeting of viral glycoproteins results from traffic out of the ER to the Golgi apparatus via ERES. To address this question, we coinfiltrated an ERES-marker, CFP-Sec24 (30), with NSvc2-N-YFP or NSvc2-YFP proteins into N. benthamiana leaf cells. As shown in Fig. 6A to C and G to I, the NSvc2-N-YFP or NSvc2-YFP bodies colocalized with CFP-Sec24 fluorescence at the ERES. These results suggest that NSvc2-N is able to redirect NSvc2-C from the ER to the ERES, from where they subsequently comigrate, most likely as a heterodimer, to the Golgi apparatus.
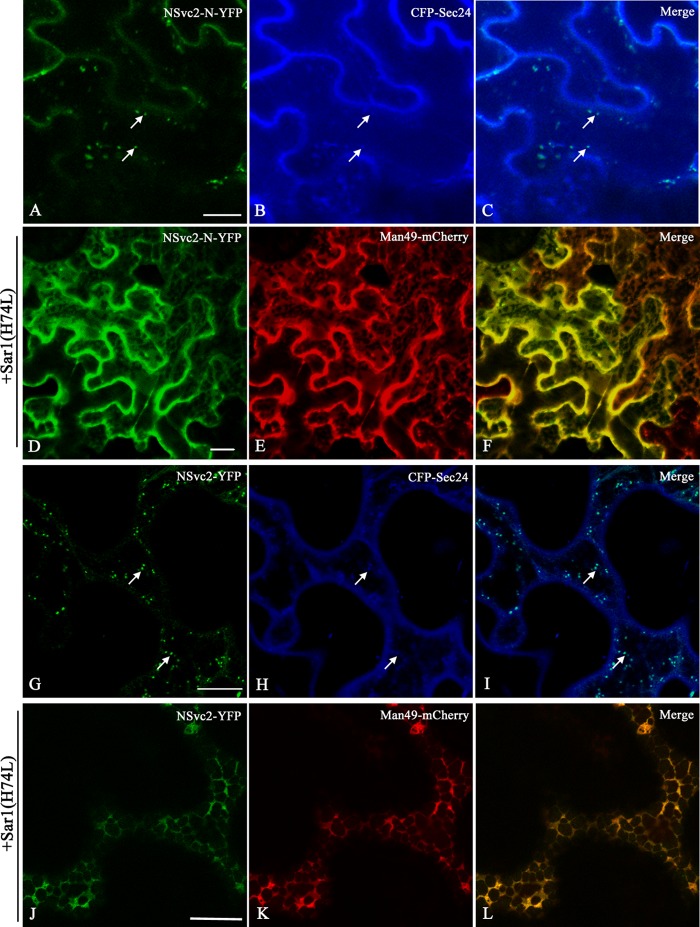
FIG 6.
ER-to-Golgi targeting of RSV NSvc2 glycoproteins depends on a functional COP II complex. (A to C) Confocal images of Nicotiana benthamiana epidermal cells coexpressing NSvc2-N-YFP (A) and the COP II marker CFP-Sec24 at 36 hpi (B). (C) Merged image of panels A and B. The arrows mark colocalization of NSvc2-N-YFP bodies with the ERES labeled with CFP-Sec24. (D to F) Coexpression of the dominant-negative mutant Sar1 (H74L) causes the redistribution of NSvc2-N-YFP (D), as well as the Golgi apparatus (E), back to the ER. (F) Merged image of panels D and E. (G to I) Cells coexpressing NSvc2-N and NSvc2-C-YFP (from their common precursor NSvc2-YFP) (G) and the ERES labeled with CFP-Sec24 at 48 hpi (H). (I) Merged image of panels G and I. (J to L) Coexpression of the dominant-negative mutant Sar1 (H74L) inhibits the transport of NSvc2-N and NSvc2-C-YFP (coexpressed from their common precursor NSvc2-YFP) to the Golgi complex. Scale bars, 20 μm.
The COP II complex is responsible for anterograde traffic out of the ER to the Golgi apparatus (31). To test whether COP II vesicles are involved in ER-to-Golgi transport of RSV NSvc2 glycoproteins, wild-type Sar1 or its dominant-negative mutant (H74L) (32) was coinfiltrated with NSvc2-N-YFP or NSvc2-YFP together with the Golgi marker Man49-mCherry into N. benthamiana. As shown in Fig. 6D to F and J to L, upon coexpression of NSvc2-N-YFP or NSvc2-YFP with Sar1 (H74L), the fluorescence of NSvc2-N-YFP or NSvc2-YFP, as well as of the Golgi bodies, was retrieved back to the ER network, whereas coexpression with wild-type Sar1 did not cause the NSvc2-N-YFP or NSvc2-YFP bodies to redistribute back to the ER (data not shown). These results suggest that the accumulation of the RSV glycoproteins at the ERES and in the Golgi bodies is dependent on a functional anterograde secretion pathway.
The accumulation of the NSvc2 glycoproteins at the Golgi bodies depends on active COP I.
To investigate whether the Golgi targeting of viral glycoproteins also involves retrograde traffic, we coinfiltrated Arf1 tagged with CFP, a COP I vesicle marker (33), with NSvc2-N-YFP or NSvc2-YFP in N. benthamiana. As shown in Fig. 7A to C and G to I, the NSvc2-N-YFP or NSvc2-YFP bodies colocalized with COP I vesicles labeled by Arf1-CFP.
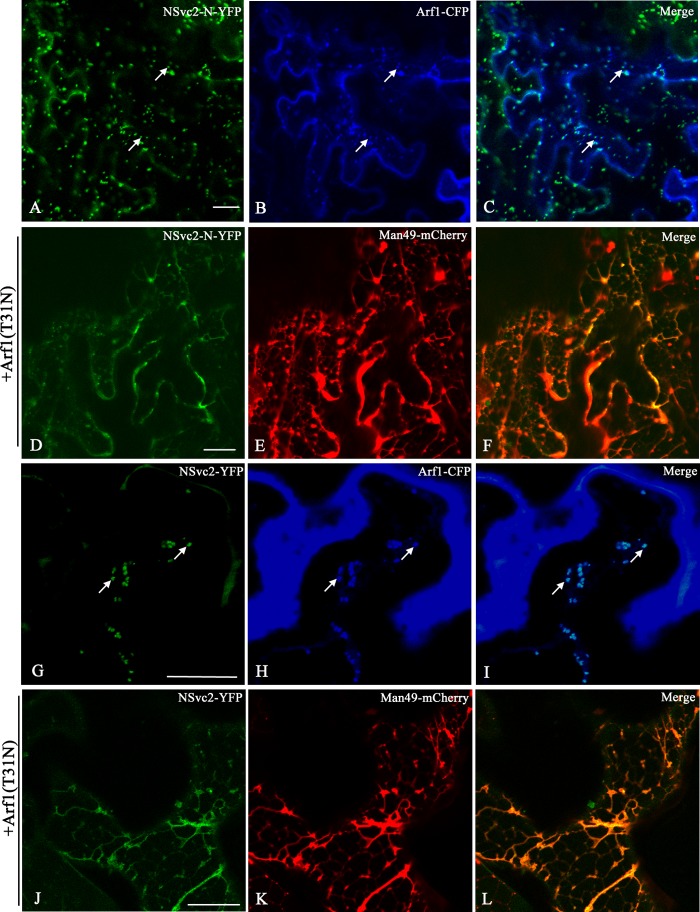
FIG 7.
ER-to-Golgi targeting of RSV NSvc2 glycoproteins depends on an active COP I complex. (A to C) Confocal images of N. benthamiana epidermal cells coexpressing NSvc2-N-YFP (A) and the COP I marker labeled with Arf1-CFP at 36 hpi (B). (C) Merged image of panels A and B. (D to F) Coexpression of the dominant-negative mutant Arf1 (T31N) led to the retention of NSvc2-N-YFP (D), as well as the Golgi apparatus (E), in the ER at 48 hpi. (F) Merged image of panels D and E. (G to I) Cells coexpressing NSvc2-YFP (G) and the COP I marker labeled with Arf1-CFP (H). (I) Merged image of panels G and H. (J to L) Coexpression of the dominant-negative Arf1 (T31N) blocks transport of NSvc2-N and NSvc2-C-YFP (coexpressed from their common precursor NSvc2-YFP) to the Golgi complex. Scale bars, 20 μm.
To determine the dependency of the ER-to-Golgi transport of RSV NSvc2 glycoproteins on active COP I, wild-type Arf1 or Arf1 (T31N), a dominant-negative mutant of COP I (33, 34), was coinfiltrated with NSvc2-N-YFP or NSvc2-YFP along with the Golgi marker Man49-mCherry into N. benthamiana. We found that NSvc2-N-YFP or NSvc2-YFP, as well as Man49-mCherry-labeled Golgi bodies redistributed back to the ER membrane in the presence of the dominant-negative Arf1 (T31N) (Fig. 7D to F and J to L). However, the coexpression of wild-type Arf1 has no such effect (data not shown). These data demonstrate that the Golgi targeting of RSV glycoproteins is also dependent on an active retrograde export route.
The Golgi targeting signal resides in a region of NSvc2-N encompassing a transmembrane domain and the 24 adjacent amino acids of the cytosolic tail.
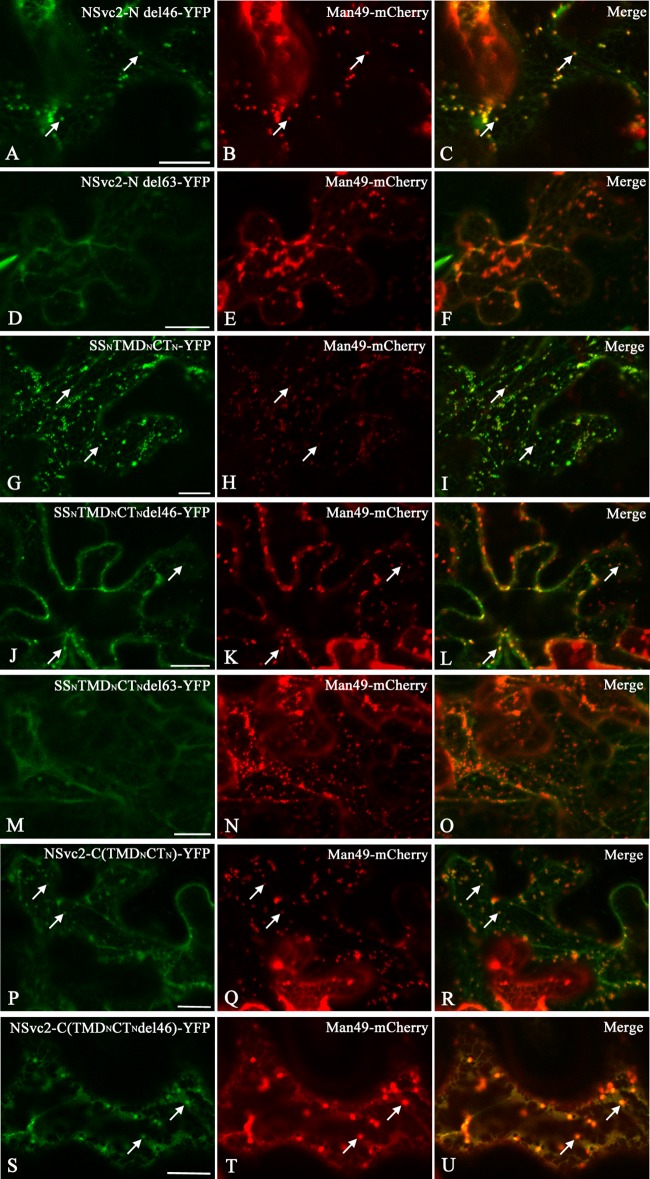
Both the NSvc2-N-YFP and NSvc2-YFP expressed in N. benthamiana localized to the Golgi complex, indicating that the Golgi retention signal resides in the N terminus of the NSvc2 protein. To map the domain responsible for the Golgi targeting of RSV NSvc2-N, a truncated NSvc2-N del46-YFP protein, where 46 amino acids at the C-terminal end of NSvc2-N within the cytosolic tail were deleted and fused with YFP (Fig. 1), was constructed and transiently expressed in N. benthamiana. The intracellular localization of this protein was determined by confocal fluorescence analysis after 48 hpi. As illustrated in Fig. 8A to C, the truncated NSvc2-N del46-YFP protein was still capable of targeting to the Golgi complex. Subsequently, 63 amino acids of the C-terminal end of the NSvc2-N protein within the cytosolic tail were deleted (Fig. 1). This truncated NSvc2-N del63-YFP protein was no longer targeted to the Golgi apparatus (Fig. 8D to F), suggesting that the amino acids in the cytosolic tail are required for entering into the Golgi apparatus.
FIG 8.
Golgi targeting signal analysis of truncated and chimeric NSvc2-N proteins. (A to U) Confocal images of N. benthamiana epidermal cells coexpressing Man49-mCherry with the truncated or chimeric proteins NSvc2-N del46-YFP (A to C), NSvc2-N del63-YFP (D to F), SSNTMDNCTN-YFP (G to I), SSNTMDN-CTNdel46-YFP (J to L), SSNTMDNCTNdel63-YFP (M to O), NSvc2-C(TMDNCTN)-YFP (P to R), and NSvc2-C(TMDNCTNdel46)-YFP (S to U), respectively, at 48 hpi. Scale bars, 20 μm.
To determine the minimum region required for Golgi targeting, the predicted transmembrane domain (amino acids 269 to 291) and the entire cytosolic domain (amino acids 292 to 361) of NSvc2-N were fused with its signal peptide sequence (amino acids 1 to 23) (Fig. 1). When this chimeric SSNTMDNCTN-YFP construct was expressed in N. benthamiana leaf cells, we found that it accumulated in the Golgi apparatus (Fig. 8G to I). Subsequently, the transmembrane domain and the 24 adjacent amino acids (CTdel46, amino acids 292 to 315) were fused with its signal peptide (Fig. 1). The resulting SSNTMDNCTNdel46-YFP construct also localized to the Golgi apparatus (Fig. 8J to L). Lastly, the transmembrane domain and the 7 adjacent amino acids (CTdel63, amino acids 292 to 298) were fused with its signal peptide (Fig. 1). As shown in Fig. 8M to O, this SSNTMDNCTNdel63-YFP construct was incapable of targeting to the Golgi complex. These analyses suggest that both the transmembrane domain (amino acids 269 to 291) and the 24 adjacent amino acids in the cytosolic tail of the NSvc2-N protein are required for Golgi targeting.
To substantiate the observation that the Golgi retention signal is located within the TMD and CT domains of NSvc2-N, the transmembrane domain (amino acids 269 to 291) and the entire cytosolic domain (amino acids 292 to 361) of NSvc2-N were swapped with those of NSvc2-C (Fig. 1). The resulting NSvc2-C(TMDNCTN)-YFP construct was coexpressed with mCherry-HDEL and Man49-mCherry separately in N. benthamiana. As shown in Fig. 8P to R, the chimeric NSvc2-C(TMDNCTN)-YFP construct was capable of targeting to the Golgi apparatus, suggesting that the transmembrane domain and the cytosolic domain of NSvc2-N was sufficient to direct NSvc2-C-YFP to the Golgi complex (Fig. 8P to R). To analyze the requirement for the Golgi targeting signal further, the transmembrane domain and the 24 adjacent amino acids in the cytosolic domain of NSvc2-N were swapped with the corresponding domain of NSvc2-C. As illustrated in Fig. 8S to U, this chimeric NSvc2-C(TMDNCTNdel46)-YFP protein was also capable of localizing to the Golgi apparatus. Taken together, these data suggest that the ER-to-Golgi targeting signal resides in the C-terminal region (amino acids 269 to 315) of NSvc2-N, encompassing the 23-amino-acid transmembrane domain and 24 adjacent amino acids in the cytosolic tail.
DISCUSSION
In this study, using N. benthamiana as a model system we demonstrated here for the first time that the glycoproteins from an unenveloped Tenuivirus could target into Golgi bodies in plant cells. The RSV NSvc2-N glycoprotein alone targeted to the Golgi apparatus, whereas the NSvc2-C glycoprotein accumulated in the ER membrane in the absence of NSvc2-N. Upon coexpression, NSvc2-N was able to redirect NSvc2-C from the ER to the Golgi apparatus. Using the Sar1 or Arf1 dominant-negative mutants, we demonstrated that the targeting of NSvc2 glycoproteins to the Golgi apparatus was dependent on an active COP I or COP II secretion pathway. We further revealed that the Golgi targeting signal mapped to a region of the NSvc2-N protein (amino acids 269 to 315) encompassing the 23-amino-acids transmembrane domain (TMD) and the adjacent 24 amino acids of the cytosolic tail.
The targeting of viral glycoproteins to the Golgi apparatus plays a pivotal role in the formation of enveloped spherical particles for the viruses (animal and plant infecting) in the Bunyaviridae family (15, 17, 35–41). Although RSV particle adopt long filamentous morphology (20, 21), the subcellular targeting to the Golgi apparatus seems to be a conserved mechanism between the unenveloped Rice stripe tenuivirus and the enveloped viruses in Bunyaviridae. Why RSV glycoproteins do not facilitate the formation of an enveloped spherical particle remains to be extensively investigated in the future. It is interesting that despite the common glycoprotein characteristics shared by RSV and viruses in the Bunyaviridae, all of the viruses in the Bunyaviridae have larger size of glycoproteins than are found in RSV.
For TSWV, the type member of Tospovirus which is the only genus containing plant-infecting viruses in the family Bunyaviridae, the glycoproteins forming the surface spikes of the mature viral particle play an important role in insect transmission (42). The key step where the virus enters the insect midgut cells is mediated by these glycoproteins (42). RSV particles must also enter the midgut cells of L. striatellus to complete their circulative-propagative transmission. The RSV-encoded glycoproteins were predicted to have a similar role in vector transmission. Although the NSvc2 protein was not detected in the filamentous RSV particle, this protein may function as a bridge between the virus particle and recognition sites on the insect cell, as is seen, for example, with helper component-proteinase (Hc-Pro) of potyvirus (43). The targeting of RSV NSvc2 proteins to the Golgi apparatus could be an essential process for glycoprotein modification and maturation, allowing the attachment of the RSV RNP particle and subsequent vector transmission.
Zhao et al. (21) reported that all of the RSV NSvc2 glycoproteins, including NSvc2-N, NSvc2-C, and the full-length NSvc2, localized exclusively to the ER membrane in Sf9 insect cells. Our findings on the Golgi targeting of NSvc2 glycoproteins in N. benthamiana cells were different from those reported by Zhao et al. in Sf9 insect cells. The RSV NSvc2 glycoproteins may have different subcellular localization patterns in different systems. The NSvc2 glycoproteins target to the Golgi apparatus in plant cells, whereas they were arrested in the ER membrane in insect cells. These two different findings together lead to an interesting new concept that the acquisition of an RSV viral particle from a plant host by L. striatellus insect vector may require glycoproteins which need to obtain glycosylation or similar modification in the Golgi apparatus, whereas transmission of an RSV viral particle from an insect vector back into a plant host may not require glycoproteins.
The leaf Golgi complex functions as a motile system that acquires products from a relatively stationary ER system (28, 31). The glycoproteins of TSWV have shown to target the Golgi body using a tobacco protoplast system (18). However, the movement of the viral glycoproteins in the plant cell has not been shown previously. We demonstrated here that the targeted NSvc2 glycoproteins moved together with the Golgi stacks along the ER/actin network in N. benthamiana epidermal cells. The movement of the NSvc2-N glycoprotein, together with the synchronous movement of the Golgi stacks in the N. benthamiana epidermal cells, gives rise to an interesting hypothesis that the NSvc2-N could be acting as a mobile system for picking up NSvc2-C from the ER and transport it into the Golgi stacks. This hypothesis is consistent with the finding that the NSvc2-N protein accumulated in the Golgi stacks as early as 24 hpi, whereas the NSvc2-C protein alone remained consistently localized in the ER. NSvc2-C only began to accumulate in the Golgi apparatus at 48 hpi in the presence of NSvc2-N. The constant movement of NSvc2-N will continue to pick up NSvc2-C in the Golgi stacks over time.
RSV NSvc2-N was able to facilitate NSvc2-C transport from the ER to the Golgi apparatus. Export of proteins from the ER in plant cells has been suggested to occur through different routes (44–47). For ER-to-Golgi transport, a widely accepted pathway is based on the sequential action of COP II and COP I complexes (27). Our results showed that RSV NSvc2-N and the NSvc2-N/NSvc2-C complex migrate to the Golgi apparatus via the ERES and that Golgi targeting was strictly dependent on a functional anterograde traffic out of the ER to the Golgi bodies or a retrograde transport route from the Golgi apparatus, since overexpression of Sar1 (H74L) and Arf1 (T31N) aborted NSvc2-N as well as NSvc2-N::NSvc2-C complex trafficking to the Golgi bodies. In the mammalian system, it has been demonstrated that the COP II coat recognizes and selects export cargo into ERES vesicles (48). Our finding that the targeting of NSvc2 protein into Golgi bodies via ERES suggests that COP II machineries, such as Sar1 or Sec23-Sec24 complex, may be involved in selecting NSvc2 glycoproteins to target into Golgi.
For viruses in the Bunyaviridae family, intracellular maturation and budding in the Golgi complex is mediated by the targeting and accumulation of the viral glycoproteins in this cellular compartment (17, 18, 35, 38–40). Previous work has shown that the Golgi targeting signal of the TSWV and BUNV glycoproteins resides in the transmembrane domain of the Gn protein, allowing for sufficient ER exit and transport to the Golgi bodies (35, 36). However, the Golgi localization signal of RVFV was mapped to a 48-amino-acid region of Gn containing the transmembrane domain and the adjacent 28 amino acids of the cytosolic tail (17). Although UUKV is also a phlebovirus, the Golgi localization signal for the UUKV glycoproteins resides in the cytosolic tail of Gn (15, 49). In the present study, we have mapped the Golgi targeting signal of RSV to a region encompassing the transmembrane domain and the 24 adjacent amino acids of the cytosolic tail of the N terminus of NSvc2. Although the tenuiviruses have a very close relationship to the phleboviruses, our finding support that the Golgi targeting motif of the RSV glycoprotein is more closely related to that of RVFV, instead of UUKV glycoprotein.
In summary, our results presented here reveal that Rice stripe tenuivirus glycoproteins were able to target into Golgi apparatus in plant cells. Targeting of RSV glycoproteins into Golgi apparatus is mediated by the N-Terminal transmembrane domain and the adjacent cytosolic 24 amino acids of NSvc2 in a COP I- and COP II-dependent manner. In light of the evidence from viruses in Bunyaviridae that targeting into Golgi apparatus is important for the viral particle assembly and vector transmission, we propose that targeting of RSV glycoproteins into Golgi apparatus in plant cells represents a physiologically relevant mechanism in the maturation of RSV particle complex for insect vector transmission.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the Program for New Century Excellent Talents in the University (NCET-12-0888), the National Natural Science Foundation of China (31222045, 31171813, and 31170142), the Special Fund for Agro-Scientific Research in the Public Interest (201303021 and 201003031), and the National Program on Key Basic Research Project of China (973 Program, 2014CB138400).
We thank Taiyun Wei for kindly providing the Sar1 (H74L) dominant-negative mutant. We also thank three anonymous referees for their valuable comments on an earlier version of this paper.
Footnotes
Published ahead of print 3 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03006-13.
REFERENCES
- 1.Toriyama S. 2000. Rice stripe virus. Descriptions of plant viruses no. 375. http://www.dpvweb.net/. [Google Scholar]
- 2.Falk BW, Tsai JH. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 36:139–163. 10.1146/annurev.phyto.36.1.139 [DOI] [PubMed] [Google Scholar]
- 3.Ramirez BC, Haenni AL. 1994. Molecular biology of tenuiviruses, a remarkable group of plant viruses. J. Gen. Virol. 75(Pt 3):467–475 [DOI] [PubMed] [Google Scholar]
- 4.Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A. 1994. Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 75(Pt 12):3569–3579 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi M, Toriyama S, Hamamatsu C, Ishihama A. 1993. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 74:769–773. 10.1099/0022-1317-74-4-769 [DOI] [PubMed] [Google Scholar]
- 6.Xiong R, Wu J, Zhou Y, Zhou X. 2009. Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387:29–40. 10.1016/j.virol.2009.01.045 [DOI] [PubMed] [Google Scholar]
- 7.Kakutani T, Hayano Y, Hayashi T, Minobe Y. 1991. Ambisense segment 3 of rice stripe virus: the first instance of a virus containing two ambisense segments. J. Gen. Virol. 72(Pt 2):465–468 [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, Hayakawa T, Toriyama S, Takahashi M. 1991. Complete nucleotide sequence of RNA 3 of rice stripe virus: an ambisense coding strategy. J. Gen. Virol. 72:763–767. 10.1099/0022-1317-72-4-763 [DOI] [PubMed] [Google Scholar]
- 9.Xiong RY, Wu JX, Zhou YJ, Zhou XP. 2008. Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82:12304–12311. 10.1128/JVI.01696-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott RM. 1990. Molecular biology of the Bunyaviridae. J. Gen. Virol. 71(Pt 3):501–522 [DOI] [PubMed] [Google Scholar]
- 11.Elliott RM. 1996. The Bunyaviridae. Plenum Press, Inc, New York, NY [Google Scholar]
- 12.Rusu M, Bonneau R, Holbrook MR, Watowich SJ, Birmanns S, Wriggers W, Freiberg AN. 2012. An assembly model of Rift Valley fever virus. Front. Microbiol. 3:254. 10.3389/fmicb.2012.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldbach R, Peters D. 1996. Molecular and biological aspects of tospoviruses, p 129–157 In Elliott RM. (ed), The Bunyaviridae. Plenum Press, Inc, New York, NY [Google Scholar]
- 14.Elliott RM. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3:572–577 [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson AM, Melin L, Persson R, Raschperger E, Wikstrom L, Pettersson RF. 1997. Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J. Virol. 71:218–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuoka Y, Chen SY, Holland CE, Compans RW. 1996. Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch. Biochem. Biophys. 336:184–189. 10.1006/abbi.1996.0547 [DOI] [PubMed] [Google Scholar]
- 17.Gerrard SR, Nichol ST. 2002. Characterization of the Golgi retention motif of Rift Valley fever virus GN glycoprotein. J. Virol. 76:12200–12210. 10.1128/JVI.76.23.12200-12210.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro D, Foresti O, Denecke J, Wellink J, Goldbach R, Kormelink RJM. 2008. Tomato spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and Golgi-derived pleomorphic membrane structures in plant cells. J. Gen. Virol. 89:1811–1818. 10.1099/vir.0.2008/001164-0 [DOI] [PubMed] [Google Scholar]
- 19.Toriyama S. 1982. Characterization of rice stripe virus: a heavy component carrying infectivity. J. Gen. Virol. 61:187–195. 10.1099/0022-1317-61-2-187 [DOI] [Google Scholar]
- 20.Toriyama S. 1986. An RNA-dependent RNA-polymerase associated with the filamentous nucleoproteins of rice stripe virus. J. Gen. Virol. 67:1247–1255. 10.1099/0022-1317-67-7-1247 [DOI] [Google Scholar]
- 21.Zhao S, Zhang G, Dai X, Hou Y, Li M, Liang J, Liang C. 2012. Processing and intracellular localization of rice stripe virus Pc2 protein in insect cells. Virology 429:148–154. 10.1016/j.virol.2012.04.018 [DOI] [PubMed] [Google Scholar]
- 22.Yao M, Zhang T, Zhou T, Zhou Y, Zhou X, Tao X. 2012. Repetitive prime-and-realign mechanism converts short capped RNA leaders into longer ones that may be more suitable for elongation during rice stripe virus transcription initiation. J. Gen. Virol. 93:194–202. 10.1099/vir.0.033902-0 [DOI] [PubMed] [Google Scholar]
- 23.Pang SZ, DeBoer DL, Wan Y, Ye GB, Layton JG, Neher MK, Armstrong CL, Fry JE, Hinchee MAW, Fromm ME. 1996. An improved green fluorescent protein gene as a vital marker in plants. Plant Physiol. 112:893–900. 10.1104/pp.112.3.893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson BK, Cai X, Nebenfuhr A. 2007. A multicolored set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants. Plant J. 51:1126–1136. 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- 25.Wei T, Wang A. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82:12252–12264. 10.1128/JVI.01329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peremyslov VV, Pan YW, Dolja VV. 2004. Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 78:3704–3709. 10.1128/JVI.78.7.3704-3709.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DaSilva LLP, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16:1753–1771. 10.1105/tpc.022673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15:441–447. 10.1046/j.1365-313X.1998.00208.x [DOI] [PubMed] [Google Scholar]
- 29.Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. 2009. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 149:1005–1016. 10.1104/pp.108.131755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F. 2007. De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 143:1640–1650. 10.1104/pp.106.094110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nebenfuhr A, Staehelin LA. 2001. Mobile factories: Golgi dynamics in plant cells. Trends Plant Sci. 6:160–167. 10.1016/S1360-1385(01)01891-X [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. 2000. A dominant-negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23:517–525. 10.1046/j.1365-313x.2000.00823.x [DOI] [PubMed] [Google Scholar]
- 33.Stefano G, Renna L, Chatre L, Hanton SL, Moreau P, Hawes C, Brandizzi F. 2006. In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46:95–110. 10.1111/j.1365-313X.2006.02675.x [DOI] [PubMed] [Google Scholar]
- 34.Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I. 2002. ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129:1507–1520. 10.1104/pp.003624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X, Lappin DF, Elliott RM. 2004. Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J. Virol. 78:10793–10802. 10.1128/JVI.78.19.10793-10802.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribeiro D, Goldbach R, Kormelink R. 2009. Requirements for ER-arrest and sequential exit to the Golgi of tomato spotted wilt virus glycoproteins. Traffic 10:664–672. 10.1111/j.1600-0854.2009.00900.x [DOI] [PubMed] [Google Scholar]
- 37.Chen SY, Compans RW. 1991. Oligomerization, transport, and Golgi retention of Punta Toro virus glycoproteins. J. Virol. 65:5902–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen SY, Matsuoka Y, Compans RW. 1991. Golgi complex localization of the Punta Toro virus G2 protein requires its association with the G1 protein. Virology 183:351–365. 10.1016/0042-6822(91)90148-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruusala A, Persson R, Schmaljohn CS, Pettersson RF. 1992. Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology 186:53–64. 10.1016/0042-6822(92)90060-3 [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Elliott RM. 2002. Golgi localization of Hantaan virus glycoproteins requires coexpression of G1 and G2. Virology 300:31–38. 10.1006/viro.2002.1414 [DOI] [PubMed] [Google Scholar]
- 41.Kikkert M, Van Lent J, Storms M, Bodegom P, Kormelink R, Goldbach R. 1999. Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73:2288–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sin SH. 2005. Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. Proc. Natl. Acad. Sci. 102:5168–5173. 10.1073/pnas.0407354102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maia IG, Haenni A, Bernardi F. 1996. Potyviral HC-Pro: a multifunctional protein. J. Gen. Virol. 77:1335–1341. 10.1099/0022-1317-77-7-1335 [DOI] [PubMed] [Google Scholar]
- 44.Hanton SL, Matheson LA, Brandizzi F. 2006. Seeking a way out: export of proteins from the plant endoplasmic reticulum. Trends Plant Sci. 11:335–343. 10.1016/j.tplants.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 45.Oufattole M, Park JH, Poxleitner M, Jiang L, Rogers JC. 2005. Selective membrane protein internalization accompanies movement from the endoplasmic reticulum to the protein storage vacuole pathway in Arabidopsis. Plant Cell 17:3066–3080. 10.1105/tpc.105.035212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi H, Saito Y, Kitagawa T, Morita S, Masumura T, Tanaka K. 2005. A novel vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar storage proteins in rice endosperm. Plant Cell Physiol. 46:245–249. 10.1093/pcp/pci019 [DOI] [PubMed] [Google Scholar]
- 47.Tormakangas K, Hadlington JL, Pimpl P, Hillmer S, Brandizzi F, Teeri TH, Denecke J. 2001. A vacuolar sorting domain may also influence the way in which proteins leave the endoplasmic reticulum. Plant Cell 13:2021–2032. 10.1105/tpc.13.9.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlowe C. 2003. Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13:295–300. 10.1016/S0962-8924(03)00082-5 [DOI] [PubMed] [Google Scholar]
- 49.Overby AK, Popov VL, Pettersson RF, Neve EPA. 2007. The cytoplasmic tails of uukuniemi virus (Bunyaviridae) GN and GC glycoproteins are important for intracellular targeting and the budding of virus-like particles. J. Virol. 81:11381–11391. 10.1128/JVI.00767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.