ABSTRACT
RIG-I-like receptors (RLRs) MDA5 and RIG-I are key players in the innate antiviral response. Upon recognition of viral RNA, they interact with MAVS, eventually inducing type I interferon production. The interferon induction pathway is commonly targeted by viruses. How enteroviruses suppress interferon production is incompletely understood. MDA5 has been suggested to undergo caspase- and proteasome-mediated degradation during poliovirus infection. Additionally, MAVS is reported to be cleaved during infection with coxsackievirus B3 (CVB3) by the CVB3 proteinase 3Cpro, whereas MAVS cleavage by enterovirus 71 has been attributed to 2Apro. As yet, a detailed examination of the RLR pathway as a whole during any enterovirus infection is lacking. We performed a comprehensive analysis of crucial factors of the RLR pathway, including MDA5, RIG-I, LGP2, MAVS, TBK1, and IRF3, during infection of CVB3, a human enterovirus B (HEV-B) species member. We show that CVB3 inhibits the RLR pathway upstream of TBK1 activation, as demonstrated by limited phosphorylation of TBK1 and a lack of IRF3 phosphorylation. Furthermore, we show that MDA5, MAVS, and RIG-I all undergo proteolytic degradation in CVB3-infected cells through a caspase- and proteasome-independent manner. We convincingly show that MDA5 and MAVS cleavages are both mediated by CVB3 2Apro, while RIG-I is cleaved by 3Cpro. Moreover, we show that proteinases 2Apro and 3Cpro of poliovirus (HEV-C) and enterovirus 71 (HEV-A) exert the same functions. This study identifies a critical role of 2Apro by cleaving MDA5 and MAVS and shows that enteroviruses use a common strategy to counteract the interferon response in infected cells.
IMPORTANCE Human enteroviruses (HEVs) are important pathogens that cause a variety of diseases in humans, including poliomyelitis, hand, foot, and mouth disease, viral meningitis, cardiomyopathy, and more. Like many other viruses, enteroviruses target the host immune pathways to gain replication advantage. The MDA5/MAVS pathway is responsible for recognizing enterovirus infections in the host cell and leads to expression of type I interferons (IFN-I), crucial antiviral signaling molecules. Here we show that three species of HEVs all employ the viral proteinase 2A (2Apro) to proteolytically target MDA5 and MAVS, leading to an efficient blockade upstream of IFN-I transcription. These observations suggest that MDA5/MAVS antagonization is an evolutionarily conserved and beneficial mechanism of enteroviruses. Understanding the molecular mechanisms of enterovirus immune evasion strategies will help to develop countermeasures to control infections with these viruses in the future.
INTRODUCTION
Type I interferons (IFNs; alpha/beta interferon [IFN-α/β]) are key players in the innate antiviral response against virus infections. Initially produced and secreted by infected cells, IFN-α/β can bind to the type I IFN receptor (IFNAR) in autocrine and paracrine manners, thereby initiating the JAK/STAT pathway. Activation of this pathway leads to the expression of hundreds of interferon-stimulated genes (ISGs), which together induce a so-called antiviral state that restricts virus replication (reviewed in reference 1). The initiation of the IFN-α/β response relies on specialized pathogen recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs), molecular motifs bearing non-self-signatures to the host cell. Some members of the Toll-like receptor (TLR) family, such as TLR3 and TLR7/8, are known to play crucial roles in virus recognition. TLRs are mostly expressed in macrophages, dendritic cells, and other immune cell types, where they detect PAMPs at the cell surface or in endosomes (2). Another family of PRRs is the RIG-I-like receptors, including RIG-I (3), MDA5 (4–6), and LGP2 (3, 7). These receptors are ubiquitously expressed and monitor the cytoplasm of virtually all nucleated cells. Upon activation by viral RNA, RIG-I and MDA5 interact with MAVS, an adaptor molecule localized at the outer membrane of mitochondria (8–11). MAVS then initiates signaling cascades via TANK-binding kinase 1 (TBK1) and IκB kinase (IKK) complexes, leading to activation of IRF3 and NF-κB, transcription factors required for transcription activation of IFN-α/β and other proinflammatory cytokine genes (2). RIG-I and MDA5 have nonredundant roles in detecting invading viruses. While RIG-I is crucial in detecting many negative-strand RNA viruses (e.g., vesicular stomatitis virus and influenza virus) and some flaviviruses (e.g., hepatitis C virus and Japanese encephalitis virus) (12–15), MDA5 is important for the recognition of members of the Picornavirus, Coronavirus, and Calicivirus families (16–18). The molecular motifs that activate RIG-I and MDA5 also vary. RIG-I is activated by 5′-triphosphate (5′ppp)-containing double-stranded RNAs (dsRNAs) as well as double-stranded regions within single-stranded RNA molecules, such as the panhandle structure formed by genomic RNAs of negative-strand RNA viruses (2, 19, 20). MDA5 requires long dsRNA duplexes for potent activation, as exemplified by the replicative form of picornaviruses (16, 21).
The Picornaviridae is a large family of nonenveloped, positive-strand RNA viruses. This family includes many important human and animal pathogens, and members of the Enterovirus genus are particularly important. Poliovirus (PV), the causative agent of poliomyelitis, is the subject of a multi-billion-dollar eradication campaign from the World Health Organization. Enterovirus 71 (EV71) continues to cause outbreaks of hand, foot, and mouth disease associated with neurological complications in Southeast Asia. Coxsackieviruses (CVs) and echoviruses are the leading causes of viral meningitis and cardiomyopathy. Rhinoviruses cause common colds and are frequently associated with asthma exacerbations and chronic obstructive pulmonary disease. Upon entry, the viral genome is immediately translated into a single polyprotein, which is proteolytically processed into mature peptides by viral proteinases 2Apro and 3Cpro. These viral proteins then facilitate viral RNA replication and, eventually, production of progeny virion particles. Like most viruses, picornaviruses have evolved to actively suppress the host IFN-α/β response to gain a replication advantage. In fact, many reports have demonstrated that picornaviruses efficiently suppress IFN-α/β at the transcription level in cultured cells (16, 22, 23). However, the mechanism by which picornaviruses interfere with IFN-α/β induction is not completely understood.
Many studies have been performed to investigate how picornaviruses interfere with the RIG-I like receptor (RLR)-mediated IFN-α/β induction pathway. MDA5, the receptor responsible for recognizing picornavirus RNA, is reported to be degraded during infection of PV (24) and EV71 (25). MDA5 degradation was shown to be dependent on caspases and, in the case of PV, also proteasome activities (24). Surprisingly, other closely related enteroviruses, such as human rhinovirus 16 (HRV16) and echovirus 1, did not induce MDA5 degradation (24), suggesting that there may be variations among enteroviruses in their strategies to suppress IFN-α/β induction. Besides MDA5, RIG-I is also targeted by several enteroviruses, such as PV, echovirus, and HRV16 and -1A, most likely via their 3Cpros (26), though it remains to be elucidated why these viruses would target an RNA sensor that does not participate in their recognition. In addition, the downstream adaptor molecule MAVS is targeted by several enteroviruses, including HRV1A (27), coxsackievirus B3 (CVB3) (23), and EV71 (28). However, different mechanisms regarding how these viruses accomplish MAVS inactivation have been proposed. MAVS is reported to be cleaved by 2Apro during EV71 infection (28) and by 3Cpro during CVB3 infection (23), whereas both of these viral proteinases, as well as caspase 3, were implicated in HRV1A-induced MAVS cleavage (27). This diversity in MAVS inactivation mechanisms is rather uncommon for enteroviruses, as they often utilize the same strategies to target a particular host factor. For instance, eukaryotic initiation factor 4G (eIF4G) is cleaved by the 2Apros of various enteroviruses and rhinoviruses (29, 30), while G3BP is cleaved by the 3Cpro of PV (31) as well as by that of CVB3 (this study). Importantly, each of the studies focused on a single factor (MDA5, MAVS, or RIG-I) and used a different group of viruses. Hence, it is yet impossible to paint a complete picture of the IFN-α/β antagonization strategy of any individual virus, and it is challenging to conclude with confidence whether different enteroviruses truly employ diverse strategies to inactivate these host factors or whether the differences merely result from the use of different reagents and/or assays in different studies.
To gain a comprehensive overview of the fate of the important components of the RLR signaling pathway during enterovirus infection, we examined multiple factors along the signaling cascade during infection of a model virus, CVB3. We show that RIG-I is targeted by 3Cpro of CVB3, PV, and EV71, resulting in similar cleavage products previously reported for PV (26). Importantly, we report that cleavage of MDA5 occurs in a caspase- and proteasome-independent manner and relies on viral proteinase 2Apro. Furthermore, in contrast to a previous report (23), we demonstrate that MAVS cleavage during CVB3 infection is also (primarily) mediated by 2Apro and not 3Cpro. Moreover, we show that 2Apros from other enteroviruses, namely, PV and EV71, also target MDA5 and MAVS for cleavage, suggesting that enteroviruses likely share common strategies to target the RLR-mediated IFN-α/β induction pathway.
MATERIALS AND METHODS
Cells and viruses.
HeLa R19 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 100 U/ml penicillin-streptomycin in a humidified incubator in the presence of 5% CO2. Coxsackievirus B3, mengovirus, and a mengovirus which lost its IFN-suppressing activities due to substitutions in the Zn finger domain of the leader protein (mengo-Zn) have been described previously (16). M-2A(CVB3/PV/EV71) and M-3C(CVB3/PV/EV71) cDNAs were generated by cloning the 2Apro- or 3Cpro-coding region of the indicated viruses, respectively, upstream of the leader-coding region in the mengovirus infectious clone pM16.1. To ensure proper maturation of the 2Apro or 3Cpro from the mengovirus polyprotein, we also added glutamine-glycine codons after the inserted sequences, to allow cleavage by mengovirus 3Cpro during polyprotein processing. Viruses were produced by directly transfecting in vitro-transcribed RNAs from the infectious clones of M-2A(CVB3/PV/EV71) and M-3C(CVB3/PV/EV71) in BHK-21 or HeLa R19 cells.
Plasmids.
pcDNA3-FLAG-MAVS-HA was generated by inserting the hemagglutinin (HA) tag in pcDNA3-FLAG-MAVS, provided by Z. Chen (University of Texas Southwestern Medical Center, Texas). The Q148A mutation was generated by site-directed mutagenesis.
Reagents.
Antibodies against MDA5 have been previously described (32). Antibodies against RIG-I were purchased from Abgent, MAVS was purchased from Alexis-Biochemicals, IRF3 was purchased from Santa Cruz Biotechnology, p-IRF3 (S396), TBK1, and phosphorylated TBK1 (p-TBK1; phosphorylation at S172) were purchased from Cell Signaling Technology, LGP2 was purchased from Abcam, actin and the FLAG tag were purchased from Sigma-Aldrich, eIF4G was purchased from Bethyl Laboratories, the HA tag was purchased from Covance, G3BP was purchased from BD Biosciences, and procyclic acidic repetitive protein (PARP) was purchased from Roche Applied Sciences. Q-VD-OPh (QVD) was purchased from BioVision, MG132 was purchased from Sigma-Aldrich, and staurosporine (STS) was purchased from Roche Applied Sciences. Recombinant CVB3 2Apro has been previously described (33), and recombinant CVB3 3Cpro was a kind gift from Rolf Hilgenfeld (University of Lübeck, Lübeck, Germany).
Quantitative real-time PCR was performed as previously described (16).
Immunoblotting.
Cells were lysed in TEN lysis buffer (40 mM Tris-HCl, 150 mM NaCl, 10 mM EDTA, 1% NP-40), and lysate was cleared by centrifugation at 16,000 × g at 4°C for 5 min. Total protein concentrations were determined using Bio-Rad protein assay dye reagent concentrate according to the manufacturer's protocols. Lysates were subjected to SDS-PAGE and immunoblotting using the indicated antibodies. Native PAGE (for IRF3 dimerization assay) was performed as described previously (22).
In vitro cleavage with recombinant 2Apro or 3Cpro.
Cells were resuspended in a 2× volume of the cell pellet of hypotonic buffer [20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM Mg(CH3COO)2, 1 mM dithiothreitol (DTT)], incubated on ice for 10 min, and lysed by repeated passage through a thin needle at 4°C. A 0.1× volume of buffer A [200 mM HEPES, pH 7.4, 1.2 M KCH3COO, 40 mM Mg(CH3COO)2, 50 mM DTT] was added to the cell lysate, and this was cleared by centrifugation at 2,500 × g for 5 min at 4°C. Unless indicated otherwise, 0.5 μg 2Apro or 0.75 μg 3Cpro was incubated with 150 μg lysate at room temperature overnight. The reaction was terminated by addition of Laemmli sample buffer and heat treatment at 95°C for 10 min.
RESULTS
CVB3 interrupts the RLR pathway upstream of TBK1 phosphorylation.
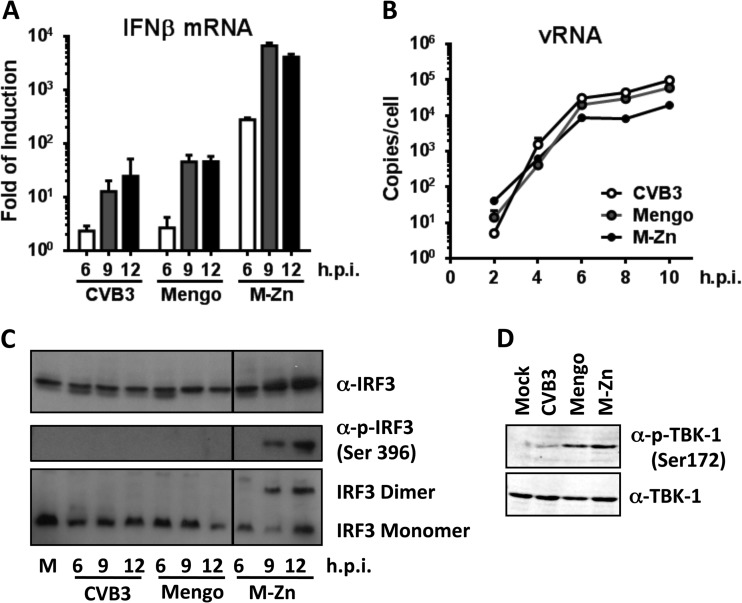
It is known that picornaviruses induce little IFN-α/β response in infected cells (23, 28, 34). In cells infected with wild-type (wt) CVB3 or mengovirus (a strain of encephalomyocarditis virus [EMCV] that was used as a control), little IFN-β mRNA was detected, and only at very late stages of infection (i.e., at 9 and 12 h postinfection [p.i.]) (Fig. 1A). In contrast, infection with a mutant mengovirus which lost its IFN-suppressing activities due to substitutions in the Zn finger domain of the leader protein (mengo-Zn) (22) already induced strong IFN-β mRNA upregulation at 6 h p.i., and this further dramatically increased and eventually reached a plateau at 9 h p.i. (Fig. 1A). This increased IFN-α/β response induced by mengo-Zn was not due to differences in virus replication, as viral RNA accumulated with kinetics and at levels similar to those for wt mengovirus and CVB3 (Fig. 1B).
FIG 1.
CVB3 antagonizes the RLR pathway upstream of TBK1, while mengovirus does so downstream of TBK1 activation. HeLa R19 cells were mock infected or infected with CVB3, mengovirus (mengo), or mengo-Zn (M-Zn) at a multiplicity of infection of 50, and cells were harvested at the indicated times (h) p.i. Total RNA was isolated and subjected to reverse transcription-quantitative PCR analysis for IFN-β mRNA (A) and viral RNA (vRNA) (B). Data were first normalized against those for actin mRNA, and then the fold induction compared to that for the mock-infected sample was calculated and is presented as the mean ± SD. (C) Infection was carried out as described for panel A, and cells were lysed at the indicated times (h) p.i. Lysates were then subjected to SDS-PAGE, followed by immunoblotting using the indicated antibodies. IRF3 dimerization analysis was carried out under native conditions. Lane M, mock infection. (D) Infection was carried out as described for panel A, and cells were lysed at 9 h p.i. Cell lysates were analyzed by SDS-PAGE, followed by immunoblotting using the indicated antibodies. Data are representative of those from at least 3 independent experiments.
To gain a first insight into how the viruses interfere with IFN-α/β induction pathways, we first asked whether the key transcription factor required for IFN-α/β transcription activation, namely, IRF3, is activated in infected cells. HeLa R19 cells were infected with CVB3 and harvested at 6, 9, and 12 h p.i. Cell lysates were subjected to immunoblotting to determine the IRF3 expression level, as well as the activation status of IRF3, as indicated by phosphorylation at serine 396 and dimerization of this transcription factor. We also performed parallel experiments with mengovirus or mengo-Zn as negative and positive controls, respectively. Infection with mengo-Zn, but not wt mengovirus, induced efficient IRF3 phosphorylation and dimerization (Fig. 1C). CVB3 infection did not induce any detectable IRF3 phosphorylation or dimerization (Fig. 1C), consistent with the lack of a significant IFN-β mRNA induction (Fig. 1A). The absence of IRF3 activation was not due to changes in IRF3 expression levels, since these remained unchanged throughout the CVB3 (and mengovirus) infection (Fig. 1C). Our data clearly indicate that the IFN-α/β induction pathway is severely inhibited upstream of IRF3 phosphorylation during wt CVB3 infection.
It is well established that IRF3 phosphorylation relies on the activity of TBK1, which is itself activated by phosphorylation at serine 172 (35). We therefore asked whether TBK1 expression and/or activation is affected in CVB3-infected cells. Cells were infected with CVB3, wt mengovirus, and mengo-Zn for 9 h, and cell lysates were analyzed by immunoblotting using a polyclonal antibody against TBK1 and a monoclonal antibody against S172-phosphorylated TBK1. As shown in Fig. 1D, the basal level of TBK1 expression did not change during infection with any of the three viruses. As expected, S172 phosphorylation of TBK1 was readily detectable in mengo-Zn-infected cells. wt mengovirus infection also resulted in TBK1 phosphorylation to the same extent as mengo-Zn infection, but this was significantly reduced in CVB3-infected cells (Fig. 1D). These results indicate that CVB3 targets the RLR pathway upstream of TBK1 phosphorylation, while mengovirus does so between TBK1 activation and IRF3 phosphorylation.
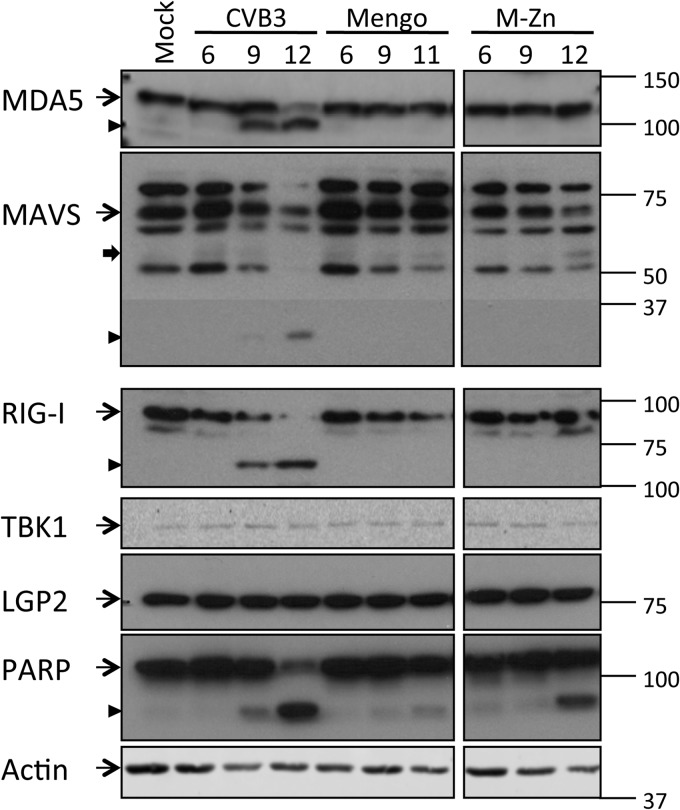
CVB3 induces MDA5, MAVS, and RIG-I cleavage in infected cells.
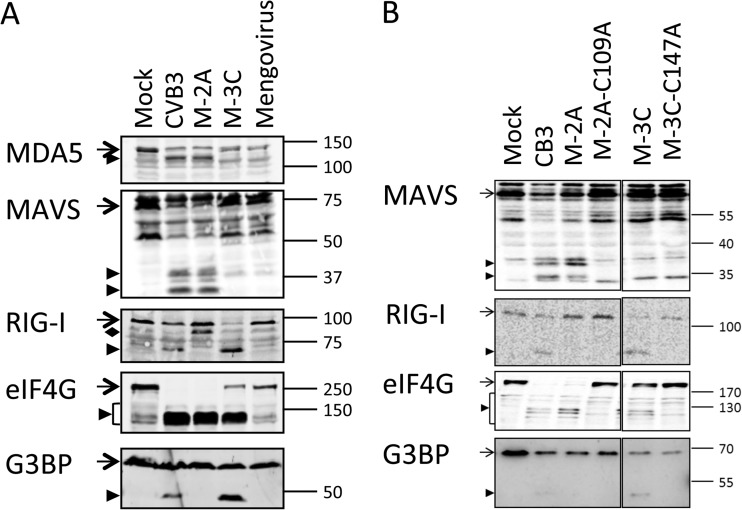
To further investigate where CVB3 interferes with the RLR pathway, we examined the crucial factors involved in this signaling cascade by immunoblotting during a course of infection. From 9 h p.i. onwards, we observed a decrease in the full-length MDA5 signal accompanied by the appearance of a smaller product of about 100 kDa (Fig. 2), suggesting that MDA5 is cleaved during CVB3 infection. Also, RIG-I and MAVS, but not TBK1 or LGP2, another member of the RLR family, were cleaved from 9 h p.i. (Fig. 2). RIG-I cleavage was accompanied by the appearance of a band at about 70 kDa, similar to the cleavage product observed in PV-infected cells (26). We also observed a putative cleavage product (<37 kDa) of MAVS (Fig. 2) which was significantly smaller than the reported cleavage product (∼50 kDa) produced by CVB3 3Cpro (23), suggesting an alternative MAVS cleavage. None of these factors were cleaved during infection of mengovirus or mengo-Zn (Fig. 2), in agreement with our observation that the RLR pathway upstream of TBK1 phosphorylation remains intact in cells infected with these viruses (Fig. 1).
FIG 2.
Fate of multiple factors of the RLR signaling pathway during infection. HeLa R19 cells were mock infected or infected with CVB3, mengovirus (mengo), or mengo-Zn (M-Zn) at a multiplicity of infection of 50, and cells were lysed at the indicated times (h) p.i. Cell lysates were analyzed by SDS-PAGE, followed by immunoblotting using the indicated antibodies. Marker bands of the indicated size (in kDa) are indicated on the right side. Arrow with a narrow tail, full-length protein; arrowhead, putative cleavage products; arrow with a wide tail, known MAVS cleavage product from caspase-mediated cleavage. Data are representative of those from at least 3 independent experiments.
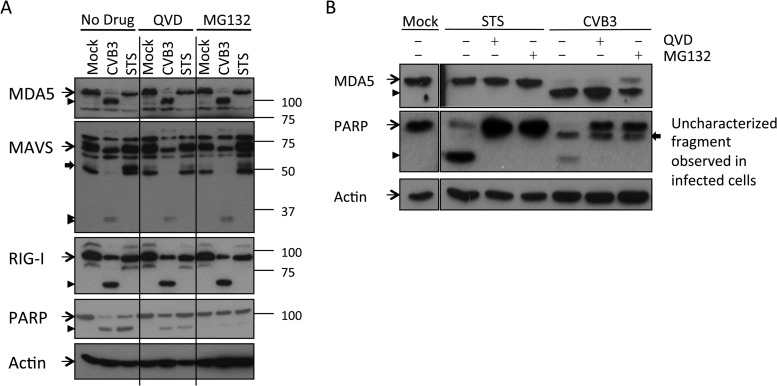
CVB3-induced MDA5, MAVS, and RIG-I cleavages are independent of caspase or proteasome activities.
We noticed that the cleavage of RIG-I, MDA5, and MAVS in CVB3-infected cells coincided with PARP cleavage, resulting in an 89-kDa cleavage product (Fig. 2), the hallmark of apoptosis. It has been suggested that poliovirus and enterovirus 71 induce caspase-dependent MDA5 degradation (24, 25). Therefore, we asked whether caspases could be responsible for cleaving MDA5, MAVS, and/or RIG-I in CVB3-infected cells. To address this question, we chemically induced apoptosis in mock-infected cells using staurosporine (STS), a general kinase inhibitor. MDA5, MAVS, and RIG-I in mock- or STS-treated cells were analyzed by immunoblotting. STS induced potent caspase activation, as shown by PARP cleavage, as well as the appearance of the known caspase 3-mediated MAVS cleavage product of about 50 kDa (27). However, it did not lead to any detectable cleavage of MDA5, MAVS, or RIG-I like that observed in CVB3-infected cells (Fig. 3A), suggesting that caspases or the proteasome may not catalyze the infection-induced cleavage events reported here. To further demonstrate that MDA5 is not cleaved by caspases, we infected cells with CVB3 in the presence of a broad-spectrum caspase inhibitor, Q-VD-OPh (QVD). We also included a proteasome inhibitor, MG132, in a parallel experiment because efficient caspase activation like the one that we observed in CVB3-infected cells (Fig. 2) often leads to protein degradation via the proteasome system. QVD and MG132 both inhibited apoptosis, as demonstrated by significantly decreased PARP cleavage in treated cells (Fig. 3A). Under these conditions, CVB3 infection still induced MDA5, MAVS, and RIG-I cleavage to an extent similar to that in nontreated cells (Fig. 3A). Also, in an independent experiment where apoptosis was completely inhibited by QVD, efficient MDA5 cleavage induced by CVB3 infection was observed (Fig. 3B). Together with our STS data, these results indicate that the cleavage events of MDA5, MAVS, and RIG-I during CVB3 infection are not mediated by caspases or the proteasome.
FIG 3.
Caspases and proteasomal proteases are not responsible for CVB3-induced MDA5, MAVS, or RIG-I cleavage. (A) HeLa R19 cells were mock infected, infected with CVB3 at a multiplicity of infection of 50 for 9 h, or treated with 2 μM staurosporine (STS) for 6 h in the presence or absence of 10 μM QVD or 10 μM MG132. Cells were lysed, and the indicated proteins were analyzed by SDS-PAGE, followed by immunoblotting. Arrow with a narrow tail, full-length protein; arrowhead, putative cleavage products; arrow with a wide tail, known MAVS cleavage product from caspase-mediated cleavage. Marker bands of the indicated size (in kDa) are indicated on the right side. (B) HeLa R19 cells were mock treated, infected with CVB3 at a multiplicity of infection of 50 for 9 h, or treated with 2 μM STS for 6 h in the presence or absence of 10 μM QVD or 10 μM MG132. Cells were lysed, and MDA5, PARP, and actin were analyzed by SDS-PAGE, followed by immunoblotting. Arrow with a narrow tail, full-length protein; arrowhead, putative cleavage products; arrow with a wide tail, additional PARP-derived fragment that was observed in CVB3-infected but not STS-treated cells and that persisted throughout CVB3 infection. Data are representative of those from at least 5 experiments.
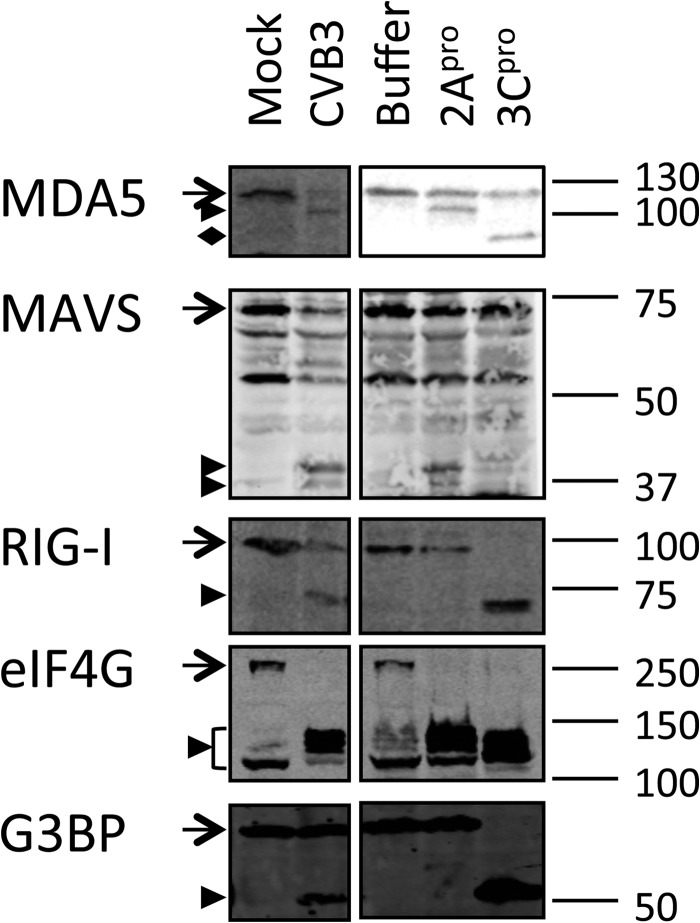
Recombinant CVB3 proteinase 2Apro induces MDA5 and MAVS cleavage, and 3Cpro induces RIG-I cleavage.
Next, we asked whether the two viral proteinases 2Apro and 3Cpro could be responsible for the cleavage events that we observed during CVB3 infection. Since these proteinases efficiently shut off host mRNA translation by cleaving eIF4G (29, 36, 37), their overexpression results in poor expression levels and severe cytotoxicity. Therefore, we chose to address this question using recombinant 2Apro and 3Cpro. Native cell lysate was treated with recombinant 2Apro or 3Cpro and subjected to immunoblotting analysis using antibodies against MDA5, MAVS, and RIG-I. To control for the activities of 2Apro and 3Cpro, we also probed for eIF4G, which is known to be cleaved by 2Apro- and 3Cpro-activated caspase 3 (29, 37, 38), and G3BP, which is cleaved by 3Cpro only (31). As clearly shown in Fig. 4, both proteinases were very active in our in vitro cleavage assay. Under these conditions, MDA5 and MAVS cleavages were observed in 2Apro-treated samples, whereas RIG-I was cleaved in 3Cpro-treated lysate (Fig. 4). The cleavage products seen in the in vitro cleavage assay showed electrophoretic mobility identical to the mobility of the cleavage products from CVB3-infected cells (Fig. 4), demonstrating the relevance of our in vitro findings. In 3Cpro-treated samples, a putative MDA5 cleavage product of approximately 100 kDa was detected; however, this protein fragment was not consistently detected in in vitro cleavage assays with 3Cpro. More importantly, we never observed this cleavage product in CVB3-infected cells. These results suggest that the CVB3-induced cleavage of MDA5 and MAVS resulted from 2Apro activity, whereas that of RIG-I resulted from 3Cpro activity.
FIG 4.
Recombinant CVB3 2Apro cleaves MDA5 and MAVS, while 3Cpro cleaves RIG-I. A native lysate of HeLa R19 cells was treated with recombinant CVB3 2Apro or 3Cpro at room temperature overnight. The reaction mixtures were subjected to SDS-PAGE, followed by immunoblotting using antibodies against the indicated proteins. As controls, lysates from mock- and CVB3-infected (multiplicity of infection, 50; 8 h) cells were also included. Arrow, full-length protein; arrowhead, putative cleavage products; filled diamond, product obtained by cleavage of MDA5 by 3Cpro that is not seen in CVB3-infected cells. Marker bands of the indicated size (in kDa) are indicated on the right side. Data are representative of those from at least 3 experiments.
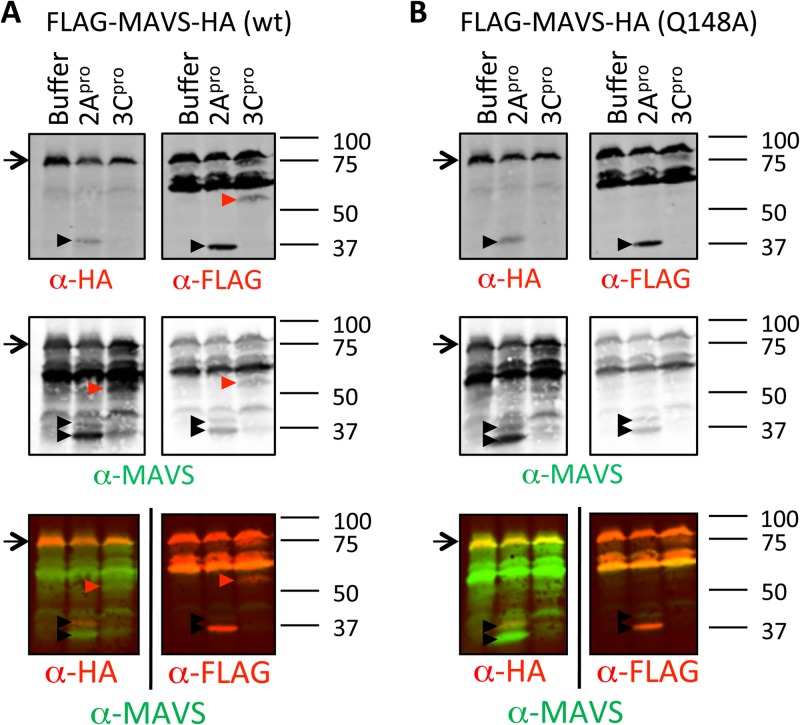
As mentioned before, it has been reported that CVB3 3Cpro cleaves MAVS, resulting in an N-terminal cleavage product of 50 kDa, and a single amino acid substitution at position 148 (Q148A) was sufficient to prevent this cleavage (23). We also observed a 3Cpro-induced cleavage product of approximately 50 kDa when we prepared lysates from cells overexpressing a FLAG- and HA-tagged MAVS protein (FLAG-MAVS-HA) (Fig. 5A). This cleavage product was also detectable using an antibody against the N-terminal FLAG tag but not one against the C-terminal HA tag, confirming that it is an N-terminal fragment of MAVS. When the reported cleavage-resistant mutant (Q148A) was overexpressed, the 3Cpro-mediated cleavage was no longer detectable (Fig. 5B), indicating that it is likely the same cleavage product reported by Mukherjee et al. (23). However, we did not observe this cleavage fragment from endogenous MAVS during CVB3 infection (Fig. 2 to 4), even when using the same antibody used in the previously published study (23) (data not shown).
FIG 5.
Both 2Apro and 3Cpro of CVB3 can cleave MAVS, but at different sites. Plasmids carrying FLAG-MAVS-HA (A) or FLAG-MAVS-HA (Q148A) (B) were transfected into cells and lysed 24 h later under native conditions. The native lysates were treated with recombinant 2Apro or 3Cpro at room temperature overnight and analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. Arrow, full-length protein; arrowhead, putative cleavage product. Marker bands of the indicated size (in kDa) are indicated on the right side of each panel. Data are representative of those from at least 2 experiments.
The 2Apro-mediated cleavage was readily detectable from both overexpressed MAVS (Fig. 5) and endogenous MAVS (Fig. 2 to 4), releasing two cleavage products (approximately 30 and 40 kDa) (Fig. 5). Using antibodies against tags at either terminus of overexpressed MAVS, we saw that the smaller cleavage product represented an N-terminal fragment, whereas the larger product was a C-terminal fragment (Fig. 5). These data suggest that MAVS is primarily cleaved by 2Apro in CVB3-infected cells.
2Apro mediates cleavage of MDA5 and MAVS, while 3Cpro cleaves RIG-I in infected cells.
We further pursued experiments to confirm these results in the context of an infection. As 2Apro and 3Cpro are both essential for CVB3 polyprotein processing, deletion mutations of these proteinases are lethal. Instead, we took advantage of the fact that wt mengovirus does not induce cleavage of RIG-I, MDA5, or MAVS (Fig. 2). By inserting CVB3 2Apro or 3Cpro in front of the leader-coding region (i.e., at the extreme 5′ terminus of the polyprotein-coding region) in the mengovirus genome, we could study the role of the CVB3 proteinases in the cleavage events during a normal picornavirus infection. These viruses, named M-2A(CVB3) and M-3C(CVB3), were used to infect cells, and RIG-I, MDA5, and MAVS cleavages were examined by immunoblotting. As shown before, wt mengovirus infection did not induce any changes in the integrity of these factors, whereas M-2A(CVB3) induced MDA5 and MAVS cleavage, and M-3C(CVB3) induced RIG-I cleavage (Fig. 6A). The cleavage products observed in M-2A(CVB3)- and M-3C(CVB3)-infected cells were exactly the same as those seen in CVB3-infected cells (Fig. 6A). We observed an additional band at about 90 kDa with the anti-RIG-I antibody in M-2A(CVB3)-infected cells (Fig. 6A). We never observed a cleavage product of this electrophoretic mobility in wt CVB3-infected cells, and this 90-kDa band was also absent in M-2A(CVB3)-infected cells in other experiments (Fig. 6B). As additional controls to confirm that the cleavages were the result of the proteinase activities of 2Apro and 3Cpro, we also produced mutant mengoviruses carrying the catalytically inactive forms of 2Apro (2A-C109A) and 3Cpro (3C-C147A). Infection with these viruses did not lead to cleavage of targets of 2Apro or 3Cpro (Fig. 6B). These results, combined with our in vitro cleavage data, convincingly show that CVB3 induces MDA5 and MAVS cleavage via 2Apro activity and RIG-I cleavage via 3Cpro activity.
FIG 6.
MDA5, MAVS, and RIG-I cleavage in the context of an infection. (A) HeLa R19 cells were mock infected or infected with CVB3, mengovirus expressing CVB3 2Apro or 3Cpro (M-2A and M-3C, respectively), or wt mengovirus at a multiplicity of infection of 50 for 8 h. Cells were lysed, and the indicated proteins were analyzed by SDS-PAGE, followed by immunoblotting. Arrow, full-length protein; arrowhead, putative cleavage products; filled diamond, product obtained by cleavage RIG-I by 2Apro that is not seen in CVB3-infected cells. (B) HeLa R19 cells were mock infected or infected with CVB3, M-2A(CVB3), M-3C(CVB3), M-2A-C109A(CVB3), or M-3C-C147A(CVB3) at a multiplicity of infection of 50 for 8 h. Cells were lysed, and the indicated proteins were analyzed by SDS-PAGE followed by immunoblotting. Arrow, full-length protein; arrowhead, putative cleavage products. Marker bands of the indicated size (in kDa) are indicated on the right side of each panel. Data are representative of those from at least 3 experiments.
Other enteroviruses also target MDA5 and MAVS by their 2Apros and RIG-I by their 3Cpros.
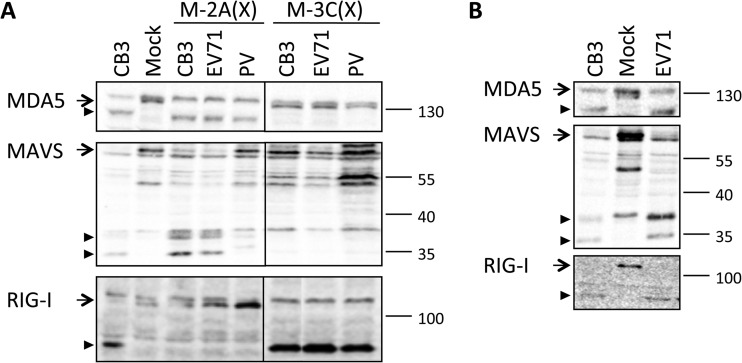
We further asked whether the cleavage events that we observed here were unique to CVB3, a human enterovirus B (HEV-B) member, or are common to other enterovirus species as well. To this end, we produced mengoviruses carrying 2Apro or 3Cpro from enterovirus 71 (EV71) and poliovirus (PV), belonging to HEV-A and HEV-C, respectively. Infections with these mutant viruses revealed that the 2Apro and 3Cpros from both viruses induced MDA5 and RIG-I cleavage, respectively (Fig. 7A). The cleavage products found with these viruses were identical to the corresponding cleavage products observed in CVB3-infected cells (Fig. 6A). M-2A(EV71) and M-2A(PV) infections also induced MAVS cleavage, though M-2A(PV) infection resulted in a less prominent MAVS cleavage and a MAVS cleavage product pattern slightly different from that of CVB3 infection. The larger cleavage product (∼40 kDa) seemed to be identical to that from M-2A(CVB3)- and M-2A(EV71)-infected cells, but the smaller product (∼35 kDa) had a slower electrophoretic mobility than that from CVB3-infected cells (∼30 kDa) (Fig. 7A).
FIG 7.
MDA5, MAVS, and RIG-I cleavage by other enteroviruses. (A) HeLa R19 cells were mock infected or infected with CVB3, M-2A(CVB3), M-2A(EV71), M-2A(PV), M-3C(CVB3), M-3C(EV71), or M-3C(PV) at a multiplicity of infection of 50 for 8 h. Cell lysates were analyzed by SDS-PAGE, followed by immunoblotting using antibodies against the indicated proteins. (B) HeLa R19 cells were mock infected or infected with CVB3 or EV71 at a multiplicity of infection of 50 for 8 h. Cells lysates were analyzed as described for panel A. Arrow, full-length protein; arrowhead, putative cleavage product. Marker bands of the indicated size (in kDa) are indicated on the right side of each panel. Data are representative of those from at least 2 experiments.
To investigate whether these cleavages also occur during a normal infection of enteroviruses, we infected cells with EV71 (strain BrCr) and analyzed MDA5, MAVS, and RIG-I by immunoblotting. Infection with EV71 induced the cleavage of all three factors similar to that seen in CVB3-infected cells (Fig. 7B). Together with data from the M-2A viruses, our results suggest that cleavage of RIG-I, MDA5, and MAVS may be a common phenomenon among enteroviruses.
DISCUSSION
The RLR signaling pathway is a crucial part of the innate antiviral response. Consequently, this pathway is targeted by numerous viruses from various families, including picornaviruses. However, our knowledge on how picornaviruses interfere with this pathway remains somewhat fragmented, since the reported studies each used a different panel of viruses and focused on only one factor of the pathway at a time. Here we performed the first comprehensive analysis of key factors of the RLR signaling pathway during enterovirus infections. Our data suggest that enteroviruses utilize their 2Apros to target MDA5 and MAVS and their 3Cpros to cleave RIG-I.
Consistent with these results, several studies have reported that IRF3 activation is severely inhibited during enterovirus infection (Fig. 1C) (23, 25, 27). Thus, cleavage of upstream factors of the RLR pathway is likely an evolutionarily conserved and advantageous mechanism to suppress IFN type I gene transcription. Meanwhile, it is worth noting that RLR pathway antagonization may not be the only evasion mechanism of enteroviruses. It is known that enteroviruses induce a shutoff of host mRNA transcription and translation, which likely further limits the production of, among other factors, virus-induced cytokines, such as IFNs. To what extent the host shutoff contributes to suppressing IFN production is difficult to investigate, since enterovirus-induced host shutoff is also mediated by 2Apro and 3Cpro (38–40), the same proteinases that cleave factors of the RLR-mediated IFN induction pathway, making it difficult to separately investigate the role of these phenomena.
MDA5, the receptor responsible for sensing picornavirus infection (16, 21), is cleaved during infection of CVB3 and EV71. Using recombinant CVB3 2Apro and our M-2A viruses carrying 2Apro of CVB3, EV71, or PV, we demonstrate that this cleavage can be mediated by all these 2Apros, indicating that this may be a common strategy by which enteroviruses inactivate MDA5. This seems to contradict previous reports that PV and EV71 induce caspase-mediated (and, in the case of PV, also proteasome-mediated) MDA5 degradation. However, in those studies, cells were first transfected with either poly(I·C) (24) or viral RNA (25) in order to upregulate MDA5 expression to a detectable level before infections were carried out. Pretreatments with these triggers induce IFN-α/β production, which, in turn, upregulates expression of interferon-stimulated genes, including MDA5. It is important to realize that IFN-α/β itself is known to potentiate cells to virus-induced apoptosis (41). Therefore, it is likely that in IFN-α/β-primed virus-infected cells, MDA5 is degraded in a caspase-dependent manner. In this study, we investigated the fate of endogenous MDA5 during enterovirus infection of naive cells. We show convincingly that MDA5 cleavage during infection is not attributed to caspases or the proteasome but is attributed to the viral proteinase 2Apro.
We also set out to identify the 2Apro cleavage site in MDA5 by infecting cells overexpressing MDA5 mutants carrying substitutions at potential 2Apro recognition sites. However, infection following protein overexpression proved to be very inefficient in our hands (i.e., viruses preferably infect nontransfected cells) (data not shown). Another approach to test potential cleavage-resistant MDA5 mutants was to use lysates from cells overexpressing tagged MDA5 mutants in our in vitro cleavage assay. However, under conditions where we did observe cleavage of endogenous MDA5 or MAVS or overexpressed MAVS (e.g., in the Q148A mutant), we did not find any cleavage product of the overexpressed MDA5 (as detected by antibodies against terminal epitope tags) (data not shown). In addition, no cleavage of recombinant MDA5 purified from mammalian cells was observed (data not shown). It eludes us why overexpressed MDA5 was so poorly cleaved by recombinant 2Apro, and unfortunately, it did not allow us to further investigate and identify the 2Apro cleavage site in MDA5.
In addition to MDA5, the downstream adaptor molecule MAVS was also cleaved in a 2Apro-dependent manner during enterovirus infection. CVB3 and EV71 induced identical MAVS cleavage products. These products were also reasonably similar to what was previously observed when in vitro-translated MAVS was treated with CVB3 2Apro (27), further supporting our conclusion that this is a 2Apro-mediated cleavage. PV 2Apro also led to MAVS cleavage, suggesting a similar cleavage mechanism, though the cleavage products were slightly different from those induced by CVB3. While this paper was in preparation, it was shown in another study that EV71 2Apro cleaves MAVS at three distinct positions, namely, Q209, Q251, and Q265, leading to the production of two cleavage products of 30 to 40 kDa (28). We show here that both CVB3 and EV71 induce identical MAVS cleavage patterns, yielding two cleavage products similar in size to those reported in that study, leading us to conclude that CVB3 2Apro likely also cleaves MAVS at these positions. Furthermore, 2Apro of PV also targeted MAVS, though the size of one of the cleavage products was slightly different from that observed in CVB3- or EV71-infected cells. It is not at odds with the finding that 2Apros from different enteroviruses cleave the same target protein at various positions. It has previously been shown that 2Apros of human rhinoviruses cleave nucleoporins at different sites, most likely due to sequence diversity in 2Apro (42). Moreover, caspase-mediated MAVS cleavage has been suggested for HRV1A (27). We did not observe the caspase-mediate MAVS cleavage product during CVB3 infection in HeLa R19 cells or cells of other human cell lines, such as HeLa Kyoto and Huh7 (data not shown). Together, our data show that the 2Apros from three different enterovirus species all target not only MDA5 but also MAVS during infection. The evolutionary conservation of these activities suggests that this is probably highly advantageous for enterovirus replication.
Although RIG-I is dispensable in sensing picornaviruses (16), it has been reported to be cleaved during infection of a few enteroviruses, such as PV, echovirus, and HRV16, mostly likely via their 3Cpros. Here we provide data that two additional enteroviruses, namely, CVB3 and EV71, also cleave RIG-I via their 3Cpros. Unlike the various 2Apro cleavage sites in MAVS, the yet unidentified site in RIG-I cleaved by 3Cpro seems to be well conserved across this panel of different enteroviruses, each yielding a putative cleavage product of approximately 70 kDa (26).
Cleavage of RIG-I, MDA5, and MAVS does not appear to be common to all picornaviruses. In this study, we also examined the fate of factors involved in the RLR pathway during infection with mengovirus, a species of EMCV of the Cardiovirus genus. We show that MDA5, MAVS, RIG-I, LGP2, and TBK1 all remained intact in cells infected with either wt mengovirus, which effectively suppresses the IFN-α/β response, or mengo-Zn, which induces high levels of IFN-α/β. Furthermore, equal levels of TBK1 activation were observed in cells infected with both wt mengovirus and mengo-Zn, indicating that factors upstream of TBK1 are not targeted by mengovirus. These results may seem to contradict earlier reports that MDA5 and RIG-I are also cleaved in mengovirus-infected cells (24). However, MDA5 cleavage was observed only in cells pretreated with poly(I·C) (24), which, as mentioned before, potentiates cells for virus-induced apoptosis and likely results in caspase-mediated MDA5 cleavage. In agreement with our observation that MDA5 is not cleaved in mengovirus-infected cells, MDA5 also remained intact during EMCV infection without poly(I·C) pretreatment in an independent study (43). Two reports have shown that RIG-I, which is dispensable for EMCV RNA recognition (16), is targeted by EMCV. Barral et al. reported that RIG-I is cleaved in EMCV-infected cells and implicated 3Cpro as the responsible proteinase since the cleavage product (of approximately 70 kDa) was similar to the cleavage product released by 3Cpro of PV (26). Another study showed a gradual decrease of RIG-I during EMCV infection which could be prevented by a caspase inhibitor, indicating that this is caspase-mediated degradation of RIG-I (43). These authors also demonstrated that recombinant EMCV 3Cpro can cleave RIG-I in vitro, resulting in a putative cleavage product of approximately 50 kDa. However, this RIG-I fragment was not observed during a normal EMCV infection (43), suggesting that it may be an artifact of in vitro study, similar to the MDA5 cleavage that we observed with CVB3 3Cpro in our own in vitro cleavage experiments.
The exact mechanism that EMCV uses to suppress the IFN-α/β response remains to be clarified. As indicated above, this blockade likely lies between TBK1 activation and IRF3 phosphorylation. It is known that TBK1 must form a complex with other kinases, such as the noncanonical IκB kinase IKK-epsilon, in order to phosphorylate its substrate, IRF3 (44–46). The assembly of a functional TBK1 complex requires not only phosphorylation of TBK1 at serine 172 but also other posttranslational modifications of IKK-epsilon and the assistance of scaffold proteins, such as NAP1 and SINTBAD (47). It is possible that mengovirus interferes with the correct assembly of the TBK1 complex to terminate the signal transduction. Alternatively, IFN-α/β shutdown may be a secondary effect of the so-called nuclear-cytoplasmic trafficking disorder that is induced by mengovirus leader protein (48). This causes an unregulated cargo transport between the nucleus and cytoplasm and possibly interferes with IRF3 activation, since this transcription factor must shuttle between these two compartments. Intriguingly, the same mutations in the leader protein (e.g., those in the Zn finger domain) simultaneously inactivate its activities in both IFN-α/β suppression (16, 22, 34) and the induction of the nuclear-cytoplasmic trafficking disorder (48), suggesting that there may be a link between these two phenomena. Future research is called upon to investigate the exact mechanism of IFN antagonization by mengovirus.
In short, our data show that several enteroviruses target MDA5, MAVS, and RIG-I in infected cells and that all do so via the same mechanisms (i.e., 2Apro targeting MDA5 and MAVS and 3Cpro targeting RIG-I). These cleavage events likely account for the lack of an IFN-α/β response in enterovirus-infected cells. Our data that TBK1 phosphorylations and, thus, activation are inhibited in CVB3-infected cells indicate that the pathway is shut down upstream of TBK1. The fact that the RLR pathway is targeted at multiple steps during infection makes it technically challenging to demonstrate the biological consequence of these cleavage events. One would have to simultaneously reconstitute a minimum of three factors, MDA5, MAVS, and RIG-I, to, it is hoped, study the effect of an intact RLR signaling pathway on virus replication. Additionally, enteroviruses may also employ additional mechanisms to ensure effective IFN-α/β suppression. For instance, enterovirus 2Apro is also known to cause nuclear-cytoplasmic trafficking disorder (49), possibly interfering with IRF3 activation, and, thereby, further ensure a total shut off of the IFN-α/β response in infected cells. Although a clear contribution of these phenomena to IFN-α/β inhibition remains to be established, this study provides important new insights into the potential roles of enterovirus proteinases in suppressing the RLR-mediated antiviral pathway.
ACKNOWLEDGMENTS
We acknowledge Rolf Hilgenfeld (University of Lübeck) for the kind gift of recombinant CVB3 3Cpro.
Q.F. and S.V.H. are supported by Mosaic grants (NWO-017.006.043 and NWO-017.002.025, respectively), M.A.L. is supported by a Rubicon grant (NWO-825.11.022), and F.J.M.V.K. is supported by an ECHO grant (NWO-CW-700.59.007), all from the Netherlands Organization for Scientific Research (NWO). P.B.F. was funded by NIH/NCI grant R01 CA097318 and the VCU MCC. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research in the MCC.
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Stark GR, Darnell JEJ. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. 10.1016/j.immuni.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis E, Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 4.Kang D, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 99:637–642. 10.1073/pnas.022637199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang D-C, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, Pestka S, Fisher PB. 2004. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23:1789–1800. 10.1038/sj.onc.1207300 [DOI] [PubMed] [Google Scholar]
- 6.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269. 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 http://www.jimmunol.org/content/175/5/2851.full.pdf [DOI] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea C-KK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 9.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 11.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- 12.Baum A, Sachidanandam R, García-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308. 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang T-H, Liao C-L, Lin Y-L. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 8:157–171. 10.1016/j.micinf.2005.06.014 [DOI] [PubMed] [Google Scholar]
- 14.Plumet S, Herschke F, Bourhis J-M, Valentin H, Longhi S, Gerlier D. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One 2:e279. 10.1371/journal.pone.0000279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr, Silverman RH. 2010. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 16:2108–2119. 10.1261/rna.2244210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJM. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2:1187–1196. 10.1016/j.celrep.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108. 10.1371/journal.ppat.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82:9829–9838. 10.1128/JVI.01199-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 20.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34. 10.1016/j.immuni.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triantafilou K, Vakakis E, Kar S, Richer E, Evans GL, Triantafilou M. 2012. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J. Cell Sci. 125:4761–4769. 10.1242/jcs.103887 [DOI] [PubMed] [Google Scholar]
- 22.Hato SV, Ricour C, Schulte BM, Lanke KH, de Bruijni M, Zoll J, Melchers WJ, Michiels T, van Kuppeveld FJ. 2007. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 9:2921–2930. 10.1111/j.1462-5822.2007.01006.x [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 7:e1001311. 10.1371/journal.ppat.1001311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barral PM, Morrison JM, Drahos J, Gupta P, Sarkar D, Fisher PB, Racaniello VR. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81:3677–3684. 10.1128/JVI.01360-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo R-L, Kao L-T, Lin S-J, Wang RY-L, Shih S-R. 2013. MDA5 plays a crucial role in enterovirus 71 RNA-mediated IRF3 activation. PLoS One 8:e63431. 10.1371/journal.pone.0063431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barral PM, Sarkar D, Fisher PB, Racaniello VR. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171–176. 10.1016/j.virol.2009.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drahos J, Racaniello VR. 2009. Cleavage of IPS-1 in cells infected with human rhinovirus. J. Virol. 83:11581–11587. 10.1128/JVI.01490-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, Jin Q, Zhao Z. 2013. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathog. 9:e1003231. 10.1371/journal.ppat.1003231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebig HD, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry 32:7581–7588. 10.1021/bi00080a033 [DOI] [PubMed] [Google Scholar]
- 30.Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, Krystek E, Blaas D, Kuechler E, Liebig HD. 1994. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4 gamma via a common recognition motif. Virology 198:741–745. 10.1006/viro.1994.1089 [DOI] [PubMed] [Google Scholar]
- 31.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]
- 32.Barral PM, Sarkar D, Su Z, Barber GN, DeSalle R, Racaniello VR, Fisher PB. 2009. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 124:219–234. 10.1016/j.pharmthera.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062–2074. 10.1128/jvi.76.5.2062-2074.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoll J, Melchers WJG, Galama JMD, van Kuppeveld FJM. 2002. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-κB. J. Virol. 76:9664–9672. 10.1128/JVI.76.19.9664-9672.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W. 2013. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 587:542–548. 10.1016/j.febslet.2013.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chau DHW, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. 2007. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12:513–524. 10.1007/s10495-006-0013-0 [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, Morace G, Gauss-Müller V, Kusov Y. 2007. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res. 35:5975–5984. 10.1093/nar/gkm645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marissen WE, Lloyd RE. 1998. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18:7565–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yalamanchili P, Harris K, Wimmer E, Dasgupta A. 1996. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J. Virol. 70:2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J. Virol. 74:1513–1523. 10.1128/JVI.74.3.1513-1523.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watters K, Palmenberg AC. 2011. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J. Virol. 85:10874–10883. 10.1128/JVI.00718-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318. 10.1016/j.virol.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 44.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. 2004. The roles of two I B kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650. 10.1084/jem.20040520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-MM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- 46.Sharma S, ten Oever BR, Grandvaux N, Zhou G-P, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151. 10.1126/science.1081315 [DOI] [PubMed] [Google Scholar]
- 47.Chau T-L, Gioia R, Gatot J-S, Patrascu F, Carpentier I, Chapelle J-P, O'Neill L, Beyaert R, Piette J, Chariot A. 2008. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33:171–180. 10.1016/j.tibs.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 48.Lidsky PV, Hato S, Bardina MV, Aminev AG, Palmenberg AC, Sheval EV, Polyakov VY, van Kuppeveld FJ, Agol VI. 2006. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 80:2705–2717. 10.1128/JVI.80.6.2705-2717.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belov G, Lidsky P, Mikitas O, Egger D, Lukyanov K, Bienz K, Agol V. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78:10166–10177. 10.1128/JVI.78.18.10166-10177.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]