ABSTRACT
A modified vaccinia virus Ankara poxvirus vector expressing the HIV-1 Env, Gag, Pol, and Nef antigens from clade B (MVA-B) is currently being tested in clinical trials. To improve its immunogenicity, we have generated and characterized the immune profile of MVA-B containing a deletion of the vaccinia viral gene N2L, which codes for an inhibitor of IRF3 (MVA-B ΔN2L). Deletion of N2L had no effect on virus growth kinetics or on the expression of HIV-1 antigens; hence, the N2 protein is not essential for MVA replication. The innate immune responses triggered by MVA-B ΔN2L revealed an increase in beta interferon, proinflammatory cytokines, and chemokines. Mouse prime-boost protocols showed that MVA-B ΔN2L improves the magnitude and polyfunctionality of HIV-1-specific CD4+ and CD8+ T cell adaptive and memory immune responses, with most of the HIV-1 responses mediated by CD8+ T cells. In the memory phase, HIV-1-specific CD8+ T cells with an effector phenotype were predominant and in a higher percentage with MVA-B ΔN2L than with MVA-B. In both immunization groups, CD4+ and CD8+ T cell responses were directed mainly against Env. Furthermore, MVA-B ΔN2L in the memory phase enhanced levels of antibody against Env. For the vector immune responses, MVA-B ΔN2L induced a greater magnitude and polyfunctionality of VACV-specific CD8+ T memory cells than MVA-B, with an effector phenotype. These results revealed the immunomodulatory role of N2L, whose deletion enhanced the innate immunity and improved the magnitude and quality of HIV-1-specific T cell adaptive and memory immune responses. These findings are relevant for the optimization of poxvirus vectors as vaccines.
IMPORTANCE On the basis of the limited efficacy of the RV144 phase III clinical trial, new optimized poxvirus vectors as vaccines against HIV/AIDS are needed. Here we have generated and characterized a new HIV/AIDS vaccine candidate on the basis of the poxvirus MVA vector expressing HIV-1 Env, Gag, Pol, and Nef antigens (MVA-B) and containing a deletion in the vaccinia virus N2L gene. Our findings revealed the immunomodulatory role of N2L and proved that its deletion from the MVA-B vector triggered an enhanced innate immune response in human macrophages and monocyte-derived dendritic cells. Furthermore, in immunized mice, MVA-B ΔN2L induced improvements in the magnitude and quality of adaptive and memory HIV-1-specific CD4+ and CD8+ T cell immune responses, together with an increase in the memory phase of levels of antibody against Env. Thus, the selective deletion of the N2L viral immunomodulatory gene is important for the optimization of MVA vectors as HIV-1 vaccines.
INTRODUCTION
Finding a safe and effective HIV/AIDS vaccine that is able to induce protective humoral and cellular immune response to HIV-1 is one of the major research goals in fighting this pandemic affecting the human population worldwide. Currently, only one HIV-1 vaccine tested in a phase III clinical trial (RV144) in Thailand has shown some level of protection against HIV-1, and it is based on a combination of recombinant poxvirus vector ALVAC and the HIV-1 gp120 protein used in a prime-boost protocol that showed 31.2% protection against HIV-1 infection (1). Since the poxvirus vector appeared to have played a significant role in the protective immune response in the combined protocol, in spite of the poor immunogenicity of the ALVAC vector (2), a main interest in improving the immunogenicity of attenuated poxvirus vectors as future HIV-1 vaccine candidates has emerged (3–5).
Among poxviruses, the highly attenuated vaccinia virus (VACV) strain modified VACV Ankara (MVA) is one of the most encouraging vectors, as it has been extensively used in preclinical and clinical trials as a prototype vaccine against HIV-1, infectious diseases, and cancer (6, 7). Numerous MVA vectors expressing different HIV-1 antigens have been produced and tested in human clinical trials (8–25), revealing that MVA vectors are safe and elicit humoral and cellular immune responses to HIV-1 antigens (for reviews, see references 3, 6, and 7), regardless of its limited replication in human and most mammalian cell types. However, MVA still contains several immunomodulatory VACV genes that counteract the host antiviral innate immune response, particularly those genes encoding proteins that inhibit the Toll-like receptor (TLR) signaling pathway (26), an important route that plays a fundamental role in the defense against pathogens through the induction of proinflammatory cytokines and type I interferon (IFN) but also in modeling adaptive immune responses to pathogens (27–29). Hence, the deletion of these immunomodulatory VACV genes is a promising approach to the generation of improved MVA-based vaccines with increasing magnitude, breadth, polyfunctionality, and durability of the antigen-specific cellular and humoral immune responses.
An attractive target for this strategy is the VACV N2L gene. The VACV N2L gene is present in the genome of VACV strains Western Reserve (WR) (VACV-WR_029), Copenhagen (N2L), and MVA (MVA 021L) but absent from NYVAC. N2L encodes a 175-amino-acid protein with a predicted molecular mass of 20.8 kDa (www.poxvirus.org). The VACV N2L gene belongs to the VACV B cell lymphoma 2 (Bcl-2) family (30), a family of intracellular proteins that are important inhibitors of the TLR signaling pathway, acting at different levels of the route, such as A46 (31–35), A52 (31, 36–39), B14 (named B15R in MVA) (36, 39–41), C6 (42–44), K7 (45–48), and N1 (49–54). Old reports showed that N2L is transcribed early during infection (55) and that a single mutation in its 5′-untranslated region is responsible for resistance to the inhibitor of RNA polymerase II (alpha-amanitin) and for the temperature-sensitive phenotype (56, 57), but as yet there is no explanation for these observations. Moreover, a yeast two-hybrid assay revealed that N2 binds to importin alpha 1, valosin-containing protein (p97)/p47 complex interacting protein 1 (VCPIP1), and phospholipid scramblase 4 (PLSCR4) (58), but the biological significance of these interactions is unknown. Recently, it has been reported that the VACV N2L gene encodes a nonessential protein expressed early during infection and located in the nucleus that inhibits the activation of the IFN-β promoter by inhibiting IFN regulatory factor 3 (IRF3) activation (59), although the exact molecular mechanism of action remains unknown. Furthermore, the VACV N2 protein is a virulence factor since a VACV WR strain with N2L deleted was attenuated in murine intranasal and intradermal models (59). However, the VACV N2L deletion mutant was not more immunogenic than the parental strain after the immunization of mice and the protection afforded against a challenge with virulent WR was indistinguishable from that of control viruses (59).
Therefore, taking into consideration the role of N2L in blocking the IFN signaling pathway through IRF3, it was of interest to establish its immunomodulatory function in vivo. Hence, in this investigation, we characterized how N2L impacts innate, adaptive, and memory immune responses in mice by examining specific immune responses to HIV-1 and VACV antigens. This was done by deleting the VACV N2L gene from the vector backbone of MVA-B, an HIV/AIDS vaccine candidate based on an attenuated recombinant MVA vector expressing HIV-1 Env and Gag-Pol-Nef (GPN) antigens from clade B (60). MVA-B has been extensively studied in vitro and in different animal models (42, 60–66), where it triggers strong and durable immune responses to HIV-1 antigens. Furthermore, MVA-B was safe and highly immunogenic when tested in a phase I clinical trial with healthy human volunteers, inducing humoral responses to Env and HIV-1-specific CD4+ and CD8+ T cell responses that were high, broad, polyfunctional, and long-lasting (12, 14). Moreover, a phase I clinical trial of HIV-1-infected patients on highly active antiretroviral therapy with MVA-B as an HIV/AIDS therapeutic vaccine is ongoing.
Here we have examined the innate immune responses (in human macrophages and monocyte-derived dendritic cells [moDCs]) and evaluated the adaptive and memory HIV-1-specific cellular and humoral immune responses of mice inoculated with MVA-B and the MVA-B ΔN2L deletion mutant. Our results demonstrate that the VACV N2 protein has an immunomodulatory role and that deleting N2L improves innate, adaptive, and memory HIV-1-specific immune responses. Thus, deletion of N2L from poxvirus vectors could be a general strategy to potentiate the vaccine utility of these vectors.
MATERIALS AND METHODS
Ethics statement.
The animal studies described here were approved by the Ethical Committee of Animal Experimentation (CEEA-CNB) of Centro Nacional de Biotecnologia (CNB-CSIC, Madrid, Spain) in accordance with national and international guidelines and with the Royal Decree (RD 1201/2005) (permit no. 12055).
Studies with peripheral blood mononuclear cells (PBMCs) from healthy blood donors recruited by the Centro de Transfusión de la Comunidad de Madrid (Madrid, Spain) were approved by the Ethical Committee of Centro de Transfusión de la Comunidad de Madrid (Madrid, Spain). Written informed consent was obtained from each donor before blood collection for the purpose of this investigation according to a collaborative agreement between the Centro de Transfusión de la Comunidad de Madrid and the CNB-CSIC. All information was kept confidential.
Cells and viruses.
DF-1 cells (a spontaneously immortalized chicken embryo fibroblast (CEF) cell line, ATCC catalog no. CRL-12203) and primary CEF cells (obtained from specific-pathogen-free 11-day-old eggs; Intervet, Salamanca, Spain) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). The THP-1 human monocytic cell line (ATCC catalog no. TIB-202) was cultured in RPMI 1640 medium containing 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 100 IU/ml penicillin, 100 μg/ml streptomycin (complete medium; all from Invitrogen, San Diego, CA) and 10% heat-inactivated FCS (Sigma-Aldrich, St. Louis, MO) as previously described (42, 62, 67). THP-1 cells were differentiated into macrophages by treatment with 0.5 mM phorbol 12-myristate 13-acetate (Sigma-Aldrich) for 24 h before use. PBMCs from buffy coats of healthy donors (recruited by the Centro de Transfusión de la Comunidad de Madrid, Madrid, Spain) were obtained by Ficoll gradient separation on Ficoll-Paque (GE Healthcare). Next, CD14+ monocytes were purified by depletion with the Dynabeads Untouched human monocytes kit (Invitrogen Dynal AS, Oslo, Norway) in accordance with the manufacturer's protocol. moDCs were then obtained as previously described (62), after the cultivation of purified monocytes for 7 days in complete RPMI 1640 medium containing 10% heat-inactivated FCS and supplemented with 50 ng/ml granulocyte-macrophage colony-stimulating factor and 20 ng/ml interleukin-4 (IL-4) (both from Gibco-Life Technologies). Cell cultures were maintained at 37°C (CEFs, THP-1 cells, and moDCs) or 39°C (DF-1 cells) in a humidified incubator containing 5% CO2. Cell lines were infected with viruses as previously described (60, 67). Virus infection of all of the cell types was performed with 2% FCS.
The poxvirus strains used in this study included the attenuated wild-type (WT) MVA (MVA-WT) obtained from the Ankara strain after 586 serial passages in CEF cells (derived from clone F6 at passage 585, kindly provided by G. Sutter) and the recombinant MVA-B expressing HIV-1IIIB GPN as an intracellular polyprotein and the HIV-1BX08 gp120 protein as a cell-released product from HIV-1 clade B isolates (60). MVA-B was used as the parental virus for the generation of the MVA-B ΔN2L deletion mutant. All viruses were grown in primary CEF cells, purified by centrifugation through two 36% (wt/vol) sucrose cushions in 10 mM Tris-HCl (pH 9), and titrated in DF-1 cells by plaque immunostaining assay as previously described (68). Determinations of the titers of the different viruses were performed at least two times. All viruses were free of contamination with mycoplasmas or other bacteria.
Construction of plasmid transfer vector pGem-RG-N2L wm.
Plasmid transfer vector pGem-RG-N2L wm was used for the construction of the MVA-B ΔN2L deletion mutant from which the VACV N2L gene has been deleted (N2L in the Copenhagen strain of VACV is equivalent to MVA 021L in MVA. For simplicity, throughout this work, we use the open reading frame nomenclature of the Copenhagen strain to refer to the MVA genes). pGem-RG-N2L wm was obtained by sequential cloning of N2L flanking sequences into plasmid pGem-RG wm (4,540 bp), whose generation was previously described (63), and contains the genes for DsRed2 and red-shifted green fluorescent protein (rsGFP) under the control of the synthetic early/late (sE/L) promoter. The MVA-B genome was used as the template to amplify the right flank of the N2L gene (474 bp) with oligonucleotides RFN2L-AatII-F (5′-GTCTAGGACGTCTAATTTTCATATGG-3′) (AatII site underlined) and RFN2L-XbaI-R (5′-AAGTATTCTAGAAATTAATGATGC-3′) (XbaI site underlined). The right flank was digested with AatII and XbaI and cloned into plasmid pGem-RG wm, which had previously been digested with the same restriction enzymes, to generate pGem-RG-RFsN2L wm (4,981 bp). The repeated right flank of the N2L gene (474 bp) was amplified by PCR from the MVA-B genome with oligonucleotides RF′N2L-EcoRI-F (5′-GTCTAGGAATTCTAATTTTCATATGG-3′) (EcoRI site underlined) and RF′N2L-ClaI-R (5′-AAGTATATCGATAATTAATGATGC-3′) (ClaI site underlined), digested with EcoRI and ClaI, and inserted into EcoRI/ClaI-digested pGem-RG-RFsN2L wm to generate pGem-RG-RFdN2L wm (5,414 bp). The left flank of the N2L gene (431 bp) was amplified by PCR from the MVA-B genome with oligonucleotides LFN2L-ClaI-F (5′-AGAGGAATCGATGATCGCATCC-3′) (ClaI site underlined) and LFN2L-BamHI-R (5′-AGCCATGGATCCACTAATCAGATCTATTAG-3′) (BamHI site underlined), digested with ClaI and BamHI, and inserted into ClaI/BamHI-digested pGem-RG-RFdN2L wm. The resulting plasmid, pGem-RG-N2L wm (5,815 bp), was confirmed by DNA sequence analysis and directs the deletion of the N2L gene from the MVA-B genome.
Construction of MVA-B ΔN2L.
The MVA-B ΔN2L deletion mutant generated in this work contains a deletion of the VACV gene N2L and was constructed by transfecting plasmid transfer vector pGem-RG-N2L wm into DF-1 cells infected with the parental virus MVA-B and then screening for transient Red2-GFP coexpression with the genes for DsRed2 and rsGFP as the transiently selectable markers as previously described (42, 63). MVA-B ΔN2L was finally obtained after six consecutive rounds of plaque purification in DF-1 cells.
PCR analysis of MVA-B ΔN2L.
To verify that the VACV gene N2L deletion in MVA-B ΔN2L was correctly generated, viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell with MVA-WT, MVA-B, or MVA-B ΔN2L as previously described (60). Primers RFN2L-AatII-F and LFN2L-BamHI-R (described above), spanning N2L flanking regions, were used for PCR analysis of the N2L locus. The amplification protocol was previously described (63). PCR products were run in 1% agarose gel and visualized by SYBR Safe staining (Invitrogen). The N2L deletion was also confirmed by DNA sequence analysis.
Expression of HIV-1IIIB GPN and HIV-1BX08 gp120 proteins by MVA-B ΔN2L.
To check the expression of the HIV-1IIIB GPN and HIV-1BX08 gp120 proteins by MVA-B ΔN2L, monolayers of DF-1 cells were mock infected or infected at 5 PFU/cell with MVA-WT, MVA-B, or MVA-B ΔN2L. At 24 h postinfection, cells were lysed in Laemmli buffer, cells extracts were fractionated by 8% SDS-PAGE, and evaluated by Western blotting with rabbit polyclonal anti-gp120 antibody against IIIB (Centro Nacional de Biotecnología; diluted 1:3,000) or polyclonal anti-gag p24 serum (ARP 432, diluted 1:1,000; National Institute for Biological Standards and Control [NIBSC] Centre for AIDS Reagents) to analyze the expression of the gp120 and GPN proteins, respectively. As loading controls, we used rabbit anti-α-tubulin (Cell Signaling, diluted 1:1,000), rabbit anti-β-actin (diluted 1:1,000; Cell Signaling), and rabbit anti-VACV E3 (diluted 1:1,000; Centro Nacional de Biotecnología) antibodies. A horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (diluted 1:5,000; Sigma) was used as the secondary antibody. The immunocomplexes were detected with an HRP-luminol enhanced-chemiluminescence system (ECL Plus; GE Healthcare).
Analysis of virus growth.
To study the virus growth profile of MVA-B ΔN2L in comparison to that of parental MVA-B, monolayers of DF-1 cells were infected in duplicate at 0.01 PFU/cell with MVA-B or MVA-B ΔN2L. Following virus adsorption for 60 min at 37°C, the inoculum was removed. The infected cells were washed with DMEM and incubated with fresh DMEM containing 2% FCS at 37°C in a 5% CO2 atmosphere. At different times (0, 24, 48, and 72 h) postinfection, cells were collected by scraping, freeze-thawed three times, and briefly sonicated. Virus titers in cell lysates were determined by immunostaining plaque assay as previously described (68).
RNA analysis by quantitative real-time PCR.
Total RNA was isolated from THP-1 cells and moDCs mock infected or infected with MVA-WT, MVA-B, or MVA-B ΔN2L with the RNeasy kit (Qiagen GmbH, Hilden, Germany). Reverse transcription of at least 500 ng of RNA was performed with the QuantiTect reverse transcription kit (Qiagen GmbH, Hilden, Germany). Quantitative PCR was performed with a 7500 Real-Time PCR system (Applied Biosystems) and Power SYBR green PCR Master Mix (Applied Biosystems) as previously described (67). The expression levels of the genes for IFN-β, tumor necrosis factor alpha (TNF-α), MIP-1α, RANTES, RIG-I, MDA-5, and hypoxanthine phosphoribosyltransferase (HPRT) were analyzed by real-time PCR with specific oligonucleotides (sequences are available upon request). Specific gene expression was expressed relative to the expression of the gene for HPRT in arbitrary units. All samples were tested in duplicate, and two different experiments were performed with each cell type.
DNA vectors.
DNA plasmids expressing HIV-1IIIB GPN fusion protein (pcDNA-IIIBGPN) and HIV-1BX08 gp120 (pCMV-BX08gp120) and the empty plasmid (pcDNA-ϕ) have been previously described (60) and were purified with the EndoFree Plasmid Mega kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's protocol and diluted for injection in endotoxin-free phosphate-buffered saline (PBS). pcDNA-ϕ (termed DNA-ϕ) or a mixture of pcDNA-IIIBGPN and pCMV-BX08gp120 (termed DNA-B) have been used in the immunization protocol as a prime.
Peptides.
HIV-1 peptide pools, with each purified peptide at 1 mg/ml per vial, were provided by BEI Resources, NIH. The peptides covered the Env, Gag, Pol, and Nef proteins present in the consensus sequence of HIV-1 clade B (gp120 from isolate BX08 and GPN from isolate IIIB) as consecutive 15-mers overlapping by 11 amino acids. The HIV-1BX08 gp120 protein was spanned by the Env-1 and Env-2 peptide pools. The HIV-1IIIB GPN fusion protein was spanned by the Gag-1, Gag-2, GPN-1, GPN-2, GPN-3, and GPN-4 pools. For immunological analysis, we grouped the peptides into three main pools, Env, Gag, and GPN. The Env pool comprises Env-1 and Env-2, the Gag pool comprises Gag-1 and Gag-2, and the GPN pool comprises GPN-1, GPN-2, GPN-3, and GPN-4.
Mouse immunization schedule.
Female BALB/c mice (6 to 8 weeks old) were purchased from Harlan Laboratories and stored in a pathogen-free barrier area of the CNB in accordance to the recommendations of the Federation of European Laboratory Animal Science Associations. A DNA prime-MVA boost immunization protocol was performed as previously described (60, 63) to assay the immunogenicity of MVA-WT, MVA-B, and MVA-B ΔN2L. Groups of animals (n = 8) received 100 μg of DNA-B (50 μg of pCMV-BX08gp120 plus 50 μg of pCDNA-IIIBGPN) or 100 μg of DNA-ϕ (100 μg of pcDNA-ϕ) by the intramuscular route and 2 weeks later received an intraperitoneal (i.p.) inoculation of 1 × 107 PFU of the corresponding recombinant VACV (MVA-WT, MVA-B, or MVA-B ΔN2L) in 200 μl of PBS. Mice primed with sham DNA (DNA-ϕ) and boosted with nonrecombinant MVA-WT were used as a control group. At 10 and 52 days after the last immunization, four mice in each group were sacrificed with carbon dioxide (CO2) and their spleens were processed to measure the adaptive and memory immune responses to VACV and HIV-1 antigens, respectively, by intracellular cytokine staining (ICS) assay. Two independent experiments were performed.
ICS assay.
The magnitude, polyfunctionality, and phenotypes of the VACV and HIV-1-specific T cell adaptive and memory responses were analyzed by ICS as previously described (42, 62, 63), with some modifications. After an overnight rest, 4 × 106 splenocytes (depleted of red blood cells) were seeded onto M96 plates and stimulated for 6 h in complete RPMI 1640 medium supplemented with 10% FCS containing 1 μl/ml Golgiplug (BD Biosciences) to inhibit cytokine secretion; monensin 1× (eBioscience), anti-CD107a–Alexa 488 (BD Biosciences); and the different HIV-1 Env, Gag, and GPN pools of peptides (5 μg/ml) or MVA-infected A20 cells (4 × 105 A20 cells; 1:10 ratio of MVA-infected A20 cells to splenocytes). Then cells were washed, stained for the surface markers, fixed, permeabilized (Cytofix/Cytoperm kit; BD Biosciences), and stained intracellularly with the appropriate fluorochromes. Dead cells were excluded with the violet LIVE/DEAD stain kit (Invitrogen). The fluorochrome-conjugated antibodies used for functional analyses were CD3-phycoerythrin (PE)-CF594, CD4-allophycocyanin (APC)-Cy7, CD8-V500, IFN-γ–PE-Cy7, TNF-α–PE, and IL-2–APC. In addition, the antibodies used for phenotypic analyses were CD62L-Alexa 700 and CD127-peridinin chlorophyll protein-Cy5.5. All antibodies were from BD Biosciences. Cells were acquired with a Gallios flow cytometer (Beckman Coulter). Analyses of the data were performed with the FlowJo software version 8.5.3 (Tree Star, Ashland, OR). The number of lymphocyte-gated events was 106. After gating, Boolean combinations of single functional gates were created with the FlowJo software to determine the frequency of each response based on all possible combinations of cytokine expression or all possible combinations of differentiation marker expression. Background responses detected in negative-control samples were subtracted from those detected in stimulated samples for every specific functional combination.
Antibody measurements by ELISA.
The total IgG, IgG1, IgG2a, and IgG3 anti-HIV-1 gp120 MN envelope protein antibodies in pooled sera from immunized mice were measured by enzyme-linked immunosorbent assay (ELISA) as previously described (60, 63).
Statistical procedures.
The statistical significance (*, P < 0.05; **, P < 0.005; ***, P < 0.001) of differences between groups was determined by Student's t test (two tailed, type 3). Statistical analysis of the ICS assay results was realized as previously described (63, 69), by an approach that corrects measurements for the medium response (RPMI), calculating confidence intervals and P values. Only antigen response values significantly larger than the corresponding RPMI are presented. Background values were subtracted from all of the values used to allow analysis of proportionate representation of responses.
RESULTS
Generation and in vitro characterization of MVA-B ΔN2L.
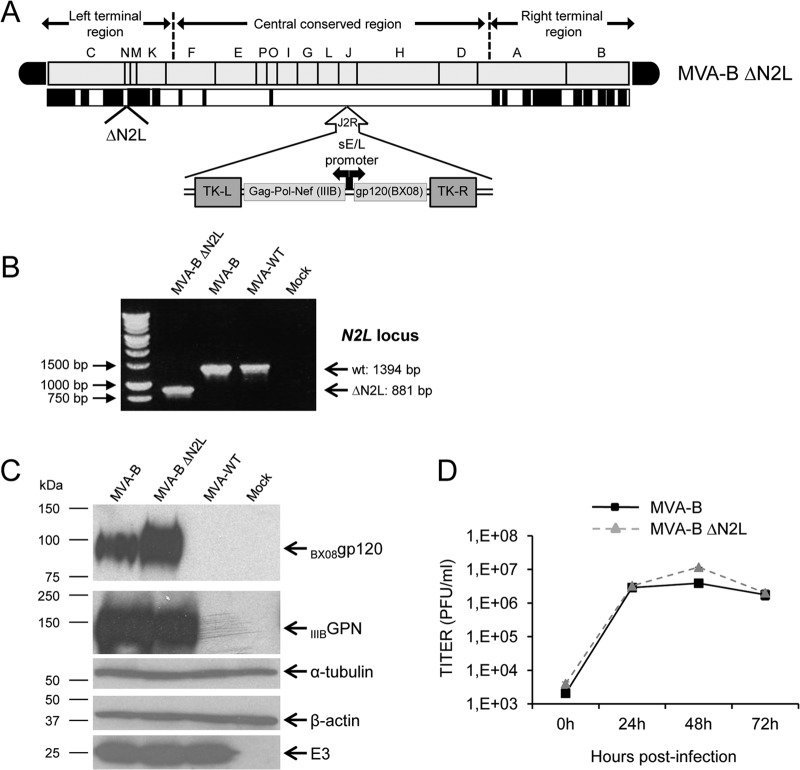
To determine whether the VACV gene N2L, encoding an inhibitor of IRF3 (59), might have an immunomodulatory role that, in turn, could influence the immunogenicity profile of antigens delivered from a poxvirus vector, we deleted N2L from the HIV/AIDS vaccine candidate MVA-B (expressing HIV-1 Env, Gag, Pol, and Nef antigens from clade B) (60), generating the MVA-B deletion mutant termed MVA-B ΔN2L (see Materials and Methods) and analyzed various properties. Figure 1A shows a diagram of MVA-B ΔN2L. PCR analysis with primers annealing within N2L flanking regions confirmed the deletion of N2L from MVA-B ΔN2L (Fig. 1B). The occurrence of the deletion was confirmed by DNA sequencing of the N2L viral locus (data not shown). Furthermore, the mutant virus was plaque purified several times to obtain a viral stock free of WT contamination. Analysis by Western blotting demonstrated that MVA-B ΔN2L expressed antigens HIV-1BX08 gp120 and HIV-1IIIB GPN as did the parental virus MVA-B (Fig. 1C). Moreover, the growth kinetics of parental MVA-B and deletion mutant MVA-B ΔN2L in cultured cells were similar (Fig. 1D), confirming that the VACV N2 protein is not required for MVA replication.
FIG 1.
Generation and in vitro characterization of MVA-B ΔN2L deletion mutant. (A) Scheme of the MVA-B ΔN2L deletion mutant genome map (adapted from references 90 and 93). The different regions are indicated by capital letters. The right and left terminal regions are shown. Below the map, the deleted or fragmented genes are depicted as black boxes. The deleted N2L gene is indicated. The HIV-1 GPN (from isolate IIIB) and gp120 (from isolate BX08) clade B sequences driven by the sE/L virus promoter inserted within the TK viral locus (J2R) are indicated (adapted from reference 60). (B) PCR analysis of the N2L locus. Viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell with MVA-WT, MVA-B, or MVA-B ΔN2L. Primers spanning N2L flanking regions were used for PCR analysis of the N2L locus. DNA products with sizes (base pairs) corresponding to the parental virus (wt) and the N2L deletion mutant are indicated on the right. Molecular size markers (1-kb ladder) with the corresponding sizes (base pairs) are indicated on the left. (C) Expression of HIV-1BX08 gp120 and HIV-1IIIB GPN proteins. DF-1 cells were mock infected or infected at 5 PFU/cell with MVA-WT, MVA-B, or MVA-B ΔN2L. At 24 h postinfection, cells were lysed in Laemmli buffer, fractionated by 8% SDS-PAGE, and analyzed by Western blotting with rabbit polyclonal anti-gp120 antibody or polyclonal anti-gag p24 serum. Rabbit anti-α-tubulin and rabbit anti-β-actin antibodies were used as protein loading controls. Rabbit anti-VACV early E3 protein antibody was used as a VACV loading control. Arrows on the right indicate the positions of the HIV-1BX08 gp120 and HIV-1IIIB GPN proteins, α-tubulin, β-actin, and the VACV E3 protein. The sizes (in kDa) of standards (Precision Plus protein standards; Bio-Rad Laboratories) are indicated on the left. (D) Viral growth kinetics in DF-1 cells. DF-1 cells were infected at 0.01 PFU/cell with MVA-B or MVA-B ΔN2L. At different times (0, 24, 48, and 72 h) postinfection, cells were collected and virus titers of cell lysates were quantified by plaque immunostaining assay with anti-WR antibodies. The mean of two independent experiments is shown.
MVA-B ΔN2L triggers a strong innate immune response in human macrophages and dendritic cells, upregulating type I IFN, proinflammatory cytokine, and chemokine expression.
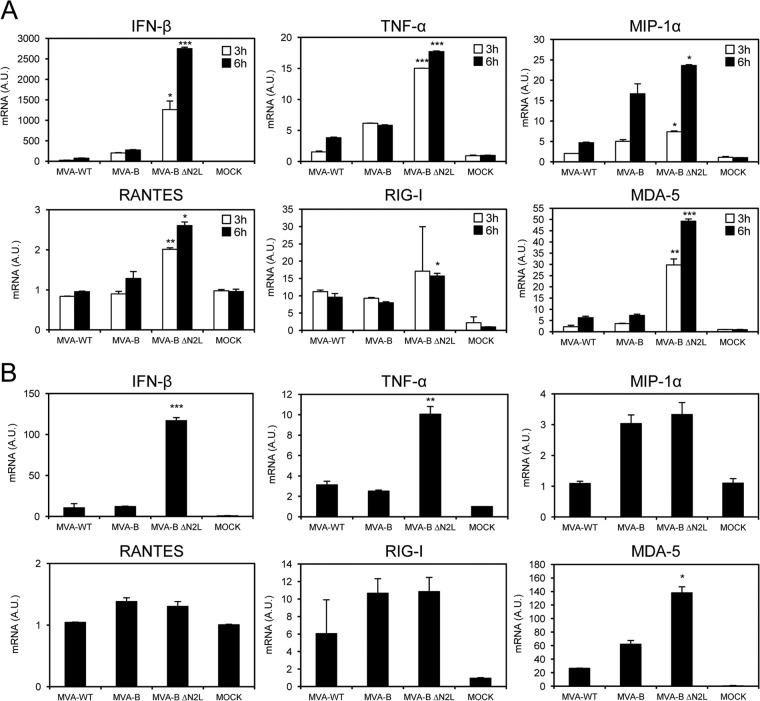
It has been recently reported that VACV N2 in the virulent WR strain blocks IRF3 and therefore, in response to WR infection, inhibits the IFN-I signaling pathway of the cell (59). Thus, to determine if similar behavior occurs in MVA, a virus known to markedly enhance the type I IFN response (70, 71), we used real-time PCR to analyze the expression of type I IFN (IFN-β), the proinflammatory cytokine TNF-α, chemokines (MIP-1α and RANTES), and some key cytosolic sensors (RIG-I and MDA-5) that lead to antiviral IFN production in human THP-1 macrophages mock infected or infected for 3 and 6 h with MVA-WT, MVA-B, and MVA-B ΔN2L at 5 PFU/cell (Fig. 2A). The results showed that, compared to MVA-WT and MVA-B, MVA-B ΔN2L significantly upregulated the mRNA levels of IFN-β, TNF-α, MIP-1α, RANTES, as well as RIG-I and MDA-5 (Fig. 2A), clearly affecting the IFN-I signaling pathway.
FIG 2.
MVA-B ΔN2L triggers a strong innate immune response in human macrophages and dendritic cells, upregulating type I IFN, proinflammatory cytokine, and chemokine expression. Human THP-1 macrophages (A) and moDCs (B) were mock infected or infected with MVA-WT, MVA-B, or MVA-B ΔN2L (5 PFU/cell in panel A and 1 PFU/cell in panel B). At different times postinfection (3 and 6 h in panel A, 6 h in panel B), RNA was extracted, and IFN-β, TNF-α, MIP-1α, RANTES, RIG-I, MDA-5, and HPRT mRNA levels were analyzed by RT-PCR. Results are expressed as the ratio of the gene of interest to HPRT mRNA levels. A.U., arbitrary units. P values indicate significant response differences between the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups (*, P < 0.05; **, P < 0.005; ***, P < 0.001). Data are means ± standard deviations of duplicate samples from one experiment and are representative of two independent experiments.
Furthermore, to verify that the VACV N2 protein impairs the IFN-I signaling pathway in innate immune cells, we infected human moDCs with MVA-WT, MVA-B, and MVA-B ΔN2L at 1 PFU/cell and measured IFN-β, TNF-α, MIP-1α, RANTES, RIG-I, and MDA-5 mRNA levels at 6 h postinfection (Fig. 2B). Similar to the results obtained with human THP-1 cells, MVA-B ΔN2L strongly increased IFN-β, TNF-α, and MDA-5 expression in moDCs, in contrast to MVA-WT and MVA-B (Fig. 2B).
Thus, deletion of N2L from the MVA-B genome promotes a strong innate immune response in human macrophages and moDCs, with high expression of type I IFN, proinflammatory cytokines, chemokines, and some key cytosolic sensors that lead to antiviral IFN production, confirming the role of the VACV N2 protein in blocking the IFN-I signaling pathway.
MVA-B ΔN2L enhances the magnitude and polyfunctionality of HIV-1-specific T cell adaptive immune responses.
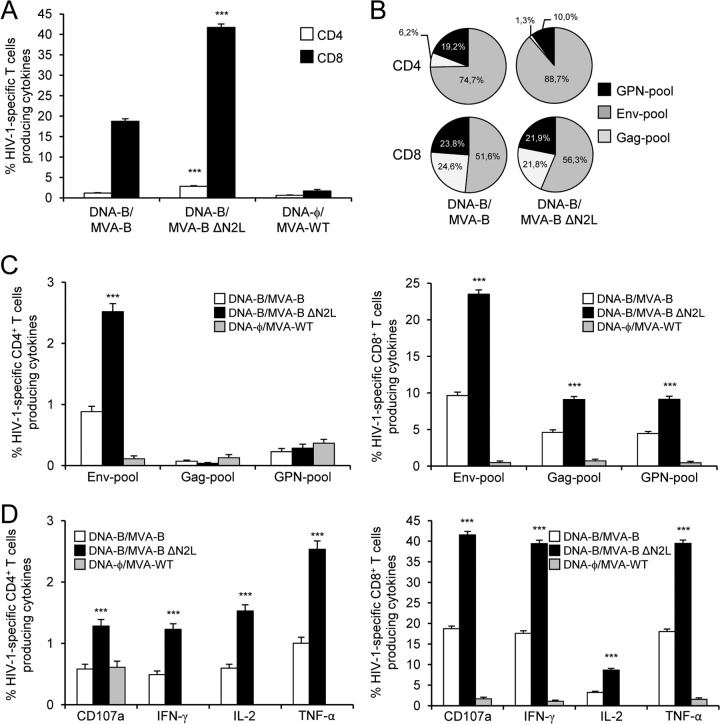
Given the immunomodulatory role of the VACV N2 protein in impairing the innate immune responses in human macrophages and DCs, we next asked if deletion of VACV N2L from MVA-B may have an impact on the immunogenicity of the vector. Therefore, to study in vivo the effect of N2L deletion on the cellular immunogenicity of the MVA vector expressing HIV-1 antigens, we analyzed the HIV-1-specific immune responses induced by MVA-B ΔN2L in BALB/c mice with a DNA prime (100 μg of DNA-B)-MVA boost (1 × 107 PFU) immunization protocol (see Materials and Methods). Animals primed with sham DNA (DNA-ϕ) and boosted with nonrecombinant MVA-WT were used as a control group. Adaptive HIV-1-specific T cell immune responses elicited by the different immunization groups (DNA-B–MVA-B, DNA-B–MVA-B ΔN2L, and DNA-ϕ–MVA-WT) were measured 10 days after the boost by polychromatic ICS assay, after the stimulation of splenocytes with pools of peptides (Env, Gag, and GPN pools) that spanned the HIV-1 Env, Gag, Pol, and Nef regions from an HIV-1 clade B consensus sequence.
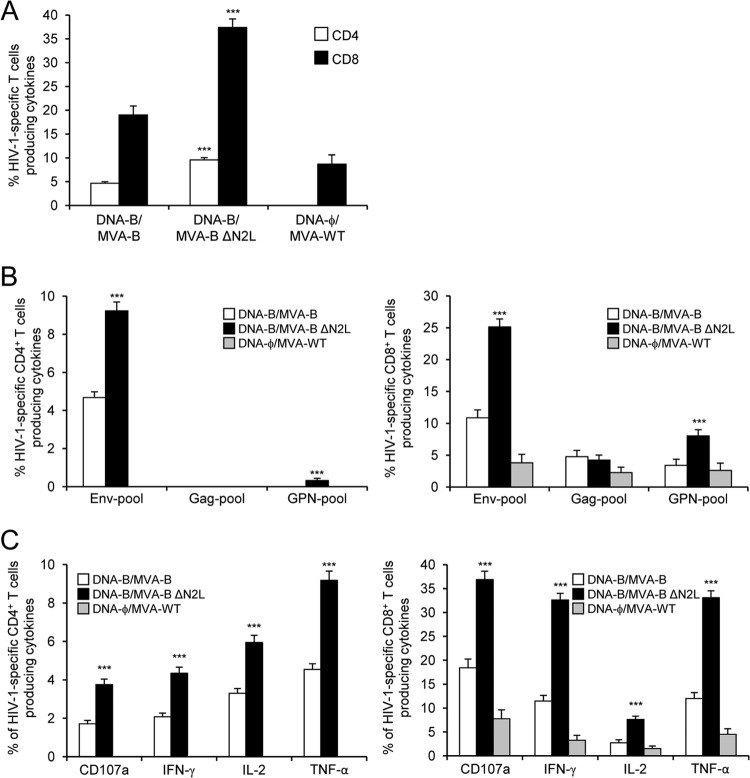
The magnitude of the total HIV-1-specific CD4+ and CD8+ T cell adaptive immune responses (determined as the sum of the individual responses producing IFN-γ, TNF-α, and/or IL-2 cytokines, as well as the expression of CD107a on the surface of activated T cells as an indirect marker of cytotoxicity) obtained for the Env, Gag, and GPN peptide pools was significantly greater in the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups than in the DNA-ϕ–MVA-WT control group (P < 0.001) (Fig. 3A). The DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups triggered an overall HIV-1-specific immune response mediated mainly by CD8+ T cells (80%) (Fig. 3A). Interestingly, immunization with DNA-B–MVA-B ΔN2L elicited a significantly greater magnitude of total HIV-1-specific CD4+ and CD8+ T cell responses than DNA-B–MVA-B, with increases of 2.04- and 1.96-fold, respectively (P < 0.001) (Fig. 3A).
FIG 3.
Immunization with MVA-B ΔN2L enhances the magnitude of HIV-1-specific CD4+ and CD8+ T cell adaptive immune responses. Splenocytes were collected from mice (n = 4 per group) immunized with DNA-ϕ–MVA-WT, DNA-B–MVA-B, or DNA-B–MVA-B ΔN2L 10 days after the last immunization. Next, HIV-1-specific CD4+ and CD8+ T cell adaptive immune responses triggered by the different immunization groups were measured by ICS assay following the stimulation of splenocytes with different HIV-1 peptide pools (Env, Gag, and GPN pools). Values from unstimulated controls were subtracted in all cases. P values indicate significant response differences between the DNA-B–MVA-B ΔN2L and DNA-B–MVA-B immunization groups (***, P < 0.001). Data are from one experiment representative of two independent experiments. (A) Overall percentages of HIV-1-specific CD4+ and CD8+ T cells. The values represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, and GPN peptide pools. (B) Percentages of Env, Gag, and GPN HIV-1-specific CD4+ and CD8+ T cells. Frequencies represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, or GPN peptide pools. (C) Percentages of HIV-1-specific CD4+ and CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against Env, Gag, and GPN peptide pools.
The pattern of HIV-1-specific T cell adaptive immune responses showed that CD4+ and CD8+ T cell responses were directed mainly against the Env pool in both the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups, with CD8+ T cell responses broadly distributed among Env, GPN, and Gag. However, DNA-B–MVA-B ΔN2L significantly enhanced the magnitude of Env- and GPN-specific CD4+ and CD8+ T cell responses (P < 0.001) (Fig. 3B).
Furthermore, HIV-1-specific CD4+ T cells producing TNF-α or IL-2 and HIV-1-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α are the most induced populations in both immunization groups (Fig. 3C). However, DNA-B–MVA-B ΔN2L induced a significantly greater magnitude of HIV-1-specific CD4+ and CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2 than DNA-B–MVA-B did (P < 0.001) (Fig. 3C).
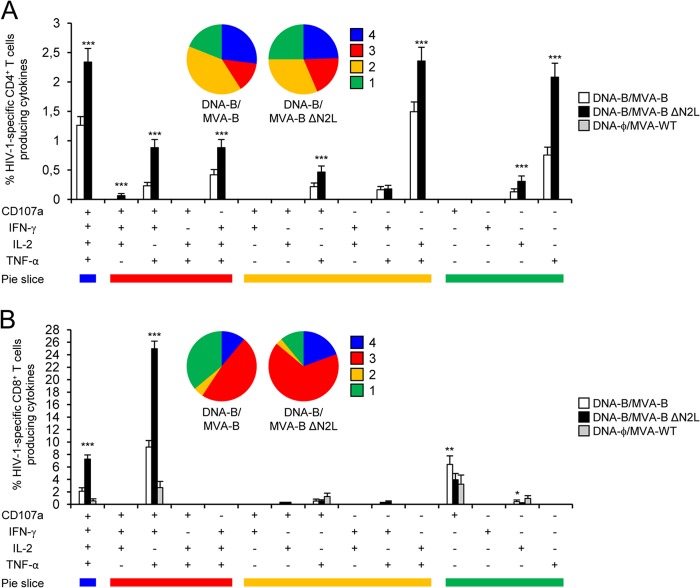
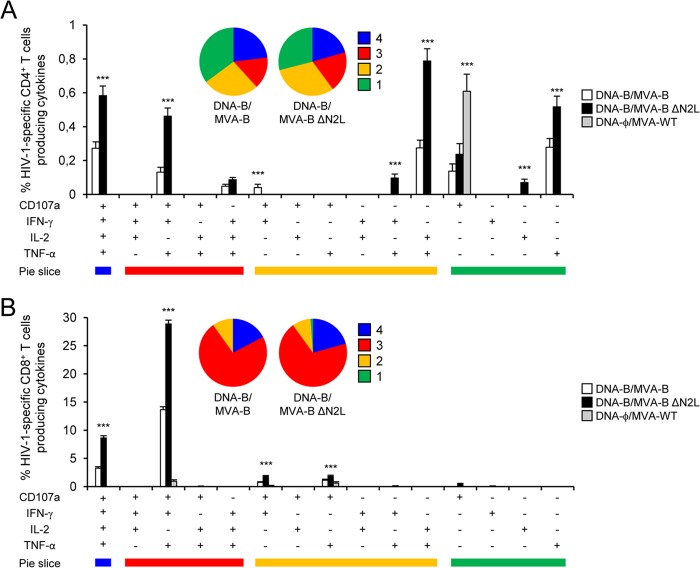
The quality of the HIV-1-specific T cell adaptive immune response was characterized in part by the pattern of cytokine production and its cytotoxic potential. Thus, on the basis of the production of CD107a, IFN-γ, TNF-α, and IL-2 from HIV-1-specific CD4+ and CD8+ T cells, 15 different HIV-1-specific CD4+ and CD8+ T cell populations could be identified (Fig. 4). As shown in Fig. 4A (pie charts), HIV-1-specific CD4+ T cell responses were similarly polyfunctional in groups immunized with DNA-B–MVA-B and DNA-B–MVA-B ΔN2L, with 41 and 44% of the CD4+ T cells exhibiting three and four functions, respectively. CD4+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2, TNF-α and IL-2, or only TNF-α were the most induced populations elicited by parental MVA-B and MVA-B ΔN2L. However, DNA-B–MVA-B ΔN2L induced a significantly greater percentage of most of the CD4+ T cells exhibiting four functions, three functions, two functions, or one function than MVA-B did (P < 0.001) (Fig. 4A, bars). On the other hand, as shown in Fig. 4B, DNA-B–MVA-B ΔN2L significantly enhanced the polyfunctionality profile of HIV-1-specific CD8+ T cell responses, with 86% of the CD8+ T cells exhibiting three or four functions, compared to DNA-B–MVA-B, where 59% of the CD8+ T cells exhibited three or four functions (Fig. 4B). CD8+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2 or CD107a, IFN-γ, and TNF-α were the most abundant populations elicited by parental MVA-B and MVA-B ΔN2L. However, DNA-B–MVA-B ΔN2L induced a significantly greater increase in the percentage of each of these populations than DNA-B–MVA-B did (P < 0.001) (Fig. 4B).
FIG 4.
Immunization with MVA-B ΔN2L enhances the polyfunctionality of HIV-1-specific CD4+ and CD8+ T cell adaptive immune responses. Functional profiles of HIV-1-specific CD4+ (A) and CD8+ (B) T cell adaptive immune responses elicited by immunization with DNA-ϕ–MVA-WT, DNA-B–MVA-B, and DNA-B–MVA-B ΔN2L. Values from unstimulated controls were subtracted in all cases. All of the possible combinations of responses are shown on the x axis (15 different T cell populations), while the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, and GPN peptide pools are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (four, three, two, or one). The pie charts summarize the data. Each slice corresponds to the proportion of the total HIV-1-specific CD4+ or CD8+ T cells exhibiting one, two, three, or four functions (CD107a and/or IFN-γ and/or TNF-α and/or IL-2) within the total HIV-1-specific CD4+ or CD8+ T cells. P values indicate significant response differences between the DNA-B–MVA-B ΔN2L and DNA-B–MVA-B immunization groups (*, P < 0.05; **, P < 0.005; ***, P < 0.001).
In summary, these results demonstrate that deletion of VACV gene N2L from the MVA-B genome enhanced the magnitude and quality of HIV-1-specific CD4+ and CD8+ T cell adaptive immune responses. Similar findings were obtained in two independent experiments.
MVA-B and MVA-B ΔN2L induce similar magnitudes and polyfunctionalities of vector VACV-specific T cell adaptive immune responses.
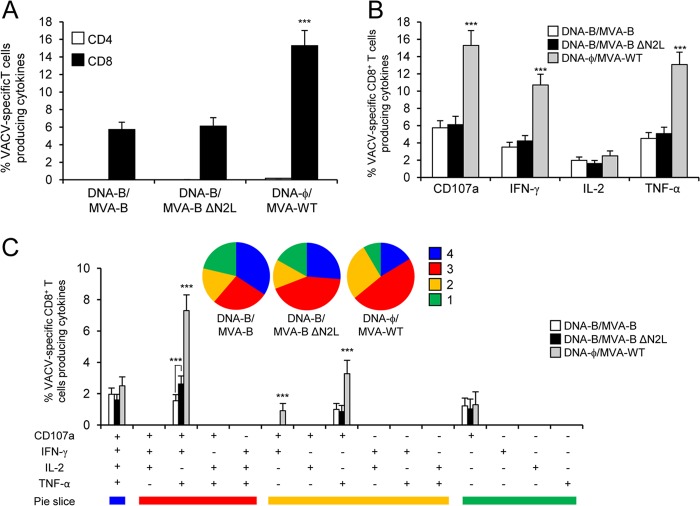
To further characterize the immune responses elicited in mice by the different immunization groups (DNA-B–MVA-B, DNA-B–MVA-B ΔN2L, and DNA-ϕ–MVA-WT), it was of interest to analyze the responses to the MVA vector. Thus, next we measured the VACV-specific T cell adaptive immune responses induced by MVA-WT, MVA-B, and MVA-B ΔN2L 10 days after the boost by ICS assay (similarly to the protocol followed in the analysis of HIV-1-specific T cell adaptive immune responses) and after the stimulation of splenocytes with MVA-infected A20 cells (a line of B cells that act as antigen-presenting cells).
All of the immunization groups triggered an overall VACV-specific adaptive immune response mediated only by CD8+ T cells (determined as the sum of the individual responses producing IFN-γ, TNF-α, and/or IL-2 cytokines, as well as the expression of CD107a on the surface of activated T cells as an indirect marker of cytotoxicity) and obtained with MVA-infected A20 cells (Fig. 5A). However, the magnitude of VACV-specific CD8+ T cell adaptive immune responses was significantly greater in the DNA-ϕ–MVA-WT immunization group than that elicited by DNA-B–MVA-B and DNA-B–MVA-B ΔN2L, with an increase of 2.5- to 2.6-fold (P < 0.001) (Fig. 5A) and no differences between the MVA-B and MVA-B ΔN2L HIV/AIDS vaccine candidates. This is likely due to the expression of HIV-1 antigens acting as competitors for immune dominance of MVA antigens.
FIG 5.
VACV-specific T cell adaptive immune responses. Splenocytes were collected from mice (n = 4 per group) immunized with DNA-ϕ–MVA-WT, DNA-B–MVA-B, or DNA-B–MVA-B ΔN2L 10 days after the last immunization. Next, VACV-specific CD4+ and CD8+ T cell adaptive immune responses triggered by the different immunization groups were measured by ICS assay following the stimulation of splenocytes with MVA-infected A20 cells. Values from unstimulated controls were subtracted in all cases. P values indicate significantly greater responses of the DNA-ϕ–MVA-WT immunization group than the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups or comparisons between DNA-B–MVA-B and DNA-B–MVA-B ΔN2L (***, P < 0.001). Data are from one experiment representative of two independent experiments. (A) Overall percentages of VACV-specific CD4+ and CD8+ T cells. The values represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected A20 cells. (B) Percentages of VACV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against MVA-infected A20 cells. (C) Functional profiles of VACV-specific CD8+ T cell adaptive immune responses. All of the possible combinations of responses are shown on the x axis (15 different T cell populations), while the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected A20 cells are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (four, three, two, or one). The pie charts summarize the data. Each slice corresponds to the proportion of the total VACV-specific CD8+ T cells exhibiting one, two, three, or four functions (CD107a and/or IFN-γ and/or TNF-α and/or IL-2) within the total VACV-specific CD8+ T cells.
Moreover, VACV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α were the most induced populations in all of the immunization groups (Fig. 5B). However, DNA-ϕ–MVA-WT induced a significantly greater magnitude of VACV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α than DNA-B–MVA-B and DNA-B–MVA-B ΔN2L did (P < 0.001) (Fig. 5B).
The quality of VACV-specific CD8+ T cell adaptive immune responses was characterized by the production of CD107a, IFN-γ, TNF-α, and/or IL-2, where 15 distinct VACV-specific CD8+ T cell populations could be identified (Fig. 5C). VACV-specific CD8+ T cell responses were similarly polyfunctional in all of the immunization groups, with 61 to 69% of the CD8+ T cells exhibiting three or four functions. CD8+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2; CD107a, IFN-γ, and TNF-α; CD107a and TNF-α; or only CD107a were the most induced populations elicited by all of the immunization groups. However, DNA-ϕ–MVA-WT induced a significantly greater percentage of most of these populations than DNA-B–MVA-B and DNA-B–MVA-B ΔN2L did (P < 0.001) (Fig. 5C).
Overall, these results showed that MVA-B and MVA-B ΔN2L elicited similar magnitudes and qualities of VACV-specific CD8+ T cell adaptive immune responses but with a lower percentage than those elicited by MVA-WT. Similar findings were obtained in two independent experiments.
MVA-B ΔN2L improved HIV-1-specific T cell memory immune responses.
Memory T cell responses might be critical for protection against HIV-1 infection (72–75), and the durability of a vaccine-induced T cell response is an important feature since long-term protection is a requirement for prophylactic vaccination. Thus, we analyzed HIV-1-specific T cell memory immune responses elicited by the different immunization groups 52 days after the boost by using the ICS assay and a protocol similar to that used in the adaptive phase.
The magnitude of HIV-1-specific CD4+ and CD8+ T cell memory immune responses, determined as the sum of the individual responses producing CD107a, IFN-γ, TNF-α, and/or IL-2 obtained with the Env, Gag, and GPN peptide pools, was significantly greater in the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups than that elicited by the DNA-ϕ–MVA-WT control group (P < 0.001) (Fig. 6A). Similar to the results obtained in the adaptive phase, the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups triggered an overall HIV-1-specific immune response mediated mainly by CD8+ T cells (>93%) (Fig. 6A). However, immunization with DNA-B–MVA-B ΔN2L again elicited a significantly greater magnitude of HIV-1-specific CD4+ and CD8+ T cell responses than DNA-B–MVA-B did, with increases of 2.4- and 2.23-fold, respectively (P < 0.001) (Fig. 6A).
FIG 6.
Immunization with MVA-B ΔN2L enhances the magnitude of HIV-1-specific CD4+ and CD8+ T cell memory immune responses. Splenocytes were collected from mice (n = 4 per group) immunized with DNA-ϕ–MVA-WT, DNA-B–MVA-B, or DNA-B–MVA-B ΔN2L 52 days after the last immunization. HIV-1-specific CD4+ and CD8+ T cell memory immune responses triggered by the different immunization groups were measured by ICS as described in the legend to Fig. 3. Values from unstimulated controls were subtracted in all cases. P values indicate significant response differences between the DNA-B–MVA-B ΔN2L and DNA-B–MVA-B immunization groups (***, P < 0.001). Data are from one experiment representative of two independent experiments. (A) Overall percentages of HIV-1-specific CD4+ and CD8+ T cells. The values represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, and GPN peptide pools. (B) Pattern of Env, Gag, and GPN HIV-1-specific CD4+ and CD8+ T cell memory immune responses. Frequencies were calculated by reporting the number of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 to the total number of CD4+ and CD8+ T cells in the different immunization groups. (C) Percentages of Env, Gag, and GPN HIV-1-specific CD4+ and CD8+ T cells. Frequencies represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, or GPN peptide pools. (D) Percentages of HIV-1-specific CD4+ and CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against Env, Gag, and GPN peptide pools.
The pattern of HIV-1-specific T cell memory immune responses showed that CD4+ and CD8+ T cell responses were directed mainly against the Env pool in both the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups, with CD8+ T cell responses broadly distributed among Env, GPN, and Gag (Fig. 6B), similar to the results obtained in the adaptive phase. However, again DNA-B–MVA-B ΔN2L significantly enhanced the magnitude of Env-specific CD4+ T cell memory responses and Env-, Gag- and GPN-specific CD8+ T cell memory responses (P < 0.001) (Fig. 6C).
Furthermore, HIV-1-specific CD4+ T memory cells producing TNF-α or IL-2 and HIV-1-specific CD8+ T memory cells producing CD107a, IFN-γ, or TNF-α immunomodulatory molecules are the most induced populations in both immunization groups (Fig. 6D), similar to the results obtained in the adaptive phase. However, again DNA-B–MVA-B ΔN2L induced a significantly greater magnitude of HIV-1-specific CD4+ and CD8+ T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2 than DNA-B–MVA-B did (P < 0.001) (Fig. 6D).
The quality of the HIV-1-specific T cell memory immune responses was characterized by analyzing the simultaneous production of CD107a, IFN-γ, TNF-α, and/or IL-2 from HIV-1-specific CD4+ and CD8+ T cells (Fig. 7), where 15 distinct HIV-1-specific CD4+ and CD8+ T cell populations could be identified. As shown in Fig. 7A, HIV-1-specific CD4+ T cell memory responses were similarly polyfunctional in groups immunized with DNA-B–MVA-B and DNA-B–MVA-B ΔN2L, with 38 and 40% of the CD4+ T cells exhibiting three or four functions, respectively. Again, CD4+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2; TNF-α and IL-2; or only TNF-α were the most induced populations elicited by parental MVA-B and MVA-B ΔN2L. However, DNA-B–MVA-B ΔN2L induced a significantly greater percentage of most of the CD4+ T cells exhibiting four functions, three functions, two functions, or one function than MVA-B did (P < 0.001) (Fig. 7A), similar to the results obtained in the adaptive phase. On the other hand, as shown in Fig. 7B, HIV-1-specific CD8+ T cell memory responses were highly and similarly polyfunctional in groups immunized with DNA-B–MVA-B and DNA-B–MVA-B ΔN2L (90% of the CD8+ T cells exhibiting three or four functions in both groups). Again, CD8+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2 or CD107a, IFN-γ, and TNF-α were the most abundant populations elicited by parental MVA-B and MVA-B ΔN2L. However, DNA-B–MVA-B ΔN2L induced a significantly greater increase in the percentage of each of these populations than DNA-B–MVA-B did (P < 0.001) (Fig. 7B).
FIG 7.
Immunization with MVA-B ΔN2L enhances the polyfunctionality of HIV-1-specific CD4+ and CD8+ T cell memory immune responses. Functional profiles of HIV-1-specific CD4+ (A) and CD8+ (B) T cell memory immune responses elicited by immunization with DNA-ϕ–MVA-WT, DNA-B–MVA-B, and DNA-B–MVA-B ΔN2L. Values from unstimulated controls were subtracted in all cases. All of the possible combinations of responses are shown on the x axis (15 different T cell populations), while the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against Env, Gag, and GPN peptide pools are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (four, three, two, or one). The pie charts summarize the data. Each slice corresponds to the proportion of the total HIV-1-specific CD4+ or CD8+ T cells exhibiting one, two, three, or four functions (CD107a and/or IFN-γ and/or TNF-α and/or IL-2) within the total HIV-1-specific CD4+ or CD8+ T cells. P values indicate significant response differences between the DNA-B–MVA-B ΔN2L and DNA-B–MVA-B immunization groups (***, P < 0.001).
Moreover, we also determined the phenotype of the Env-, Gag-, and GPN-specific CD4+ and CD8+ T memory cells by measuring the expression of the CD127 and CD62L surface markers, which allow the definition of the different memory subpopulations: central memory (TCM, CD127+/CD62L+), effector memory (TEM, CD127+/CD62L−), and effector (TE, CD127-/CD62L−) T cells (76) (Fig. 8). The results showed that in the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups, HIV-1-specific CD4+ and CD8+ T memory cells (determined as the sum of the individual responses producing CD107a, IFN-γ, TNF-α, and/or IL-2 obtained for the Env, Gag, and GPN peptide pools) were mainly of the TEM phenotype (Fig. 8, bottom part), but immunization with MVA-B ΔN2L induced a significantly greater increase in the percentage of HIV-1-specific CD4+ and CD8+ TEM and TE cells than immunization with MVA-B did (P < 0.001) (Fig. 8, upper part).
FIG 8.
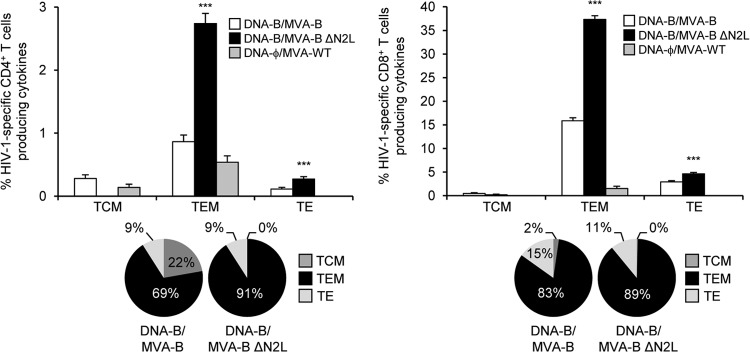
Phenotypic profile of memory HIV-1-specific CD4+ and CD8+ T cells. Percentages of TCM, TEM, and TE HIV-1-specific CD4+ and CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of TCM, TEM, and TE memory populations producing CD107a, IFN-γ, TNF-α, or IL-2 against Env, Gag, and GPN peptide pools. Values from unstimulated controls were subtracted in all cases. The pie charts summarize the data. Each slice corresponds to the proportion of TCM, TEM, and TE memory populations within the total HIV-1-specific CD4+ or CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. P values indicate significant response differences between the DNA-B–MVA-B ΔN2L and DNA-B–MVA-B immunization groups (***, P < 0.001).
In summary, these results demonstrate that deletion of VACV gene N2L from the MVA-B genome enhanced the magnitude and quality of HIV-1-specific CD4+ and CD8+ T cell memory immune responses, with an increase in the percentage of TEM and TE cells. Similar findings were obtained in two independent experiments.
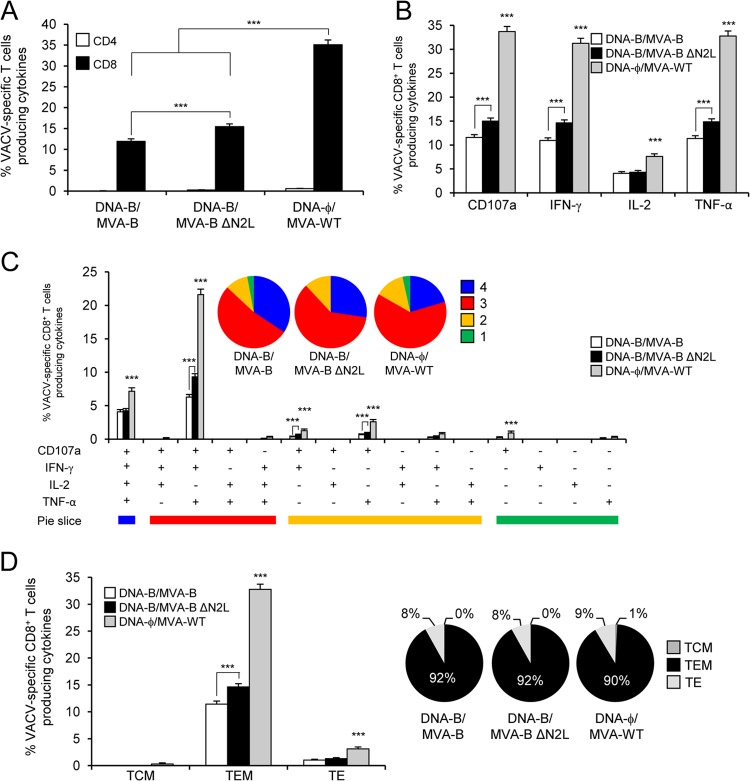
MVA-B ΔN2L improved vector VACV-specific T cell memory immune responses.
The VACV-specific T cell memory immune responses elicited by the different immunization groups were measured by ICS assay 52 days after the boost. Similar to the results obtained in the adaptive phase, all of the immunization groups triggered an overall VACV-specific memory immune responses mediated mainly by CD8+ T cells, determined as the sum of the individual responses producing CD107a, IFN-γ, TNF-α, and/or IL-2 obtained with MVA-infected A20 cells (Fig. 9A). However, the magnitude of VACV-specific CD8+ T cell memory immune responses in the DNA-ϕ–MVA-WT immunization group was significantly greater than that elicited by DNA-B–MVA-B and DNA-B–MVA-B ΔN2L, with increases of 2.94- and 2.27-fold, respectively (P < 0.001) (Fig. 9A). Furthermore, DNA-B–MVA-B ΔN2L induced a significantly greater magnitude of VACV-specific CD8+ T cell memory immune responses than DNA-B–MVA-B, with an increase of 1.3-fold (P < 0.001) (Fig. 9A).
FIG 9.
VACV-specific T cell memory immune responses. Splenocytes were collected from mice (n = 4 per group) immunized with DNA-ϕ–MVA-WT, DNA-B–MVA-B, or DNA-B–MVA-B ΔN2L 52 days after the last immunization. Next, VACV-specific CD4+ and CD8+ T cell memory immune responses triggered by the different immunization groups were measured by ICS assay following the stimulation of splenocytes with MVA-infected A20 cells. Values from unstimulated controls were subtracted in all cases. P values indicate significantly greater responses of the DNA-ϕ–MVA-WT immunization group than the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups or comparisons between DNA-B–MVA-B and DNA-B–MVA-B ΔN2L (***, P < 0.001). Data are from one experiment representative of two independent experiments. (A) Overall percentages of VACV-specific CD4+ and CD8+ T memory cells. The values represent the sum of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected A20 cells. (B) Percentage of VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2 against MVA-infected A20 cells. (C) Functional profiles of VACV-specific CD8+ T cell memory immune responses. Responses are grouped and color coded on the basis of the number of functions (four, three, two, or one). All of the possible combinations of responses are shown on the x axis (15 different T cell populations), while the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected A20 cells are shown on the y axis. The pie charts summarize the data. Each slice corresponds to the proportion of VACV-specific CD8+ T memory cells exhibiting one, two, three, or four functions (CD107a and/or IFN-γ and/or TNF-α and/or IL-2) within the total VACV-specific CD8+ T cells. (D) Percentages of TCM, TEM, and TE VACV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of TCM, TEM, and TE memory populations producing CD107a, IFN-γ, TNF-α, or IL-2 against MVA-infected A20 cells. The pie charts summarize the data. Each slice corresponds to the proportion of TCM, TEM, and TE memory populations within the total VACV-specific CD4+ or CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2.
Moreover, VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, or TNF-α are the most induced populations in all of the immunization groups (Fig. 9B). However, DNA-ϕ–MVA-WT induced a significantly greater magnitude of VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2 than DNA-B–MVA-B and DNA-B–MVA-B ΔN2L did (P < 0.001) (Fig. 9B). Furthermore, DNA-B–MVA-B ΔN2L induced a significantly greater magnitude of VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, or TNF-α than DNA-B–MVA-B did (P < 0.001) (Fig. 9B).
The quality of VACV-specific CD8+ T cell memory immune responses was characterized by the simultaneous production of CD107a, IFN-γ, TNF-α, and/or IL-2, similar to the protocol followed in the adaptive phase, where 15 distinct VACV-specific CD8+ T cell populations could be identified (Fig. 9C). VACV-specific CD8+ T cell responses were highly and similarly polyfunctional in all of the immunization groups, with 83 to 88% of the CD8+ T cells exhibiting three or four functions. CD8+ T cells producing CD107a, IFN-γ, TNF-α, and IL-2; CD107a, IFN-γ, and TNF-α; or CD107a and TNF-α were the most induced populations elicited by all of the immunization groups. However, DNA-ϕ–MVA-WT induced a significantly greater percentage of these populations than DNA-B–MVA-B and DNA-B–MVA-B ΔN2L did (P < 0.001) (Fig. 9C). Furthermore, DNA-B–MVA-B ΔN2L induced a significantly greater magnitude of triple and double VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2 than DNA-B–MVA-B did (P < 0.001) (Fig. 9C).
Additionally, the analysis of the memory phenotype of VACV-specific CD8+ T memory cells by measuring the expression of the CD127 and CD62L surface markers showed that all of the immunization groups elicited mainly VACV-specific CD8+ T memory cells of the TEM phenotype (Fig. 9D), but immunization with MVA-WT induced a significantly greater percentage of VACV-specific CD8+ TEM and TE cells than MVA-B and MVA-B ΔN2L did (P < 0.001) (Fig. 9D). Furthermore, MVA-B ΔN2L elicited a significantly higher percentage of VACV-specific CD8+ TEM cells than MVA-B did (P < 0.001) (Fig. 9D).
In summary, these results show that MVA-B ΔN2L elicited a greater magnitude and quality of VACV-specific CD8+ T cell memory immune responses than MVA-B did, with also an increase in the percentage of TEM cells, but both immunization groups elicited a lower percentage than MVA-WT did. Similar findings were obtained in two independent experiments.
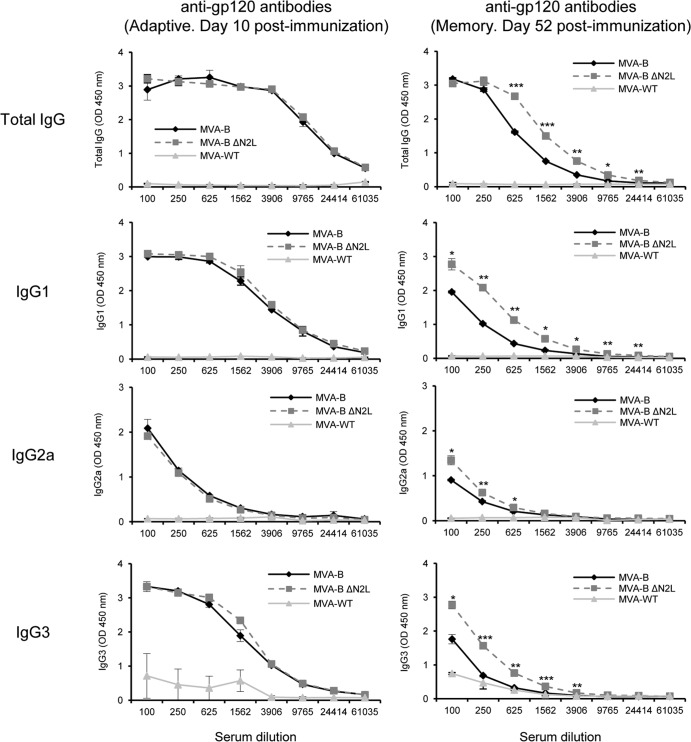
MVA-B ΔN2L enhances the levels of antibodies against HIV-1 gp120 in the memory phase.
Since cells infected with MVA-B release monomeric gp120 (60) and both the cellular and humoral arms of the immune system are thought to be necessary to control HIV-1 infection (77), we analyzed the humoral responses elicited after immunization with DNA-B–MVA-B and DNA-B–MVA-B ΔN2L, quantifying by ELISA the total IgG and subclass IgG1, IgG2a, and IgG3 levels of antibodies against HIV-1 Env (clade B) in pooled sera obtained from mice 10 and 52 days postboost (Fig. 10). The results showed that in the group boosted with MVA-B ΔN2L, the levels of total IgG, IgG1, IgG2a, and IgG3 anti-gp120 antibodies were significantly higher than those obtained in animals immunized with parental MVA-B only in the memory phase. These results indicate that MVA-B and MVA-B ΔN2L induced antibodies against HIV-1 gp120, and deletion of VACV gene N2L produces in mice an increase of around 2-fold in the humoral memory immune response to HIV-1 Env.
FIG 10.
Humoral immune responses elicited by MVA-B and MVA-B ΔN2L against HIV-1 gp120 protein. Levels of gp120-specific total IgG, IgG1, IgG2a, and IgG3 binding antibodies were measured by ELISA in pooled sera from mice immunized with DNA-B–MVA-B, DNA-B–MVA-B ΔN2L or DNA-ϕ–MVA-WT (n = 4 at each time point) 10 days (left panels) or 52 days (right panels) after the last immunization. Mean absorbance values (measured at 450 nm) and standard deviations of duplicate pooled serum dilutions are presented. P values indicate significant differences in antibody levels between the DNA-B–MVA-B and DNA-B–MVA-B ΔN2L immunization groups (*, P < 0.05; **, P < 0.005; ***, P < 0.001). Data are from one experiment representative of two independent experiments.
DISCUSSION
Poxvirus vectors are promising HIV/AIDS vaccine candidates, and several recombinant poxvirus vectors (such as the MVA, NYVAC, canarypox, and fowlpox viruses) expressing different HIV-1 antigens have been widely used in many human clinical trials, proving that they are safe and immunogenic, inducing HIV-1-specific humoral and cellular immune responses (for reviews, see references 3–5 and 7). In fact, the canarypox ALVAC combined with HIV-1 gp120 protein is actually the only HIV/AIDS vaccine candidate that is able to trigger a 31.2% protective effect (1). However, this efficacy is modest, and the immunogenicity against HIV-1 antigens induced by all of the poxvirus vector-based HIV vaccines tested in human clinical trials is limited and restricted. Thus, new, more efficient, and optimized poxvirus vector-based HIV vaccines able to enhance humoral and cellular immunogenicity against HIV-1 antigens are needed. Different approaches have been developed to improve vector immunogenicity, such as the use of attenuated, replication-competent poxvirus vectors; combination of heterologous vectors; the use of costimulatory molecules, and removal of VACV viral genes encoding immunosuppressive molecules (reviewed in reference 3). The latter is one of the most promising strategies actually developed.
Several MVA deletion mutants containing deletions in different immunomodulatory VACV genes have been generated and analyzed in mice (42, 62, 63, 78–80) and nonhuman primates (81, 82). These studies revealed that MVA vectors with deletions of single VACV genes encoding inhibitors of the type I IFN signaling pathway (C6L [42]), apoptosis (F1L [80]), and IL-18 binding protein (C12L [79]) or the uracyl-DNA glycosylase gene (UDG [82]) enhanced the overall immune responses to HIV-1 antigens. The HIV-1-specific CD4+ and CD8+ T cell immune responses were further enhanced by MVA vectors with deletions of two (secreted CC-chemokine binding protein [A41L] and secreted IL-1β receptor [B16R] [63] or inhibitors of the type I IFN signaling pathway C6L and K7R [62]) or four (IL-18 binding protein [C12L], Toll/IL-1 receptor homolog [A46R], CC-chemokine binding protein [B7R], and the secreted IL-1β receptor [B16R]) VACV immunomodulatory genes (81), but an additional fifth deletion of the uracil-DNA glycosylase gene (MVA101R) decreased the responses (81). Moreover, NYVAC vectors with a single deletion of a VACV gene encoding a TLR inhibitor (A46R [83]) and with single or double deletions in VACV genes encoding type I and II IFN binding proteins (B19R and B8R, respectively [84–86]) were also able to enhance the immune responses to HIV-1 antigens in mice.
Thus, as a way to try to improve the immune responses to HIV-1 antigens by selective deletion of VACV immunomodulatory genes in a poxvirus vector-based HIV vaccine candidate, we focused this work on VACV N2L, a gene whose product we predicted by bioinformatic comparison to belong to a family of VACV proteins with a Bcl-2 fold and with an immunomodulatory role (30). Recently, it has been demonstrated that VACV N2L encodes a nuclear inhibitor of IRF3 that blocks the IFN I signaling pathway at the intracellular level (59). As part of our effort to find out the immunomodulatory role of the VACV Bcl-2 family of proteins (30), here we deleted the VACV N2L gene from an MVA vector-based HIV/AIDS vaccine candidate termed MVA-B, which expresses HIV-1 Env, Gag, Pol, and Nef antigens from clade B (60) and has been well characterized in vitro (61, 65), in mice (42, 60, 62–64), in nonhuman primates (66), and in a phase I clinical trial in humans (12, 14), showing that MVA-B is a promising HIV/AIDS vaccine candidate that is safe, highly immunogenic, and able to induce broad, polyfunctional, and durable CD4+ and CD8+ T cell responses to HIV-1 antigens, together with humoral responses to Env.
To examine the immunomodulatory role of N2L, we used mice immunized in DNA prime-MVA boost protocols to compare the innate, adaptive, and memory immune responses to HIV-1 antigens induced by parental MVA-B and the deletion mutant MVA-B ΔN2L. First, our results showed that deletion of the VACV N2L gene from the MVA-B genome has no effect on the replication capacity of the virus in cultured cells, showing that N2L is not essential for viral replication; similar behavior was observed when N2L was deleted from the WR strain (59). Moreover, this deletion has no effect on the expression of gp120 and GPN HIV-1 antigens from MVA-B. Furthermore, MVA-B ΔN2L prompts the upregulation in infected human macrophages and moDCs of type I IFN (IFN-β), the proinflammatory cytokine TNF-α, chemokines (MIP-1α and RANTES), and some key cytosolic sensors that lead to antiviral IFN production (RIG-I and MDA-5), revealing that the innate immune responses are enhanced by selective deletion of the VACV N2L gene and confirming the role of the VACV N2 protein in blocking the IFN-I signaling pathway (59). Remarkably, compared to MVA-B, the deletion mutant MVA-B ΔN2L significantly enhanced the adaptive and memory HIV-1-specific T cell immune responses. In both immunization groups, HIV-1-specific CD4+ and CD8+ T cells were activated, but with a higher frequency of HIV-1-specific CD8+ T cells, similar to previous results with MVA or NYVAC vectors containing deletions in VACV genes (42, 62, 63, 80, 84). However, MVA-B ΔN2L significantly enhanced the magnitude of the overall adaptive and memory HIV-1-specific CD4+ and CD8+ T cell responses compared to MVA-B. Furthermore, both immunization groups elicited similar polyfunctional profiles of adaptive and memory HIV-1-specific CD4+ and CD8+ T cells, but MVA-B ΔN2L significantly enhanced the magnitude of these polyfunctional HIV-1-specific CD4+ and CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. In the memory phase, both immunization groups elicited an HIV-1-specific T cell response mainly due to the TEM phenotype, but MVA-B ΔN2L significantly enhanced the magnitude of these TEM cells, an important and relevant feature because the presence of TEM has been correlated with protection in the macaque-simian immunodeficiency virus model (87, 88). Interestingly, both immunization groups were able to induce HIV-1-specific CD4+ and CD8+ T cell responses of TEM and TE phenotypes in the adaptive phase (data not shown), with again MVA-B ΔN2L significantly enhancing the magnitude of CD4+ and CD8+ TEM and TE cells. The fast acquisition of TEM and TE phenotypes in the adaptive phase could be important in developing the T cell memory responses and mounting more effective immunity during a primary pathogen encounter. Moreover, adaptive and memory CD4+ T cell immune responses were directed mainly against Env in both immunization groups. In contrast to other MVA-B deletion mutants previously characterized, where a pattern of GPN-specific CD8+ T cell immune responses was induced (42, 62, 63), MVA-B ΔN2L triggered CD8+ T cell immune responses preferentially directed against Env. The biological relevance of this shift is not known. Moreover, a similar CD8+ T cell immune response to Gag and GPN pools was observed after immunization with MVA-B and MVA-B ΔN2L in the memory phase, indicating that the response to the GPN region is directed mainly to Gag. Furthermore, this enhanced HIV-1-specific CD4+ and CD8+ T cell response elicited after the immunization of mice with MVA-B ΔN2L is similar to what we have previously reported after deleting from the MVA-B genome other VACV genes that block the IFN-I signaling pathway at the intracellular level, such as C6L (42, 62) or K7R (62), or when we deleted other VACV genes, such as A41L and B16R (63). These findings once more reinforce the deletion strategy as a useful approach to improving the HIV-1-specific immune responses elicited by poxvirus vector-based HIV/AIDS vaccine candidates.
On B cell immune activation, MVA-B ΔN2L induced higher levels of total IgG, IgG1, IgG2a, and IgG3 anti-gp120 humoral responses than MVA-B did in the memory phase. This enhancement may be mediated by the increase in innate immune responses and HIV-1-specific CD4+ T helper cells triggered by the MVA-B ΔN2L deletion mutant. Thus, the generation of antibodies against gp120 by MVA-B ΔN2L is a positive immune parameter. In fact, a humoral response to V1/V2 loops of HIV-1 gp120 has been correlated with efficacy in the RV144 phase III clinical trial (89).
In addition to an evaluation of the immune responses to HIV-1 antigens, we have also analyzed the adaptive and memory specific immune responses directed against the poxvirus vector. The results showed that in the adaptive phase, immunization with MVA-B and MVA-B ΔN2L elicited similar VACV-specific T cell immune responses due mainly to CD8+ T cells, although the values were significantly lower than those elicited in the control group by MVA-WT. However, in the memory phase, MVA-B ΔN2L induced a significantly greater magnitude of VACV-specific CD8+ T cell responses than MVA-B did, but again, the response was significantly weaker than that elicited by MVA-WT. Similar results were also obtained by analyzing the response to VACV peptides E3 and F2 (data not shown). The fact that immunization with MVA-WT elicited greater adaptive and memory VACV-specific immune responses than MVA-B and MVA-B ΔN2L did could be related to competition between the HIV-1 and VACV antigens, diminishing the overall VACV-specific immune responses. On the other hand, the increase in memory VACV-specific immune responses by MVA-B ΔN2L, compared to MVA-B, could reflect a long-term role of the VACV N2L gene, increasing both VACV and HIV-1 memory immune responses.
How does the deletion of N2L affect the immune responses elicited by MVA-B? As recently reported, N2L is a nuclear inhibitor of IRF3 (59) and therefore acts on the IFN signaling pathway, inhibiting this important route involved in the host antiviral response (27–29). Thus, once N2L is deleted from the MVA genome, the IFN signaling pathway should be reestablished upon viral infection, and consequently an upregulation of type I IFN, proinflammatory cytokines, and chemokines should be produced, which will finally activate the antigen-specific T cells. As predicted, this phenomenon was clearly observed in our study, where enhanced expression of type I IFN, proinflammatory cytokines, and chemokines was observed after the infection of human macrophages and moDCs with MVA with the VACV N2L gene deleted. Moreover, the enhanced adaptive and memory HIV-1-specific T cell immune responses elicited in mice by the MVA-B ΔN2L deletion mutant could be explained by the biological role of N2L. Thus, after the i.p. inoculation of mice with MVA-B ΔN2L, the enhanced secretion of type I IFN, proinflammatory cytokines, and chemokines induced by MVA-B ΔN2L-infected macrophages could prompt a greater recruitment of immature DCs and lymphocytes, producing a suitable environment for the uptake and presentation of the HIV-1 antigens to T cells. Immature MVA-B ΔN2L-infected DCs then migrate to the lymph nodes, maturing en route, and then activate HIV-1-specific T cells, increasing immunogenicity against HIV-1 antigens. In agreement with this, it has been recently reported that on days 5 and 7 postinfection, the total number of cells in the bronchoalveolar lavage fluid of mice immunized intranasally with a VACV WR N2L deletion mutant was higher than in that of mice immunized with the parental virus (59).
While the WR strain with N2L deleted is attenuated in mice and is not more immunogenic than the parental virus (59), however, the MVA-B ΔN2L deletion mutant generated in this study is able to enhance immunogenicity against HIV-1 and VACV antigens. This could be due to the difference between the two vectors in the number of immunomodulatory genes (90–92).
Overall, our findings have revealed the immunomodulatory role of N2L, proving that its deletion from the MVA-B vector led to an enhanced innate immune response and improvements in the magnitude and quality of adaptive and memory HIV-1-specific CD4+ and CD8+ T cell immune responses. The selective deletion of the N2L viral immunomodulatory gene is important for the optimization of MVA vectors as HIV-1 vaccines.
ACKNOWLEDGMENTS
This investigation was supported by grants from Spain, SAF2008-02036, FIPSE 36-0731-09, and Red Sida RD12/0017/0038.
We are grateful to NIBSC (United Kingdom) for Gag antibodies. We thank Victoria Jimenez for tissue culture and virus growth.
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220. 10.1056/NEJMoa0908492 [DOI] [PubMed] [Google Scholar]
- 2.Russell ND, Graham BS, Keefer MC, McElrath MJ, Self SG, Weinhold KJ, Montefiori DC, Ferrari G, Horton H, Tomaras GD, Gurunathan S, Baglyos L, Frey SE, Mulligan MJ, Harro CD, Buchbinder SP, Baden LR, Blattner WA, Koblin BA, Corey L. 2007. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J. Acquir. Immune Defic. Syndr. 44:203–212. 10.1097/01.qai.0000248356.48501.ff [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gómez CE, Perdiguero B, García-Arriaza J, Esteban M. 2012. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum. Vaccin. Immunother. 8:1192–1207. 10.4161/hv.20778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell RJ, Kim JH, Corey L, Michael NL. 2012. Human immunodeficiency virus vaccine trials. Cold Spring Harb. Perspect. Med. 2:a007351. 10.1101/cshperspect.a007351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pantaleo G, Esteban M, Jacobs B, Tartaglia J. 2010. Poxvirus vector-based HIV vaccines. Curr. Opin. HIV AIDS 5:391–396. 10.1097/COH.0b013e32833d1e87 [DOI] [PubMed] [Google Scholar]
- 6.Cottingham MG, Carroll MW. 2013. Recombinant MVA vaccines: dispelling the myths. Vaccine 31:4247–4251. 10.1016/j.vaccine.2013.03.021 [DOI] [PubMed] [Google Scholar]
- 7.Gómez CE, Perdiguero B, García-Arriaza J, Esteban M. 2013. Clinical applications of attenuated MVA poxvirus strain. Expert Rev. Vaccines 12:1395–1416. 10.1586/14760584.2013.845531 [DOI] [PubMed] [Google Scholar]
- 8.Aboud S, Nilsson C, Karlen K, Marovich M, Wahren B, Sandström E, Gaines H, Biberfeld G, Godoy-Ramírez K. 2010. Strong HIV-specific CD4+ and CD8+ T-lymphocyte proliferative responses in healthy individuals immunized with an HIV-1 DNA vaccine and boosted with recombinant modified vaccinia virus Ankara expressing HIV-1 genes. Clin. Vaccine Immunol. 17:1124–1131. 10.1128/CVI.00008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakari M, Aboud S, Nilsson C, Francis J, Buma D, Moshiro C, Aris EA, Lyamuya EF, Janabi M, Godoy-Ramírez K, Joachim A, Polonis VR, Brave A, Earl P, Robb M, Marovich M, Wahren B, Pallangyo K, Biberfeld G, Mhalu F, Sandström E. 2011. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine 29:8417–8428. 10.1016/j.vaccine.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cebere I, Dorrell L, McShane H, Simmons A, McCormack S, Schmidt C, Smith C, Brooks M, Roberts JE, Darwin SC, Fast PE, Conlon C, Rowland-Jones S, McMichael AJ, Hanke T. 2006. Phase I clinical trial safety of DNA- and modified virus Ankara-vectored human immunodeficiency virus type 1 (HIV-1) vaccines administered alone and in a prime-boost regime to healthy HIV-1-uninfected volunteers. Vaccine 24:417–425. 10.1016/j.vaccine.2005.08.041 [DOI] [PubMed] [Google Scholar]
- 11.Currier JR, Ngauy V, de Souza MS, Ratto-Kim S, Cox JH, Polonis VR, Earl P, Moss B, Peel S, Slike B, Sriplienchan S, Thongcharoen P, Paris RM, Robb ML, Kim J, Michael NL, Marovich MA. 2010. Phase I safety and immunogenicity evaluation of MVA-CMDR, a multigenic, recombinant modified vaccinia Ankara-HIV-1 vaccine candidate. PLoS One 5:e13983. 10.1371/journal.pone.0013983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García F, Bernaldo de Quiros JC, Gómez CE, Perdiguero B, Nájera JL, Jimenez V, García-Arriaza J, Guardo AC, Perez I, Diaz-Brito V, Conde MS, González N, Alvarez A, Alcami J, Jimenez JL, Pich J, Arnaiz JA, Maleno MJ, Leon A, Munoz-Fernandez MA, Liljestrom P, Weber J, Pantaleo G, Gatell JM, Plana M, Esteban M. 2011. Safety and immunogenicity of a modified pox vector-based HIV/AIDS vaccine candidate expressing Env, Gag, Pol, and Nef proteins of HIV-1 subtype B (MVA-B) in healthy HIV-1-uninfected volunteers: A phase I clinical trial (RISVAC02). Vaccine 29:8309–8316. 10.1016/j.vaccine.2011.08.098 [DOI] [PubMed] [Google Scholar]
- 13.Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, Hural J, DeRosa SC, DeFawe OD, Tomaras GD, Montefiori DC, Xu Y, Lai L, Kalams SA, Baden LR, Frey SE, Blattner WA, Wyatt LS, Moss B, Robinson HL. 2011. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J. Infect. Dis. 203:610–619. 10.1093/infdis/jiq105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez CE, Nájera JL, Perdiguero B, García-Arriaza J, Sorzano CO, Jimenez V, González-Sanz R, Jimenez JL, Munoz-Fernandez MA, Lopez Bernaldo de Quiros JC, Guardo AC, García F, Gatell JM, Plana M, Esteban M. 2011. The HIV/AIDS vaccine candidate MVA-B administered as a single immunogen in humans triggers robust, polyfunctional, and selective effector memory T cell responses to HIV-1 antigens. J. Virol. 85:11468–11478. 10.1128/JVI.05165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goonetilleke N, Moore S, Dally L, Winstone N, Cebere I, Mahmoud A, Pinheiro S, Gillespie G, Brown D, Loach V, Roberts J, Guimarães-Walker A, Hayes P, Loughran K, Smith C, De Bont J, Verlinde C, Vooijs D, Schmidt C, Boaz M, Gilmour J, Fast P, Dorrell L, Hanke T, McMichael AJ. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J. Virol. 80:4717–4728. 10.1128/JVI.80.10.4717-4728.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudmundsdotter L, Nilsson C, Brave A, Hejdeman B, Earl P, Moss B, Robb M, Cox J, Michael N, Marovich M, Biberfeld G, Sandström E, Wahren B. 2009. Recombinant modified vaccinia Ankara (MVA) effectively boosts DNA-primed HIV-specific immune responses in humans despite pre-existing vaccinia immunity. Vaccine 27:4468–4474. 10.1016/j.vaccine.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimarães-Walker A, Mackie N, McCormack S, Hanke T, Schmidt C, Gilmour J, Barin B, McMichael A, Weber J, Legg K, Babiker A, Hayes P, Gotch F, Smith C, Dally L, Dorrell L, Cebere I, Kay R, Winstone N, Moore S, Goonetilleke N, Fast P. 2008. Lessons from IAVI-006, a phase I clinical trial to evaluate the safety and immunogenicity of the pTHr.HIVA DNA and MVA.HIVA vaccines in a prime-boost strategy to induce HIV-1 specific T-cell responses in healthy volunteers. Vaccine 26:6671–6677. 10.1016/j.vaccine.2008.09.016 [DOI] [PubMed] [Google Scholar]
- 18.Jaoko W, Nakwagala FN, Anzala O, Manyonyi GO, Birungi J, Nanvubya A, Bashir F, Bhatt K, Ogutu H, Wakasiaka S, Matu L, Waruingi W, Odada J, Oyaro M, Indangasi J, Ndinya-Achola J, Konde C, Mugisha E, Fast P, Schmidt C, Gilmour J, Tarragona T, Smith C, Barin B, Dally L, Johnson B, Muluubya A, Nielsen L, Hayes P, Boaz M, Hughes P, Hanke T, McMichael A, Bwayo J, Kaleebu P. 2008. Safety and immunogenicity of recombinant low-dosage HIV-1 A vaccine candidates vectored by plasmid pTHr DNA or modified vaccinia virus Ankara (MVA) in humans in East Africa. Vaccine 26:2788–2795. 10.1016/j.vaccine.2008.02.071 [DOI] [PubMed] [Google Scholar]
- 19.Keefer MC, Frey SE, Elizaga M, Metch B, De Rosa SC, Barroso PF, Tomaras G, Cardinali M, Goepfert P, Kalichman A, Philippon V, McElrath MJ, Jin X, Ferrari G, Defawe OD, Mazzara GP, Montefiori D, Pensiero M, Panicali DL, Corey L. 2011. A phase I trial of preventive HIV vaccination with heterologous poxviral-vectors containing matching HIV-1 inserts in healthy HIV-uninfected subjects. Vaccine 29:1948–1958. 10.1016/j.vaccine.2010.12.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehendale S, Thakar M, Sahay S, Kumar M, Shete A, Sathyamurthi P, Verma A, Kurle S, Shrotri A, Gilmour J, Goyal R, Dally L, Sayeed E, Zachariah D, Ackland J, Kochhar S, Cox JH, Excler JL, Kumaraswami V, Paranjape R, Ramanathan VD. 2013. Safety and immunogenicity of DNA and MVA HIV-1 subtype C vaccine prime-boost regimens: a phase I randomised trial in HIV-uninfected Indian volunteers. PLoS One 8:e55831. 10.1371/journal.pone.0055831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mwau M, Cebere I, Sutton J, Chikoti P, Winstone N, Wee EG, Beattie T, Chen YH, Dorrell L, McShane H, Schmidt C, Brooks M, Patel S, Roberts J, Conlon C, Rowland-Jones SL, Bwayo JJ, McMichael AJ, Hanke T. 2004. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 85:911–919. 10.1099/vir.0.19701-0 [DOI] [PubMed] [Google Scholar]
- 22.Peters BS, Jaoko W, Vardas E, Panayotakopoulos G, Fast P, Schmidt C, Gilmour J, Bogoshi M, Omosa-Manyonyi G, Dally L, Klavinskis L, Farah B, Tarragona T, Bart PA, Robinson A, Pieterse C, Stevens W, Thomas R, Barin B, McMichael AJ, McIntyre JA, Pantaleo G, Hanke T, Bwayo J. 2007. Studies of a prophylactic HIV-1 vaccine candidate based on modified vaccinia virus Ankara (MVA) with and without DNA priming: effects of dosage and route on safety and immunogenicity. Vaccine 25:2120–2127. 10.1016/j.vaccine.2006.11.016 [DOI] [PubMed] [Google Scholar]
- 23.Ramanathan VD, Kumar M, Mahalingam J, Sathyamoorthy P, Narayanan PR, Solomon S, Panicali D, Chakrabarty S, Cox J, Sayeed E, Ackland J, Verlinde C, Vooijs D, Loughran K, Barin B, Lombardo A, Gilmour J, Stevens G, Smith MS, Tarragona-Fiol T, Hayes P, Kochhar S, Excler JL, Fast P. 2009. A phase 1 study to evaluate the safety and immunogenicity of a recombinant HIV type 1 subtype C-modified vaccinia Ankara virus vaccine candidate in Indian volunteers. AIDS Res. Hum. Retroviruses 25:1107–1116. 10.1089/aid.2009.0096 [DOI] [PubMed] [Google Scholar]
- 24.Sandström E, Nilsson C, Hejdeman B, Brave A, Bratt G, Robb M, Cox J, Vancott T, Marovich M, Stout R, Aboud S, Bakari M, Pallangyo K, Ljungberg K, Moss B, Earl P, Michael N, Birx D, Mhalu F, Wahren B, Biberfeld G. 2008. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J. Infect. Dis. 198:1482–1490. 10.1086/592507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasan S, Schlesinger SJ, Chen Z, Hurley A, Lombardo A, Than S, Adesanya P, Bunce C, Boaz M, Boyle R, Sayeed E, Clark L, Dugin D, Boente-Carrera M, Schmidt C, Fang Q, LeiBa Huang Y, Zaharatos GJ, Gardiner DF, Caskey M, Seamons L, Ho M, Dally L, Smith C, Cox J, Gill D, Gilmour J, Keefer MC, Fast P, Ho DD. 2010. Phase 1 safety and immunogenicity evaluation of ADMVA, a multigenic, modified vaccinia Ankara-HIV-1 B′/C candidate vaccine. PLoS One 5:e8816. 10.1371/journal.pone.0008816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911–922. 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowie AG, Haga IR. 2005. The role of Toll-like receptors in the host response to viruses. Mol. Immunol. 42:859–867. 10.1016/j.molimm.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 29.Perdiguero B, Esteban M. 2009. The interferon system and vaccinia virus evasion mechanisms. J. Interferon Cytokine Res. 29:581–598. 10.1089/jir.2009.0073 [DOI] [PubMed] [Google Scholar]
- 30.González JM, Esteban M. 2010. A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history. Virol. J. 7:59. 10.1186/1743-422X-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O'Neill LA. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 97:10162–10167. 10.1073/pnas.160027697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lysakova-Devine T, Keogh B, Harrington B, Nagpal K, Halle A, Golenbock DT, Monie T, Bowie AG. 2010. Viral inhibitory peptide of TLR4, a peptide derived from vaccinia protein A46, specifically inhibits TLR4 by directly targeting MyD88 adaptor-like and TRIF-related adaptor molecule. J. Immunol. 185:4261–4271. 10.4049/jimmunol.1002013 [DOI] [PubMed] [Google Scholar]
- 33.Oda S, Franklin E, Khan AR. 2011. Poxvirus A46 protein binds to TIR domain-containing Mal/TIRAP via an alpha-helical sub-domain. Mol. Immunol. 48:2144–2150. 10.1016/j.molimm.2011.07.014 [DOI] [PubMed] [Google Scholar]
- 34.Stack J, Bowie AG. 2012. Poxviral protein A46 antagonizes Toll-like receptor 4 signaling by targeting BB loop motifs in Toll-IL-1 receptor adaptor proteins to disrupt receptor:adaptor interactions. J. Biol. Chem. 287:22672–22682. 10.1074/jbc.M112.349225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stack J, Haga IR, Schröder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201:1007–1018. 10.1084/jem.20041442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham SC, Bahar MW, Cooray S, Chen RA, Whalen DM, Abrescia NG, Alderton D, Owens RJ, Stuart DI, Smith GL, Grimes JM. 2008. Vaccinia virus proteins A52 and B14 share a Bcl-2-like fold but have evolved to inhibit NF-kappaB rather than apoptosis. PLoS Pathog. 4:e1000128. 10.1371/journal.ppat.1000128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW, Smith GL, Bowie A, O'Neill LA. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197:343–351. 10.1084/jem.20021652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maloney G, Schröder M, Bowie AG. 2005. Vaccinia virus protein A52R activates p38 mitogen-activated protein kinase and potentiates lipopolysaccharide-induced interleukin-10. J. Biol. Chem. 280:30838–30844. 10.1074/jbc.M501917200 [DOI] [PubMed] [Google Scholar]
- 39.McCoy SL, Kurtz SE, Macarthur CJ, Trune DR, Hefeneider SH. 2005. Identification of a peptide derived from vaccinia virus A52R protein that inhibits cytokine secretion in response to TLR-dependent signaling and reduces in vivo bacterial-induced inflammation. J. Immunol. 174:3006–3014 http://www.jimmunol.org/content/174/5/3006.long [DOI] [PubMed] [Google Scholar]
- 40.Chen RA, Jacobs N, Smith GL. 2006. Vaccinia virus strain Western Reserve protein B14 is an intracellular virulence factor. J. Gen. Virol. 87:1451–1458. 10.1099/vir.0.81736-0 [DOI] [PubMed] [Google Scholar]
- 41.Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. 2008. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 4:e22. 10.1371/journal.ppat.0040022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Arriaza J, Nájera JL, Gómez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244. 10.1371/journal.pone.0024244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumner RP, Ren H, Smith GL. 2013. Deletion of immunomodulator C6 from vaccinia virus strain Western Reserve enhances virus immunogenicity and vaccine efficacy. J. Gen. Virol. 94(Part 5):1121–1126. 10.1099/vir.0.049700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unterholzner L, Sumner RP, Baran M, Ren H, Mansur DS, Bourke NM, Randow F, Smith GL, Bowie AG. 2011. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 7:e1002247. 10.1371/journal.ppat.1002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benfield CT, Ren H, Lucas SJ, Bahsoun B, Smith GL. 2013. Vaccinia virus protein K7 is a virulence factor that alters the acute immune response to infection. J. Gen. Virol. 94:1647–1657. 10.1099/vir.0.052670-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalverda AP, Thompson GS, Vogel A, Schröder M, Bowie AG, Khan AR, Homans SW. 2009. Poxvirus K7 protein adopts a Bcl-2 fold: biochemical mapping of its interactions with human DEAD box RNA helicase DDX3. J. Mol. Biol. 385:843–853. 10.1016/j.jmb.2008.09.048 [DOI] [PubMed] [Google Scholar]
- 47.Oda S, Schröder M, Khan AR. 2009. Structural basis for targeting of human RNA helicase DDX3 by poxvirus protein K7. Structure 17:1528–1537. 10.1016/j.str.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 48.Schröder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27:2147–2157. 10.1038/emboj.2008.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoyagi M, Zhai D, Jin C, Aleshin AE, Stec B, Reed JC, Liddington RC. 2007. Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein. Protein Sci. 16:118–124. 10.1110/ps.062454707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartlett N, Symons JA, Tscharke DC, Smith GL. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83:1965–1976 http://vir.sgmjournals.org/content/83/8/1965.long [DOI] [PubMed] [Google Scholar]
- 51.Cooray S, Bahar MW, Abrescia NG, McVey CE, Bartlett NW, Chen RA, Stuart DI, Grimes JM, Smith GL. 2007. Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J. Gen. Virol. 88:1656–1666. 10.1099/vir.0.82772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, Arvikar S, Latz E, Fitzgerald KA, Marshall WL. 2004. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by Toll-like receptors. J. Biol. Chem. 279:36570–36578. 10.1074/jbc.M400567200 [DOI] [PubMed] [Google Scholar]
- 53.Jacobs N, Bartlett NW, Clark RH, Smith GL. 2008. Vaccinia virus lacking the Bcl-2-like protein N1 induces a stronger natural killer cell response to infection. J. Gen. Virol. 89:2877–2881. 10.1099/vir.0.2008/004119-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maluquer de Motes C, Cooray S, Ren H, Almeida GM, McGourty K, Bahar MW, Stuart DI, Grimes JM, Graham SC, Smith GL. 2011. Inhibition of apoptosis and NF-kappaB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathog. 7:e1002430. 10.1371/journal.ppat.1002430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan JR, Roberts BE. 1984. Organization of RNA transcripts from a vaccinia virus early gene cluster. J. Virol. 51:283–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamin A, Esposito J, Hruby D. 1991. A single nucleotide substitution in the 5′-untranslated region of the vaccinia N2L gene is responsible for both alpha-amanitin-resistant and temperature-sensitive phenotypes. Virology 182:393–396. 10.1016/0042-6822(91)90688-8 [DOI] [PubMed] [Google Scholar]
- 57.Tamin A, Villarreal EC, Weinrich SL, Hruby DE. 1988. Nucleotide sequence and molecular genetic analysis of the vaccinia virus HindIII N/M region encoding the genes responsible for resistance to alpha-amanitin. Virology 165:141–150. 10.1016/0042-6822(88)90667-8 [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Villa NY, Rahman MM, Smallwood S, Shattuck D, Neff C, Dufford M, Lanchbury JS, Labaer J, McFadden G. 2009. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J. Proteome Res. 8:4311–4318. 10.1021/pr900491n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferguson BJ, Benfield CT, Ren H, Lee VH, Frazer GL, Strnadova P, Sumner RP, Smith GL. 2013. Vaccinia virus protein N2 is a nuclear IRF3 inhibitor that promotes virulence. J. Gen. Virol. 94:2070–2081. 10.1099/vir.0.054114-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gómez CE, Nájera JL, Jimenez EP, Jimenez V, Wagner R, Graf M, Frachette MJ, Liljestrom P, Pantaleo G, Esteban M. 2007. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1(IIIB) Gag-Pol-Nef proteins of clade B. Vaccine 25:2863–2885. 10.1016/j.vaccine.2006.09.090 [DOI] [PubMed] [Google Scholar]
- 61.Climent N, Guerra S, García F, Rovira C, Miralles L, Gómez CE, Pique N, Gil C, Gatell JM, Esteban M, Gallart T. 2011. Dendritic cells exposed to MVA-based HIV-1 vaccine induce highly functional HIV-1-specific CD8(+) T cell responses in HIV-1-infected individuals. PLoS One 6:e19644. 10.1371/journal.pone.0019644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García-Arriaza J, Arnaez P, Gómez CE, Sorzano CO, Esteban M. 2013. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate MVA-B by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS One 8:e66894. 10.1371/journal.pone.0066894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.García-Arriaza J, Nájera JL, Gómez CE, Sorzano CO, Esteban M. 2010. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS One 5:e12395. 10.1371/journal.pone.0012395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gómez CE, Nájera JL, Sanchez R, Jimenez V, Esteban M. 2009. Multimeric soluble CD40 ligand (sCD40L) efficiently enhances HIV specific cellular immune responses during DNA prime and boost with attenuated poxvirus vectors MVA and NYVAC expressing HIV antigens. Vaccine 27:3165–3174. 10.1016/j.vaccine.2009.03.049 [DOI] [PubMed] [Google Scholar]
- 65.Guerra S, González JM, Climent N, Reyburn H, Lopez-Fernandez LA, Nájera JL, Gómez CE, García F, Gatell JM, Gallart T, Esteban M. 2010. Selective induction of host genes by MVA-B, a candidate vaccine against HIV/AIDS. J. Virol. 84:8141–8152. 10.1128/JVI.00749-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mooij P, Balla-Jhagjhoorsingh SS, Koopman G, Beenhakker N, van Haaften P, Baak I, Nieuwenhuis IG, Kondova I, Wagner R, Wolf H, Gómez CE, Nájera JL, Jimenez V, Esteban M, Heeney JL. 2008. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J. Virol. 82:2975–2988. 10.1128/JVI.02216-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gómez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480. 10.1371/journal.ppat.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Ramírez JC, Gherardi MM, Esteban M. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923–933. 10.1128/JVI.74.2.923-933.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nájera JL, Gómez CE, García-Arriaza J, Sorzano CO, Esteban M. 2010. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One 5:e11406. 10.1371/journal.pone.0011406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Büttner M, Czerny CP, Lehner KH, Wertz K. 1995. Interferon induction in peripheral blood mononuclear leukocytes of man and farm animals by poxvirus vector candidates and some poxvirus constructs. Vet. Immunol. Immunopathol. 46:237–250. 10.1016/0165-2427(94)05357-X [DOI] [PubMed] [Google Scholar]
- 71.Waibler Z, Anzaghe M, Ludwig H, Akira S, Weiss S, Sutter G, Kalinke U. 2007. Modified vaccinia virus Ankara induces Toll-like receptor-independent type I interferon responses. J. Virol. 81:12102–12110. 10.1128/JVI.01190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, Forster R, Rowland-Jones S, Sekaly RP, McMichael AJ, Pantaleo G. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106–111. 10.1038/35065118 [DOI] [PubMed] [Google Scholar]
- 73.Sallusto F, Geginat J, Lanzavecchia A. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22:745–763. 10.1146/annurev.immunol.22.012703.104702 [DOI] [PubMed] [Google Scholar]
- 74.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- 75.Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247–258. 10.1038/nri2274 [DOI] [PubMed] [Google Scholar]
- 76.Bachmann MF, Wolint P, Schwarz K, Oxenius A. 2005. Recall proliferation potential of memory CD8+ T cells and antiviral protection. J. Immunol. 175:4677–4685 http://www.jimmunol.org/content/175/7/4677.long [DOI] [PubMed] [Google Scholar]
- 77.McElrath MJ, Haynes BF. 2010. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 33:542–554. 10.1016/j.immuni.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cottingham MG, Andersen RF, Spencer AJ, Saurya S, Furze J, Hill AV, Gilbert SC. 2008. Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA). PLoS One 3:e1638. 10.1371/journal.pone.0001638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Falivene J, Del Medico Zajac MP, Pascutti MF, Rodriguez AM, Maeto C, Perdiguero B, Gómez CE, Esteban M, Calamante G, Gherardi MM. 2012. Improving the MVA vaccine potential by deleting the viral gene coding for the IL-18 binding protein. PLoS One 7:e32220. 10.1371/journal.pone.0032220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perdiguero B, Gómez CE, Nájera JL, Sorzano CO, Delaloye J, González-Sanz R, Jimenez V, Roger T, Calandra T, Pantaleo G, Esteban M. 2012. Deletion of the viral anti-apoptotic gene F1L in the HIV/AIDS vaccine candidate MVA-C enhances immune responses against HIV-1 antigens. PLoS One 7:e48524. 10.1371/journal.pone.0048524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garber DA, O'Mara LA, Gangadhara S, McQuoid M, Zhang X, Zheng R, Gill K, Verma M, Yu T, Johnson B, Li B, Derdeyn CA, Ibegbu C, Altman JD, Hunter E, Feinberg MB. 2012. Deletion of specific immune-modulatory genes from modified vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in rhesus macaques. J. Virol. 86:12605–12615. 10.1128/JVI.00246-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garber DA, O'Mara LA, Zhao J, Gangadhara S, An I, Feinberg MB. 2009. Expanding the repertoire of modified vaccinia Ankara-based vaccine vectors via genetic complementation strategies. PLoS One 4:e5445. 10.1371/journal.pone.0005445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perdiguero B, Gómez C, Di Pilato M, Sánchez Sorzano C, Delaloye J, Roger T, Calandra T, Pantaleo G, Esteban M. 2013. Deletion of the vaccinia virus gene A46R, encoding for an inhibitor of TLR signalling, is an effective approach to enhance the immunogenicity of the HIV/AIDS vaccine candidate NYVAC-C. PLoS One 8:e74831. 10.1371/journal.pone.0074831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gómez CE, Perdiguero B, Nájera JL, Sorzano CO, Jimenez V, González-Sanz R, Esteban M. 2012. Removal of vaccinia virus genes that block interferon type I and II pathways improves adaptive and memory responses of the HIV/AIDS vaccine candidate NYVAC-C in mice. J. Virol. 86:5026–5038. 10.1128/JVI.06684-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kibler KV, Gómez CE, Perdiguero B, Wong S, Huynh T, Holechek S, Arndt W, Jimenez V, González-Sanz R, Denzler K, Haddad EK, Wagner R, Sekaly RP, Tartaglia J, Pantaleo G, Jacobs BL, Esteban M. 2011. Improved NYVAC-based vaccine vectors. PLoS One 6:e25674. 10.1371/journal.pone.0025674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quakkelaar ED, Redeker A, Haddad EK, Harari A, McCaughey SM, Duhen T, Filali-Mouhim A, Goulet JP, Loof NM, Ossendorp F, Perdiguero B, Heinen P, Gómez CE, Kibler KV, Koelle DM, Sekaly RP, Sallusto F, Lanzavecchia A, Pantaleo G, Esteban M, Tartaglia J, Jacobs BL, Melief CJ. 2011. Improved innate and adaptive immunostimulation by genetically modified HIV-1 protein expressing NYVAC vectors. PLoS One 6:e16819. 10.1371/journal.pone.0016819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. 10.1038/nature10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M, Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293–299. 10.1038/nm.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286. 10.1056/NEJMoa1113425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Antoine G, Scheiflinger F, Dorner F, Falkner FG. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365–396. 10.1006/viro.1998.9123 [DOI] [PubMed] [Google Scholar]
- 91.Blanchard TJ, Alcami A, Andrea P, Smith GL. 1998. Modified vaccinia virus Ankara undergoes limited replication in human cells and lacks several immunomodulatory proteins: implications for use as a human vaccine. J. Gen. Virol. 79(Part 5):1159–1167 [DOI] [PubMed] [Google Scholar]
- 92.Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, Hausmann J. 2007. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J. Gen. Virol. 88:3249–3259. 10.1099/vir.0.83156-0 [DOI] [PubMed] [Google Scholar]
- 93.Nájera JL, Gómez CE, Domingo-Gil E, Gherardi MM, Esteban M. 2006. Cellular and biochemical differences between two attenuated poxvirus vaccine candidates (MVA and NYVAC) and role of the C7L gene. J. Virol. 80:6033–6047. 10.1128/JVI.02108-05 [DOI] [PMC free article] [PubMed] [Google Scholar]