ABSTRACT
There is a need to develop a single and highly effective vaccine against the emerging chikungunya virus (CHIKV), which causes a severe disease in humans. Here, we have generated and characterized the immunogenicity profile and the efficacy of a novel CHIKV vaccine candidate based on the highly attenuated poxvirus vector modified vaccinia virus Ankara (MVA) expressing the CHIKV C, E3, E2, 6K, and E1 structural genes (termed MVA-CHIKV). MVA-CHIKV was stable in cell culture, expressed the CHIKV structural proteins, and triggered the cytoplasmic accumulation of Golgi apparatus-derived membranes in infected human cells. Furthermore, MVA-CHIKV elicited robust innate immune responses in human macrophages and monocyte-derived dendritic cells, with production of beta interferon (IFN-β), proinflammatory cytokines, and chemokines. After immunization of C57BL/6 mice with a homologous protocol (MVA-CHIKV/MVA-CHIKV), strong, broad, polyfunctional, and durable CHIKV-specific CD8+ T cell responses were elicited. The CHIKV-specific CD8+ T cells were preferentially directed against E1 and E2 proteins and, to a lesser extent, against C protein. CHIKV-specific CD8+ memory T cells of a mainly effector memory phenotype were also induced. The humoral arm of the immune system was significantly induced, as MVA-CHIKV elicited high titers of neutralizing antibodies against CHIKV. Remarkably, a single dose of MVA-CHIKV protected all mice after a high-dose challenge with CHIKV. In summary, MVA-CHIKV is an effective vaccine against chikungunya virus infection that induced strong, broad, highly polyfunctional, and long-lasting CHIKV-specific CD8+ T cell responses, together with neutralizing antibodies against CHIKV. These results support the consideration of MVA-CHIKV as a potential vaccine candidate against CHIKV.
IMPORTANCE We have developed a novel vaccine candidate against chikungunya virus (CHIKV) based on the highly attenuated poxvirus vector modified vaccinia virus Ankara (MVA) expressing the CHIKV C, E3, E2, 6K, and E1 structural genes (termed MVA-CHIKV). Our findings revealed that MVA-CHIKV is a highly effective vaccine against chikungunya virus, with a single dose of the vaccine protecting all mice after a high-dose challenge with CHIKV. Furthermore, MVA-CHIKV is highly immunogenic, inducing strong innate responses: high, broad, polyfunctional, and long-lasting CHIKV-specific CD8+ T cell responses, together with neutralizing antibodies against CHIKV. This work provides a potential vaccine candidate against CHIKV.
INTRODUCTION
Chikungunya virus (CHIKV) is an alphavirus of the family Togaviridae that is transmitted by mosquitoes of the genus Aedes (1). The virus causes chikungunya fever in humans, a disease characterized by skin rash, high fever, headache, vomiting, myalgia, and, mainly, polyarthralgia (1–6). Most of the symptoms resolve after 10 days, but the polyarthralgia can persist for months or years (4, 6, 7), and severe symptoms, such as encephalitis, hemorrhagic disease, and mortality, have also been described (5, 8, 9).
CHIKV contains a positive, single-stranded RNA genome of around 11.8 kb which encodes four nonstructural and five structural proteins (10, 11). The nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) are required for virus replication. The structural proteins are cleaved by capsid (C) autoproteinase and signalases from a polyprotein precursor to generate the C and envelope (E3, E2, 6K, and E1) proteins (10–12). Virions are 70-nm enveloped particles containing 240 heterodimers of E1/E2 glycoproteins on their surfaces (13).
CHIKV infection was first described in 1952 in Tanzania, and the virus was isolated in 1953 (14). In 2005, CHIKV reemerged as an outbreak on La Réunion Island (15) and has spread to different places in Africa, islands in the Indian Ocean, India, Southeast Asia, and southern Europe, affecting millions of people (3, 16–23), revealing that the virus is a public threat that could cause a worldwide epidemic (4, 6, 24, 25). Thus, the development of a prophylactic CHIKV vaccine is a high priority that has been moving forward to control CHIKV infection (26). Several vaccine approaches against CHIKV, such as a formalin-inactivated CHIKV (27–29), a live attenuated CHIKV (30, 31), a recombinant E2 protein-based vaccine (32), chimeric alphavirus vectors (33–35), an adenovirus vector (36), a virus-like particle vaccine (37–39), DNA vaccines (40, 41), an internal ribosome entry site (IRES)-based live attenuated CHIKV vaccine (42–44), and a recombinant measles vaccine (45), have been developed. However, currently there are no licensed CHIKV vaccines or effective antiviral therapies that could control the disease (26).
Modified vaccinia virus Ankara (MVA) is a highly attenuated poxvirus strain that has been widely used in several preclinical and clinical trials as a vaccine vector against many infectious diseases and cancer (46–49), showing that MVA vectors are safe, express high levels of heterologous antigens, and are strongly immunogenic. Thus, the use of MVA as a vector to generate a vaccine candidate against CHIKV could be a useful approach to counteract the disease.
In this study, we have generated an MVA-based CHIKV vaccine candidate (termed MVA-CHIKV) expressing the CHIKV C-E3-E2-6K-E1 structural genes, and we have characterized (i) the innate immune responses that it elicits in human macrophages and monocyte-derived dendritic cells (moDCs), (ii) the adaptive and memory cellular immunogenicities that it elicits in mice, (iii) its ability to induce neutralizing antibodies against CHIKV, and (iv) its efficacy after a challenge with CHIKV. The results showed that MVA-CHIKV promotes a robust innate immune response characterized by the expression of beta interferon (IFN-β), proinflammatory cytokines, and chemokines, and that it is highly immunogenic to activating T and B cells, inducing strong, broad, polyfunctional, and durable CHIKV-specific T cell responses, together with high titers of neutralizing antibodies against CHIKV. Significantly, just one dose of the vaccine protected all mice from a CHIKV challenge.
MATERIALS AND METHODS
Ethics statement.
The immunogenicity animal studies were approved by the Ethical Committee of Animal Experimentation (CEEA-CNB) of the Centro Nacional de Biotecnología (CNB-CSIC, Madrid, Spain) in accordance with national and international guidelines and with the Royal Decree (RD 1201/2005). Experiments were conducted at the CNB, Madrid, Spain (permit number 13002).
The pathogenicity animal studies were approved by the Stockholms Norra djurförsöksetiska nämnd (permit number N74/11) and were conducted at the Astrid Fagraeus Laboratory (AFL) at the Karolinska Institutet, Stockholm, Sweden.
Studies with peripheral blood mononuclear cells (PBMCs) from healthy blood donors recruited by the Centro de Transfusión de la Comunidad de Madrid (Madrid, Spain) were approved by the Ethical Committee of the Centro de Transfusión de la Comunidad de Madrid (Madrid, Spain). Written informed consent was obtained from each donor before blood collection for the purpose of this investigation according to a collaborative agreement between the Centro de Transfusión de la Comunidad de Madrid and the CNB-CSIC. All information was kept confidential.
Cells and viruses.
HeLa cells (a human epithelial cervix adenocarcinoma; ATCC CCL-2), DF-1 cells (a spontaneously immortalized chicken embryo fibroblast cell line; ATCC CRL-12203), and primary chicken embryo fibroblasts (CEFs) (obtained from specific-pathogen-free 11-day-old eggs; Intervet, Salamanca, Spain) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with penicillin (100 units/ml; Sigma-Aldrich), streptomycin (100 μg/ml; Sigma-Aldrich), l-glutamine (2 mM; Sigma-Aldrich), nonessential amino acids (0.1 mM; Sigma-Aldrich), gentamicin (50 μg/ml; Sigma-Aldrich), amphotericin B (Fungizone, 0.5 μg/ml; Gibco-Life Technologies), and 10% heat-inactivated fetal calf serum (FCS). The human monocytic THP-1 cell line (ATCC TIB-202) was cultured in RPMI 1640 medium containing 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin (complete medium; all from Sigma-Aldrich), and 10% heat-inactivated FCS (Sigma-Aldrich, St. Louis, MO), as previously described (50–52). THP-1 cells were differentiated into macrophages by treatment with 0.5 mM phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) for 24 h before usage. Freshly isolated PBMCs from buffy coats of healthy donors (recruited by the Centro de Transfusión de la Comunidad de Madrid, Madrid, Spain) were obtained by Ficoll gradient separation on Ficoll-Paque (GE Healthcare). Next, CD14+ monocytes were purified by depletion using a Dynabeads untouched human monocyte kit (Invitrogen Dynal AS, Oslo, Norway), by following the manufacturer's protocol. Then, human moDCs were obtained as previously described (51), after cultivation of purified monocytes for 7 days in six-well culture plates (3 × 106 cells/well at 1 × 106 cells/ml) in complete RPMI 1640 medium containing 10% FCS and supplemented with 50 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 20 ng/ml interleukin 4 (IL-4) (both from Gibco-Life Technologies). The purity of sorted DC populations was >99%. Cell cultures were maintained at 37°C (HeLa, CEF, and THP-1 cells and moDCs) or 39°C (DF-1 cells) in a humidified incubator containing 5% CO2. Cell lines were infected with viruses as previously described (50, 53). Virus infections were performed in the presence of 2% FCS for all cell types.
The poxvirus strain used in this study as the parental virus for the generation of the recombinant MVA-CHIKV is a modified vaccinia virus Ankara (MVA) strain (obtained from the Ankara strain after 586 serial passages in CEFs; this strain is derived from clone F6 at passage 585, kindly provided by G. Sutter, Germany). We modified this virus by inserting the gene encoding the green fluorescent protein (GFP) into the thymidine kinase (TK) locus and by deleting the immunomodulatory vaccinia virus (VACV) genes C6L, K7R, and A46R. The GFP cassette in the TK locus was replaced by the CHIKV cassette to generate MVA-CHIKV. We also used in this study the parental attenuated MVA (referred to as wild-type MVA [MVA-WT]). All viruses were grown in primary CEFs, purified by centrifugation through two 36% (wt/vol) sucrose cushions in 10 mM Tris-HCl, pH 9, and titrated in DF-1 cells by a plaque immunostaining assay, as previously described (54). The titer determinations of the different viruses were performed at least three times. All viruses were free of contamination with mycoplasmas, fungi, or bacteria.
Construction of the infectious CHIKV clone LR2006-OPY1 has previously been described (55). Briefly, a plasmid for production of CHIKV was generated by cloning the CHIKV cDNA under the control of the SP6 RNA polymerase promoter. CHIKV was subsequently produced in BHK-21 cells as previously described (56, 57).
Construction of the plasmid transfer vector pCyA20-ICRES1.
The plasmid transfer vector pCyA-ICRES1 was generated and used for the construction of recombinant MVA-CHIKV, in which CHIKV structural genes (those for C, E3, E2, 6K, and E1) instead of the GFP gene were inserted into the TK locus of the parental MVA-GFP. In detail, a 3.7-kbp DNA fragment encoding the CHIKV structural proteins (C, E3, E2, 6K, and E1) was amplified with oligonucleotides ICRES1 upper (5′-ATATAGTTTAAACATGGAGTTCATCCCAACCCAAAC-3′; PmeI site underlined) and ICRES1 lower (5′-TATATAGATCTTTAGTGCCTGCTGAACGACACG-3′; BglII site underlined) from plasmid pSP6-ICRES1, which contains the structural genes from the CHIKV genome (CHIKV isolate LR2006-OPY1). The amplification reaction was performed with Platinum Pfx DNA polymerase (Invitrogen) according to the manufacturer's recommendations. Then, this fragment was digested with PmeI and BglII and cloned into the VACV insertion vector pCyA-20 (7,582 bp), previously digested with the same restriction enzymes and dephosphorylated by incubation with shrimp alkaline phosphatase, to generate the plasmid transfer vector pCyA-ICRES1 (11,324 bp). The plasmid pCyA-20, used for the generation of plasmid pCyA-ICRES1, was previously described (58) and contains a synthetic band containing the viral early/late promoter and a multiple-cloning site in the plasmid pLZAW1 (provided by Sanofi-Pasteur). The correct generation of pCyA-ICRES1 was further confirmed by DNA sequence analysis. This plasmid was used in transient-infection and transfection assays for the insertion of CHIKV structural genes (those for C, E3, E2, 6K, and E1) into the TK locus of the MVA genome under the transcriptional control of the viral synthetic early/late (sE/L) promoter. It contains a β-galactosidase (β-Gal) reporter gene sequence between two repetitions of the left TK-flanking arm, which allows the reporter gene to be deleted from the final recombinant virus by homologous recombination after successive passages.
Construction of recombinant MVA-CHIKV.
DF-1 cells (3 × 106 cells) were infected with parental MVA-GFP at a multiplicity of infection (MOI) of 0.05 PFU/cell and transfected 1 h later with 10 μg of DNA of plasmid pCyA-ICRES1, using Lipofectamine reagent according to the manufacturer's recommendations (Invitrogen). At 48 h postinfection (hpi), the cells were harvested, lysed by freeze-thaw cycling, sonicated, and used for recombinant-virus screening. Recombinant MVAs containing the 3.7-kb CHIKV DNA fragment (encoding the structural proteins of CHIKV, C, E3, E2, 6K, and E1), with the GFP gene deleted, and transiently coexpressing the β-Gal marker gene (MVA-CHIKV, X-Gal+) were selected by consecutive rounds of plaque purification in DF-1 cells stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 400 μg/ml, for three passages in total). In the following plaque purification steps, recombinant MVAs containing the 3.7-kb CHIKV DNA fragment and with the β-Gal gene deleted by homologous recombination from between the left TK arm and the short left TK arm repeat flanking the marker (MVA-CHIKV, X-Gal−) were isolated by three additional consecutive rounds of plaque purification screening for nonstaining viral foci in DF-1 cells in the presence of X-Gal (400 μg/ml). In each round of purification, the isolated plaques were expanded in DF-1 cells for 3 days, and the crude viruses obtained were used for the next plaque purification round. The resulting recombinant virus, MVA-CHIKV (passage 2 [P2] stock), was grown in CEFs, purified through two 36% (wt/vol) sucrose cushions, and titrated by plaque immunostaining assay.
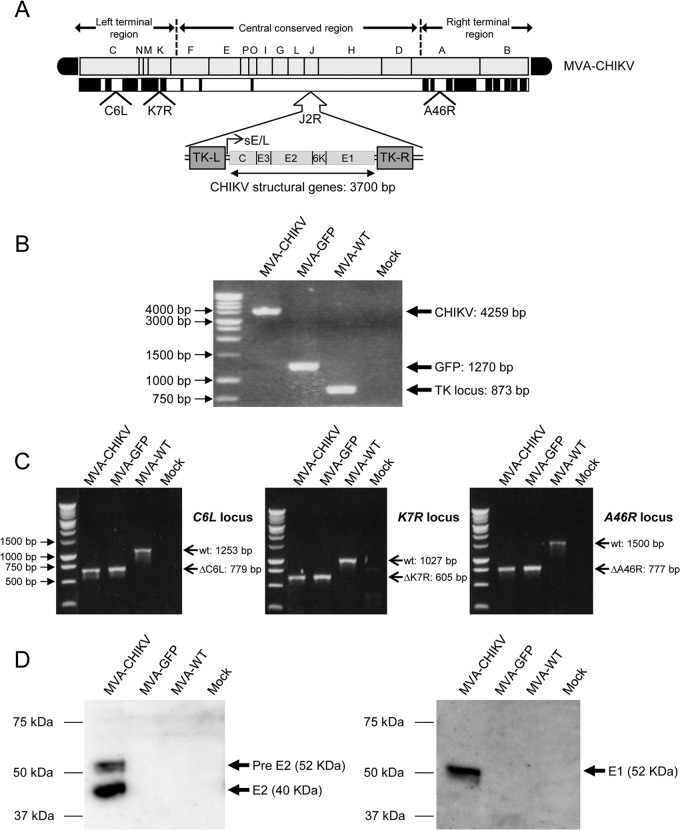
PCR analysis of recombinant MVA-CHIKV.
The purity of recombinant MVA-CHIKV was confirmed by PCR with primers spanning the junction regions of the CHIKV insert and by DNA sequence analysis. Thus, to test the identity and purity of the recombinant MVA-CHIKV, viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell with MVA-WT, MVA-GFP, or MVA-CHIKV, as previously described (53). Primers TK-L and TK-R (previously described [58]), annealing in the TK gene-flanking regions, were used for PCR analysis of the TK locus. Furthermore, to verify that deletion of the VACV genes C6L, K7R, and A46R from MVA-CHIKV was correctly done, primers RFC6L-AatII-F and LFC6L-BamHI-R (previously described [52]), LFK7R-AatII-F and RFK7R-BamHI-R (previously described [51]), and LFA46R-AatII-F and RFA46R-BamHI-R (sequences will be provided upon request), spanning the VACV C6L-, K7R-, and A46R-flanking regions, respectively, were used for PCR analysis of the C6L, K7R, and A46R loci, respectively. All the amplification reactions were performed with Platinum Taq DNA polymerase (Invitrogen) according to the manufacturer's recommendations, and the amplification protocol was previously described (59). PCR products were run in 1% agarose gel and visualized by SYBR Safe staining (Invitrogen). The 3.7-kb CHIKV DNA insertion and C6L, K7R, and A46R deletions were also confirmed by DNA sequence analysis. No mutations were included in the 3.7-kb CHIKV DNA insert during the PCR.
Analysis of virus growth.
To determine the virus growth profile of MVA-CHIKV, in comparison to that of MVA-WT, monolayers of DF-1 cells grown in 12-well plates were infected in duplicate at 0.01 PFU/cell with MVA-WT or recombinant MVA-CHIKV. Following virus adsorption for 60 min at 37°C, the inoculum was removed, and the infected cells were washed with DMEM and incubated with fresh DMEM containing 2% FCS at 37°C in a 5% CO2 atmosphere. At different times postinfection (0, 24, 48, and 72 h), cells were harvested by being scraped from plates (lysates at 5 × 105 cells/ml), frozen and thawed three times, and briefly sonicated. Virus titers in cell lysates were determined by a plaque immunostaining assay in DF-1 cells, as previously described (54), using rabbit polyclonal anti-vaccinia virus strain WR (Centro Nacional de Biotecnología; diluted 1:1,000), followed by anti-rabbit-horseradish peroxidase (HRP) (Sigma; diluted 1:1,000).
Expression of CHIKV proteins from recombinant MVA-CHIKV by Western blotting.
To check the correct expression of CHIKV antigens by the recombinant MVA-CHIKV, monolayers of DF-1 cells were mock infected or infected at 5 PFU/cell with MVA-WT, MVA-GFP, or MVA-CHIKV. At 24 hpi, cells were lysed in Laemmli buffer, and cells extracts were fractionated in 10% SDS-PAGE and analyzed by Western blotting with mouse polyclonal antibody against E2 (antibody 3E4, kindly provided by Philippe Desprès from the Pasteur Institute, Paris, France; diluted 1:500), mouse monoclonal antibody against E1-E2 (antibody D3-62; kindly provided by Marc Lecuit and Therese Couderc from the Pasteur Institute, Paris, France; diluted 1:500), or rabbit polyclonal antibody against E1 (kindly provided by Lisa F. P. Ng from the Singapore Immunology Network, Agency for Science, Technology and Research, Biopolis, Singapore; diluted 1:1,000) to evaluate the expression of the different CHIKV structural proteins. The anti-rabbit HRP-conjugated antibody (Sigma; diluted 1:5,000), or anti-mouse-HRP-conjugated antibody (Sigma; diluted 1:2,000), were used as secondary antibodies. The immunocomplexes were detected using an enhanced HRP-luminol chemiluminescence system (ECL Plus; GE Healthcare).
Genetic stability of recombinant MVA-CHIKV by expression analysis.
The genetic stability of recombinant MVA-CHIKV was analyzed as previously described (58), with some modifications. Monolayers of DF-1 cells were infected at 0.05 PFU/cell with recombinant MVA-CHIKV (P2 stock). At 72 hpi, cells were collected by being scraped. After three freeze-thaw cycles and a brief sonication, the cellular extract was centrifuged at 1,500 rpm for 5 min, and the supernatant was used for a new round of infection at a low MOI. The same procedure was repeated nine times. Expression of CHIKV proteins at all nine passages was detected by Western blotting after infection of DF-1 cells with virus stocks from each passage, using a mouse polyclonal antibody against E2 (antibody 3E4; diluted 1:500), mouse monoclonal antibody against E1 and E2 (antibody D3-62; diluted 1:500), or rabbit polyclonal antibody against E1 (diluted 1:1,000) to evaluate the expression of the different CHIKV proteins. As loading controls, we used rabbit anti-β-actin (Cell Signaling; diluted 1:1,000) and rabbit anti-VACV E3 (Centro Nacional de Biotecnología; diluted 1:1,000) antibodies. An anti-rabbit HRP-conjugated antibody (Sigma; diluted 1:5,000) or an anti-mouse HRP-conjugated antibody (Sigma; diluted 1:2,000) was used as the secondary antibody. The immunocomplexes were detected using an enhanced HRP-luminol chemiluminescence system (ECL Plus; GE Healthcare).
Furthermore, the genetic stability of MVA-CHIKV (P2 stock, P3 stock, and passage 9) was analyzed by quantifying the number of individual plaques expressing CHIKV and VACV antigens after a plaque immunostaining assay of DF-1 cells, as previously described (54). We used a mouse polyclonal antibody against CHIKV E2 (antibody 3E4; diluted 1:500), mouse monoclonal antibody against CHIKV E1 and E2 (antibody D3-62; diluted 1:500), or a rabbit polyclonal anti-vaccinia virus strain WR antibody (Centro Nacional de Biotecnología; diluted 1:1,000), followed by anti-rabbit-horseradish peroxidase (HRP) (Sigma; diluted 1:1,000) or an anti-mouse HRP-conjugated antibody (Sigma; diluted 1:1,000).
Expression of CHIKV proteins from recombinant MVA-CHIKV by immunofluorescence microscopy analysis.
Immunofluorescence studies were done as previously described (58). Briefly, HeLa cells cultured on glass coverslips to a confluence of 50% were mock infected or infected with MVA-WT or MVA-CHIKV at an MOI of 0.5 PFU/cell. At 6 and 16 hpi, cells were washed with phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde (PFA) in PBS at room temperature (RT) for 15 min. Then, cells were quenched for 15 min in the presence of 50 mM NH4Cl and permeabilized and blocked with 0.05% saponin and 5% FCS in PBS. Mouse polyclonal antibodies against CHIKV protein E2 (antibodies 3E4, D3-62, and K9-1, kindly provided by Marc Lecuit and Therese Couderc from the Pasteur Institute, Paris, France; diluted 1:2,000, 1:1,000, and 1:3,000, respectively, in PBS containing 0.05% saponin-5% FCS) and the endoplasmic reticulum (ER) marker rabbit polyclonal anti-calnexin antibody (BioNova) were used to stain the cells for 1 h at RT in the presence of 0.05% saponin-5% FCS in PBS. Then, coverslips were washed with PBS and cells were blocked again with 0.05% saponin-5% FCS in PBS for 15 min and stained for 1 h in the dark at RT with specific mouse or rabbit secondary antibodies conjugated with the fluorochromes Alexa Fluor 488 (green) or Alexa Fluor 594 (red) (Invitrogen). Coverslips were then washed with PBS, and cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma) in PBS for 15 min, along with the high-affinity F-actin phalloidin probe conjugated to the red-orange fluorescent dye tetramethylrhodamine (TRITC) (Sigma) or the trans-Golgi marker wheat germ agglutinin (WGA) probe conjugated to the red fluorescent dye Alexa Fluor 594 (Invitrogen), when required. After being stained, coverslips were mounted on glass slides and conserved in ProLong Gold antifade reagent (Invitrogen). Images of sections of the cells were acquired using a Leica TCS SP5 microscope and were recorded and processed by the specialized software LasAF (Leica Microsystems).
Electron microscopy analysis.
Electron microscopy analysis was done as previously described (58). Briefly, confluent HeLa cells grown in DMEM supplemented with 10% newborn calf serum (NCS) were infected with MVA-WT or MVA-CHIKV at an MOI of 5 PFU/cell. At 6 and 16 hpi, cell monolayers were fixed in situ in a plastic dish for 1 h with 2% glutaraldehyde-1% tannic acid in 0.4 M HEPES buffer (pH 7.5) at RT. After fixation, monolayers of cells were carefully scraped, centrifuged to eliminate the fixative, and maintained in HEPES buffer at 4°C. Afterwards, cells were processed by conventional embedding in the epoxy resin EML-812 (Taab Laboratories, Adermaston, Berkshire, United Kingdom). Processing consists of postfixation in a mixture of 1% osmium tetroxide-0.8% potassium ferricyanide in distilled water for 1 h at 4°C, followed by staining with 2% uranyl acetate and progressive dehydration in increasing concentrations of acetone (50, 70, 90, and 100%). Then infiltration in the epoxy resin was carried out at RT for 1 day and maintained at 60°C for 3 days to allow resin polymerization. Ultrathin sections (70 nm thick) of the samples were sliced using a Leica EM UC6 ultramicrotome and collected on copper grids. The cells sections were finally stained with saturated uranyl acetate and lead citrate by ordinary methods. Sections of infected cells were studied in a JEOL 1011 transmission electron microscope operating at 100 kV. Digital images were recorded with an ES1000W Erlangshen charge-coupled-device (CCD) camera (Gatan, CA, USA).
RNA analysis by quantitative real-time PCR.
Total RNA was isolated from both THP-1 cells and moDCs mock infected or infected with MVA-WT, MVA-GFP, or MVA-CHIKV, using the RNeasy kit (Qiagen GmbH, Hilden, Germany). THP-1 cells were mock infected or infected at 5 PFU/cell, and total RNA was obtained at 3 and 6 hpi. moDCs were mock infected or infected at 1 PFU/cell, and total RNA was obtained at 6 hpi. Reverse transcription of at least 500 ng of RNA was performed using the QuantiTect reverse transcription kit (Qiagen GmbH, Hilden, Germany). Quantitative PCR was performed with a 7500 real-time PCR system (Applied Biosystems) using the Power SYBR green PCR master mix (Applied Biosystems), as previously described (50). Expression levels of the IFN-β, TNF-α, MIP-1α, RANTES, IP-10, IFIT1, IFIT2, RIG-I, MDA-5, and HPRT genes were analyzed by real-time PCR using specific oligonucleotides (the sequences will be provided upon request). Gene-specific expression was expressed relative to the expression of HPRT in arbitrary units (AU). All samples were tested in duplicate, and two different experiments were performed with each cell type.
Peptides.
CHIKV peptides representative of the capsid (C), E1, and E2 CHIKV proteins were used in the immunological analysis. Peptides sequences are as follows: ACLVGDJVM (capsid), HSMTNAAVTI (E1), and IILYYYELY (E2).
C57BL/6 mouse immunogenicity study.
Female C57BL/6 mice (6 to 8 weeks old) were purchased from Harlan Laboratories and stored in a pathogen-free barrier area of the CNB (Madrid, Spain) in accordance with recommendations of the Federation of European Laboratory Animal Science Associations. A homologous MVA prime-MVA boost immunization protocol was performed to assay the immunogenicity of the recombinant MVA-CHIKV. Groups of animals (n = 10) were immunized with 1 × 107 PFU of MVA-WT or MVA-CHIKV by the intraperitoneal (i.p.) route in 200 μl of PBS. Two weeks later, animals received 2 × 107 PFU of MVA-WT or MVA-CHIKV by the i.p. route in 200 μl of PBS. At 10 and 52 days after the last immunization, five mice in each group were sacrificed using carbon dioxide (CO2), and their spleens were processed to measure the adaptive and memory immune responses, respectively, to the CHIKV and VACV antigens by an intracellular cytokine staining (ICS) assay. Two independent experiments were performed.
The ability of MVA-CHIKV to induce antibody responses was studied in mice at the AFL at Karolinska Institutet, Stockholm, Sweden. Groups of 5- to 6-week-old female C57BL/6 mice (n = 5) were immunized with 1 × 107 PFU of MVA-WT or MVA-CHIKV by the i.p. route in 200 μl PBS. MVA-WT was administered once, whereas MVA-CHIKV was administered once or twice with a 3-week immunization interval. Serum was collected by tail bleedings 3 weeks after the first immunization and 6 weeks after the second immunization, and antibody responses were analyzed by enzyme-linked immunosorbent assay (ELISA) and a neutralization assay.
CHIKV challenge study.
A challenge study to evaluate the efficacy of the recombinant MVA-CHIKV was performed in a biosafety level 3 laboratory at the AFL at Karolinska Institutet, Stockholm, Sweden. Female C57BL/6 mice previously immunized with MVA-WT or MVA-CHIKV to study antibody responses were used (see “C57BL/6 mouse immunogenicity study” above [5 mice/group]). Seven weeks after the last immunization, mice were challenged with a total of 106 PFU of CHIKV (CHIKV isolate LR2006-OPY1) via the subcutaneous route in the dorsal side of each hind foot. Blood samples were collected at days 1 to 3 postchallenge from tail bleedings, and viremia was analyzed by plaque assay (PFU/ml). Foot swelling (height by breadth) was measured using a digital caliper before challenge and days 4 to 8 after challenge. Two independent experiments were performed.
ICS assay.
The magnitudes, polyfunctionality, and phenotypes of the VACV- and CHIKV-specific adaptive and memory T cell responses were analyzed by ICS, as previously described (51, 52, 58, 59), with some modifications. After an overnight rest, 4 × 106 splenocytes (depleted of red blood cells) were seeded on M96 plates and stimulated for 6 h in complete RPMI 1640 medium supplemented with 10% FCS containing 1 μl/ml GolgiPlug (BD Biosciences) (to inhibit cytokine secretion), anti-CD107a–Alexa Fluor 488 (BD Biosciences), Monensin (1×; eBioscience), and 1 μg/ml of the different CHIKV peptides (C, E1, or E2) or MVA-infected EL4 cells (4 × 105 EL4 cells in 1 × 106 splenocytes; the ratio of MVA-infected EL4 cells to splenocytes was equal to 1:2.5). Then, cells were washed, stained for the surface markers, fixed, permeabilized (Cytofix/Cytoperm kit; BD Biosciences), and stained intracellularly with the appropriate fluorochromes. Dead cells were excluded using the violet LIVE/DEAD stain kit (Invitrogen). For functional analyses, the following fluorochrome-conjugated antibodies were used: CD3-phycoerythrin (PE)-CF594, CD4-allophycocyanin (APC)-Cy7, CD8-V500, IFN-γ–PE–Cy7, tumor necrosis factor alpha (TNF-α)–PE, and interleukin 2 (IL-2)–APC. In addition, for phenotypic analyses, the following antibodies were used: CD62L-Alexa 700 and CD127-PerCP-Cy5.5. All antibodies were from BD Biosciences. Cells were acquired using a Gallios flow cytometer (Beckman Coulter). Analyses of the data were performed using FlowJo software version 8.5.3 (Tree Star, Ashland, OR). The number of lymphocyte-gated events was 106. After gating, Boolean combinations of single functional gates were then created using FlowJo software to determine the frequency of each response based on all possible combinations of cytokine.
ELISA.
Levels of binding of antibodies to CHIKV present in the sera of immunized mice were determined using ELISA. Ninety-six-well Nunc MaxiSorp plates were coated with 50 μl of purified recombinant CHIKV E1-p62 protein (13) at a concentration of 2 μg/ml in PBS at 4°C overnight. Plates were washed with PBS supplemented with 0.05% Tween 20 (PBS-Tween) and blocked with 1% BSA (Gibco, Life Technologies) in PBS for 1 h at RT. Sera were diluted in PBS-Tween, added to plates, and incubated at 4°C overnight. Plates were then washed, and a secondary HRP-conjugated goat-anti-mouse IgG (Southern Biotech, AL, USA; diluted 1:5,000 in PBS-Tween) was added and incubated for 1.5 h at RT. Plates were washed, and the o-phenylenediamine dihydrochloride (OPD) substrate (Sigma-Aldrich) was added. One molar HCl was added after 15 min to stop the color development. Absorbance was read at 490 nm.
Neutralizing antibodies.
Titers of neutralizing antibodies against CHIKV present in the sera of immunized mice collected 6 weeks after the last immunization were determined using CHIKV viral-replicon particles (VRPs) expressing Gaussia luciferase (Gluc), as previously described (60). Briefly, VRPs (MOI, 0.5) were preincubated with 2-fold serial dilutions of sera from immunized mice for 1 h at 37°C before the mixture was added to BHK-21 cells seeded in 24-well plates. After incubation for 1 h at 37°C, the inoculum was removed, cells were washed with PBS, and medium was added. A readout of secreted Gluc was performed at 24 hpi using the Renilla luciferase assay system (Promega, WI, USA). Neutralization potency was determined as a percentage of measured Gluc activity and compared to the Gluc readout after VRP application without antibody/serum, and results are presented as 50% neutralization titers (NT50).
Data analysis and statistical procedures.
The statistical significance (*, P < 0.05; **, P < 0.005; ***, P < 0.001) of differences between groups was determined by Student's t test (two tailed, type 2). Statistical analysis of the ICS data was realized as previously described (59, 61), using an approach that corrects measurements for the medium response (RPMI 1640), with calculation of confidence intervals and P values. Only antigen response values significantly larger than the corresponding RPMI 1640 values are represented. Background values were subtracted from all values used to allow analysis of proportionate representation of responses.
RESULTS
Generation and in vitro characterization of an MVA recombinant expressing CHIKV structural genes (MVA-CHIKV).
To generate a vaccine against CHIKV, we constructed an MVA-based candidate vaccine expressing the CHIKV C, E3, E2, 6K, and E1 structural genes (termed MVA-CHIKV) in the vector backbone of the parent, MVA-WT, which also contains deletions in the VACV immunomodulatory genes C6L, K7R, and A46R (termed MVA-GFP) (see Materials and Methods). The deletions were introduced into the genome of MVA, as we have previously described the benefit of enhancing cellular and humoral immune responses to human immunodeficiency virus type 1 (HIV-1) antigens from MVA vectors lacking selected viral immunomodulatory genes (51, 52, 59). In Fig. 1A, a diagram of the MVA-CHIKV genome is shown with the corresponding VACV deletions and the CHIKV C, E3, E2, 6K, and E1 structural genes inserted into the VACV TK locus under the transcriptional control of the synthetic early/late viral (sE/L) promoter driving the constitutive expression of the structural CHIKV proteins. The correct insertion and purity of recombinant MVA-CHIKV was confirmed by PCR and DNA sequence analysis. PCR using primers annealing in the VACV TK gene-flanking regions confirmed the presence of the CHIKV structural genes in MVA-CHIKV, with no wild-type contamination in the preparation (Fig. 1B); in the parental virus MVA-GFP, the GFP was present, and in the MVA-WT, the VACV TK locus was amplified (Fig. 1B). Moreover, PCRs using primers annealing in the C6L-, K7R-, and A46R-flanking regions confirmed the deletion of the VACV genes C6L, K7R, and A46R, respectively, from MVA-CHIKV (Fig. 1C), also with no wild-type contamination in the preparation. The proper insertion and correct sequence of the CHIKV structural genes in the TK locus and the correct deletions of the VACV genes C6L, K7R, and A46R were also confirmed by DNA sequencing (data not shown).
FIG 1.
Generation and in vitro characterization of recombinant MVA-CHIKV. (A) Scheme of the MVA-CHIKV genome map. The different regions are indicated by capital letters. The right and left terminal regions are shown. Below the map, the deleted or fragmented genes are depicted as black boxes. The CHIKV C, E3, E2, 6K, and E1 structural genes (from CHIKV isolate LR2006-OPY1), driven by the synthetic early/late (sE/L) virus promoter and inserted within the VACV TK viral locus (J2R), are indicated. The deleted VACV C6L, K7R, and A46R genes are also indicated. TK-L, TK left; TK-R, TK right. (B) PCR analysis of the VACV TK locus. Viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell with MVA-CHIKV, MVA-GFP, or MVA-WT. Primers spanning the TK locus-flanking regions were used for PCR analysis of the CHIKV genes inserted within the TK locus. DNA products with their corresponding sizes (base pairs) are indicated by an arrow on the right. A molecular size marker (1-kb ladder) with the corresponding sizes (base pairs) is indicated on the left. (C) PCR analysis of the C6L, K7R, and A46R loci. Viral DNA was extracted from DF-1 cells mock infected or infected at 5 PFU/cell with MVA-CHIKV, MVA-GFP, or MVA-WT. Primers spanning C6L-, K7R-, and A46R-flanking regions were used for PCR analysis of the C6L, K7R, and A46R loci, respectively. DNA products with the corresponding size (base pairs) of the parental virus (wt) or the virus with the C6L, K7R, and A46R deletions are indicated by an arrow on the right. A molecular size marker (1-kb ladder) with the corresponding sizes (base pairs) is indicated on the left. (C) Expression of CHIKV E1 and E2 proteins. DF-1 cells were mock infected or infected at 5 PFU/cell with MVA-CHIKV, MVA-GFP, or MVA-WT. At 24 h postinfection, cells were lysed in Laemmli buffer, fractionated by 10% SDS-PAGE, and analyzed by Western blotting using rabbit polyclonal antibody against E1 or mouse polyclonal antibody against E2. Arrows on the right indicate the positions of the CHIKV E1 and E2 proteins, with the corresponding sizes in kDa. The sizes of standards (in kDa) (Precision Plus protein standards; Bio-Rad Laboratories) are indicated on the left.
Additionally, to confirm that MVA-CHIKV constitutively expresses and correctly processes the CHIKV structural polyprotein C-E3-E2-6K-E1, we carried out a Western blot analysis of DF-1 cells infected with MVA-CHIKV, MVA-GFP, or MVA-WT using specific antibodies that recognize the CHIKV E1 and E2 proteins. The results demonstrated that the viral polyprotein was correctly processed and that MVA-CHIKV expressed the CHIKV E1 and E2 proteins (Fig. 1D). Expression of the C, E3, and 6K proteins is not shown, since we did not have specific antibodies. However, the proper processing of the polyprotein indicates that the other proteins should be correctly expressed by MVA-CHIKV.
MVA-CHIKV replicates in cell culture.
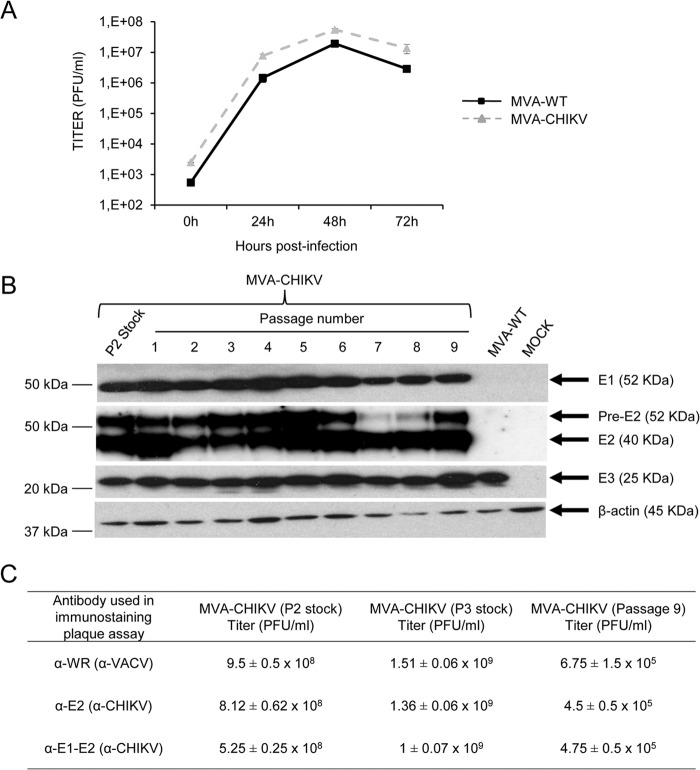
To further determine whether expression of CHIKV structural proteins affects MVA replication in cell culture, we next analyzed the growth of MVA-CHIKV and MVA-WT in DF-1 cells. The results showed that the kinetics of viral growth were similar between the two viruses (Fig. 2A), indicating that the constitutive expression of CHIKV structural proteins does not impair vector replication under permissive conditions.
FIG 2.
Viral growth kinetics and stability of MVA-CHIKV. (A) Viral growth kinetics. Monolayers of DF-1 cells were infected at 0.01 PFU/cell with MVA-WT or MVA-CHIKV. At different times postinfection (0, 24, 48, and 72 h), cells were collected and virus titers in cell lysates were quantified by a plaque immunostaining assay with anti-WR antibodies. The means of results from two independent experiments are shown. (B) Stability of MVA-CHIKV. MVA-CHIKV (P2 stock) was continuously grown to passage 9 in DF-1 cells. Then, DF-1 cells were mock infected or infected with MVA-CHIKV from the different passages or with MVA-WT. At 24 h postinfection, cells were lysed in Laemmli buffer, fractionated by 10% SDS-PAGE, and analyzed by Western blotting using rabbit polyclonal antibody against E1 or mouse polyclonal antibody against E2. Rabbit anti-β-actin antibody was used as a protein loading control. Rabbit anti-VACV early E3 protein antibody was used as a VACV loading control. Arrows on the right indicate the positions of the CHIKV E1 and E2 proteins, β-actin, and the VACV E3 protein, with their corresponding sizes in kDa. The sizes of standards (in kDa) (Precision Plus protein standards; Bio-Rad Laboratories) are indicated on the left. (C) Genetic stability of MVA-CHIKV, as determined by plaque immunostaining. Virus titers of MVA-CHIKV (P2 stock, P3 stock, and passage 9) were quantified by a plaque immunostaining assay with anti-VACV and anti-CHIKV antibodies. The means and standard deviations of results from two independent determinations are shown. It should be noted that the P3 stock represents virus purified with two sucrose cushions, while P2 and passage 9 stocks are virus titers from crude cell lysates.
MVA-CHIKV is stable in cell culture.
To ensure that MVA-CHIKV can be maintained in cultured cells without the loss of the CHIKV transgene, a stability test was performed. Here, the recombinant MVA-CHIKV was further grown in DF-1 cells infected at a low multiplicity during 9 consecutive passages, and expression of the CHIKV E1 and E2 proteins during the different passages was determined by Western blotting (Fig. 2B). The results showed that MVA-CHIKV efficiently expresses the CHIKV E1 and E2 proteins after successive passages, indicating that the recombinant MVA-CHIKV is genetically stable. Moreover, we also analyzed the genetic stability of MVA-CHIKV at passage 9 by a plaque immunostaining assay (Fig. 2C). The results showed that MVA-CHIKV titers (in the P2 stock, in the P3 stock, and at passage 9) were similar when plaques expressing VACV antigens or plaques expressing CHIKV antigens were analyzed, confirming the high genetic stability of MVA-CHIKV.
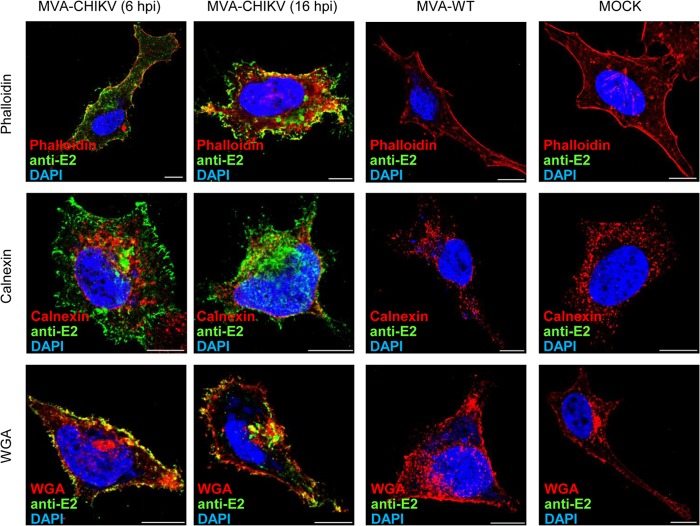
MVA-CHIKV expresses the CHIKV E2 protein in the cytoplasm and in the cell membrane.
The expression and intracellular localization of the structural CHIKV E2 protein was also analyzed by immunofluorescence in human HeLa cells infected with MVA-WT or MVA-CHIKV using antibodies specific to E2 and specific probes or antibodies for different proteins and compartments, such as actin (phalloidin), ER (anticalnexin), and the Golgi apparatus or Golgi apparatus-derived membranes, as well as for the plasma membrane (WGA). As shown in Fig. 3, the CHIKV E2 protein is expressed mainly at the plasma membrane and throughout the cytoplasm, where it is localized in discrete accumulations at the viral factories. Moreover, no colocalization between E2 and the ER was observed. Nevertheless, partial colocalization between E2 and actin was found in discrete areas and spots, and it was also detected between E2 and the plasma membrane and between E2 and the Golgi apparatus or Golgi apparatus-derived membranes.
FIG 3.
Immunofluorescence analysis of CHIKV E2 protein produced in HeLa cells infected with MVA-CHIKV. Subconfluent HeLa cells mock infected or infected with MVA-CHIKV or MVA-WT were fixed at 6 and 16 hpi (MVA-WT- and mock-infected cells are represented at 16 hpi), labeled with the corresponding primary antibodies or probes, monitored for appropriate fluorescence from protein-conjugated secondary antibodies, and visualized by confocal microscopy. The antibodies or probes used were anti-E2 (D3-62; to detect CHIKV E2 protein, shown in green), phalloidin (to stain actin, shown in red), anti-calnexin (to detect the ER, shown in red), WGA (to detect Golgi apparatus membranes and the plasma membrane, shown in red), and DAPI (to mark DNA, shown in blue). Bar, 10 μm.
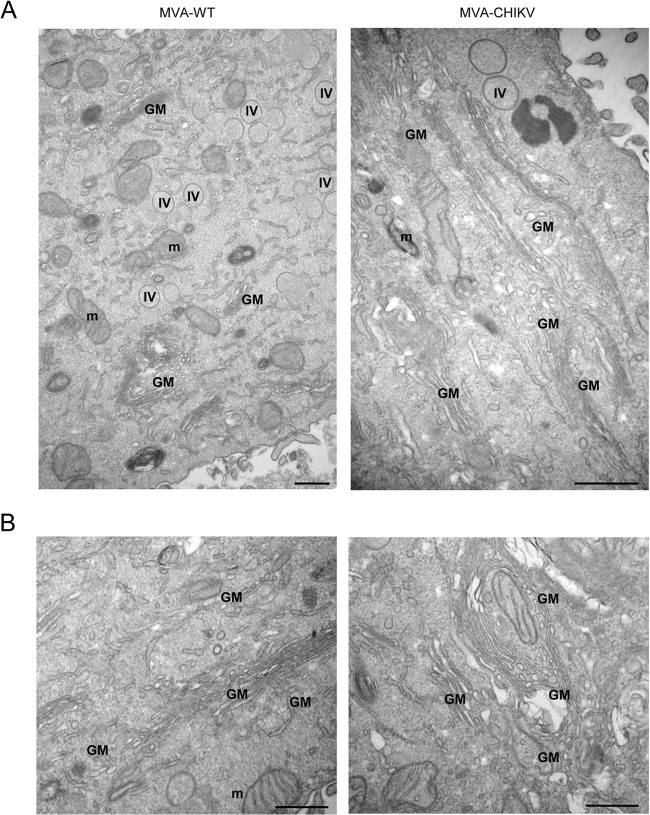
MVA-CHIKV induces specific morphological cell alterations, with the formation and accumulation of Golgi apparatus-derived membranes.
We next examined the impact of CHIKV structural proteins constitutively expressed by MVA-CHIKV on HeLa cell architecture using transmission electron microscopy analysis. Thus, ultrathin sections of HeLa cells infected with MVA-WT or MVA-CHIKV were visualized by electron microscopy at low and high magnifications (Fig. 4). Both in MVA-WT- and in MVA-CHIKV-infected cells, the assembly of immature virus (IV) forms of MVA was detected, and remarkably, in MVA-CHIKV-infected cells, an atypical growth and accumulation of Golgi apparatus-like membranes was observed.
FIG 4.
Architecture of HeLa cells following infection with MVA-CHIKV or MVA-WT. HeLa cells infected with MVA-WT or MVA-CHIKV were chemically fixed at 6 hpi and then processed for conventional embedding in an epoxy resin and observed by electron microscopy. (A) General overview of a cell infected with MVA-WT (left) and MVA-CHIKV (right); (B) two high-magnification panels of a cell infected with MVA-CHIKV. m, mitochondria; IV, immature virus; GM, Golgi apparatus membranes. Bars: 500 nm.
MVA-CHIKV triggers an innate immune response in human macrophages and dendritic cells, inducing type I IFN, proinflammatory cytokines, and chemokine expression.
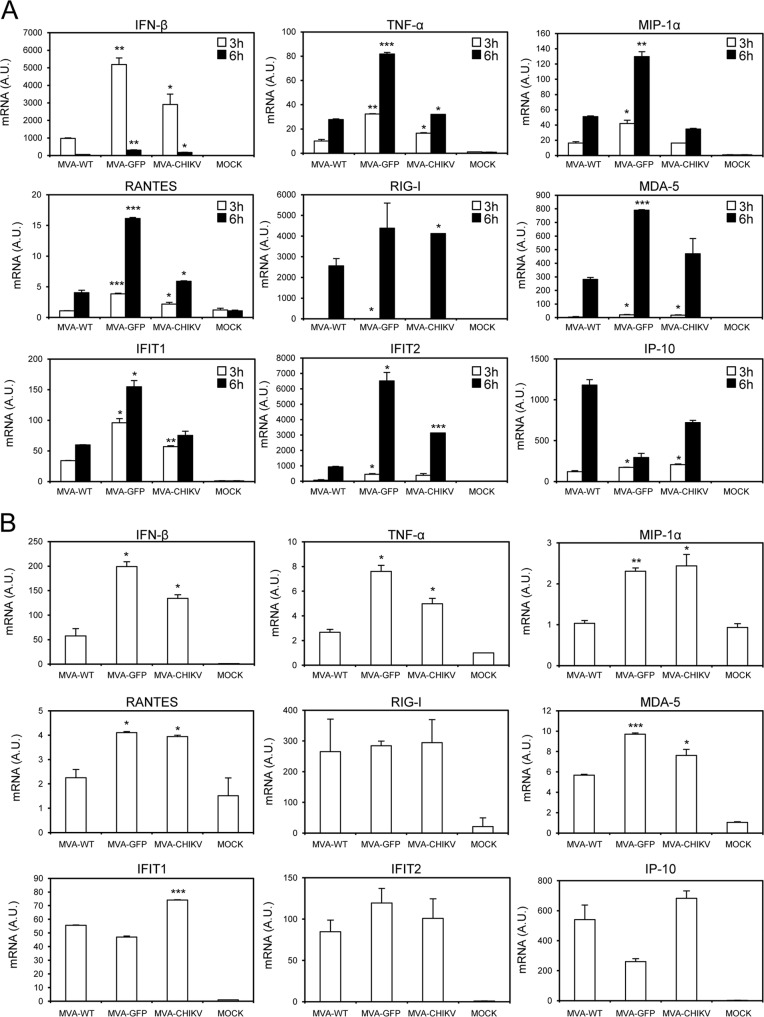
Type I IFN innate immune responses play a critical role in controlling CHIKV viral replication (62–66). Thus, to evaluate whether the presence of the CHIKV structural genes in the MVA genome is able to impair the response of innate immune cells to MVA infection, we analyzed by real-time PCR the expression of type I IFN (IFN-β), proinflammatory cytokines (TNF-α), chemokines (MIP-1α, IP-10, and RANTES), IFN-inducible genes (IFIT1 and IFIT2), and some key cytosolic sensors that lead to antiviral IFN production (RIG-I and MDA-5) by human THP-1 macrophages that had been mock infected or infected for 3 and 6 h with 5 PFU/cell of MVA-WT, MVA-GFP, and MVA-CHIKV (Fig. 5A). The results showed that, compared to MVA-WT, MVA-CHIKV significantly upregulated the mRNA levels of most of these genes (Fig. 5A).
FIG 5.
MVA-CHIKV triggers an innate immune response in human macrophages and dendritic cells, upregulating type I IFN, proinflammatory cytokines, and chemokine expression. Human THP-1 macrophages (A) and moDCs (B) were mock infected or infected with MVA-WT, MVA-GFP, or MVA-CHIKV at 5 PFU/cell (A) and 1 PFU/cell (B). At different times postinfection (3 h and 6 h in panel A, 6 h in panel B), RNA was extracted and the mRNA levels of IFN-β, TNF-α, MIP-1α, RANTES, IP-10, IFIT1, IFIT2, RIG-I, MDA-5, and HPRT were analyzed by reverse transcription-PCR. Results are expressed as the ratio of the gene of interest to the hypoxanthine phosphoribosyltransferase (HPRT) mRNA level. A.U., arbitrary units. P values indicate significantly higher responses in comparisons of MVA-WT to MVA-GFP or MVA-CHIKV at the same hour (*, P < 0.05; **, P < 0.005; ***, P < 0.001). Data are means ± standard deviations of results with duplicate samples from one experiment and are representative of two independent experiments.
To verify these results, we infected human moDCs with 1 PFU/cell of MVA-WT, MVA-GFP, and MVA-CHIKV and measured IFN-β, TNF-α, MIP-1α, IP-10, RANTES, IFIT1, IFIT2, RIG-I, and MDA-5 mRNA levels at 6 h postinfection (Fig. 5B). In the same way as in human THP-1 cells, in moDCs, MVA-CHIKV strongly increased the mRNA levels of most of these genes, compared to levels in MVA-WT-infected cells (Fig. 5B).
In conclusion, MVA-CHIKV promotes a robust innate immune response in human macrophages and moDCs by inducing the expression of type I IFN (IFN-β), proinflammatory cytokines (TNF-α), chemokines (MIP-1α, IP-10, and RANTES), IFN-inducible genes (IFIT1 and IFIT2), and some key cytosolic sensors that lead to antiviral IFN production (RIG-I and MDA-5).
MVA-CHIKV induces strong, broad, and polyfunctional adaptive CHIKV-specific T cell immune responses.
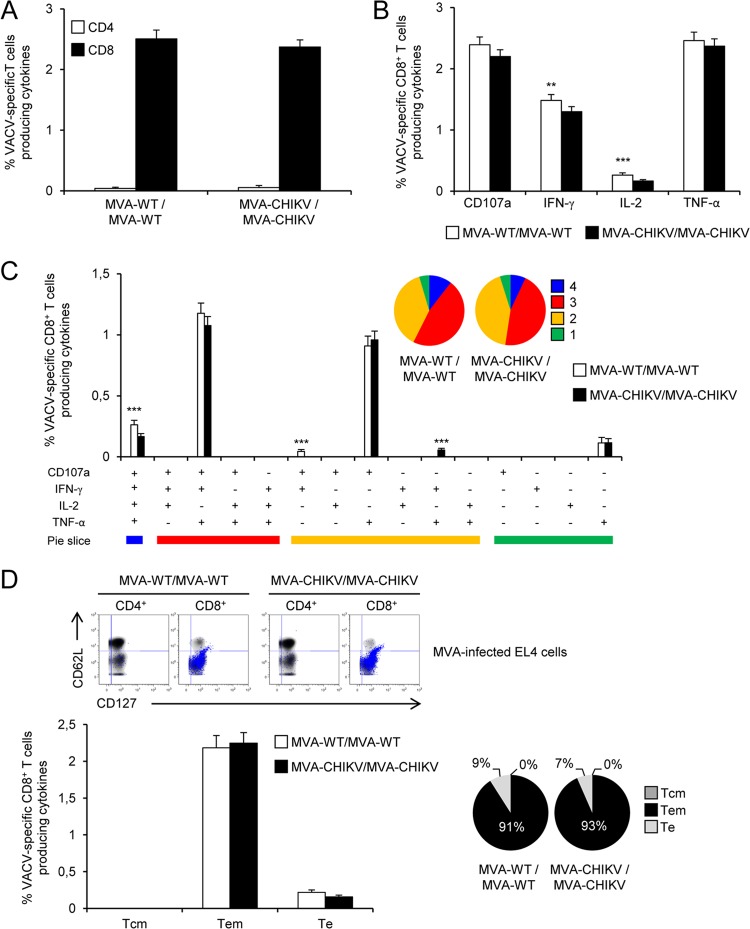
The role of T cell responses in controlling CHIKV infection is not well known (67, 68). Thus, to study in vivo the effect of the presence of the CHIKV structural genes in the MVA genome on cellular immunogenicity against CHIKV antigens, we next analyzed the CHIKV-specific immune responses induced by MVA-CHIKV in C57BL/6 mice by using a homologous MVA-CHIKV prime (1 × 107 PFU) and MVA-CHIKV boost (2 × 107 PFU) immunization protocol (see Materials and Methods). Animals primed with nonrecombinant MVA-WT and boosted with MVA-WT were used as a control group. Adaptive, CHIKV-specific T cell immune responses elicited by both immunization groups (MVA-WT/MVA-WT and MVA-CHIKV/MVA-CHIKV) were measured 10 days after the last immunization by a polychromatic intracellular cytokine staining (ICS) assay, after the stimulation of splenocytes with specific peptides representative of the C, E1, and E2 CHIKV proteins.
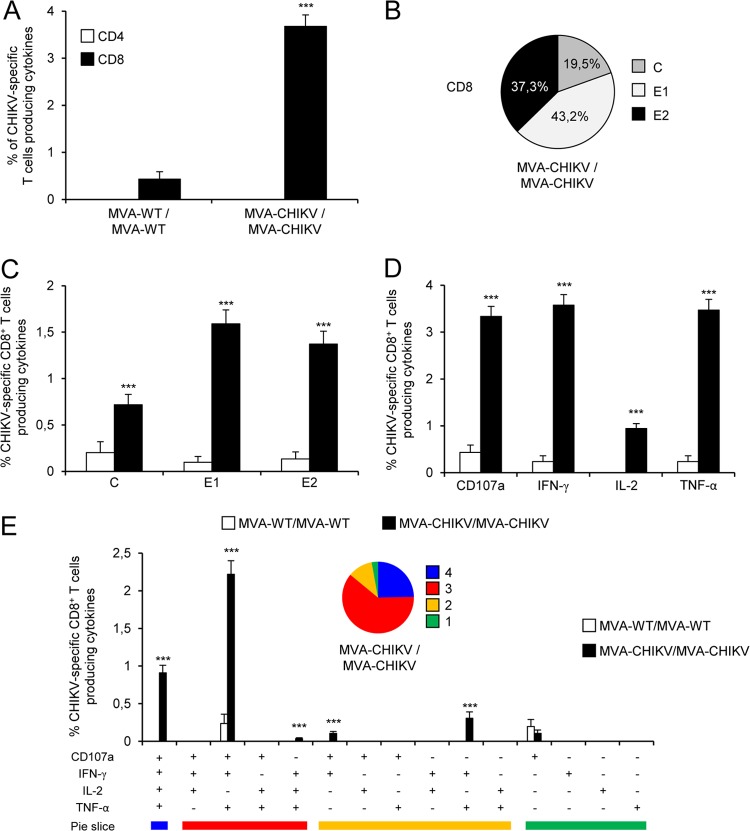
Immunization with MVA-CHIKV/MVA-CHIKV elicited robust adaptive CHIKV-specific CD8+ T cell immune responses (determined as the sum of the individual responses obtained for the C, E1, and E2 CHIKV peptides, producing IFN-γ, TNF-α, and/or IL-2 cytokines, as well as the expression of CD107a on the surfaces of activated T cells as an indirect marker of cytotoxicity) (Fig. 6A). The MVA-CHIKV/MVA-CHIKV immunization group triggered an overall CHIKV-specific immune response mediated only by CD8+ T cells, with no CHIKV-specific CD4+ T cells detected, indicating the selectivity for CD8+ T cells of the peptides used in the ICS assay (Fig. 6A).
FIG 6.
Immunization with MVA-CHIKV induces strong, broad, and polyfunctional adaptive CHIKV-specific T cell immune responses. Splenocytes were collected from mice (5 per group) immunized with MVA-WT/MVA-WT or MVA-CHIKV/MVA-CHIKV 10 days after the last immunization. Next, adaptive, CHIKV-specific CD4+ and CD8+ T cell immune responses triggered by both immunization groups were measured by the ICS assay following stimulation of splenocytes with different CHIKV peptides (C, E1, and E2). Values from unstimulated controls were subtracted in all cases. P values indicate significantly higher responses in comparisons of MVA-WT/MVA-WT to MVA-CHIKV/MVA-CHIKV (***, P < 0.001). Data are from one experiment and are representative of data from two independent experiments. (A) Overall magnitudes of CHIKV-specific CD4+ and CD8+ T cells. The values represent the sums of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against the C, E1, and E2 peptides. (B) Pattern of adaptive, CHIKV-specific CD8+ T cell immune responses to C, E1, and E2. Frequencies were calculated by reporting the number of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 and are specific for each peptide to the total number of CD8+ T cells in the MVA-CHIKV/MVA-CHIKV immunization group. (C) Magnitudes of C, E1, and E2 CHIKV-specific CD8+ T cells. Frequencies represent the sums of the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against the C, E1, or E2 peptide. (D) Magnitudes of CHIKV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against the C, E1, and E2 peptides. (E) Polyfunctional profile of adaptive, CHIKV-specific CD8+ T cell immune responses. All the possible combinations of the responses are shown on the x axis (15 different T cell populations), while the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against the C, E1, and E2 peptides are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (4, 3, 2, or 1). The pie charts summarize the data. Each slice corresponds to the proportion of CHIKV-specific CD8+ T cells exhibiting one, two, three, or four functions within the total population of CHIKV-specific CD8+ T cells.
The pattern of adaptive, CHIKV-specific T cell immune responses showed that CD8+ T cell responses induced by MVA-CHIKV/MVA-CHIKV were broad, with most of the responses directed mainly against the E1 and E2 peptides (80%) and to a lesser extent against C (Fig. 6B). Furthermore, compared to MVA-WT/MVA-WT, MVA-CHIKV/MVA-CHIKV significantly enhanced the magnitude of C-, E1- and E2-specific CD8+ T cell responses (P < 0.001) (Fig. 6C).
Moreover, CHIKV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α are the populations most induced by the MVA-CHIKV/MVA-CHIKV immunization group (Fig. 6D); levels were also of a significantly higher magnitude than those induced by MVA-WT/MVA-WT (P < 0.001) (Fig. 6D).
The quality of the adaptive, CHIKV-specific T cell immune response was characterized in part by the pattern of cytokine production and its cytotoxic potential. Thus, on the basis of the production of CD107a, IFN-γ, TNF-α, and IL-2 from CHIKV-specific CD8+ T cells, 15 different CHIKV-specific CD8+ T cell populations could be identified (Fig. 6E). MVA-CHIKV/MVA-CHIKV induced a high polyfunctional profile, with 97% of CD8+ T cells exhibiting two, three, or four functions (Fig. 6E). Furthermore, CD8+ T cells producing CD107a plus IFN-γ plus TNF-α plus IL-2, CD107a plus IFN-γ plus TNF-α, or IFN-γ plus TNF-α were the most abundant populations elicited by MVA-CHIKV/MVA-CHIKV, which, compared to MVA-WT/MVA-WT, also induced significantly higher increases in the percentages of most of the populations (P < 0.001) (Fig. 6E).
Collectively, these results demonstrate that MVA-CHIKV induced strong, broad, and polyfunctional adaptive CHIKV-specific CD8+ T cell immune responses. Similar findings were observed in two independent experiments.
MVA-WT and MVA-CHIKV induce similar magnitudes and polyfunctionalities of adaptive, VACV-specific T cell immune responses.
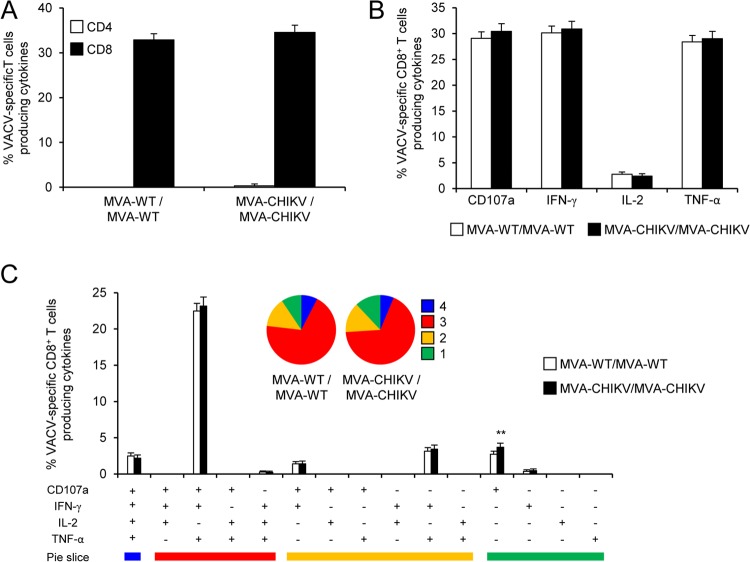
To further characterize the immune responses elicited in mice by both immunization groups (MVA-WT/MVA-WT and MVA-CHIV/MVA-CHIKV), it was of interest to analyze the responses to the MVA vector. Thus, we next measured adaptive, VACV-specific T cell immune responses induced by MVA-WT and MVA-CHIKV 10 days after the boost by an ICS assay (similar to the protocol followed in the analysis of adaptive CHIKV-specific T cell immune responses) after the stimulation of splenocytes with MVA-infected EL4 cells.
Both immunization groups triggered an overall adaptive, VACV-specific immune response mediated only by CD8+ T cells (determined as the sum of the individual responses producing IFN-γ, TNF-α, and/or IL-2 cytokines, as well as the expression of CD107a on the surfaces of activated T cells as an indirect marker of cytotoxicity); this experiment was carried out with MVA-infected EL4 cells, and the magnitudes of adaptive, VACV-specific CD8+ T cell immune responses were similar in the two immunization groups (Fig. 7A). Moreover, VACV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α were the most induced populations in both immunization groups, and they were induced at similar magnitudes (Fig. 7B).
FIG 7.
Adaptive, VACV-specific T cell immune responses. Splenocytes were collected from mice (5 per group) immunized with MVA-WT/MVA-WT or MVA-CHIKV/MVA-CHIKV 10 days after the last immunization. Next, adaptive, VACV-specific CD4+ and CD8+ T cell immune responses triggered by both immunization groups were measured by ICS assay following stimulation of splenocytes with MVA-infected EL4 cells. Values from unstimulated controls were subtracted in all cases. Data are from one experiment that is representative of two independent experiments. (A) Overall magnitude of VACV-specific CD4+ and CD8+ T cells. The values represent the sums of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected EL4 cells. (B) Percentages of VACV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against MVA-infected EL4 cells. (C) Functional profiles of adaptive, VACV-specific CD8+ T cell immune responses. All the possible combinations of the responses are shown on the x axis (15 different T cell populations), while the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected EL4 cells are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (4, 3, 2, or 1). The pie charts summarize the data. Each slice corresponds to the proportion of VACV-specific CD8+ T cells exhibiting one, two, three, or four functions within the total population of VACV-specific CD8+ T cells.
The quality of adaptive, VACV-specific CD8+ T cell immune responses was characterized by the production of CD107a, IFN-γ, TNF-α, and/or IL-2, and 15 distinct VACV-specific CD8+ T cell populations could be identified (Fig. 7C). VACV-specific CD8+ T cell responses were similarly polyfunctional in the two immunization groups, with 88 to 91% of the CD8+ T cells exhibiting two, three, or four functions. CD8+ T cells producing CD107a plus IFN-γ plus TNF-α plus IL-2, CD107a plus IFN-γ plus TNF-α, IFN-γ plus TNF-α, CD107a plus IFN-γ, or only CD107a were the most induced populations elicited by both immunization groups (at similar percentages) (Fig. 7C).
In summary, these results showed that MVA-WT and MVA-CHIKV elicited similar magnitudes and levels of quality of adaptive, VACV-specific CD8+ T cell immune responses. Similar findings were observed in two independent experiments.
MVA-CHIKV induces strong, broad, polyfunctional, and durable CHIKV-specific memory T cell immune responses.
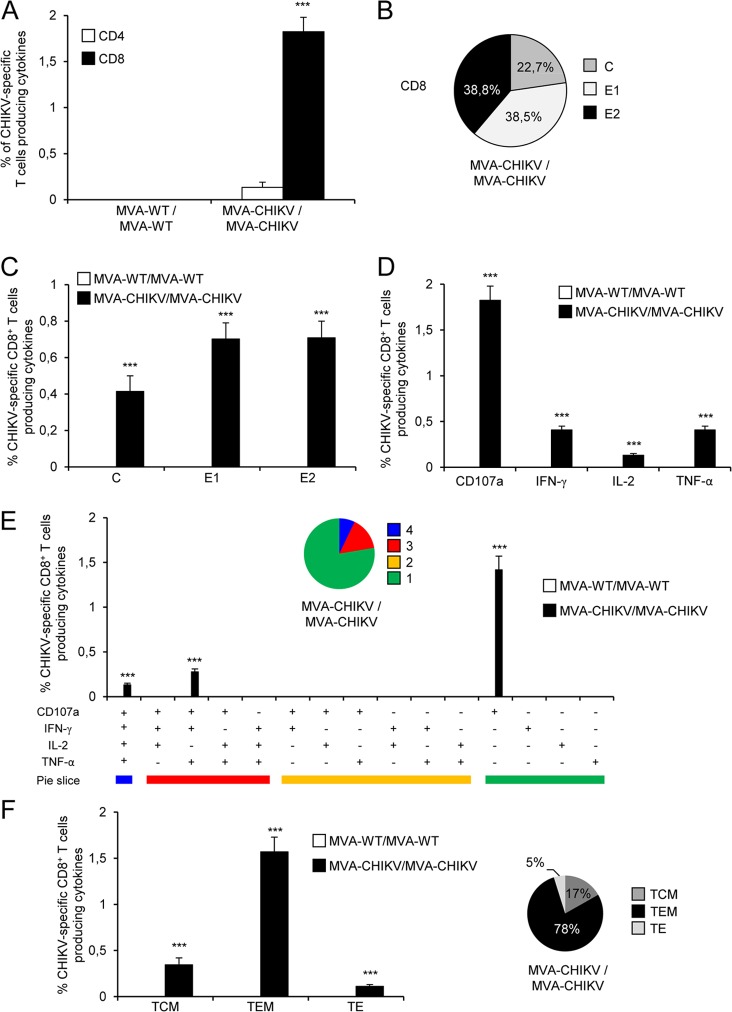
The durability of a vaccine-induced T cell response is an important feature, since long-term protection is a requirement for prophylactic vaccination. Thus, we analyzed CHIKV-specific memory T cell immune responses elicited by both immunization groups 52 days after the last immunization by the ICS assay in a manner similar to that of the protocol followed in the adaptive phase.
Immunization with MVA-CHIKV/MVA-CHIKV elicited robust CHIKV-specific CD8+ memory T cell immune responses (determined as the sum of the individual responses to the CHIKV C, E1, and E2 peptides in splenocytes producing IFN-γ, TNF-α, and/or IL-2 cytokines, and the expression of CD107a on the surfaces of activated T cells was used as an indirect marker of cytotoxicity) (Fig. 8A). As with the results obtained in the adaptive phase, the MVA-CHIKV/MVA-CHIKV immunization group triggered an overall CHIKV-specific memory immune response mediated only by CD8+ T cells, with no CHIKV-specific CD4+ T cells detected, indicating the selectivity for CD8+ T cells of the peptides used in the ICS assay (Fig. 8A).
FIG 8.
Immunization with MVA-CHIKV induces strong, broad, and polyfunctional CHIKV-specific memory T cell immune responses. Splenocytes were collected from mice (5 per group) immunized with MVA-WT/MVA-WT or MVA-CHIKV/MVA-CHIKV 52 days after the last immunization. CHIKV-specific CD4+ and CD8+ memory T cell immune responses triggered by both immunization groups were measured by ICS assay as described in the legend to Fig. 6. Values from unstimulated controls were subtracted in all cases. P values indicate significantly higher responses in comparisons of MVA-WT/MVA-WT to MVA-CHIKV/MVA-CHIKV (***, P < 0.001). Data are from one experiment that is representative of two independent experiments. (A) Overall magnitudes of CHIKV-specific CD4+ and CD8+ T cells. The values represent the sums of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against C plus E1 plus E2. (B) Pattern of C, E1, and E2 CHIKV-specific CD8+ memory T cell immune responses. Frequencies were calculated by reporting the number of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 and are specific for each peptide to the total number of CD8+ T cells in the MVA-CHIKV/MVA-CHIKV immunization group. (C) Magnitudes of C, E1, and E2 CHIKV-specific CD8+ T cells. Frequencies represent the sums of the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against the C, E1, or E2 peptide. (D) Magnitudes of CHIKV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of CD8+ T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against C plus E1 plus E2. (E) Polyfunctional profile of CHIKV-specific CD8+ memory T cell immune responses. All the possible combinations of the responses are shown on the x axis (15 different T cell populations), while the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against C plus E1 plus E2 are shown on the y axis. Responses are grouped and color coded on the basis of the number of functions (4, 3, 2, or 1). The pie charts summarize the data. Each slice corresponds to the proportion of CHIKV-specific CD8+ T cells exhibiting one, two, three, or four functions within the total population of CHIKV-specific CD8+ T cells. (F) Phenotypic profile of CHIKV-specific CD8+ memory T cells. CD127 and CD62L expression was used to identify central memory (TCM, CD127+ CD62L+), effector memory (TEM, CD127+ CD62L−), and effector (TE, CD127− CD62L−) subpopulations. Magnitudes of TCM, TEM, and TE CHIKV-specific CD8+ T cells are represented. The values represent percentages of TCM, TEM, and TE populations producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against the C plus E1 plus E2 peptides. The pie charts summarize the data. Each slice corresponds to the proportion of the TCM, TEM, or TE population within the total CHIKV-specific CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2. P values indicate significantly higher responses in comparisons of MVA-WT/MVA-WT to MVA-CHIKV/MVA-CHIKV (***, P < 0.001).
The pattern of CHIKV-specific memory T cell immune responses showed that CD8+ T cell responses induced by MVA-CHIKV/MVA-CHIKV were broad, with most of the responses being directed mainly against the E1 and E2 peptides (77%) and, to a lesser extent, against C (Fig. 8B), as with the results obtained in the adaptive phase. Furthermore, compared to MVA-WT/MVA-WT, MVA-CHIKV/MVA-CHIKV significantly enhanced the magnitudes of C-, E1-, and E2-specific CD8+ T cell responses (P < 0.001) (Fig. 8C).
Furthermore, CHIKV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α are the populations most induced by the MVA-CHIKV/MVA-CHIKV immunization group (Fig. 8D), and their levels are also of a significantly higher magnitude than those induced by MVA-WT/MVA-WT (P < 0.001) (Fig. 8D), results similar to those obtained in the adaptive phase.
The quality of the CHIKV-specific memory T cell immune responses was characterized by analyzing the simultaneous production of CD107a, IFN-γ, TNF-α, and/or IL-2 from CHIKV-specific CD8+ T cells (Fig. 8E), where 15 distinct CHIKV-specific CD8+ T cell populations could be identified. MVA-CHIKV/MVA-CHIKV induced a polyfunctional profile, with 22% of CD8+ T cells exhibiting two, three, or four functions (Fig. 8E). Furthermore, again, CD8+ T cells producing CD107a plus IFN-γ plus TNF-α plus IL-2, CD107a plus IFN-γ plus TNF-α, or CD107a were the most abundant populations elicited by MVA-CHIKV/MVA-CHIKV, which also induced increases in the percentages of most of the populations that were significantly higher than those induced by MVA-WT/MVA-WT (P < 0.001) (Fig. 8E), results similar to those obtained in the adaptive phase.
Moreover, we also determined the phenotype of the C-, E1-, and E2-specific memory T cells by measuring the expression of CD127 and CD62L surface markers, which allowed us to define the different memory subpopulations: central memory (TCM; CD127+/CD62L+), effector memory (TEM; CD127+/CD62L−), and effector (TE; CD127−/CD62L−) T cells (69) (Fig. 8F). The results showed that MVA-CHIKV/MVA-CHIKV induced CHIKV-specific CD8+ memory T cells (determined as the sum of the individual responses, i.e., the levels of CD107a, IFN-γ, TNF-α, and/or IL-2 produced against the C plus E1 plus E2 peptides), mainly of the TEM phenotype (78%) (Fig. 8F, right). Furthermore, immunization with MVA-CHIKV/MVA-CHIKV induced significantly higher increases in the percentages of CHIKV-specific CD8+ TCM, TEM, and TE cells than immunization with MVA-WT (P < 0.001) (Fig. 8F, left).
In summary, these results demonstrate that MVA-CHIKV induced strong, broad, polyfunctional, and durable CHIKV-specific CD8+ memory T cell immune responses. Similar findings were observed in two independent experiments.
MVA-WT and MVA-CHIKV induce similar magnitudes and polyfunctionalities of VACV-specific memory T cell immune responses.
VACV-specific memory T cell immune responses elicited by both immunization groups (MVA-WT/MVA-WT and MVA-CHIV/MVA-CHIKV) were measured 52 days after the boost by ICS assay, which is similar to the protocol followed in the adaptive phase.
Similarly to the results obtained in the adaptive phase, both immunization groups triggered overall VACV-specific memory immune responses mediated only by CD8+ T cells, determined as the sums of the individual levels of production of CD107a, IFN-γ, TNF-α, and/or IL-2 obtained in MVA-infected EL4 cells (Fig. 9A), with the magnitudes of VACV-specific CD8+ memory T cell immune responses being similar in the two immunization groups (Fig. 9A). Moreover, VACV-specific CD8+ T cells producing CD107a, IFN-γ, or TNF-α are the most induced populations in both immunization groups, and these populations were also produced at similar magnitudes (Fig. 9B).
FIG 9.
VACV-specific memory T cell immune responses. Splenocytes were collected from mice (5 per group) immunized with MVA-WT/MVA-WT or MVA-CHIKV/MVA-CHIKV 52 days after the last immunization. Next, VACV-specific CD4+ and CD8+ memory T cell immune responses triggered by both immunization groups were measured by ICS assay following stimulation of splenocytes with MVA-infected EL4 cells. Values from unstimulated controls were subtracted in all cases. P values indicate significantly higher responses in comparisons of MVA-WT/MVA-WT to MVA-CHIKV/MVA-CHIKV (***, P < 0.001). Data are from one experiment that is representative of two independent experiments. (A) Overall magnitude of VACV-specific CD4+ and CD8+ memory T cells. The values represent the sums of the percentages of T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected EL4 cells. (B) Percentage of VACV-specific CD8+ T memory cells producing CD107a, IFN-γ, TNF-α, or IL-2. Frequencies represent percentages of CD8+ memory T cells producing CD107a, IFN-γ, TNF-α, or IL-2 against MVA-infected EL4 cells. (C) Functional profiles of VACV-specific CD8+ memory T cell immune responses. Responses are grouped and color coded on the basis of the number of functions (4, 3, 2, or 1). All possible combinations of the responses are shown on the x axis (15 different T cell populations), while the percentages of CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected EL4 cells are shown on the y axis. The pie charts summarize the data. Each slice corresponds to the proportion of VACV-specific CD8+ memory T cells exhibiting one, two, three, or four functions within the total population of VACV-specific CD8+ T cells. (D) Phenotypic profile of VACV-specific CD4+ and CD8+ memory T cells. Fluorescence-activated cell sorter (FACS) plots of MVA-infected EL4 cells are represented. CD127 and CD62L expression was used to identify central memory (TCM, CD127+ CD62L+), effector memory (TEM, CD127+ CD62L−), and effector (TE, CD127− CD62L−) subpopulations. The memory T cell subpopulations are depicted as density plots. Blue dots represent T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2. The graph represents the magnitudes of TCM, TEM, and TE VACV-specific CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2. Frequencies represent percentages of TCM, TEM, and TE populations producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2 against MVA-infected EL4 cells. The pie charts summarize the data. Each slice corresponds to the proportion of the TCM, TEM, or TE population within the total population of VACV-specific CD8+ T cells producing CD107a and/or IFN-γ and/or TNF-α and/or IL-2.
The quality of VACV-specific CD8+ memory T cell immune responses was characterized as the simultaneous production of CD107a, IFN-γ, TNF-α, and/or IL-2 and was assessed by using a protocol like that followed in the adaptive phase, where 15 distinct VACV-specific CD8+ T cell populations could be identified (Fig. 9C). VACV-specific CD8+ T cell responses were similarly polyfunctional in the two immunization groups, with 95% of CD8+ T cells exhibiting two, three, or four functions. CD8+ T cells producing CD107a plus IFN-γ plus TNF-α plus IL-2, CD107a plus IFN-γ plus TNF-α, IFN-γ plus TNF-α, CD107a plus IFN-γ, or only CD107a were the most induced populations elicited by both immunization groups, and they were also induced at similar percentages (Fig. 9C).
Additionally, the analysis of the memory phenotype of VACV-specific CD8+ memory T cells by measurement of the expression of CD127 and CD62L surface markers showed that both immunization groups elicited mainly VACV-specific CD8+ memory T cells of the TEM phenotype and did so at similar magnitudes (Fig. 9D).
In summary, these results showed that MVA-WT and MVA-CHIKV elicited similar magnitudes and levels of quality of VACV-specific CD8+ memory T cell immune responses. Similar findings were observed in two independent experiments.
MVA-CHIKV induces high titers of neutralizing antibodies against CHIKV.
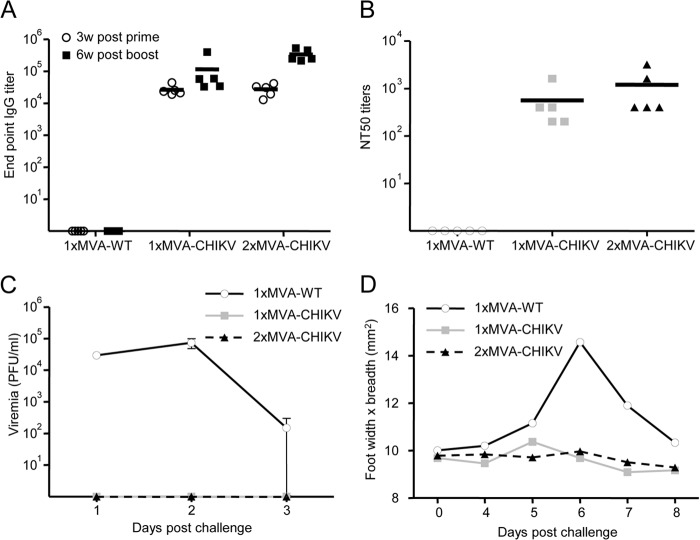
Antibodies against CHIKV are crucial to control CHIKV infection (37, 42, 44, 45, 70–78). Thus, to study the ability of MVA-CHIKV to elicit humoral immune responses against CHIKV, we analyzed the levels of CHIKV envelope-specific antibodies present in the sera of C57BL/6 mice immunized with one or two doses of MVA-CHIKV (see Materials and Methods). Animals immunized with nonrecombinant MVA-WT were used as a control group. The results show that one immunization with MVA-CHIKV elicited high titers of IgG antibodies against CHIKV that were further enhanced by the second immunization (Fig. 10A). Nevertheless, a single immunization with MVA-CHIKV was enough to induce high levels of IgG antibody responses to CHIKV. Next, we analyzed the titers of neutralizing antibodies against CHIKV present in the sera of immunized mice at 6 weeks postboost. The results showed that in contrast to infection with MVA-WT, immunization with one or two doses of MVA-CHIKV resulted in the production of high titers of CHIKV-neutralizing antibodies (Fig. 10B). In comparison to neutralizing antibody titers after a single immunization, titers after two doses were slightly increased.
FIG 10.
Immunization with MVA-CHIKV induces neutralizing antibodies against CHIKV and protects mice against CHIKV infection. Five C57BL/6 mice per group were immunized as described in Materials and Methods. Data are from one experiment that is representative of two independent experiments. (A) Antibody titers against CHIKV E1-p62 detected by ELISA at 3 weeks after the prime or 6 weeks after the boost in sera of animals immunized with one dose of MVA-WT or one or two doses of MVA-CHIKV. The detection limit is 100. (B) CHIKV-neutralizing antibody titers (NT50) detected at 6 weeks after the last immunization in sera of animals immunized with one dose of MVA-WT or one or two doses of MVA-CHIKV. The detection limit is 100. (C) Viremia detected in animals immunized with one dose of MVA-WT or one or two doses of MVA-CHIKV and challenged with a total of 106 PFU of CHIKV subcutaneously in both feet. Blood samples were collected at days 1 to 3 postchallenge from tail bleedings, and viremia was analyzed by plaque assay (PFU/ml). The detection limit is 250 PFU/ml. (D) Foot swelling in animals immunized with one dose of MVA-WT or one or two doses of MVA-CHIKV and challenged with a total of 106 PFU of CHIKV subcutaneously in both feet. Foot swelling was analyzed on the day of challenge and days 4 to 8 following challenge.
MVA-CHIKV protects mice against CHIKV infection.
To study whether MVA-CHIKV protected mice against a CHIKV challenge, we immunized mice with one or two doses of MVA-CHIKV or one dose of MVA-WT (as a control). Seven weeks after the last immunization, mice were challenged with a high dose of CHIKV in their feet (see Materials and Methods). Protection was evaluated by determining viremia and analyzing foot swelling in CHIKV-challenged mice during the days following challenge. The results showed that no CHIKV infection was developed in MVA-CHIKV-vaccinated mice, as no viremia was detected postchallenge (Fig. 10C). However, MVA-WT-immunized mice developed CHIKV infection, with high levels of CHIKV (104 to 105 PFU/ml) detected in blood that peaked day 2 postchallenge (Fig. 10B). Moreover, MVA-CHIKV-vaccinated mice did not develop foot swelling after challenge, while MVA-WT-immunized mice developed severe foot swelling that peaked 6 days postchallenge (Fig. 10D).
In conclusion, MVA-CHIKV is a highly effective vaccine that protected mice against challenge with a high dose of CHIKV, with no viremia detected in their blood and no signs of inflammation. Remarkably, just a single dose of MVA-CHIKV protected all mice from challenge with CHIKV.
DISCUSSION
CHIKV is an important emerging virus that causes a severe disease in humans; there have been recent outbreaks of the epidemic worldwide (4, 6, 24, 25). Thus, finding and developing an effective vaccine are needed to avoid CHIKV spreading and to prevent the disease's progression (26).
In this study, we generated a new and effective CHIKV vaccine candidate based on the highly attenuated poxvirus vector MVA expressing the CHIKV C, E3, E2, 6K, and E1 structural genes (termed MVA-CHIKV). This novel vaccine candidate is highly stable in cell culture and expresses high levels of CHIKV antigens. We observed colocalization of the E2 protein expressed by MVA-CHIKV with the plasma membrane, which was highly expected given that E2 is responsible for viral attachment to the cellular surface via receptor binding prior to its entry via endocytosis (79). Moreover, as it has been reported that the viral envelope protein E1, which forms a heterodimeric complex with E2 (13), plays a direct role in the fusion of the viral membrane to the plasma membrane and to intracellular membranes (80); therefore, it is very probable that we are visualizing E2 associated with E1. In addition, MVA-CHIKV induces specific morphological cell alterations with the formation and accumulation of Golgi apparatus-derived membranes. This striking overgrowth of Golgi stacks may be due to the presence of a large amount of CHIKV structural proteins, especially E1 and E2, that interact with cellular membranes directly and that somehow might induce the profusion of certain cellular membranes. Furthermore, we have not detected virus-like particles (VLPs), other than the C, E3, E2, 6K, and E1 CHIKV proteins expressed by MVA-CHIKV (from CHIKV strain LR2006 OPY-1), after infection of HeLa cells. The lack of VLP formation after cloning and expressing this same structural CHIKV cassette has been shown to be strain dependent, with the yield of VLPs from CHIKV strain 37997 approximately 100 times more than that from CHIKV strain LR2006 OPY-1 (37–39). Moreover, MVA-CHIKV triggers a robust innate immune response with the upregulation of IFN-β, proinflammatory cytokines, and chemokines in human macrophages and moDCs, effector molecules which are likely to influence the type of immune B and T cells activated and, in turn, the vaccine's efficacy. The increases in innate immune responses triggered by MVA-CHIKV and the parental virus, MVA-GFP, were similar, revealing that this enhancement may not be due to the heterologous GFP or CHIKV antigens present in the MVA genome but rather due to the absence in MVA-GFP and MVA-CHIKV of the VACV immunomodulatory C6L, K7R, and A46R genes, whose deletion was previously described and which enhanced the innate immune responses elicited by an MVA-based HIV/AIDS vaccine candidate (51). Furthermore, MVA-CHIKV is highly immunogenic in C57BL/6 mice, inducing strong, broad, and polyfunctional CHIKV-specific CD8+ T cell adaptive and memory immune responses. Most of the responses were directed against E1 and E2 and to a lesser extent against C, and CHIKV-specific CD8+ effector memory T cells were induced. Interestingly, the vaccine induced high titers of neutralizing antibodies against CHIKV, even after a single dose. Remarkably, just a single dose of MVA-CHIKV protected all mice from challenge with CHIKV, showing a strong protective effect in this experimental model. This relevant characteristic may be important in future human clinical trials in the areas were CHIKV is endemic, where a CHIKV vaccine that induces a long-term protective effect after one or two doses is desirable.
Currently, there is not a licensed vaccine that can control CHIKV infection (26), although several CHIKV vaccine candidates have been developed using different strategies (27–45). All these approaches were immunogenic in preclinical trials (mice or nonhuman primates) or early human clinical trials, but many of them required many immunizations and will be expensive to manufacture. Thus, further improvements are needed to obtain a strong and long-lasting protective immunity in humans, with fewer immunizations, a greater safety profile, and low costs.
Poxviruses and, in particular, MVA have been extensively used as vectors against many different infectious diseases, such as HIV, malaria, tuberculosis, hepatitis B, hepatitis C, influenza, and cancer (46–49). For use as vaccine candidates against different infectious diseases, MVA vectors contain some ideal characteristics, such as a good safety profile, low cost, ease of manufacture, high expression of heterologous antigens, and high immunogenicity profile. Furthermore, it has been reported that a viral vector, such as vaccinia virus, induces high levels of memory B cells (81) that are maintained much longer than protein antigen vaccines, playing a role in the maintenance of protective immunity because of the strong innate responses during vaccination.
The high innate, adaptive, and memory cellular immunogenicity and humoral immune responses elicited by MVA-CHIKV will likely contribute to the effective protection observed in C57BL/6 mice and will be a more effective way to obtain durable protection against CHIKV. Strong innate immune responses, with high levels of type I IFN, proinflammatory cytokines, and chemokines, have been detected in acute CHIKV-infected patients (64–66, 82–90), nonhuman primates (91), and mice (62–64, 66, 92, 93) and might play a critical role in controlling virus replication. Although high levels of some proinflammatory cytokines have been associated with disease severity (82–85, 87–89), type I IFN was identified as a key mediator responsible for viral clearance, and IFN-stimulated genes (ISGs) or IFN response factors 3 and 7 have been described to be critical in the control of CHIKV infection (63, 64, 66, 90, 94, 95). Moreover, IFN-α/β knockout mice (A129 mice) exhibited severe CHIKV disease (62–64, 66, 92), confirming the key role of type I IFNs in the control of CHIKV infection, by restricting virus replication early in the infection, although their effects are not enough to eliminate CHIKV from infected hosts. In fact, it has been reported that in the absence of an IFN-α/β response, IFN-γ partially controls CHIKV replication (77). Therefore, our results showing that MVA-CHIKV promotes a strong innate immune response in human macrophages and moDCs, upregulating the expression of IFN-β, IFN-inducible genes (IFIT1 and IFIT2), and some key cytosolic sensors that lead to antiviral IFN production (RIG-I and MDA-5), could be relevant to promoting the recruitment of leukocytes to sites of infection and therefore be an efficient antiviral defense in the control of CHIKV infection after challenge.
In epidemiological studies, anti-CHIKV antibodies have been shown to play an important role in protection (70, 72–74). In addition, natural antibodies present in the sera of uninfected C57BL/6 mice were able to partially inhibit CHIKV (75). Furthermore, therapies based on passively transferred anti-CHIKV neutralizing antibodies conferred protection to A129 mice (42, 44, 45, 70, 71, 73, 76–78) and nonhuman primates (37) against CHIKV and protection to mice against more distantly related arthrogenic alphaviruses (43, 93, 96), establishing the key role of humoral immunity in the control of CHIKV replication. Interestingly, complete protection from CHIKV challenge can be obtained using adoptive transfer of antibodies induced by immunization with a CHIKV/IRES vaccine candidate and occurs in the absence of CD4+/CD8+ T cells (42); a titer of 35 for neutralizing antibodies was sufficient to protect mice from a challenge with 100 PFU of CHIKV (42). Thus, the high titers of CHIKV-neutralizing antibodies induced by MVA-CHIKV in C57BL/6 mice, even after just one dose, is an important feature that may represent one of the most relevant parameters necessary to control CHIKV infection. In fact, we obtained NT50 neutralization titers of 103 to 104, compared to titers of 35 obtained previously using a CHIKV/IRES vaccine candidate (42); these were sufficient to protect mice from a challenge with a high dose of CHIKV (106 PFU).
The role of T cell responses in controlling CHIKV infection is not well understood, although they are probably needed to maintain long-term immunity, as CD8+ T cells are important in the control of virus-infected cells and CD4+ T cells are associated with the development of humoral immune responses (67, 68). Humans infected with CHIKV have large numbers of activated CD4+ and CD8+ CHIKV-specific T cells (65, 87), and a combination of strong innate immunity and CD8+ T cells was associated with the acute phase of infection, while CD4+ T cells were developed at a later stage of infection (65). CHIKV-specific CD4+ and CD8+ T cells producing IFN-γ were induced after CHIKV infection in C57BL/6 mice, but neither CD4+ nor CD8+ T cells were needed to control CHIKV viremia in C57BL/6 mice, and CD4+ T cells are involved in the pathology detected in the footpads of infected A129 mice (97). Furthermore, a CHIKV/IRES vaccine candidate activates adaptive CD4+ and CD8+ T cell and memory immune responses in A129 mice, but the adoptive transfer of these T cells did not confer protection against CHIKV challenge (42). However, CD4 knockout mice control CHIKV infection despite having small amounts of anti-CHIKV antibodies, showing that the breath of protective antibodies upon infection of mice with CHIKV is dependent on CD4+ T cells (75). Thus, CHIKV-specific T cell immune responses occur early in infection and play an early role in the control of viral replication, prior to generation of neutralizing antibodies. Therefore, our results showing that MVA-CHIKV induced high, broad, and polyfunctional CHIKV-specific CD8+ T cells producing CD107a, IFN-γ, TNF-α, and/or IL-2 in the adaptive and memory phases could be important in the protective effect observed in C57BL/6 mice, also if we take into consideration the fact that CHIKV challenge was administered 7 weeks after the last immunization. CHIKV-specific CD8+ T cells were directed mainly against E1 and E2, in a way similar to what has been previously described for C57BL/6 mice vaccinated with other CHIKV vaccine candidates after analysis of the immune response by enzyme-linked immunosorbent spot assay (ELISPOT) (33, 41). The fact that we detected only CHIKV-specific CD8+ T cell responses, and not CD4+ T cells, is due to the use of synthetic CHIKV C, E1, and E2 peptides that contain CD8+ T cell epitopes (41). Thus, generation of CHIKV-specific T cell responses by MVA-CHIKV may complement the strong humoral immune responses induced by the vaccine, leading to protection from CHIKV challenge.
Modified A129 mice, lacking the IFN-α/β receptor, are susceptible to lethal CHIKV infection (62), and some CHIKV vaccine candidates have been tested in this animal model to assess immune protection after challenge with CHIKV (34, 42-45, 77). However, adult C57BL/6 mice can be used as a model of CHIKV disease when CHIKV challenge is administered to the footpad (93, 98). Thus, to avoid the possibility that normal humoral and cellular immune responses against CHIKV are affected by the lack of a functional IFN-α/β response, we have defined the immunogenicity and efficacy of MVA-CHIKV in C57BL/6 wild-type mice as for other CHIKV vaccine candidates (33, 41). In our study, we analyzed foot swelling and not cell infiltration or tissue damage postchallenge. However, it has been described that, in our infection model, the level of foot swelling postinfection correlates to the level of mononuclear infiltrates and proinflammatory mediators, as well as tissue damage (93, 98), supporting the notion that CHIKV arthritis is an inflammatory disease; we therefore assume that the lack of foot swelling observed in mice immunized with MVA-CHIKV reflects a negligible level of local inflammatory cell infiltration and tissue damage. We cannot exclude the possibility that viral replication occurs in the feet, as we have analyzed only CHIKV viremia. However, it would be difficult to differentiate between the 106 PFU CHIKV injected at challenge and the possibly newly produced CHIKV, since high levels of viremia are present in feet as long as 9 days after infection (93, 98).
In conclusion, in this investigation, we described a novel CHIKV vaccine candidate based on the MVA vector (MVA-CHIKV) and defined its resulting innate, adaptive, and memory cellular immunogenicity, together with its ability to induce neutralizing antibodies against CHIKV, and efficacy in a mouse model. We showed that this vector is strongly immunogenic, eliciting high innate immune responses and strong CHIKV-specific humoral and cellular B and T cell immune responses, and is highly effective in protecting all mice from challenge with CHIKV after only one dose. Thus, this viral vector, MVA-CHIKV, represents a promising vaccine candidate against CHIKV and deserves to be further tested in nonhuman primates and in phase I clinical trials.
ACKNOWLEDGMENTS
This investigation was supported by ICRES (Integrated Chikungunya Research), a collaborative project supported by the European Union under the Health Cooperation Work Programme of the 7th Framework Program (grant agreement 261202). Mariano Esteban was supported by Spanish grant SAF2008-02036 and Peter Liljeström by the Swedish Research Council.
We thank Victoria Jiménez and Cristina Sánchez for tissue culture and virus growth and Janett Wieseler for excellent technical assistance with the CHIKV neutralization assay. We thank Philippe Desprès, Marc Lecuit, Therese Couderc, and Lisa F. P. Ng for providing anti-CHIKV antibodies and Félix Rey for providing the E2-E1 cell line.
Footnotes
Published ahead of print 8 January 2014
REFERENCES
- 1.Her Z, Kam YW, Lin RT, Ng LF. 2009. Chikungunya: a bending reality. Microbes Infect. 11:1165–1176. 10.1016/j.micinf.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 2.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671. 10.1016/S0140-6736(11)60281-X [DOI] [PubMed] [Google Scholar]
- 3.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. 2007. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7:319–327. 10.1016/S1473-3099(07)70107-X [DOI] [PubMed] [Google Scholar]
- 4.Powers AM, Logue CH. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363–2377. 10.1099/vir.0.82858-0 [DOI] [PubMed] [Google Scholar]
- 5.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8:491–500. 10.1038/nrmicro2368 [DOI] [PubMed] [Google Scholar]
- 6.Simon F, Savini H, Parola P. 2008. Chikungunya: a paradigm of emergence and globalization of vector-borne diseases. Med. Clin. North Am. 92:1323–1343, ix. 10.1016/j.mcna.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 7.Dupuis-Maguiraga L, Noret M, Brun S, Le Grand R, Gras G, Roques P. 2012. Chikungunya disease: infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Negl. Trop. Dis. 6:e1446. 10.1371/journal.pntd.0001446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandak NH, Kashyap RS, Kabra D, Karandikar P, Saha SS, Morey SH, Purohit HJ, Taori GM, Daginawala HF. 2009. Neurological complications of Chikungunya virus infection. Neurol. India 57:177–180. 10.4103/0028-3886.51289 [DOI] [PubMed] [Google Scholar]
- 9.Ganesan K, Diwan A, Shankar SK, Desai SB, Sainani GS, Katrak SM. 2008. Chikungunya encephalomyeloradiculitis: report of 2 cases with neuroimaging and 1 case with autopsy findings. AJNR Am. J. Neuroradiol. 29:1636–1637. 10.3174/ajnr.A1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin D. 2006. Alphaviruses, p 1023–1067 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 12.Simizu B, Yamamoto K, Hashimoto K, Ogata T. 1984. Structural proteins of Chikungunya virus. J. Virol. 51:254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. 10.1038/nature09555 [DOI] [PubMed] [Google Scholar]
- 14.Ross RW. 1956. The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J. Hyg. (Lond.) 54:177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgherini G, Poubeau P, Staikowsky F, Lory M, Le Moullec N, Becquart JP, Wengling C, Michault A, Paganin F. 2007. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin. Infect. Dis. 44:1401–1407. 10.1086/517537 [DOI] [PubMed] [Google Scholar]
- 16.Charrel RN, de Lamballerie X, Raoult D. 2007. Chikungunya outbreaks—the globalization of vectorborne diseases. N. Engl. J. Med. 356:769–771. 10.1056/NEJMp078013 [DOI] [PubMed] [Google Scholar]
- 17.Grandadam M, Caro V, Plumet S, Thiberge JM, Souares Y, Failloux AB, Tolou HJ, Budelot M, Cosserat D, Leparc-Goffart I, Despres P. 2011. Chikungunya virus, southeastern France. Emerg. Infect. Dis. 17:910–913. 10.3201/eid1705.101873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R, Djauzi, Wandra T, Master J, Kosasih H, Hartati S, Beckett C, Sedyaningsih ER, Beecham HJ, III, Corwin AL. 2005. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans. R. Soc. Trop. Med. Hyg. 99:128–141. 10.1016/j.trstmh.2004.03.013 [DOI] [PubMed] [Google Scholar]
- 19.Mavalankar D, Shastri P, Raman P. 2007. Chikungunya epidemic in India: a major public-health disaster. Lancet Infect. Dis. 7:306–307. 10.1016/S1473-3099(07)70091-9 [DOI] [PubMed] [Google Scholar]
- 20.Poletti P, Messeri G, Ajelli M, Vallorani R, Rizzo C, Merler S. 2011. Transmission potential of chikungunya virus and control measures: the case of Italy. PLoS One 6:e18860. 10.1371/journal.pone.0018860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. 2007. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840–1846. 10.1016/S0140-6736(07)61779-6 [DOI] [PubMed] [Google Scholar]
- 22.Schaffner F, Bellini R, Petric D, Scholte EJ, Zeller H, Rakotoarivony LM. 2013. Development of guidelines for the surveillance of invasive mosquitoes in Europe. Parasit. Vectors 6:209. 10.1186/1756-3305-6-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel MP, Brehin AC, Cubito N, Despres P, Kunst F, Rey FA, Zeller H, Brisse S. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263. 10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staples JE, Breiman RF, Powers AM. 2009. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin. Infect. Dis. 49:942–948. 10.1086/605496 [DOI] [PubMed] [Google Scholar]
- 25.Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. 2011. Chikungunya virus: evolution and genetic determinants of emergence. Curr. Opin. Virol. 1:310–317. 10.1016/j.coviro.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver SC, Osorio JE, Livengood JA, Chen R, Stinchcomb DT. 2012. Chikungunya virus and prospects for a vaccine. Expert Rev. Vaccines 11:1087–1101. 10.1586/erv.12.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison VR, Eckels KH, Bartelloni PJ, Hampton C. 1971. Production and evaluation of a formalin-killed Chikungunya vaccine. J. Immunol. 107:643–647 [PubMed] [Google Scholar]
- 28.Tiwari M, Parida M, Santhosh SR, Khan M, Dash PK, Rao PV. 2009. Assessment of immunogenic potential of Vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine 27:2513–2522. 10.1016/j.vaccine.2009.02.062 [DOI] [PubMed] [Google Scholar]
- 29.White A, Berman S, Lowenthal JP. 1972. Comparative immunogenicities of Chikungunya vaccines propagated in monkey kidney monolayers and chick embryo suspension cultures. Appl. Microbiol. 23:951–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. 2000. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. Am. J. Trop. Med. Hyg. 62:681–685 [DOI] [PubMed] [Google Scholar]
- 31.Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE, Jr, Lupton HW. 1986. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4:157–162. 10.1016/0264-410X(86)90003-4 [DOI] [PubMed] [Google Scholar]
- 32.Kumar M, Sudeep AB, Arankalle VA. 2012. Evaluation of recombinant E2 protein-based and whole-virus inactivated candidate vaccines against chikungunya virus. Vaccine 30:6142–6149. 10.1016/j.vaccine.2012.07.072 [DOI] [PubMed] [Google Scholar]
- 33.Chattopadhyay A, Wang E, Seymour R, Weaver SC, Rose JK. 2013. A chimeric vesiculo/alphavirus is an effective alphavirus vaccine. J. Virol. 87:395–402. 10.1128/JVI.01860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang E, Kim DY, Weaver SC, Frolov I. 2011. Chimeric Chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. J. Virol. 85:9249–9252. 10.1128/JVI.00844-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang E, Volkova E, Adams AP, Forrester N, Xiao SY, Frolov I, Weaver SC. 2008. Chimeric alphavirus vaccine candidates for chikungunya. Vaccine 26:5030–5039. 10.1016/j.vaccine.2008.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Suhrbier A, Penn-Nicholson A, Woraratanadharm J, Gardner J, Luo M, Le TT, Anraku I, Sakalian M, Einfeld D, Dong JY. 2011. A complex adenovirus vaccine against chikungunya virus provides complete protection against viraemia and arthritis. Vaccine 29:2803–2809. 10.1016/j.vaccine.2011.01.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. 2010. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16:334–338. 10.1038/nm.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metz SW, Gardner J, Geertsema C, Le TT, Goh L, Vlak JM, Suhrbier A, Pijlman GP. 2013. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl. Trop. Dis. 7:e2124. 10.1371/journal.pntd.0002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metz SW, van den Doel P, Geertsema C, Osterhaus AD, Vlak JM, Martina BE, Pijlman GP. 2013. Chikungunya virus-like particles are more immunogenic in a lethal AG129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 31:6092–6096. 10.1016/j.vaccine.2013.09.045 [DOI] [PubMed] [Google Scholar]
- 40.Mallilankaraman K, Shedlock DJ, Bao H, Kawalekar OU, Fagone P, Ramanathan AA, Ferraro B, Stabenow J, Vijayachari P, Sundaram SG, Muruganandam N, Sarangan G, Srikanth P, Khan AS, Lewis MG, Kim JJ, Sardesai NY, Muthumani K, Weiner DB. 2011. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS Negl. Trop. Dis. 5:e928. 10.1371/journal.pntd.0000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthumani K, Lankaraman KM, Laddy DJ, Sundaram SG, Chung CW, Sako E, Wu L, Khan A, Sardesai N, Kim JJ, Vijayachari P, Weiner DB. 2008. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine 26:5128–5134. 10.1016/j.vaccine.2008.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu H, Das SC, Fuchs JF, Suresh M, Weaver SC, Stinchcomb DT, Partidos CD, Osorio JE. 2013. Deciphering the protective role of adaptive immunity to CHIKV/IRES, a novel candidate vaccine against Chikungunya in the A129 mouse model. Vaccine 31:3353–3360. 10.1016/j.vaccine.2013.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Partidos CD, Paykel J, Weger J, Borland EM, Powers AM, Seymour R, Weaver SC, Stinchcomb DT, Osorio JE. 2012. Cross-protective immunity against o'nyong-nyong virus afforded by a novel recombinant chikungunya vaccine. Vaccine 30:4638–4643. 10.1016/j.vaccine.2012.04.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plante K, Wang E, Partidos CD, Weger J, Gorchakov R, Tsetsarkin K, Borland EM, Powers AM, Seymour R, Stinchcomb DT, Osorio JE, Frolov I, Weaver SC. 2011. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 7:e1002142. 10.1371/journal.ppat.1002142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandler S, Ruffie C, Combredet C, Brault JB, Najburg V, Prevost MC, Habel A, Tauber E, Despres P, Tangy F. 2013. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine 31:3718–3725. 10.1016/j.vaccine.2013.05.086 [DOI] [PubMed] [Google Scholar]
- 46.Cottingham MG, Carroll MW. 2013. Recombinant MVA vaccines: dispelling the myths. Vaccine 31:4247–4251. 10.1016/j.vaccine.2013.03.021 [DOI] [PubMed] [Google Scholar]
- 47.Gomez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2013. Clinical applications of attenuated MVA poxvirus strain. Expert Rev. Vaccines 12:1395–1416. 10.1586/14760584.2013.845531 [DOI] [PubMed] [Google Scholar]
- 48.Gomez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. 2011. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr. Gene Ther. 11:189–217. 10.2174/156652311795684731 [DOI] [PubMed] [Google Scholar]
- 49.Volz A, Sutter G. 2013. Protective efficacy of modified vaccinia virus Ankara in preclinical studies. Vaccine 31:4235–4240. 10.1016/j.vaccine.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 50.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480. 10.1371/journal.ppat.1000480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Garcia-Arriaza J, Arnaez P, Gomez CE, Sorzano CO, Esteban M. 2013. Improving adaptive and memory immune responses of an HIV/AIDS vaccine candidate, MVA-B, by deletion of vaccinia virus genes (C6L and K7R) blocking interferon signaling pathways. PLoS One 8:e66894. 10.1371/journal.pone.0066894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244. 10.1371/journal.pone.0024244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez CE, Najera JL, Jimenez EP, Jimenez V, Wagner R, Graf M, Frachette MJ, Liljestrom P, Pantaleo G, Esteban M. 2007. Head-to-head comparison on the immunogenicity of two HIV/AIDS vaccine candidates based on the attenuated poxvirus strains MVA and NYVAC co-expressing in a single locus the HIV-1BX08 gp120 and HIV-1(IIIB) Gag-Pol-Nef proteins of clade B. Vaccine 25:2863–2885. 10.1016/j.vaccine.2006.09.090 [DOI] [PubMed] [Google Scholar]
- 54.Ramirez JC, Gherardi MM, Esteban M. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923–933. 10.1128/JVI.74.2.923-933.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pohjala L, Utt A, Varjak M, Lulla A, Merits A, Ahola T, Tammela P. 2011. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One 6:e28923. 10.1371/journal.pone.0028923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liljestrom P, Garoff H. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology 9:1356–1361. 10.1038/nbt1291-1356 [DOI] [PubMed] [Google Scholar]
- 57.Liljestrom P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez CE, Perdiguero B, Cepeda MV, Mingorance L, Garcia-Arriaza J, Vandermeeren A, Sorzano CO, Esteban M. 2013. High, broad, polyfunctional, and durable T cell immune responses induced in mice by a novel hepatitis C virus (HCV) vaccine candidate (MVA-HCV) based on modified vaccinia virus Ankara expressing the nearly full-length HCV genome. J. Virol. 87:7282–7300. 10.1128/JVI.03246-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia-Arriaza J, Najera JL, Gomez CE, Sorzano CO, Esteban M. 2010. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS One 5:e12395. 10.1371/journal.pone.0012395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glasker S, Lulla A, Lulla V, Couderc T, Drexler JF, Liljestrom P, Lecuit M, Drosten C, Merits A, Kummerer BM. 2013. Virus replicon particle based Chikungunya virus neutralization assay using Gaussia luciferase as readout. Virol. J. 10:235. 10.1186/1743-422X-10-235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Najera JL, Gomez CE, Garcia-Arriaza J, Sorzano CO, Esteban M. 2010. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One 5:e11406. 10.1371/journal.pone.0011406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, Despres P, Arenzana-Seisdedos F, Michault A, Albert ML, Lecuit M. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 4:e29. 10.1371/journal.ppat.0040029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, Anraku I, Kumagai Y, Loo YM, Gale M, Jr, Akira S, Khromykh AA, Suhrbier A. 2012. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J. Virol. 86:9888–9898. 10.1128/JVI.00956-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schilte C, Couderc T, Chretien F, Sourisseau M, Gangneux N, Guivel-Benhassine F, Kraxner A, Tschopp J, Higgs S, Michault A, Arenzana-Seisdedos F, Colonna M, Peduto L, Schwartz O, Lecuit M, Albert ML. 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 207:429–442. 10.1084/jem.20090851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wauquier N, Becquart P, Nkoghe D, Padilla C, Ndjoyi-Mbiguino A, Leroy EM. 2011. The acute phase of Chikungunya virus infection in humans is associated with strong innate immunity and T CD8 cell activation. J. Infect. Dis. 204:115–123. 10.1093/infdis/jiq006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werneke SW, Schilte C, Rohatgi A, Monte KJ, Michault A, Arenzana-Seisdedos F, Vanlandingham DL, Higgs S, Fontanet A, Albert ML, Lenschow DJ. 2011. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 7:e1002322. 10.1371/journal.ppat.1002322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kam YW, Ong EK, Renia L, Tong JC, Ng LF. 2009. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect. 11:1186–1196. 10.1016/j.micinf.2009.09.003 [DOI] [PubMed] [Google Scholar]
- 68.Teo TH, Lum FM, Lee WW, Ng LF. 2012. Mouse models for Chikungunya virus: deciphering immune mechanisms responsible for disease and pathology. Immunol. Res. 53:136–147. 10.1007/s12026-012-8266-x [DOI] [PubMed] [Google Scholar]
- 69.Bachmann MF, Wolint P, Schwarz K, Oxenius A. 2005. Recall proliferation potential of memory CD8+ T cells and antiviral protection. J. Immunol. 175:4677–4685 [DOI] [PubMed] [Google Scholar]
- 70.Couderc T, Khandoudi N, Grandadam M, Visse C, Gangneux N, Bagot S, Prost JF, Lecuit M. 2009. Prophylaxis and therapy for Chikungunya virus infection. J. Infect. Dis. 200:516–523. 10.1086/600381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fric J, Bertin-Maghit S, Wang CI, Nardin A, Warter L. 2013. Use of human monoclonal antibodies to treat Chikungunya virus infection. J. Infect. Dis. 207:319–322. 10.1093/infdis/jis674 [DOI] [PubMed] [Google Scholar]
- 72.Kam YW, Lee WW, Simarmata D, Harjanto S, Teng TS, Tolou H, Chow A, Lin RT, Leo YS, Renia L, Ng LF. 2012. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. J. Virol. 86:13005–13015. 10.1128/JVI.01780-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kam YW, Lum FM, Teo TH, Lee WW, Simarmata D, Harjanto S, Chua CL, Chan YF, Wee JK, Chow A, Lin RT, Leo YS, Le Grand R, Sam IC, Tong JC, Roques P, Wiesmuller KH, Renia L, Rotzschke O, Ng LF. 2012. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol. Med. 4:330–343. 10.1002/emmm.201200213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kam YW, Simarmata D, Chow A, Her Z, Teng TS, Ong EK, Renia L, Leo YS, Ng LF. 2012. Early appearance of neutralizing immunoglobulin G3 antibodies is associated with chikungunya virus clearance and long-term clinical protection. J. Infect. Dis. 205:1147–1154. 10.1093/infdis/jis033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lum FM, Teo TH, Lee WW, Kam YW, Renia L, Ng LF. 2013. An essential role of antibodies in the control of Chikungunya virus infection. J. Immunol. 190:6295–6302. 10.4049/jimmunol.1300304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, Smit JM, Fremont DH, Pierson TC, Heise MT, Diamond MS. 2013. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 9:e1003312. 10.1371/journal.ppat.1003312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Partidos CD, Weger J, Brewoo J, Seymour R, Borland EM, Ledermann JP, Powers AM, Weaver SC, Stinchcomb DT, Osorio JE. 2011. Probing the attenuation and protective efficacy of a candidate chikungunya virus vaccine in mice with compromised interferon (IFN) signaling. Vaccine 29:3067–3073. 10.1016/j.vaccine.2011.01.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Selvarajah S, Sexton NR, Kahle KM, Fong RH, Mattia KA, Gardner J, Lu K, Liss NM, Salvador B, Tucker DF, Barnes T, Mabila M, Zhou X, Rossini G, Rucker JB, Sanders DA, Suhrbier A, Sambri V, Michault A, Muench MO, Doranz BJ, Simmons G. 2013. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus e2 protects from disease. PLoS Negl. Trop. Dis. 7:e2423. 10.1371/journal.pntd.0002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708. 10.1038/nature09546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuo SC, Chen YJ, Wang YM, Tsui PY, Kuo MD, Wu TY, Lo SJ. 2012. Cell-based analysis of Chikungunya virus E1 protein in membrane fusion. J. Biomed. Sci. 19:44. 10.1186/1423-0127-19-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amanna IJ, Carlson NE, Slifka MK. 2007. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357:1903–1915. 10.1056/NEJMoa066092 [DOI] [PubMed] [Google Scholar]
- 82.Chaaitanya IK, Muruganandam N, Sundaram SG, Kawalekar O, Sugunan AP, Manimunda SP, Ghosal SR, Muthumani K, Vijayachari P. 2011. Role of proinflammatory cytokines and chemokines in chronic arthropathy in CHIKV infection. Viral Immunol. 24:265–271. 10.1089/vim.2010.0123 [DOI] [PubMed] [Google Scholar]
- 83.Chirathaworn C, Poovorawan Y, Lertmaharit S, Wuttirattanakowit N. 2013. Cytokine levels in patients with chikungunya virus infection. Asian Pac. J. Trop. Med. 6:631–634. 10.1016/S1995-7645(13)60108-X [DOI] [PubMed] [Google Scholar]
- 84.Chirathaworn C, Rianthavorn P, Wuttirattanakowit N, Poovorawan Y. 2010. Serum IL-18 and IL-18BP levels in patients with Chikungunya virus infection. Viral Immunol. 23:113–117. 10.1089/vim.2009.0077 [DOI] [PubMed] [Google Scholar]
- 85.Chow A, Her Z, Ong EK, Chen JM, Dimatatac F, Kwek DJ, Barkham T, Yang H, Renia L, Leo YS, Ng LF. 2011. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J. Infect. Dis. 203:149–157. 10.1093/infdis/jiq042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Her Z, Malleret B, Chan M, Ong EK, Wong SC, Kwek DJ, Tolou H, Lin RT, Tambyah PA, Renia L, Ng LF. 2010. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J. Immunol. 184:5903–5913. 10.4049/jimmunol.0904181 [DOI] [PubMed] [Google Scholar]
- 87.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, Tallet F, Moiton MP, Gauzere BA, Bruniquet S, Jaffar Bandjee Z, Morbidelli P, Martigny G, Jolivet M, Gay F, Grandadam M, Tolou H, Vieillard V, Debre P, Autran B, Gasque P. 2010. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J. Immunol. 184:5914–5927. 10.4049/jimmunol.0900255 [DOI] [PubMed] [Google Scholar]
- 88.Kelvin AA, Banner D, Silvi G, Moro ML, Spataro N, Gaibani P, Cavrini F, Pierro A, Rossini G, Cameron MJ, Bermejo-Martin JF, Paquette SG, Xu L, Danesh A, Farooqui A, Borghetto I, Kelvin DJ, Sambri V, Rubino S. 2011. Inflammatory cytokine expression is associated with chikungunya virus resolution and symptom severity. PLoS Negl. Trop. Dis. 5:e1279. 10.1371/journal.pntd.0001279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng LF, Chow A, Sun YJ, Kwek DJ, Lim PL, Dimatatac F, Ng LC, Ooi EE, Choo KH, Her Z, Kourilsky P, Leo YS. 2009. IL-1beta, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS One 4:e4261. 10.1371/journal.pone.0004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NK, Koh EG, Chow A, Leo YS, Merits A, Chin KC, Ng LF. 2012. Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest. 122:4447–4460. 10.1172/JCI63120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, Guigand L, Dubreil L, Lebon P, Verrier B, de Lamballerie X, Suhrbier A, Cherel Y, Le Grand R, Roques P. 2010. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J. Clin. Invest. 120:894–906. 10.1172/JCI40104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gardner CL, Burke CW, Higgs ST, Klimstra WB, Ryman KD. 2012. Interferon-alpha/beta deficiency greatly exacerbates arthritogenic disease in mice infected with wild-type chikungunya virus but not with the cell culture-adapted live-attenuated 181/25 vaccine candidate. Virology 425:103–112. 10.1016/j.virol.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya virus arthritis in adult wild-type mice. J. Virol. 84:8021–8032. 10.1128/JVI.02603-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost MC, Fsihi H, Frenkiel MP, Blanchet F, Afonso PV, Ceccaldi PE, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saib A, Rey FA, Arenzana-Seisdedos F, Despres P, Michault A, Albert ML, Schwartz O. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog. 3:e89. 10.1371/journal.ppat.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.White LK, Sali T, Alvarado D, Gatti E, Pierre P, Streblow D, Defilippis VR. 2011. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J. Virol. 85:606–620. 10.1128/JVI.00767-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holzer GW, Coulibaly S, Aichinger G, Savidis-Dacho H, Mayrhofer J, Brunner S, Schmid K, Kistner O, Aaskov JG, Falkner FG, Ehrlich H, Barrett PN, Kreil TR. 2011. Evaluation of an inactivated Ross River virus vaccine in active and passive mouse immunization models and establishment of a correlate of protection. Vaccine 29:4132–4141. 10.1016/j.vaccine.2011.03.089 [DOI] [PubMed] [Google Scholar]
- 97.Teo TH, Lum FM, Claser C, Lulla V, Lulla A, Merits A, Renia L, Ng LF. 2013. A pathogenic role for CD4+ T cells during Chikungunya virus infection in mice. J. Immunol. 190:259–269. 10.4049/jimmunol.1202177 [DOI] [PubMed] [Google Scholar]
- 98.Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, Gunn BM, Elmore SA, Heise MT. 2011. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. Am. J. Pathol. 178:32–40. 10.1016/j.ajpath.2010.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]