ABSTRACT
Recombination is currently recognized as a factor for high genetic diversity, but the frequency of such recombination events and the genome segments involved are not well known. In the present study, we initially focused on the detection of recombinant porcine reproductive and respiratory syndrome virus (PRRSV) isolates by examining previously published data sets of ORF5 sequences (genotypes 1 and 2) obtained worldwide. We then examined full-length genome sequences in order to determine potential recombination breakpoints along the viral genome. For ORF5, 11 sets of genotype 1 sequences from different geographical areas, including 2 Asian, 1 American, and 7 European regions, and three sets of genotype 2, including sets from China, Mexico, and the United States, were analyzed separately. Potential recombination breakpoints were detected in 10/11 genotype 1 sets, including 9 cases in which the clustering of at least one isolate was different before and after the breakpoints. In genotype 2, potential breakpoints and different tree clustering of at least one strain before and after the breakpoint were observed in 2 out of 3 sets. The results indicated that most of the ORF5 data sets contained at least one recombinant sequence. When the full-length genome sequences were examined, both genotype 1 and 2 sets presented breakpoints (10 and 9, respectively), resulting in significantly different topologies before and after the breakpoints. Mosaic genomes were detected in genotype 1 sequences. These results may have significant implications for the understanding of the molecular epidemiology of PRRSV.
IMPORTANCE PRRSV is one of the most important viruses affecting swine production worldwide, causing big economic losses and sanitary problems. One of the key questions on PRRSV arises from its genetic diversity, which is thought to have a direct impact on immunobiology, epidemiology, diagnosis, and vaccine efficacy. One of the causes of this genetic diversity is recombination among strains. This study provides evidence that recombinant PRRSV isolates are common in most of the countries with significant swine production, especially PRRSV genotype 1. This observation has implications in the proper characterization of PRRSV strains, in the future development of phylogenetic studies, and in the development of new PRRSV control strategies. Moreover, the present paper emphasizes the need for a deeper understanding of the mechanisms and circumstances involved in the generation of genetic diversity of PRRSV.
INTRODUCTION
In the last 20 years, porcine reproductive and respiratory syndrome virus (PRRSV) has become endemic in most countries with industrial swine production. The infection of sows with PRRSV causes stillbirths, delayed or premature farrowing, and birth of weak piglets. PRRSV is one of the key components of the porcine respiratory disease complex (PRDC) in piglets, and it contributes significantly to increased mortality rates and medication costs in herds (1). The significant impact of PRRSV infection has been emphasized recently due to the emergence of so-called hot strains causing fatality rates of >20% in Asian piglets (2, 3).
One hallmark of PRRSV is its high genetic diversity (4, 5). This diversity arises first from the unusual fact of the parallel emergence in Europe and America in the early 1990s of two different PRRSV genotypes, designated genotype 1 and genotype 2 (6), that shared only about 55% similarity. Second, the replication strategy of PRRSV involves an RNA-dependent RNA polymerase that lacks 3′ proofreading abilities, and as a result, the rate of introduction of random mutations is very high (7, 8).
The precise nature of selection pressure in PRRSV is not well understood. Chronological drift of variants is not easily observed when phylogenetic analyses are carried out for any genotype of PRRSV (4, 5). Thus, in the current panorama two genotypes are recognized, which are further divided into a wide range of clusters, each containing diverse variants (4, 5). This high degree of diversity is a hallmark of PRRSV and could be due to inter- and intragenic recombinations which may break the directionality of the genome drift (9, 10) or by the introduction of new strains from areas not sufficiently studied, such as eastern Europe or Russia (11, 12).
Interestingly, when the complete PRRSV genome is examined, it is obvious that genetic diversity arises mainly (in terms of frequency) from a limited number of highly variable regions, such as the segments of open reading frame 1a (ORF1a) coding for nonstructural proteins 1 and 2 and the structural proteins encoded by ORF3-5. The other regions of the viral genome are more conserved. Moreover, there is evidence indicating that intergenic recombination among PRRSV isolates would be possible and would generate mosaic viruses (9). In a recent work, Shi and coworkers (10) published that the novel highly pathogenic PRRS viruses were recombinant viruses, but it is still unknown whether or not this phenomenon played a role in the increase in virulence of these virus strains.
ORF5 has been widely used for phylogenetic analysis, because it contains some hypervariable segments and some less variable ones, allowing discrimination of groups of strains in a reasonably meaningful way. However, it is worth pointing out that correlation cannot be established between phylogenetic groupings using ORF5 and virulence or cross-protection (13, 14). The use of ORF5 for PRRSV phylogenetic analysis often reveals that some strains are not groupable within a clade and that ORF5 grouping does not correspond to grouping based on other genes, such as ORF7 (12). These observations indicated that recombination in ORF5 could be common and could explain, at least partially, the origin of ungroupable strains in phylogenetic analysis. The aim of the present study was to examine all publicly available ORF5 and full-length genome sequences of genotypes 1 and 2 for recombination breakpoints and the existence of mosaic PRRSV isolates.
MATERIALS AND METHODS
ORF5 sequences.
The initial study of ORF5 was conducted using 14 sets of ORF5 sequences representing PRRSV isolates of genotypes 1 and 2. For genotype 1, 11 sets of sequences (n = 564) were examined; 10 sets contained previously published sequences, and one set contained newly sequenced isolates from Spain. The published sequences of genotype 1 were isolated from Germany (n = 48) (15), Hungary (two sets, named 1 and 2; n = 39 and n = 11, respectively) (16, 17), Italy (n = 63) (18), Republic of Korea (n = 25) (19), Romania (n = 15) (20), Thailand (n = 69) (21), the United Kingdom (n = 140) (22), the United States (n = 33) (23), and eastern Europe (n = 74) (11, 12). Forty-seven genotype 1 sequences from Spain were obtained from routine diagnostic samples, which were obtained from different farms of the same company between November 2007 and October 2010 (GenBank accession numbers KF666904 to KF666950) using a previously published reverse transcription-PCR (RT-PCR) method (24). Sets of sequences for genotype 2 PRRSV strains (n = 237) corresponded to previously published sequences from China (n = 38) (25) and the United States (n = 51) (26) and a set of 148 sequences from PRRSV isolates from pig farms of Sonora (Mexico) retrieved between 2003 and 2011 (A. Burgara-Estrella, M. Reséndiz-Sandoval, M. Cortey, E. Mateu, and J. Hernández, submitted for publication). All sequences included were free of termination codons, creating truncated proteins.
Detection of potential recombination events in ORF5 sequences.
Each data set of sequences was subjected to a phylogenetic analysis in MEGA 5.1 (27) by means of the neighbor-joining (NJ) method (maximum composite likelihood model) with 1,000 iterations for bootstrapping. Sequences with similarities of ≥99% were depurated to avoid unnecessary redundancy, and the resulting set was reanalyzed by the maximum likelihood (ML) method (general time-reversible model) with 1,000 iterations for bootstrapping. In parallel, all sets of sequences were analyzed using MrBayes 3.2 (28), setting the parameters to 2 runs, 1,000,000 generations, a sampling frequency of 100, and 100 bootstrap replicates.
The different sets of sequences were then analyzed using the single-breakpoint detection utility (SPB) and the genetic algorithm recombination detection (GARD) (29) utility available online (http://www.datamonkey.org). Those utilities allowed the detection of potential recombination events in the examined sets and indicated the potential breakpoints where recombination could have taken place. For SBP, the Akaike information criterion (AIC) or the AICc (for sets of <45 sequences) were calculated (30). For GARD, the Kishino-Hasegawa (KH) test (31) was conducted in order to examine topological incongruences resulting from the comparison of trees made after splitting sequences in the set by the calculated breakpoints.
The sets of sequences for which SPB/GARD detected potential breakpoints were selected for the next step of the analysis. For each of those sets, sequences were split into two segments, one from nucleotide (nt) 1 up to each of the breakpoints detected, and the second from this point to the last nucleotide in ORF5. The phylogenetic analysis of the two segments was then performed using ML and MrBayes as described above. The resulting trees were compared in order to search for discrepancies in the clustering of isolates. When clear discrepancies were found, namely, the same strain located in different clusters for each analyzed segment, a similarity matrix was constructed with the nearest strains in each tree.
Sets made of the nearest isolates to the potential recombinant strain in each tree were examined in RDP v.4.6 (32) in order to establish potential parental strains for that isolate. An automated analysis was run using default settings for RDP, Geneconv, Chimaera, MaxChi, Bootscan, Siscan, 3seq, and LARD utilities. The significance level was set at 0.05.
Analysis of recombination events using full ORF1-7 sequences of genotype 1 and genotype 2 isolates.
In the next step, the objective was to investigate whether recombination occurred simply in ORF5 or whether it can take place in other segments of the viral genome, resulting in mosaic isolates. For that purpose, full ORF1-7 sequences of genotype 1 and 2 isolates were examined. The set of genotype 1 isolates consisted of 24 sequences retrieved from different countries of Europe, Asia, and the United States: Denmark (n = 8) (33), Spain (n = 7) (34), The Netherlands (n = 2), China (n = 2), the Republic of Korea (n = 1), Thailand (n = 1), and the United States (n = 2). Additionally, the full sequences from two commercial vaccines (Porcilis PRRS and Amervac PRRS) were included. Two genotype 2 sets were analyzed. The first one included 23 isolates, of which 13 were collected and sequenced in Denmark (35), and another 10 were isolated in other countries and included the vaccine strains. A second genotype 2 set, including 61 Asian and North American full-length sequences already analyzed by Gauger et al. in 2012 (36), including two vaccine sequences, was also analyzed. Table 1 summarizes the names and GenBank accession numbers of the isolates. For control purposes, a set of Chinese strains of genotype 2 previously reported to contain a mosaic strain (9) was included in the examination.
TABLE 1.
Genotype 1 and 2 full-length sequences used in this study
| Genotype and accession no. | Name |
|---|---|
| 1 | |
| KC862574 | DK-2012-01-05-2 |
| KC862567 | DK-2011-05-11-14 |
| KC862568 | DK-2010-10-10-3 |
| KC862573 | DK-2008-10-5-2 |
| KC862572 | DK-2003-7-2 |
| KC862566 | DK-1992-PRRS-111_92 |
| KC862571 | DK-2003-6-5 |
| KC862569 | DK-2011-05-23-9 |
| GQ461593 | SHE-CN |
| EU076704 | HKEU16 |
| AY366525 | EuroPRRS |
| DQ489311 | SD01-08 |
| FJ349261 | KNU07 |
| DQ864705 | 01CB1 |
| M96262 | Lelystad |
| AY588319 | LV4 2.1 |
| JF276430 | CReSA 2982 |
| JF276432 | CReSA 3256 |
| JF276434 | CReSA 3266 |
| JF276435 | CReSA 3267 |
| JF276433 | CReSA 3249 |
| JF276431 | CReSA 3262 |
| KC862570 | ESP-1991-Olot91 |
| GU067771 | Amervac PRRSV |
| KJ127878 | Porcillis |
| 2 | |
| Set 1 | |
| KC862577 | DK-2011-030311-1 |
| KC862578 | DK-2004-1-7-Pl |
| KC862576 | DK-1997-19407B |
| KC862581 | DK-2010-10-2-1 |
| KC862580 | DK-2010-10-7-1 |
| KC862575 | DK-2012-01-11-3 |
| KC862583 | DK-2010-10-4-1 |
| KF183946 | DK-2010-10-13-1 |
| KF183947 | DK-2011-88005-A8-Pl |
| KC862582 | DK-2008-10-1-3 |
| KC862584 | DK-2003-2-3 |
| KC862585 | DK-2004-2-1 |
| KC862579 | DK-2010-10-1-2 |
| NC_001961 | NC_001961 strain |
| AY545985 | TS |
| AY424271 | JA142TS |
| AF325691 | SAMSTS |
| AY032626 | CH1a |
| EU708726 | X143CN |
| GQ857656 | SX1CN |
| ET880437 | HN2007CN |
| T87392 | VR2332 |
| AF066183 | RespPRRS |
| Set 2 | |
| U87392 | VR-2332 |
| AY585241 | PL97-1 |
| EU360129 | rV63 |
| EF153486 | Isolate CC-1 |
| FJ899592 | Clone 20 |
| DQ056373 | 01NP1.2 |
| FJ950747 | BJSD |
| GQ914997 | SD1-100 |
| AF066183 | RespPRRS MLV |
| EF484033 | Clone pMLV |
| AF159149 | RespPRRS/Repro |
| EU360128 | rV68 |
| FJ524377 | pV7-myc |
| AF176348 | PA8 |
| AF046869 | 16244B 2/18/97(Nebraska)pass.3 |
| EF532816 | ISU-P |
| DQ473474 | Isolate LMY |
| GQ330474 | APRRS |
| AF184212 | SP |
| DQ779791 | Prime Pac |
| AB288356 | EDRD-1 |
| DQ176020 | Isolate MN184B |
| DQ176019 | Isolate MN184A |
| EF488739 | Isolate MN184C |
| EF484031 | Clone pMN184 |
| HQ699067 | NC16845 |
| DQ988080 | Isolate Ingelvac ATP |
| AY545985 | NVSL 97-7895 |
| AY424271 | JA142 |
| AF325691 | NVSL 97-7985 IA 1-4-2 |
| AF494042 | P129 |
| AY262352 | PRRSV HB-2(sh)/2002 |
| EU360130 | HB-1/3.9 |
| AY150312 | PRRSV HB-1(sh)/2002 |
| GU269541 | GD3 |
| GQ359108 | SD-CXA/2008 |
| GQ857656 | SX-1 |
| GU168567 | 09HUB7 |
| EU678352 | WUH2 |
| GU168568 | 09HUB5 |
| FJ895329 | SX2009 |
| GU454850 | GDQY2 |
| GU168569 | 08SDWF |
| GU169411 | 08HuN |
| FJ889130 | CWZ-1-F3 |
| GU232736 | TP P60 |
| GU232737 | TP P90 |
| GU232738 | YN9 |
| EU880435 | Isolate YN2008 |
| GQ374441 | GDQJ |
| FJ950746 | BJPG |
| GU232735 | KP |
| GU461292 | AH0701 |
| FJ889129 | CBB-1-F3 |
| EU880432 | Isolate JX2006 |
| EU880436 | Isolate XL2008 |
| EF488048 | Clone pJX143 |
| FJ950745 | BJBLZ |
| FJ950744 | BJSY-1 |
| EU880435 | YN2008 |
| GQ374442 | GDBY1 |
The analysis of the above-mentioned sets started with aligning the sequences. Since some sequences did not include the full 5′-untranslated region (UTR), the analysis was initiated at the earliest common nucleotide. A similar approach was taken for the 3′-UTR. The sequences were then divided into three overlapping segments: segment 1, comprising nucleotides 1 to 6,000; segment 2, comprising nucleotides 5,000 to 11,000; and segment 3, from nucleotide 10,000 until the 3′ end. Each set of segments was analyzed as described above by means of GARD for obtaining significant breakpoints in the Kishino-Hasegawa test. Once breakpoints were detected, the full-genome sequences were reexamined by segmenting them according to the breakpoints detected by GARD. The sets comprising the new segments were analyzed by the ML method (general time-reversible model) also with 1,000 iterations for bootstrapping. The resulting topologies for each segment were compared to the topology obtained from the full-genome analysis.
For isolates with significant evidence of recombination, an RDP analysis was conducted, as described above, using the closest isolates in the different clusters. In this case, because of the difficulties of RDP for managing the analysis of full-length sequences, viral genomes were again segmented, considering the breakpoints at which different topologies were observed.
RESULTS
Detection of potential recombination events in ORF5 sequences.
Evidence of recombination was found in both 9 out of 11 genotype 1 and two out of three genotype 2 sequences. Each set of sequences for which a recombination breakpoint was detected was then split into two different segments: one from nt 1 up to the breakpoint and the other from the breakpoint nt until the end of the ORF. The phylogenetic analysis using both the ML and Bayesian methods showed different topologies of some strains in ORF5 within sets of genotypes 1 and 2 before and after the breakpoint, providing strong evidence for the actual existence of recombination events. The presence of breakpoints, the significance obtained by means of SBP/GARD analysis, and the presence of different topologies before and after the breakpoint by means of phylogenetic analysis of each set of sequences are summarized in Table 2.
TABLE 2.
Summary of breakpoints and their statistical significance by analysis of the genotype 1 and 2 sets used in this studya
| Genotype and sequence set | Breakpoint(s) (nt from ORF5) | Significance byb: |
Different clustering before and after breakpoint | |
|---|---|---|---|---|
| SBP (AIC/AICc support) | GARD significance (P < 0.01) | |||
| PRRSV1 | ||||
| Germany | 181 | 0.688 | NS | Yes |
| Hungary 1 | 146 and 172 | 0.54 | NS | Yes |
| Hungary 2 | 149 | 0.777 | NS | No |
| Italy | 180 and 219 | 0.203 and 0.744 | P < 0.01 (nt 219) | Yes |
| Republic of Korea | 346 | 0.137 | P < 0.1 | Yes |
| Romania | Not found | NS | NS | No |
| Spain | 166 | 0.994 | NS | Yes |
| Thailand | 165 | 0.492 | NS | Yes |
| UK | 296 | 0.705 | NS | Yes |
| USA | 225 | 0.354 | NS | Yes |
| Eastern Europe | 175 | 0.99 | NS | Yes |
| PRRSV2 | ||||
| China | Not found | No | ||
| Mexico | 177 | 0.61 | Yes | |
| USA | 135 and 153 | 0.304 and 0.300 | P < 0.01 | Yes |
The presence of different topologies before and after the breakpoints by means of the phylogenetic analysis is also summarized in this table.
NS, not significant.
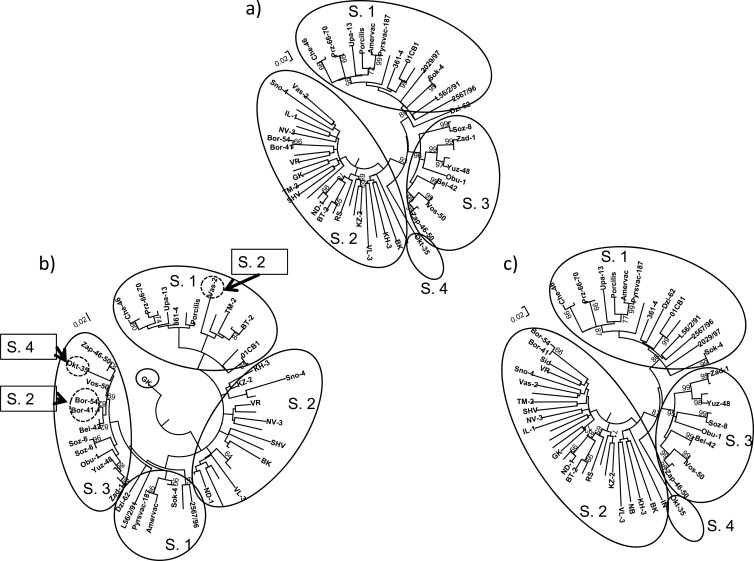
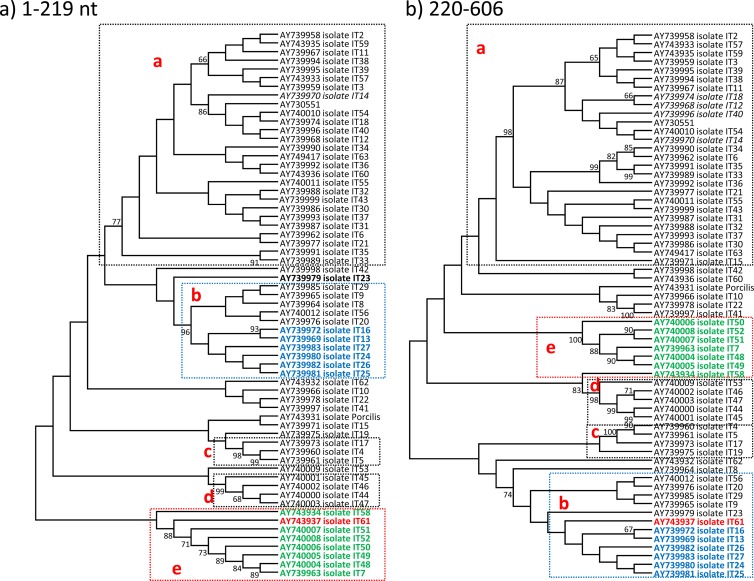
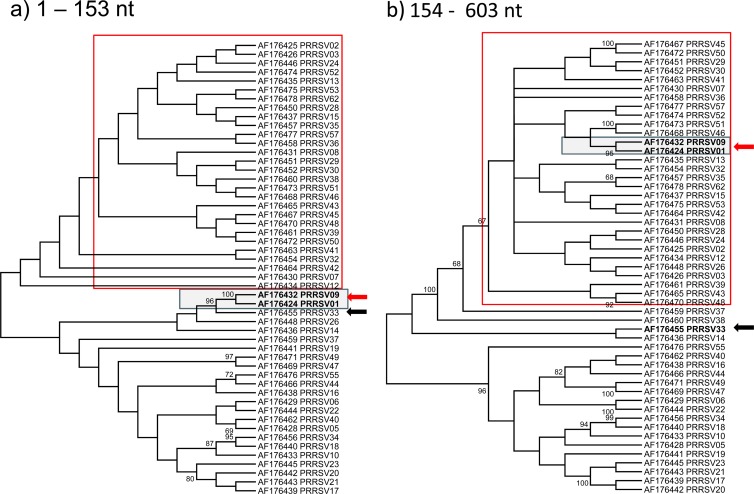
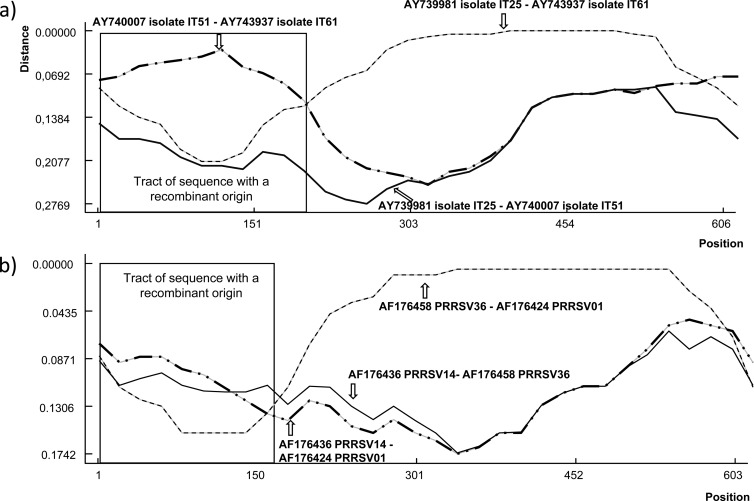
For the eastern Europe sequences, Fig. 1 shows phylogenetic grouping of the isolates considering the full ORF5 and the segments before and after the breakpoint. These results show that the OKT-35, Bor41, Bor54, and Vas-2 strains were grouping in different subtypes before and after the breakpoint, suggesting that recombination between subtypes occurred. Furthermore, ORF5 sequences obtained from Germany, Italy, Spain, the United States, the United Kingdom, and Mexico showed strong evidence of recombination events. Figures 2 and 3 show that phylogenetic grouping of Italian genotype 1 and U.S. genotype 2 strains by the ML method was different before and after the breakpoints, and similar results were obtained by NJ and Bayesian analysis (Fig. 4). RDP enabled the parental isolates for a number of recombinant isolates to be identified. Figure 5 shows the change of similarity percentages before and after a breakpoint, taking as reference the potential parental strains for each segment, where the Italian isolate IT61 (GenBank accession no. AY743937) is shown as the recombinant example for genotype 1 (Fig. 5A) and the U.S. isolate PRRSV01 (GenBank accession no. AF176424) is shown as the recombinant example for genotype 2 (Fig. 5B).
FIG 1.
Maximum likelihood (ML) consensus trees (1,000 replicates for bootstrapping) for the ORF5 gene (a) and the segments containing nt 63 to 175 (b) and nt 176 to 606 (c) of ORF5. Strains OKT-35 (subtype 4 [S. 4]; a and c), Bor41, and 54 (subtype 2; a and c) are grouped with strains of subtype 2 in the segment before the breakpoint (b) with high bootstrap values (>65), indicating recombination between subtypes. For Vas-2 (subtype 2; a and c), although its grouping before this same breakpoint is not supported by bootstrap values of >65, it seems to be phylogenetically closer to strains belonging to subtype 1 than to subtype 2, but these two subtypes are not as well defined in the ML tree shown in panel a as in the trees shown in panels a and c.
FIG 2.
ML consensus trees (1,000 replicates for bootstrapping) for ORF5 sequences from genotype 1 isolates of Italy. Only bootstrap values of >65% are shown. Panels a and b represent phylogenetic trees resulting from splitting Italian ORF5 sequences into two segments, up to the detected recombination breakpoint (nt 1 to 219) and after the recombination breakpoint (nt 220 to 606). The red strain is the recombinant strain that jumps from the e cluster (green isolates) to the b cluster (blue strains).
FIG 3.
ML consensus trees (1,000 replicates for bootstrapping) for ORF5 sequences from genotype 2 isolates of the United States. Only bootstrap values of >65% are shown. Panels a and b represent phylogenetic trees resulting from splitting American ORF5 sequences into two segments, up to the detected recombination breakpoint (nt 1 to 153) and after the recombination breakpoint (nt 154 to 603). The red arrow indicates the recombinant strains, and the black arrow indicates the parental strain before the breakpoint.
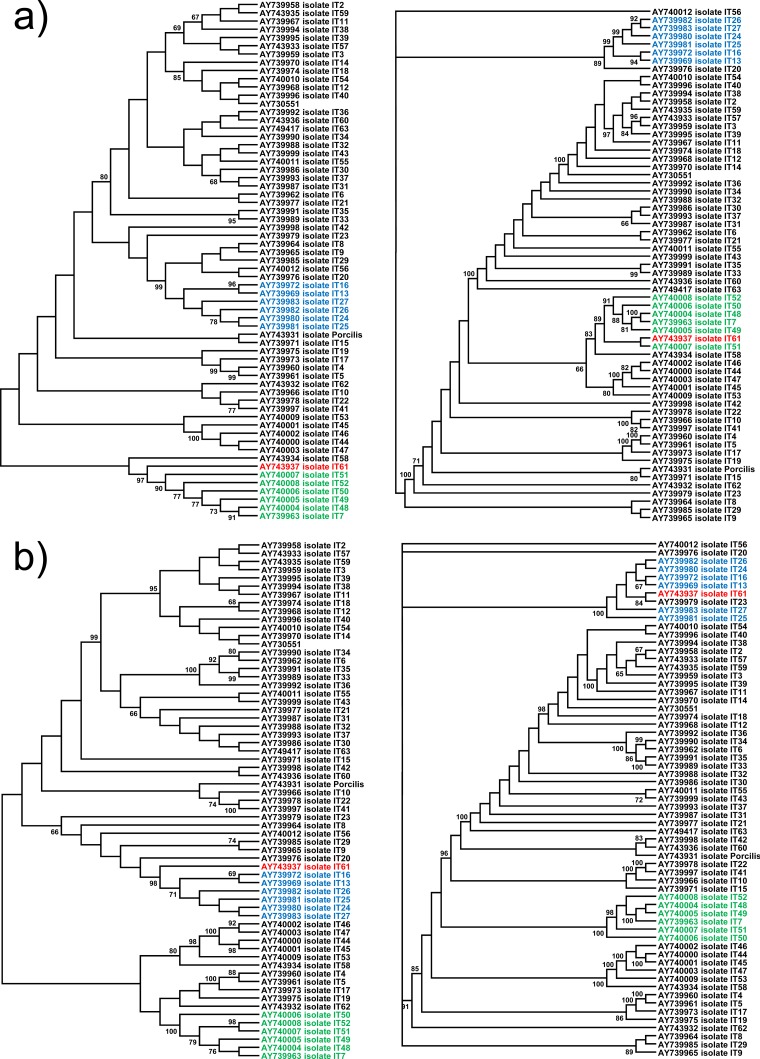
FIG 4.
Neighbor-joining and Bayesian inference consensus trees (1,000 replicates for bootstrapping) for ORF5 sequences from genotype 1 isolates of Italy. Only bootstrap values of >65% are shown. Panels a and b represent phylogenetic trees resulting from splitting Italian ORF5 sequences into two segments, up to the detected recombination breakpoint (nt 1 to 219) and after the recombination breakpoint (nt 220 to 606). The red strain is the recombinant strain that jumps from the e cluster (green isolates) to b cluster (blue strains).
FIG 5.
Distance plots for recombinant and parental strains in the Italian and U.S. sets of ORF5 sequences. The x axis shows the nucleotide position, and the y axis shows the nucleotide similarity. Each line represents similarity for a pair of isolates. The upper graph depicts Italian isolates IT25, IT51, and IT61. In that case, isolate IT61 was detected as a recombinant strain where the major parental virus was IT25 (99.3% probability) and IT52 was the minor parental virus (96.5% probability) (P = 2.37 × 10−15). The lower graph depicts the case of genotype 2 isolates, PRRSV01, PRRSV14, and PRRSV36, with PRRSV01 resulting from the recombination of strain PRRSV36 as a major parent (81.7%) and PRRSV14 (88.9%) as a minor parent (P = 6.94 × 10−11). GenBank accession numbers are shown for identification purposes.
Analysis of recombination events using full ORF1-7 sequences of genotype 1 and genotype 2 isolates.
The examination of the control set of genotype 2 previously published by Li et al. (9) showed that with the methodology of the present work, a mosaic strain was found coincidently with what has been previously reported. Thus, the present methodology was considered to be validated for the analysis of full genomes.
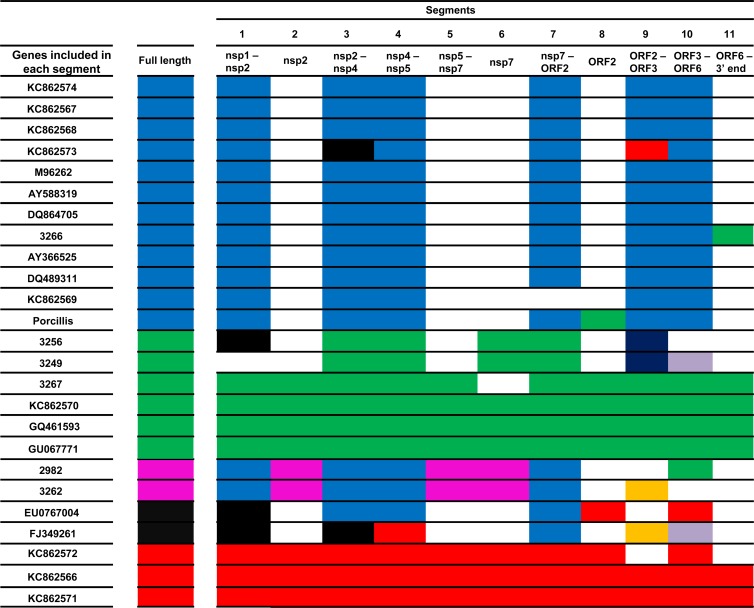
In the analysis of the genotype 1 set, 11 significant breakpoints were detected (Table 3). These breakpoints were scattered throughout the viral genome, in regions encoding both nonstructural and structural proteins. Of the 24 genotype 1 sequences, seven had significantly different topologies in different segments; that is, they jumped from one significant cluster to another significant cluster (>65% of iterations) (Fig. 5). For three additional strains (KC862569, KC862572, and JF276435), indirect evidence of recombination was seen, namely, the isolate changed from one significant cluster to another cluster; however, the threshold of 65% of iterations was not reached in this new clade (Fig. 6). Interestingly, the sequence from the genotype 1 vaccine examined also showed evidence of being a recombinant itself or, alternatively, being a parental of a recombinant strain because of the change between clusters along the genome (Fig. 6).
TABLE 3.
Fragments of the full-length sequenced genotype 1 (11) and 2 (10) strains delimited by the GARD analysis breakpointsa
| Fragment | Genotype |
||
|---|---|---|---|
| 1 | 2 |
||
| Set 1 | Set 2 | ||
| 1 | 1–2281 | 1–2245 | 1–2456 |
| 2 | 2282–2734 | 2246–4596 | 2456–3804 |
| 3 | 2735–5696 | 4597–5755 | 3804–6834 |
| 4 | 5697–6196 | 5756–6464 | 6834–7640 |
| 5 | 6197–6434 | 6465–9405 | 7640–11064 |
| 6 | 6435–6920 | 9406–12051 | 11064–12888 |
| 7 | 6921–11614 | 12052–12927 | 12888–14058 |
| 8 | 11615–12386 | 12928–13789 | 14058-end |
| 9 | 12387–12986 | 13790–15009 | |
| 10 | 12987–14638 | 15009-end | |
| 11 | 14638-end | ||
These segments were found to show different topologies according to Kishino-Hasengawa test with significant values (P < 0.05).
FIG 6.
Clustering of the full-length genotype 1 strains and the 11 different segments determined by the breakpoints obtained with the GARD analysis is shown by means of a color code. Blue, green, pink, black, and red were first determined by the clustering observed in the full-length ML tree (1,000 iterations) based on the biggest groups determined by bootstrap values of >65. Clustering of full-length genomes then was compared individually to the other 11 segments. A matching color of a strain in full-length sequence and a segment means they have the same clustering. A change of color means a change of clustering and further evidence of recombination. White indicates that no clustering was determined that was supported by bootstrap values of >65. Other colors mean new clustering not observed in the full-length tree.
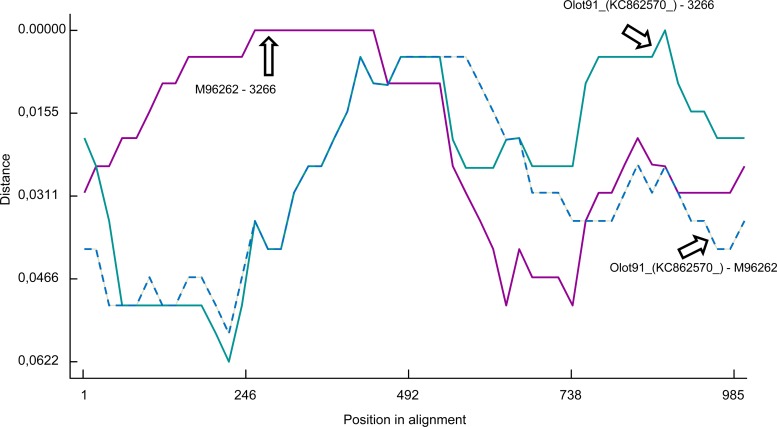
The examination of potential recombinant strains in RDP showed that the origin of recombination could be traced in some cases. For example, strain 3266 (Fig. 7) was a recombinant between a Lelystad-like virus (up to the beginning of ORF7; 97.3% probability) and an Olot 91-like virus (the 3′ end of ORF7; 99.7% probability). The vaccine strain (Porcillis) is a recombinant with an Olot 91-like or Amervac-like virus (at the end of NSP12 and the beginning of ORF2 between nt 11614 and 12386; 98% and 98.9% probability, respectively; not possible to discriminate between both potential parental strains) and a Lelystad-like strain (in the rest of the genome; probability of 99.5%), and the EU076704 strain is a recombinant between a 3262-like strain (in ORF2 between nt 12386 and 12986; probability of 93.4%) and a DK-1992-like isolate (between nt 11614 and 12286 and between nt 12986 up to ORF7; probability, 92.9%). No other parental strains were determined with other potential recombinant strains observed in the tree topologies.
FIG 7.
Distance plots for recombinant 3266 strain and parental strains Olot-91 and LV in ORF6 and ORF7 of the full-length genotype 1 data set. The x axis shows the nucleotide position, and the y axis shows the nucleotide similarity. Each line represents similarity for a pair of isolates. Strain 3266 was a recombinant between a Lelystad-like virus as the major parent (up to the beginning of ORF7; 97.3% probability) and an Olot 91-like virus (the 3′ end of ORF7; 99.7% probability) as a minor parent (P = 3.01 × 10−21).
For genotype 2, eight significant breakpoints (Table 3) were identified in set 1 and seven in set 2 by GARD. The analysis of the topologies obtained after splitting the genomes into 9 segments showed only one potential recombinant strain in set 1 (GenBank accession no. AF325691) corresponding to the isolates of the mid-1990s causing the so-called sow acute mortality syndrome, or atypical PRRS. No recombinants were found among Danish genotype 2 isolates. In set 2, one strain (DQ473474) had significantly different topologies in at least two segments. Another strain (AB288356) also showed indirect evidence of recombination in the same way as that described above for the three genotype 1 strains.
DISCUSSION
Although early studies (36, 37) provided statistical indications of the existence of recombination in PRRSV, the first clear evidence was reported by Yuan et al. (38). At that time, the perception was that the frequency of recombinant strains in the environment was low due to their failure to compete with parental strains in vitro. Recently, more evidence has evolved indicating that the existence and establishment of recombinant PRRSV genotype 2 strains in the field is a common fact for genotype 2 isolates (9, 10, 39, 40), but this has not been shown for genotype 1. The present paper provides evidence that intragenic recombination in ORF5 of genotype 1 field isolates is a common event, as indicated by the detection of recombinant isolates in many of the ORF5 sets of sequences examined. The characteristics of the recombinant isolates were generally compatible with the copy-choice recombination model (41, 42). The copy-choice model has previously been proposed as a model for recombination for other Nidovirales members, such as avian gammacoronaviruses (43), some canine coronaviruses (44), feline coronavirus type II (45), and severe acute respiratory syndrome coronavirus (46). A recent work carried out in the Republic of Korea (47) demonstrated the presence of 2 out of 11 recombinant infectious bronchitis virus strains, a proportion similar to that of PRRSV recombinants compared to the results obtained in the present work. Within ORF5, recombination breakpoints were mostly located in the 5′ half of the ORF but did not comprise ORF5a, a possible indication of the essential nature of the encoded protein (48). As mentioned before, the Mexican set was retrieved from another work (Burgara-Estrella et al., submitted) that used a different methodology. Although the methods used by Burgara-Estrella in the previous work were different from the ones in the present study, the same potential recombinant isolates were detected, reinforcing the validity of our methodology and results.
The examination of full-length genome sequences showed that recombination can take place all along the genome, resulting in mosaic strains, which in some cases jumped to different clusters in more than 3 segments (Fig. 6). The change in tree topologies was significant in 10/25 strains in genotype 1 and probably in 1/23 and 2/61 in genotype 2 strains, suggesting that recombination is a frequent phenomenon in the generation of PRRSV and should be taken into account when studying the molecular epidemiology of this virus. However, the identification of parental strains before and after a breakpoint was not possible in 3/7 recombinant genotype 1 strains when using RDP analysis, probably due to the lack of sequencing activity. The nucleotide distances between the potential parental strain and the recombinant strain are too high to properly determine significantly if they are parentally related in these cases.
Most of the currently available PRRSV sequence information is based on analysis of ORF5, while there is a significant lack of information regarding full-length genome sequences. This fact makes it very difficult to detect and trace the phylogenetic origin of recombinant strains, especially when sequence sets were not produced with such a purpose. This emphasizes the need for regularly sequencing PRRSV isolates in full in order to enhance PRRSV surveillance to construct complete databases to properly study the evolution and epidemiology of this virus. This is particularly important for Europe, where the number of full-length sequences is still scarce.
The potential impact of PRRSV recombination can be high. The critical points for controlling PRRS would require the determination of the frequency of horizontal introduction of the virus into the farms and the establishment of the epidemiological relationships for the newly introduced strains in order to ascertain the origin of the infections. This will only be possible if adequate identification and classification of the circulating virus are available. Currently, this is done by means of detection using PCR and further sequencing of ORF5. The actual circulation of recombinant strains is certainly an obstacle for this approach; therefore, this strategy must be refined. Indeed, the evidence of the existence of mosaic viruses provided in this paper supports that a single-gene sequence strategy is useful but not sufficient. Thus, a more holistic approach should be considered, including detection of recombination breakpoints for sets of sequences of related farms (geographically or by sharing animals, transport, etc.) before conclusions can be drawn. Optimally, full-genome sequencing, or at least sequencing of two nonadjacent genes showing high diversity (for example, the nsp2 coding segment of ORF1a and ORF5), should be used for phylogenetic and epidemiologic studies. This is particularly important for the assessment of the circulation of vaccine-derived recombinant strains.
The existence of recombinant strains circulating in the field further complicates the evaluation of homologous/heterologous protection, which is one of the critical points for the design of next-generation PRRSV vaccines. Full-length genome sequencing combined with careful assessment of phylogenetic trees constructed based on all genes is required to be able to make sustained conclusions on homology between the vaccine and any tested challenge strain.
In summary, the present paper provides clear evidence that recombinant genotype 1 and 2 PRRSV strains are circulating in the field and emphasizes the need for more research on the role of recombination in the generation of genetic diversity of PRRSV.
ACKNOWLEDGMENT
This work has been funded in part by project PoRRSCon, porcine reproductive and respiratory syndrome (PRRS): new generation, efficacious and safe vaccine, new control strategies, grant no. 245141 of the 7th Framework Program of the European Union.
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Holtkamp DJ, Kliebenstein JB, Neumann EJ, Zimmerman JJ, Rotto HF, Yoder TK, Chong Wang C, Yeske PE, Mowrer CL, Haley CA. 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J. Swine Health Prod. 21:72–84 [Google Scholar]
- 2.Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P. 2007. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet. J. 174:577–584. 10.1016/j.tvjl.2007.07.032 [DOI] [PubMed] [Google Scholar]
- 3.Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2(6):e526. 10.1371/journal.pone.0000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murtaugh MP, Stadejek T, Abrahante JE, Lam TT, Leung FC. 2010. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 154:18–30. 10.1016/j.virusres.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 5.Shi M, Lam TT, Hon CC, Hui RK, Faaberg KS, Wennblom T, Murtaugh MP, Stadejek T, Leung FC. 2010. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. 154:7–17. 10.1016/j.virusres.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 6.Meng XJ, Paul PS, Halbur PG, Lum MA. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the U. S. A. and Europe. Arch. Virol. 140:745–755. 10.1007/BF01309962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsberg R, Oleksiewicz MB, Petersen AM, Hein J, Bøtner A, Storgaard T. 2001. A molecular clock dates the common ancestor of European-type porcine reproductive and respiratory syndrome virus at more than 10 years before the emergence of disease. Virology 289:174–179. 10.1006/viro.2001.1102 [DOI] [PubMed] [Google Scholar]
- 8.Forsberg R. 2005. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 22:2131–2134. 10.1093/molbev/msi208 [DOI] [PubMed] [Google Scholar]
- 9.Li B, Fang L, Xu Z, Liu S, Gao J, Jiang Y, Chen H, Xiao S. 2009. Recombination in vaccine and circulating strains of porcine reproductive and respiratory syndrome viruses. Emerg. Infect. Dis. 15:2032–2035. 10.3201/eid1512.090390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi M, Holmes EC, Brar MS, Leung FC. 2013. Recombination is associated with an outbreak of novel highly pathogenic porcine reproductive and respiratory syndrome viruses in China. J. Virol. 87:10904–10907. 10.1128/JVI.01270-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadejek T, Oleksiewicz MB, Scherbakov AV, Timina AM, Krabbe JS, Chabros K, Potapchuk D. 2008. Definition of subtypes in the European genotype of porcine reproductive and respiratory syndrome virus: nucleocapsid characteristics and geographical distribution in Europe. Arch. Virol. 153:1479–1488. 10.1007/s00705-008-0146-2 [DOI] [PubMed] [Google Scholar]
- 12.Stadejek T, Mittelholzer Ch Oleksiewicz MB, Paweska J, Belák S. 2006. Highly diverse type of equine arteritis virus (EAV) from the semen of a South African donkey: short communication. Acta Vet. Hung. 54:263–270. 10.1556/AVet.54.2006.2.12 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Murtaugh MP. 2012. Dissociation of porcine reproductive and respiratory syndrome virus neutralization from antibodies specific to major envelope protein surface epitopes. Virology 433:367–376. 10.1016/j.virol.2012.08.026 [DOI] [PubMed] [Google Scholar]
- 14.Martínez-Lobo FJ, Díez-Fuertes F, Simarro I, Castro JM, Prieto C. 2011. Porcine Reproductive and respiratory syndrome virus isolates differ in their susceptibility to neutralization. Vaccine 29:6928–6940. 10.1016/j.vaccine.2011.07.076 [DOI] [PubMed] [Google Scholar]
- 15.Greiser-Wilke I, Fiebig K, Drexler C, Grosse Beilage E. 2010. Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) in selected herds in a pig-dense region of north-western Germany. Vet. Microbiol. 143:213–223. 10.1016/j.vetmic.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 16.Balka G, Hornyák A, Bálint A, Kiss I, Kecskeméti S, Bakonyi T, Rusvai M. 2008. Genetic diversity of porcine reproductive and respiratory syndrome virus strains circulating in Hungarian swine herds. Vet. Microbiol. 127:128–135. 10.1016/j.vetmic.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 17.Kiss I, Sámi L, Kecskeméti S, Hanada K. 2006. Genetic variation of the prevailing porcine respiratory and reproductive syndrome viruses occurring on a pig farm upon vaccination. Arch. Virol. 151:2269–2276. 10.1007/s00705-006-0805-0 [DOI] [PubMed] [Google Scholar]
- 18.Pesente P, Rebonato V, Sandri G, Giovanardi D, Ruffoni LS, Torriani S. 2006. Phylogenetic analysis of ORF5 and ORF7 sequences of porcine reproductive and respiratory syndrome virus (PRRSV) from PRRS-positive Italian farms: a showcase for PRRSV epidemiology and its consequences on farm management. Vet. Microbiol. 114:214–224. 10.1016/j.vetmic.2005.11.061 [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Roh IS, Choi EJ, Lee C, Lee CH, Lee KH, Lee KK, Song YK, Lee OS, Park CK. 2010. A molecular analysis of European porcine reproductive and respiratory syndrome virus isolated in South Korea. Vet. Microbiol. 143:394–400. 10.1016/j.vetmic.2009.11.039 [DOI] [PubMed] [Google Scholar]
- 20.Zaulet M, Gurau MR, Petrovan V, Buburuzan L. 2012. Genetic diversity characterization of porcine reproductive and respiratory syndrome virus isolates in Romania, based on phylogenetic analysis. Int. J. Mol. Sci. 13:12046–12061. 10.3390/ijms130912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilubol D, Tripipat T, Hoonsuwan T, Tipsombatboon P, Piriyapongsa J. 2013. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes I and II in Thailand. Arch. Virol. 158:943–953. 10.1007/s00705-012-1573-7 [DOI] [PubMed] [Google Scholar]
- 22.Frossard JP, Hughes GJ, Westcott DG, Naidu B, Williamson S, Woodger NG, Steinbach F, Drew TW. 2013. Porcine reproductive and respiratory syndrome virus: genetic diversity of recent British isolates. Vet. Microbiol. 162:507–518. 10.1016/j.vetmic.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, Schneider P, Zhang WP, Faaberg KS, Nelson EA, Rowland RR. 2007. Diversity and evolution of a newly emerged North American type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch. Virol. 152:1009–1017. 10.1007/s00705-007-0936-y [DOI] [PubMed] [Google Scholar]
- 24.Mateu E, Martín M, Vidal D. 2003. Genetic diversity and phylogenetic analysis of glycoprotein 5 of European-type porcine reproductive and respiratory virus strains in Spain. J. Gen. Virol. 84:529–534. 10.1099/vir.0.18478-0 [DOI] [PubMed] [Google Scholar]
- 25.Yin G, Gao L, Shu X, Yang G, Guo S, Li W. 2012. Genetic diversity of the ORF5 gene of porcine reproductive and respiratory syndrome virus isolates in southwest China from 2007 to 2009. PLoS One 7(3):e33756. 10.1371/journal.pone.0033756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg TL, Hahn EC, Weigel RM, Scherba G. 2000. Genetic, geographical and temporal variation of porcine reproductive and respiratory syndrome virus in Illinois. J. Gen. Virol. 81:171–179 [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher N, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3:Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 29.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901. 10.1093/molbev/msl051 [DOI] [PubMed] [Google Scholar]
- 30.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19:716–723. 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- 31.Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequences, and the branching order of Hominoidea. J. Mol. Evol. 29:170–179. 10.1007/BF02100115 [DOI] [PubMed] [Google Scholar]
- 32.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. 10.1093/bioinformatics/16.6.562 [DOI] [PubMed] [Google Scholar]
- 33.Kvisgaard LK, Hjulsager CK, Kristensen CS, Lauritsen KT, Larsen LE. 2013. Genetic and antigenic characterization of complete genomes of type 1 porcine reproductive and respiratory syndrome viruses (PRRSV) isolated in Denmark over a period of 10 years. Virus Res. 178:197–205. 10.1016/j.virusres.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 34.Darwich L, Gimeno M, Sibila M, Diaz I, de la Torre E, Dotti S, Kuzemtseva L, Martin M, Pujols J, Mateu E. 2011. Genetic and immunobiological diversities of porcine reproductive and respiratory syndrome genotype I strains. Vet. Microbiol. 150:49–62. 10.1016/j.vetmic.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 35.Kvisgaard LK, Hjulsager CK, Brar MS, Leung FC, Larsen LE. 2013. Genetic dissection of complete genomes of type 2 PRRS viruses isolated in Denmark over a period of 15 years. Vet. Microbiol. 167:334–344. 10.1016/j.vetmic.2013.09.023 [DOI] [PubMed] [Google Scholar]
- 36.Gauger PC, Faaberg KS, Guo B, Kappes MA, Opriessnig T. 2012. Genetic and phenotypic characterization of a 2006 United States porcine reproductive and respiratory virus isolate associated with high morbidity and mortality in the field. Virus Res. 163:98–107. 10.1016/j.virusres.2011.08.017 [DOI] [PubMed] [Google Scholar]
- 37.Kapur V, Elam MR, Pawlovich TM, Murtaugh MP. 1996. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 77:1271–1276. 10.1099/0022-1317-77-6-1271 [DOI] [PubMed] [Google Scholar]
- 38.Yuan S, Nelsen CJ, Murtaugh MP, Schmitt BJ, Faaberg KS. 1999. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 61:87–98. 10.1016/S0168-1702(99)00029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N, Yu X, Wang L, Wu J, Zhou Z, Ni J, Li X, Zhai X, Tian K. 2013. Two natural recombinant highly pathogenic porcine reproductive and respiratory syndrome viruses with different pathogenicities. Virus Genes 46:473–478. 10.1007/s11262-013-0892-4 [DOI] [PubMed] [Google Scholar]
- 40.Shi M, Lemey P, Singh Brar M, Suchard MA, Murtaugh MP, Carman S, D'Allaire S, Delisle B, Lambert MÈ, Gagnon CA, Ge L, Qu Y, Yoo D, Holmes EC, Chi-Ching Leung F. 2013. The spread of type 2 porcine reproductive and respiratory syndrome virus (PRRSV) in North America: a phylogeographic approach. Virology 447:146–154. 10.1016/j.virol.2013.08.028 [DOI] [PubMed] [Google Scholar]
- 41.Lai MM. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon-Loriere E, Holmes EC. 2011. Why do RNA viruses recombine? Nat. Rev. Microbiol. 9:617–626. 10.1038/nrmicro2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackwood MW, Hall D, Handel A. 2012. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 12:1305–1311. 10.1016/j.meegid.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decaro N, Mari V, Elia G, Addie DD, Camero M, Lucente MS, Martella V, Buonavoglia C. 2010. Recombinant canine coronaviruses in dogs, Europe. Emerg. Infect. Dis. 16:41–47. 10.3201/eid1601.090726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 72:4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stavrinides J, Guttman DS. 2004. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 78:76–82. 10.1128/JVI.78.1.76-82.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song JE, Jeong WG, Sung HW, Kwon HM. 2013. Sequencing, phylogenetic analysis, and potential recombination events of infectious bronchitis viruses isolated in Korea. Virus Genes 46:371–374. 10.1007/s11262-012-0856-0 [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Li Y, Liu R, Wang X, Gao F, Lin T, Huang T, Yao H, Tong G, Fan H, Wei Z, Yuan S. 2013. Porcine reproductive and respiratory syndrome virus ORF5a protein is essential for virus viability. Virus Res. 171:178–185. 10.1016/j.virusres.2012.11.005 [DOI] [PubMed] [Google Scholar]