ABSTRACT
The human immunodeficiency virus type 1 (HIV-1)-encoded virion infectivity factor (Vif) is required to inactivate the host restriction factor APOBEC3 by engaging Cullin 5 (Cul5)-RING ubiquitin ligase (CRL5). Core binding factor beta (CBF-β) is a novel regulator of Vif-CRL5 function; as yet, its mechanism of regulation remains unclear. In the present study, we demonstrate that CBF-β promotion of Vif-CRL5 assembly is independent of its influence on Vif stability and is also a conserved feature of primate lentiviral Vif proteins. Furthermore, CBF-β is critical for the formation of the Vif-ElonginB/ElonginC-Cul5 core E3 ubiquitin ligase complex in vitro. CBF-β from diverse vertebrate species supported HIV-1 Vif function, indicating the conserved nature of Vif–CBF-β interfaces. Considering the importance of the interaction between Vif and CBF-β in viral CRL5 function, disrupting this interaction represents an attractive pharmacological intervention against HIV-1.
IMPORTANCE HIV-1 encodes virion infectivity factor (Vif) to inactivate its host's antiviral APOBEC3 proteins. Vif triggers APOBEC3 degradation by forming Vif-Cullin 5 (Cul5)-RING ubiquitin ligase (CRL5). Core binding factor beta (CBF-β) is a novel regulator of Vif-CRL5 function whose mechanism of regulation remains poorly defined. In the present study, we demonstrate that the promotion of Vif-CRL5 assembly by CBF-β can be separated from its influence on Vif stability. The promotion of Vif-CRL5 assembly, but not the influence on Vif stability, is conserved among primate lentiviral Vif proteins: we found that CBF-β from diverse vertebrate species supported HIV-1 Vif function. Considering the importance of the interaction between Vif and CBF-β in viral CRL5 function and HIV-1 replication, disrupting this interaction is an attractive strategy against HIV-1.
INTRODUCTION
Most lentiviruses encode a virion infectivity factor (Vif) that is necessary for viral replication and survival in their hosts. Recent studies have indicated that lentiviral Vif is required to inactivate the host restriction factors (1, 2) present in natural target cells, such as CD4+ T cells and macrophages. These host restriction factors were identified as cytidine deaminase apolipoprotein B mRNA-editing catalytic polypeptide-like 3 (APOBEC3) proteins (3–14).
Human immunodeficiency virus type 1 (HIV-1) Vif suppresses the antiviral activity of APOBEC3 by degrading APOBEC3 as well as by inhibiting its packaging into newly synthesized virus particles. Vif interacts with cellular Cullin 5 (Cul5)-ElonginB-ElonginC (ElonginB/C) proteins to form an E3 ubiquitin ligase complex that targets selected APOBEC3 proteins for proteasomal degradation (15–23). Both the HIV and simian immunodeficiency virus (SIV) Vif proteins have evolved a conserved HX5CX17–18CX3–5H (HCCH) motif in the carboxyl-terminal region that recruits Cul5 (24–28). In contrast, HIV-1 Vif recognizes various APOBEC3 proteins through unique motifs in its amino-terminal region (17, 29–37).
We along with others have recently identified a novel cellular factor, core binding factor beta (CBF-β), a ubiquitously expressed cellular protein that is critical for the regulation of HIV-1 Vif activity (38–45). In uninfected cells, CBF-β forms stable heterodimeric transcription factors with RUNX family proteins to enhance RUNX's DNA-binding affinity. These transcription factors are also involved in diverse cellular functions, including development, hematopoiesis, and T-cell differentiation (46, 47). Previous studies have shown that during HIV infection, CBF-β specifically interacts with HIV-1 Vif to promote the degradation of APOBEC3G (A3G) and APOBEC3F (APOBEC3G/F). Depletion of endogenous CBF-β or mutations in Vif that disrupt CBF-β binding prevent Vif from suppressing the antiviral activity of APOBEC3G (41, 44). Furthermore, silencing of endogenous CBF-β with small interfering RNAs (siRNAs) inhibits HIV-1 replication in CD4+ T cells, even in the presence of Vif (41, 44).
However, the mechanism by which CBF-β regulates Vif function is not fully understood. One study has concluded that CBF-β increases the stability of Vif, resulting in increased protein levels (41), while a previous study from our group indicated that CBF-β is still required for Vif function even when Vif expression levels are not affected by CBF-β (44). Here, we report that CBF-β is required by HIV-1 Vif from three primary isolates, CCR5, CXCR4, and a dual-tropic isolate, for efficient Vif-Cul5 formation and subsequent APOBEC3G degradation. In vivo, stabilization of Vif expression by the proteasome inhibitor MG132 did not restore the Vif-Cul5 interaction in the absence of CBF-β. In vitro, purified recombinant Vif protein did not interact with Cul5 in the absence of CBF-β. Furthermore, CBF-β promoted the interaction between Cul5 and multiple SIV Vif proteins without influencing Vif stability. Thus, we suspect that CBF-β promotes the Vif-Cul5 interaction by inducing conformational changes in Vif.
MATERIALS AND METHODS
Plasmid construction.
HIV-1 Vif expression vector pVif-HA (where HA is hemagglutinin), SIVmac Vif expression vector pSIVmac Vif-HA, SIVagm Vif expression vector pSIVagm-myc, bovine immunodeficiency virus (BIV) Vif expression vector pBIV Vif-HA, feline immunodeficiency virus (FIV) Vif expression vector pFIV Vif-HA, A3G expression vector pA3G-HA, RhA3G (where Rh is rhesus macaque) vector pRhA3G-myc, RhA3C vector pRhA3C-myc, RhA3H vector pRhA3H-myc, cow A3F expression vector pCow A3F-HA, feline A3CH (fA3CH) expression vector pfA3CH-HA, and the CBF-β expression vector pCBF-β-myc have been previously described (44). Bos taurus CBF-β was constructed by site-directed mutagenesis (F88L) from human CBF-β. Mus musculus CBF-β (GenBank accession no. NM_001161458), Gallus gallus CBF-β (EMBL accession no. AAM09650.2), Danio rerio CBF-β (EMBL accession no. AAG49892.1), Drosophila melanogaster CBF-β (GenBank accession no. AAF47538.3), and Saccoglossus kowalevskii CBF-β (EMBL accession no. ACY92495.1) were synthesized by Shanghai Generay Biotech Co. and constructed in the VR1012 vector with a myc tag.
Antibodies and cell lines.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% fetal bovine serum and penicillin-streptomycin (D-10 medium) and passaged when confluent. The antibodies used in this study were anti-Vif antibody (catalog number 2221; AIDS Research and Reference Reagent Program), anti-CBF-β antibody (ab11921; Abcam), anti-Cul5 antibody (sc-13014; Santa Cruz), anti-ElonginB antibody (sc-11447; Santa Cruz), anti-ElonginC antibody (610760; BD Transduction Lab), anti-β-actin antibody (A3853; Sigma), anti-HA antibody (MMS-101R-1000; Covance), anti-myc antibody(05-724; Upstate), and anti-HA Affinity Matrix antibody (11815016001; Roche).
CBF-β silencing by RNA interference.
HEK293T cells were cotransfected with pLKO.1 or pLKO.1–CBF-β (clone TRCN0000016645, 5′-GAAGATAGAGACAGGTCTCAT-3′ [Open Biosystems]) together with pRSV-Rev (where RSV is Rous sarcoma virus), pMDLg/pRRE (where RRE is Rev-responsive element), and pCMV-VSVG (where CMV and VSVG are cytomegalovirus and vesicular stomatitis virus G protein, respectively). The assembled virus-like particles (VLPs) in the culture supernatants were used to infect fresh HEK293T cells. Three days later, HEK293T cells were selected with 5 μg/ml puromycin for 10 days. CBF-β expression was monitored by immunoblotting.
Pulldown assays.
Cul5 (residues 1 to 393 without tag), Cul5 (residues 1 to 393 with a glutathione S-transferase [GST] tag), Vif-ElonginB/C, and Vif–CBF-β–ElonginB/C were purified as described previously (45). In brief, Cul5 (residues 1 to 393) with a GST tag was expressed in the Escherichia coli BL21 (DE) strain at 16°C overnight and lysed by sonication, followed by affinity chromatography with glutathione-Sepharose 4B. At this stage, Cul5-GST in the beads was ready for use in GST pulldowns. To produce tag-free Cul5 protein, the GST tag was then removed using Prescission protease. Gel filtration chromatography was utilized for further purification. Vif-ElonginB/C and Vif–CBF-β–ElonginB/C were purified with nickel beads via His-tagged CBF-β 140 and/or His-tagged ElonginB. Purified proteins were buffer exchanged into phosphate-buffered saline (PBS) before the pulldown assays and adjusted to 0.5 mg/ml. For GST pulldown assays, GST-Cul5 beads were added to Vif-ElonginB/C and Vif–CBF-β–ElonginB/C, followed by 3 h of incubation at 4°C with shaking. For nickel bead pulldown, Cul5 (no tag) and Vif-ElonginB/C or Vif–CBF-β–ElonginB/C were mixed and incubated with nickel beads for 3 h at 4°C with shaking. The beads were then washed with PBS buffer five times, and the input and pulldown fractions were analyzed by SDS-PAGE and immunoblotting.
Gel filtration chromatography.
The purified N-terminal region of Cul5 (Cul5N) was mixed with purified Vif-ElonginB/C or Vif–CBF-β–ElonginB at a molar ratio of 1:1 and incubated at 4°C for 1 h. The protein mixture was then loaded onto a Superdex 200 10/300 GL column (GE Healthcare) with a 500-μl loop and run at a flow rate of 0.5 ml/min. The collected peak fractions were subjected to SDS-PAGE, followed by immunoblotting analysis with specific antibodies indicated in Materials and Methods. The gel filtration column was calibrated using vitamin B12 (1,370 Da), myoglobin (17,000 Da), ovalbumin (44,000 Da), gamma globulin (158,000 Da), and thyroglobulin (670,000 Da) as standards.
Transfection, immunoblot analysis, and immunoprecipitation.
Transfections and immunoblot analysis were performed as previously described (44, 48, 49). For immunoprecipitation assays, the lysate was centrifuged at 18,000 × g for 20 min at 4°C. HA or myc affinity matrix was then added to the supernatant and incubated with gentle rocking at room temperature for 2 h. After a quick spin, the beads were washed six times with washing buffer. Proteins were eluted with glycine hydrochloride (pH 2.5).
RESULTS
Separate roles for CBF-β in the promotion of HIV-1 Vif-CRL5 assembly and Vif stability.
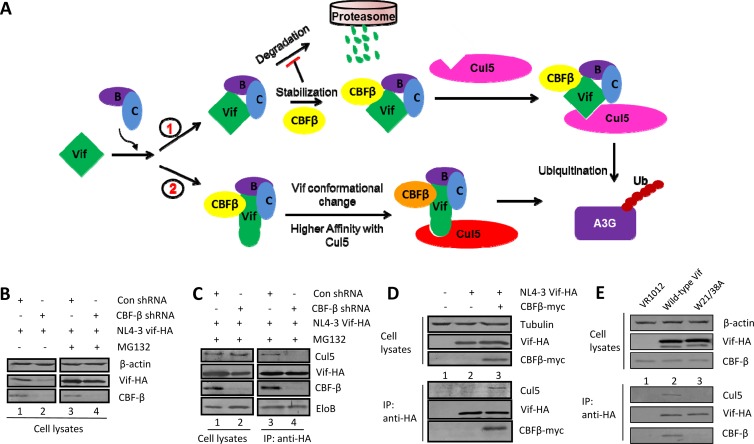
CBF-β has been shown to be a critical regulator of HIV-1 Vif function (38–45). Two possible roles for CBF-β in HIV-1 Vif regulation have been proposed (Fig. 1A). Since Vif is sensitive to proteasome-mediated degradation in the absence of CBF-β (39, 41), CBF-β may protect Vif from degradation and thus increase the amount of Vif available for Cul5 binding. Alternatively, CBF-β could induce a conformational change in Vif during binding, enabling more efficient binding of the Vif complex to Cul5 (44, 45). We first examined whether the reduced Vif-Cul5 interaction in the absence of CBF-β was a result of reduced Vif stability. When proteasome activity was inhibited by MG132 treatment, Vif expression was restored in both CBF-β-silenced HEK293T cells and control cells (Fig. 1B). However, inhibiting proteasome activity with MG132 did not restore the Vif-Cul5 interaction in CBF-β-silenced cells (Fig. 1C). In the presence of CBF-β, endogenous Cul5 was efficiently coprecipitated with HIV-1 Vif (Fig. 1C, lane 3), but in CBF-β-silenced cells, a significantly reduced amount of Cul5 was coprecipitated with HIV-1 Vif, even in the presence of MG132 (Fig. 1C, lane 4). When we coimmunoprecipitated ElonginB with Vif, we found that the interactions between Vif and ElonginB were unaffected (Fig. 1C). These results indicated that inhibiting proteasome activity did not restore the interaction of HIV-1 Vif with Cul5 in the absence of CBF-β. Furthermore, ectopically expressed CBF-β could restore the HIV-1 Vif-Cul5 interaction in CBF-β-silenced HEK293T cells (Fig. 1D). Thus, we were able to separate the unique role of CBF-β in promoting assembly of Vif-Cullin 5-RING ubiquitin ligase (Vif-CRL5) from its influence on HIV-1 Vif stability.
FIG 1.
Separate roles for CBF-β in promoting HIV-1 Vif-CRL5 assembly and Vif stability. (A) The proposed mechanisms by which CBF-β functions in Vif regulation. CBF-β could be involved either in Vif-CRL5 E3 ubiquitin (Ub) ligase complex formation (pathway 2) or in increasing Vif stability (pathway 1). B, ElonginB; C, ElonginC. (B) The stability of Vif was enhanced in CBF-β-silenced cells by the proteasome inhibitor MG132. HEK293T or CBF-β-silenced HEK293T cells were transfected with the Vif-HA (HIV-1 NL4-3) expression vector. After 36 h, cells were treated with 2 μM MG132 for 14 h. Cell lysates were then analyzed by SDS-PAGE and immunoblotting using specific antibodies as indicated. β-Actin was used as an internal control. The data shown represent one of three independent experiments. (C) The Vif-Cul5 interaction was not restored by MG132 treatment in CBF-β-silenced HEK293T cells. HEK293T cells or CBF-β-silenced HEK293T cells were transfected with an expression vector for Vif-HA and treated with MG132. Cell lysates were prepared for immunoblotting or immunoprecipitated (IP) with the anti-HA antibody, and the precipitated samples were analyzed by immunoblotting with anti-HA antibody to detect Vif-HA. Samples were also analyzed using anti-CBF-β, anti-Cul5, or anti-ElonginB antibody. The data shown represent one of three independent experiments. (D) Expression of exogenous CBF-β in CBF-β-silenced HEK293T cells restores the Vif-Cul5 interaction without affecting Vif expression. The data shown represent one of three independent experiments. (E) The CBF-β binding-defective HIV-1 Vif mutant (Trp21 to Ala and Trp38 to Ala substitutions; W21/38A) cannot interact with Cul5 even if its stability is maintained. HEK293T cells were transfected with the indicated expression vectors. Cell lysates were prepared and immunoprecipitated, followed by immunoblot analysis with specific antibodies against Cul5, CBF-β, or the HA tag (to detect Vif-HA). β-Actin was used as an internal control. The data shown represent one of three independent experiments.
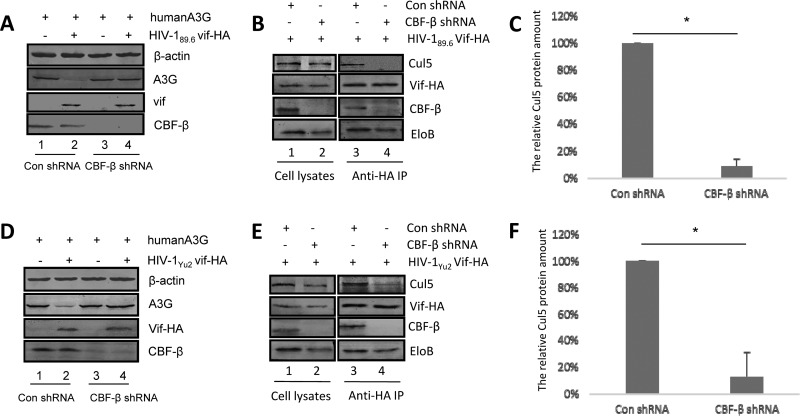
Similar results were observed for Vif proteins from the HIV-1 CCR5-tropic strain Yu2 and the dual-tropic strain 89.6 (Fig. 2). First, we observed that CBF-β is necessary for HIV-1 89.6 and HIV-1 Yu2 Vif proteins to degrade A3G (Fig. 2A and D). We then restored Vif expression with MG132, followed by immunoprecipitation. As was observed for Vif from HIV-1 NL4-3, coimmunoprecipitation of Cul5 with Vif from both HIV-1 89.6 and HIV-1 Yu2 was significantly inhibited when CBF-β was silenced, yet the association between ElonginB and Vif was unaffected under the same conditions (Fig. 2B and E). The interaction of Cul5 with Vif from both HIV-1 89.6 (Fig. 2C) and HIV-1 Yu2 (Fig. 2F) was significantly inhibited in repeated experiments when CBF-β was silenced. Taken together, these results demonstrate that CBF-β promotes the interaction of Cul5 and Vif proteins from both laboratory and primary HIV-1 strains.
FIG 2.
CBF-β is critical for the primary HIV-1 Vif-Cul5 interaction. CBF-β is important for HIV-189.6 Vif-mediated (A) and HIV-1Yu2 Vif-mediated (D) degradation of human A3G. HEK293T cells were transfected with expression vectors for HIV-189.6 Vif-HA (B) or HIV-1Yu2 Vif-HA (E) in the presence of control shRNA (lane 1) or shRNA against CBF-β (lane 2). After 36 h, cells were lysed, and the supernatant was immunoprecipitated (IP) and analyzed by immunoblotting as described in Materials and Methods (lanes 3 and 4). Coimmunoprecipitated Cul5 protein bands with HIV-189.6 Vif (B, lanes 3 and 4) and HIV-1Yu2 Vif (E, lanes 3 and 4) were quantified as previously described (51) and shown in panel C and F, respectively. Coimmunoprecipitated Cul5 from control cells was set to 100%. The data shown represent one of three independent experiments. Error bars represent the standard deviation of the mean of triplicate samples. EloB, ElonginB; Con, control.
CBF-β promotes the Vif-Cul5 interaction in vitro.
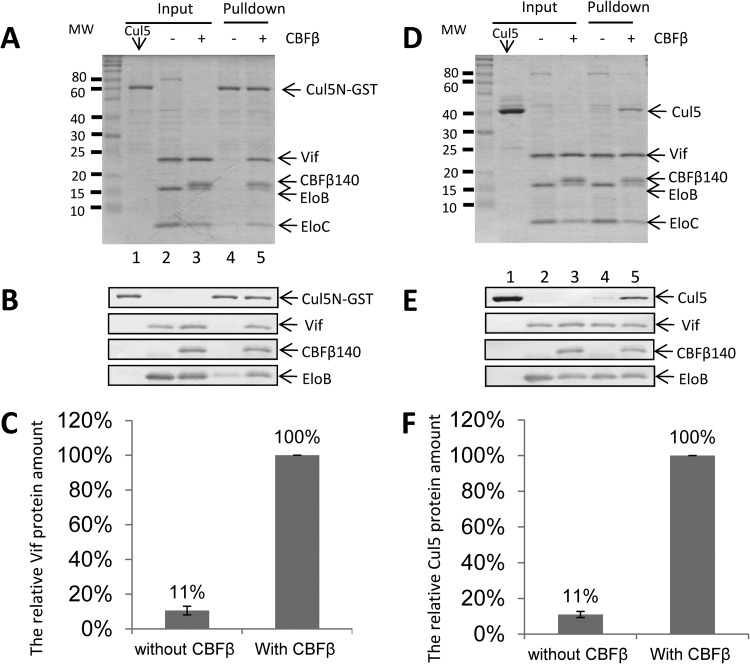
To examine whether CBF-β could promote Vif-CRL5 E3 ligase complex formation in vitro, in the absence of protease activity, we expressed and purified these proteins separately. These proteins included the N-terminal region of Cul5 (Cul5N), which is sufficient for the interaction of Vif with ElonginB/C (45), the Vif-ElonginB/C complex with a His tag, and the Vif–CBF-β–ElonginB/C complex with a His tag. Vif could be efficiently pulled down by GST-Cul5N when CBF-β was present, as determined by SDS-PAGE and Coomassie blue staining (Fig. 3A, lane 5). In contrast, Vif-ElonginB/C complexes lacking CBF-β could not be efficiently pulled down by GST-Cul5N (Fig. 3A, lane 4). This difference was further confirmed by immunoblotting with a Vif-specific antibody (Fig. 3B). Quantitative analysis of the Vif protein from the Coomassie-stained gel suggested that 90% less Vif could be pulled down by Cul5 in the absence of CBF-β than when CBF-β was present (Fig. 3C). We also examined the interaction of Vif with Cul5 in a reciprocal experiment. Cul5N could be efficiently immunoprecipitated by the Vif–CBF-β–ElonginB/C complex (Fig. 3D, lane 5) but not by the Vif-ElonginB/C complex (lane 4). This difference was also confirmed by immunoblotting with a Cul5-specific antibody (Fig. 3D). Quantitative analysis of the Cul5N protein from the Coomassie-stained gel suggested that 90% less Cul5N could be pulled down by Vif in the absence of CBF-β than in its presence (Fig. 3F). These results demonstrated that CBF-β allows the Vif-ElonginB/C complex to recruit Cul5 in a more efficient manner.
FIG 3.
CBF-β enhances the interaction of HIV-1 Vif and Cul5 in vitro. (A) Efficient pulldown of Vif–CBF-β–ElonginB/C but not Vif-ElonginB/C by Cul5N-GST. Cul5N-GST, Vif–CBF-β–ElonginB/C, and Vif-ElonginB/C proteins were expressed in E. coli and purified as described in Materials and Methods. The Vif-Cul5 interaction was examined by GST pulldown assay. The input and pulldown samples were analyzed by SDS-PAGE and Coomassie staining. (B) The input and pulldown samples were also analyzed by SDS-PAGE and immunoblotting, using specific antibodies as indicated. (C) Quantification of Vif protein bands in the pulldown fractions from the Coomassie-stained gel by ImageJ. Values are expressed as means ± standard deviations for three independent experiments. (D) Efficient pulldown of Cul5N by Vif–CBF-β–ElonginB/C but not Vif-ElonginB/C. Cul5N, His-tagged Vif–CBF-β–ElonginB/C, and His-tagged Vif-ElonginB/C proteins were expressed in E. coli and purified as described in Materials and Methods. The Vif-Cul5 interaction was examined by nickel bead pulldown assays. The input and pulldown samples were analyzed by SDS-PAGE and Coomassie staining. (E) The input and pulldown samples were also analyzed by SDS-PAGE and immunoblotting, using specific antibodies as indicated. (F) Quantification of Cul5N protein bands in the pulldown fractions from the Coomassie-stained gel by ImageJ. Values are expressed as means ± standard deviations for three independent experiments. MW, molecular weight (in thousands).
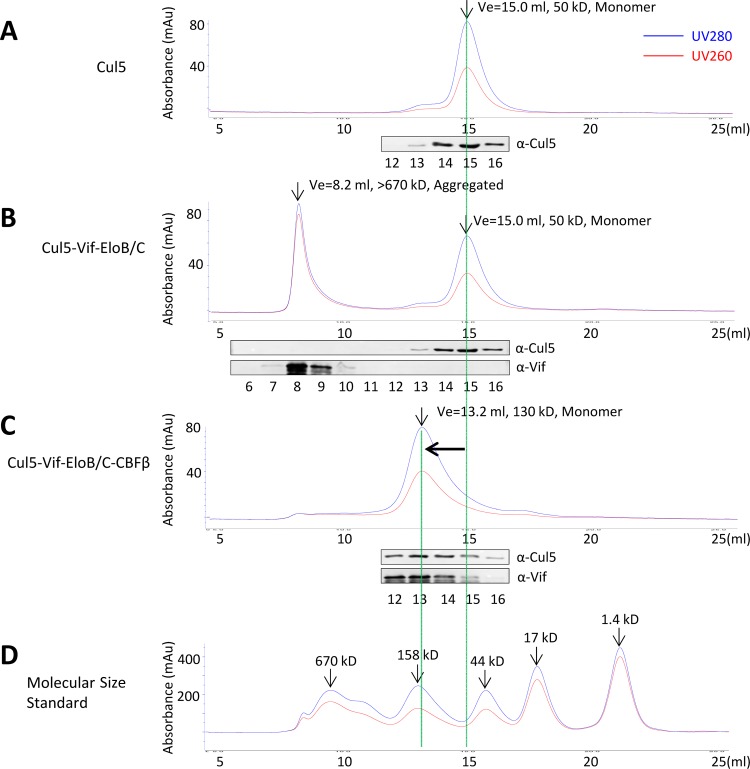
We further examined the Cul5-Vif interaction, with or without CBF-β, by size exclusion chromatography. Untagged, purified Cul5N ran as monomer in size exclusion chromatography, and it eluted at ∼15.0 ml on a Superdex 200 column, consistent with its molecular size (Fig. 4A). Subsequently, we incubated purified Cul5N with the Vif-ElonginB/C complex (Fig. 4B) or the Vif–CBF-β–ElonginB/C complex (Fig. 4C) at 4°C for 1 h and then analyzed the samples by gel filtration chromatography. The elution profiles of the molecular mass standards were also analyzed (Fig. 4D). Only when Cul5N was mixed with Vif–CBF-β–ElonginB/C was the expected Cul5N peak at 15 ml replaced by a protein complex with a slightly larger molecular size (eluting at ∼13.2 ml) (Fig. 4C). These results indicate that Cul5N can readily form complexes with Vif–CBF-β–ElonginB/C but not with Vif-ElonginB/C, and they further support a role for CBF-β in promoting a more efficient Vif-Cul5 interaction.
FIG 4.
Vif-Cul5-ElonginB/C forms stable complexes only in the presence of CBF-β. Purified Cul5N (residues 1 to 393) was mixed with purified Vif-ElonginB/C or Vif–CBF-β–ElonginB/C at a molar ratio of 1:1 and incubated at 4°C for 1 h. The protein mixture was then loaded onto a Superdex 200 10/300 GL column. The collected peak fractions were subjected to SDS-PAGE, followed by immunoblot analysis with specific antibodies indicated in Materials and Methods. Ve, elution volume; EloB/C, ElonginB/C. (A) Gel filtration profile of Cul5N alone. Cul5N is a monomer in gel filtration, with a molecular size of ∼50 kDa. (B) Gel filtration profile of Cul5N-Vif-ElonginB/C. Cul5 and Vif-ElonginB/C, which eluted in different peaks. The Cul5 peak did not change. (C) Gel filtration profile of Cul5N–Vif–Elongin/C–CBF-β. Cul5 and Vif–ElonginB/C–CBF-β eluted in the same peak. The Cul5 peak moved from 15.0 ml (∼50 kDa) to 13.2 ml (130 kDa). The theoretical molecular size of Cul5–Vif–ElonginB/C–CBF-β is 110 kDa. (D) Gel filtration profile of the molecular size standards: vitamin B12 (1,370 Da), myoglobin (17,000 Da), ovalbumin (44,000 Da), gamma globulin (158,000 Da), and thyroglobulin (670,000 Da). Au, arbitrary units; α, anti.
CBF-β is required for SIV Vif function and the assembly of the SIV Vif-CRL5 E3 ubiquitin ligase complex.
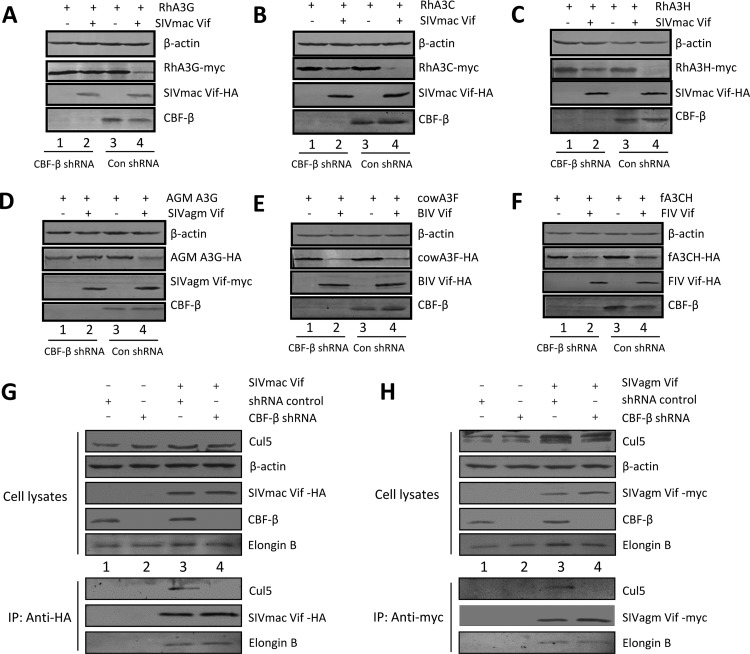
To determine whether CBF-β has a conserved effect on the activity of other lentiviral Vif proteins, we used short hairpin RNAs (shRNAs) to knock down the expression of endogenous CBF-β; CBF-β was reduced by >80% with the CBF-β-specific shRNA compared to control shRNA-treated cells. In the control shRNA-treated cells, SIVmac Vif efficiently reduced the expression of rhesus macaque A3G (RhA3G) compared to the level in the negative control (Fig. 5A). When endogenous CBF-β was silenced, the ability of SIVmac Vif to degrade RhA3G was blocked. Similarly, SIVmac Vif required CBF-β to deplete RhA3C (Fig. 5B) and RhA3H (Fig. 5C). CBF-β was also required for the SIVagm Vif-mediated depletion of African green monkey (AGM) A3G (Fig. 5D). Although CBF-β was important for the function of SIVmac and SIVagm Vif, silencing CBF-β did not affect SIVmac or SIVagm Vif expression (Fig. 5A to D).
FIG 5.
CBF-β is important for SIV Vif but not FIV or BIV Vif function. (A to C) SIVmac Vif-mediated downregulation of RhA3 requires CBF-β. HEK293T cells or CBF-β-silenced HEK293T cells were cotransfected with expression vectors for RhA3G-myc (A), RhA3C (B), or RhA3H (C) plus SIVmac Vif-HA or a control vector (pcDNA3.1). Cell lysates were prepared at 36 h posttransfection and analyzed by SDS-PAGE and immunoblotting using specific antibodies as indicated. β-Actin was used as an internal control. (D) SIVagm Vif-mediated downregulation of AGM A3G requires CBF-β. (E) BIV Vif-mediated downregulation of bovine A3F does not require CBF-β. (F) FIV Vif-mediated downregulation of fA3CH does not require CBF-β. (G and H) CBF-β is important for the SIV Vif-Cul5 interaction. HEK293T or CBF-β-silenced HEK293T cells were transfected with an expression vector for SIVmac Vif-HA (G) or SIVagm Vif-myc (H). Cell lysates were prepared for immunoblotting or were immunoprecipitated with the anti-HA antibody or anti-myc antibody. The precipitated samples were analyzed by immunoblotting with an anti-HA antibody to detect SIVmac Vif-HA (G) or with anti-myc antibody to detect SIVagm Vif-myc (H). Samples were also analyzed using anti-Cul5 or anti-ElonginB antibody.
In contrast to the results with primate lentiviral Vif proteins, the BIV Vif-induced depletion of bovine A3F (Fig. 5E) and FIV Vif-induced depletion of feline A3CH (Fig. 5F) were not affected by CBF-β silencing. Thus, our results suggest that the role of CBF-β in primate lentiviral Vif-induced APOBEC3 degradation is evolutionarily conserved. Similarly, CBF-β appears to promote SIV Vif function without affecting its stability. Interestingly, HIV-1 and diverse SIV Vif molecules contain a unique HCCH zinc-binding motif that is critical for the Cul5 interaction (24–28). This motif is not detected in nonprimate lentiviral Vif proteins, such as those from BIV, FIV, or visna virus. To determine whether CBF-β might play a role in the assembly of the SIVmac and SIVagm Vif-Cul5 E3 ubiquitin ligases, we performed a coimmunoprecipitation experiment and found that the amount of coimmunoprecipitation of Cul5 with SIVmac Vif-HA or SIVagm Vif-myc was significantly decreased when CBF-β was silenced (Fig. 5G and H). However, coprecipitation of ElonginB with SIVmac Vif-HA or SIVagm Vif-myc was not significantly affected under the same conditions. Thus, the CBF-β-mediated Vif-Cul5 interaction appears to be a conserved feature among primate lentiviruses.
CBF-β from diverse animal species can support HIV-1 Vif function.
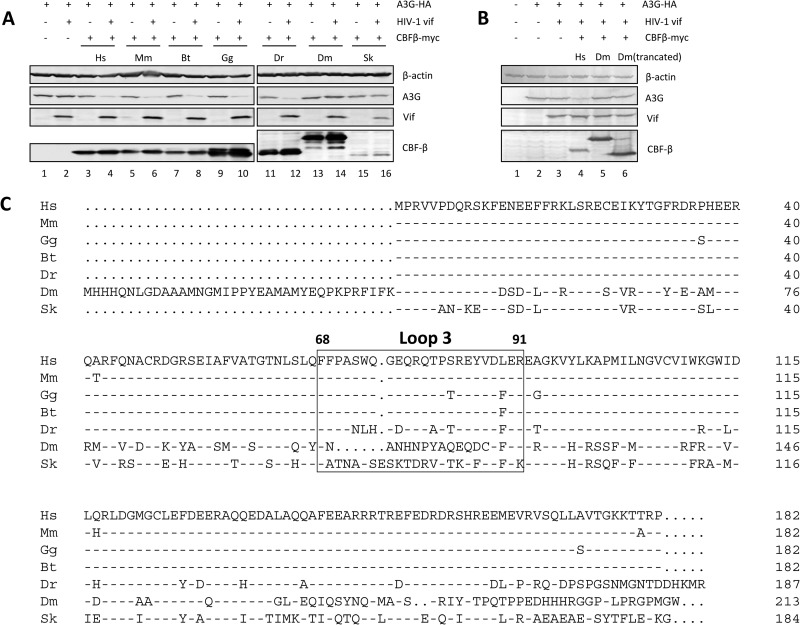
Since CBF-β homologs have been found in many vertebrate and invertebrate species, we wanted to further investigate the interactions between CBF-β proteins from diverse hosts and the HIV Vif protein. For this purpose, we constructed CBF-β expression vectors of multiple species: Mus musculus, Bos taurus, Gallus gallus, Danio rerio, Drosophila melanogaster, and Saccoglossus kowalevskii. CBF-β from these species was cotransfected with HIV-I Vif into CBF-β-silenced HEK293T cells. In CBF-β-silenced cells, HIV-1 Vif could not degrade A3G (Fig. 6A, lane 2). Expression of human CBF-β restored the ability of Vif to degrade A3G (Fig. 6A, lanes 3 and 4). Like human CBF-β, the CBF-β from Mus musculus (Fig. 6A, lanes 5 and 6), Bos taurus (lanes 7 and 8), Gallus gallus (lanes 9 and 10), and Danio rerio (lanes 11 and 12) could all restore HIV-1 Vif's ability to degrade A3G. In contrast, CBF-β from the two invertebrate species, Drosophila melanogaster and Saccoglossus kowalevskii, could not restore Vif function. Previous studies have shown that the loop 3 region of human CBF-β is important for HIV-1 Vif binding (44). The loop 3 sequences are highly conserved in the animal CBF-β proteins that were able to restore Vif function (Fig. 6C). From the sequence alignment, we could see a high degree of diversity in the loop 3 region between these two CBF-βs and the others. There was also an extra sequence at the amino terminus of Drosophila melanogaster CBF-β. To exclude the possibility that the effect we observed was caused by this extra sequence, we constructed a truncated Drosophila CBF-β with a deletion of residues 1 to 36 [CBF-β(Δ1-36)], which still could not restore the function of HIV-1 Vif (Fig. 6B, lane 6).
FIG 6.
CBF-β from multiple animal species can support HIV-1 Vif function. (A) CBF-β-silenced HEK293T cells were cotransfected with expression vectors for human A3G-HA, HIV-1 Vif, and CBF-β–myc from different animal species as indicated. The expression levels of A3G, HIV-1 Vif, and CBF-β–myc were then examined by immunoblotting. β-Actin was used as an internal control. Species names are abbreviated as follows: Hs, Homo sapiens; Mm, Mus musculus; Bt, Bos taurus; Gg, Gallus gallus; Dr, Danio rerio; Dm, Drosophila melanogaster; Sk, Saccoglossus kowalevskii. (B) Truncated Drosophila CBF-β also could not restore HIV-1 Vif function. (C) Alignment of CBF-β sequences from multiple species. Loop 3 (amino acids 68 to 91) is defined. This region is important for the interaction of CBF-β with HIV-1 Vif.
DISCUSSION
HIV-1 Vif assembles with Cul5-ElonginB/C (Vif-CRL5) to target APOBEC3 for polyubiquitination and degradation. CBF-β has recently been identified as a critical regulator of HIV-1 Vif-CRL5 function (38–45). However, the mechanism by which CBF-β regulates Vif function is not fully understood. It has been observed that CBF-β enhances the stability of HIV-1 Vif (39, 41) in virus-producing cells and increases its resistance to protease digestion in vitro (42). In the current study, we demonstrate that the promotion of Vif-CRL5 assembly by CBF-β is a separate function from its regulation of Vif stability. First, we showed that stabilization of HIV-1 Vif expression with a proteasome inhibitor did not restore the Vif-Cul5 interaction in cultured cells in the absence of CBF-β (Fig. 1C). Second, in CBF-β-silenced cells, ectopically expressed CBF-β was able to restore the interaction (Fig. 1D). Third, CBF-β binding-defective Vif mutants were expressed at the same levels as wild-type Vif proteins (Fig. 1E), yet these mutant Vif proteins interacted poorly with Cul5 compared to wild-type Vif. Next, we showed that CBF-β was also essential for the interaction of purified Vif and Cul5 in vitro (Fig. 3 and 4). By conducting thermodynamic measurements, Salter et al. have also shown that CBF-β increases the affinity of Vif for Cul5 (43). Finally, CBF-β did not influence the stability of SIVmac Vif or SIVagm Vif (Fig. 5). However, the formation of the Vif-CRL5 E3 ubiquitin ligase complex with SIVmac and SIVagm Vif did depend on the association of these SIV Vif proteins with CBF-β.
The ordered assembly of Vif-CRL5 requires CBF-β. Vif-ElonginB/C assembly, either in vivo or in vitro, was not affected by CBF-β. However, further assembly of Vif-ElonginB/C with Cul5 depends on CBF-β. Since CBF-β does not bind Cul5 directly (41, 44), it is likely that CBF-β primes Vif to enhance its interaction with Cul5. CBF-β may induce conformational changes in Vif that promote the Vif-Cul5 interaction. CBF-β-induced conformational changes in Vif could also explain the increased stability (39, 41), solubility (45), and reduced oligomerization (42, 45) of Vif molecules in the presence of CBF-β.
Most lentiviruses encode a Vif protein that is necessary for viral replication and survival in the host. Lentiviral Vif proteins have been shown to inactivate APOBEC3 antiviral factors from their natural hosts. In this study, we have demonstrated that CBF-β is a conserved cellular regulator of primate lentiviral Vif proteins from HIV-1, SIVmac, and SIVagm. CBF-β was found to be essential for SIVmac or SIVagm Vif-mediated inactivation of RhA3 or AGM A3, respectively. However, CBF-β was not required for the function of nonprimate lentiviral Vif proteins such as FIV and BIV. When endogenous CBF-β was silenced, both the FIV and BIV Vif-mediated degradation of sensitive A3 proteins could still be detected. Thus, CBF-β is a unique regulator of HIV/SIV Vif-Cul5-CRL E3 ubiquitin ligases.
Why CBF-β was not required for nonprimate lentiviral Vif (e.g., BIV Vif) function is not clear. It is unlikely that differences in mammalian CBF-β could account for this observation since CBF-β proteins from diverse mammals (including Mus musculus and Bos taurus) and nonmammalian vertebrates (including Gallus gallus and Danio rerio) were able to support Vif function. It is conceivable that nonprimate lentiviral Vif proteins have evolved to use different mechanisms for the assembly of the viral Vif-E3 ubiquitin ligase for inactivating host APOBEC3 proteins.
Although diverse vertebrate CBF-β proteins were able to support the function of HIV-1 Vif, CBF-β proteins from certain invertebrates were defective in this regard. We have demonstrated that loop 3 of human CBF-β is important for Vif binding, and this region is highly divergent among animal CBF-β proteins (Fig. 5). While these sequence differences in loop 3 may contribute to the differences seen in their abilities to promote Vif function, we along with others have also observed that hydrophobic residues throughout the HIV-1 Vif molecule are important for the interaction with CBF-β (42, 50). These data indicate that multiple regions in CBF-β are likely involved in the Vif–CBF-β interaction. Definitive identification of the critical CBF-β residues involved in Vif binding will require structural information concerning the Vif–CBF-β complex. This is a promising area for future research.
CBF-β is important for the function of both RUNX and Vif. We have demonstrated previously that Vif and RUNX can utilize different domains of CBF-β (38, 44). A similar observation has been reported by other investigators (40). Mutations in CBF-β regions that are known to be important for RUNX protein binding had little effect on the Vif–CBF-β interaction, suggesting that the interaction of Vif with CBF-β does not require RUNX proteins (44). Furthermore, regions in CBF-β that are involved in Vif binding do not affect RUNX binding. CBF-β mutants that cannot support HIV-1 Vif function are still competent for promoting RUNX function (38). Diverse Vif proteins from HIV and SIV interact with CBF-β, indicating a conserved motif(s) that may be required for this essential interaction. Since Vif–CBF-β interfaces could be disrupted without affecting CBF-β's cellular function, disrupting this interaction represents an attractive pharmacological intervention against HIV-1.
ACKNOWLEDGMENTS
We are grateful to Kun Luo and Egbert Hoiczyck for advice and helpful discussions and to Nancy A. Speck and Alex Bullock for plasmids. We thank Deborah McClellan for editorial assistance. Antibody to Vif was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (catalog number 2221; contributed by D. Gabuzda).
This work was supported in part by funding from the Chinese Ministry of Science and Technology (2012CB911100 and 2013ZX0001-005), the Chinese Ministry of Education (IRT1016), and the Key Laboratory of Molecular Virology, Jilin Province, China (20102209).
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- 2.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. 10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115. 10.1038/ni1125 [DOI] [PubMed] [Google Scholar]
- 4.Chiu YL, Greene WC. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353. 10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- 5.Cullen BR. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067–1076. 10.1128/JVI.80.3.1067-1076.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goff SP. 2004. Retrovirus restriction factors. Mol. Cell 16:849–859. 10.1016/j.molcel.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 7.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868–877. 10.1038/nri1489 [DOI] [PubMed] [Google Scholar]
- 8.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. 10.1016/j.chom.2008.04.008 [DOI] [PubMed] [Google Scholar]
- 9.Navarro F, Landau NR. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16:477–482. 10.1016/j.coi.2004.05.006 [DOI] [PubMed] [Google Scholar]
- 10.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. 2005. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 4:1281–1285. 10.4161/cc.4.9.1994 [DOI] [PubMed] [Google Scholar]
- 11.Rose KM, Marin M, Kozak SL, Kabat D. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291–297. 10.1016/j.molmed.2004.04.008 [DOI] [PubMed] [Google Scholar]
- 12.Rose KM, Marin M, Kozak SL, Kabat D. 2004. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J. Biol. Chem. 279:41744–41749. 10.1074/jbc.M406760200 [DOI] [PubMed] [Google Scholar]
- 13.Turelli P, Trono D. 2005. Editing at the crossroad of innate and adaptive immunity. Science 307:1061–1065. 10.1126/science.1105964 [DOI] [PubMed] [Google Scholar]
- 14.Niewiadomska AM, Yu XF. 2009. Host restriction of HIV-1 by APOBEC3 and viral evasion through Vif. Curr. Top. Microbiol. Immunol. 339:1–25. 10.1007/978-3-642-02175-6_1 [DOI] [PubMed] [Google Scholar]
- 15.Conticello SG, Harris RS, Neuberger MS. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009–2013. 10.1016/j.cub.2003.10.034 [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Yu X, Luo K, Yu Y, Yu XF. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072–2081. 10.1128/JVI.78.4.2072-2081.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403. 10.1038/nm946 [DOI] [PubMed] [Google Scholar]
- 18.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792–7798. 10.1074/jbc.M313093200 [DOI] [PubMed] [Google Scholar]
- 19.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407. 10.1038/nm945 [DOI] [PubMed] [Google Scholar]
- 20.Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591–601. 10.1016/S1097-2765(03)00353-8 [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. 10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- 22.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861–2866. 10.1101/gad.1249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867–2872. 10.1101/gad.1250204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444–11449. 10.1073/pnas.0502440102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259–17265. 10.1074/jbc.M602413200 [DOI] [PubMed] [Google Scholar]
- 26.Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J. 21:217–222. 10.1096/fj.06-6773com [DOI] [PubMed] [Google Scholar]
- 27.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. 2007. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 373:541–550. 10.1016/j.jmb.2007.07.029 [DOI] [PubMed] [Google Scholar]
- 28.Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290–299. 10.1016/j.virol.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 29.He Z, Zhang W, Chen G, Xu R, Yu XF. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381:1000–1011. 10.1016/j.jmb.2008.06.061 [DOI] [PubMed] [Google Scholar]
- 30.Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235–13241. 10.1128/JVI.00204-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pery E, Rajendran KS, Brazier AJ, Gabuzda D. 2009. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 83:2374–2381. 10.1128/JVI.01898-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell RA, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201–8210. 10.1128/JVI.00395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrofelbauer B, Senger T, Manning G, Landau NR. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984–5991. 10.1128/JVI.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. 10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80:3112–3115. 10.1128/JVI.80.6.3112-3115.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, He Z, Wang T, Xu R, Yu XF. 2009. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 83:8674–8682. 10.1128/JVI.00653-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang Y, Wang X, Zhou T, York IA, Zheng YH. 2009. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 83:8544–8552. 10.1128/JVI.00651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du J, Zhao K, Rui Y, Li P, Zhou X, Zhang W, Yu XF. 2013. Differential requirements for HIV-1 Vif-mediated APOBEC3G degradation and RUNX1-mediated transcription by core binding factor beta. J. Virol. 87:1906–1911. 10.1128/JVI.02199-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hultquist JF, Binka M, LaRue RS, Simon V, Harris RS. 2012. Vif proteins of human and simian immunodeficiency viruses require cellular CBFβ to degrade APOBEC3 restriction factors. J. Virol. 86:2874–2877. 10.1128/JVI.06950-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hultquist JF, McDougle RM, Anderson BD, Harris RS. 2012. HIV type 1 viral infectivity factor and the RUNX transcription factors interact with core binding factor beta on genetically distinct surfaces. AIDS Res. Hum. Retroviruses 28:1543–1551. 10.1089/aid.2012.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. 10.1038/nature10693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DY, Kwon E, Hartley PD, Crosby DC, Mann S, Krogan NJ, Gross JD. 2013. CBFbeta stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell 49:632–644. 10.1016/j.molcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salter JD, Lippa GM, Belashov IA, Wedekind JE. 2012. Core-binding factor beta increases the affinity between human Cullin 5 and HIV-1 Vif within an E3 ligase complex. Biochemistry 51:8702–8704. 10.1021/bi301244z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. 10.1038/nature10718 [DOI] [PubMed] [Google Scholar]
- 45.Zhou X, Evans SL, Han X, Liu Y, Yu XF. 2012. Characterization of the interaction of full-length HIV-1 Vif protein with its key regulator CBFbeta and CRL5 E3 ubiquitin ligase components. PLoS One 7:e33495. 10.1371/journal.pone.0033495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bruijn MF, Speck NA. 2004. Core-binding factors in hematopoiesis and immune function. Oncogene 23:4238–4248. 10.1038/sj.onc.1207763 [DOI] [PubMed] [Google Scholar]
- 47.Ito Y. 2008. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99:33–76. 10.1016/S0065-230X(07)99002-8 [DOI] [PubMed] [Google Scholar]
- 48.Dettenhofer M, Yu XF. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu XF, Dawson L, Tian CJ, Flexner C, Dettenhofer M. 1998. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J. Virol. 72:3412–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Han X, Zhao K, Du J, Evans SL, Wang H, Li P, Zheng W, Rui Y, Kang J, Yu XF. 18 December 2013. Dispersed and conserved hydrophobic residues of HIV-1 Vif are essential for CBFβ recruitment and A3G suppression. J. Virol. 10.1128/JVI.03604-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei W, Guo H, Han X, Liu X, Zhou X, Zhang W, Yu XF. 2012. A novel DCAF1-binding motif required for Vpx-mediated degradation of nuclear SAMHD1 and Vpr-induced G2 arrest. Cell. Microbiol. 14:1745–1756. 10.1111/j.1462-5822.2012.01835.x [DOI] [PubMed] [Google Scholar]