ABSTRACT
The study of cellular central carbon metabolism modulations induced by viruses is an emerging field. Human cytomegalovirus (HCMV), herpes simplex virus (HSV), Kaposi's sarcoma-associated herpesvirus (KSHV), and hepatitis C virus (HCV) have been shown recently to reprogram cell metabolism to support their replication. During HCV infection the global glucidolipidic metabolism of hepatocytes is highly impacted. It was suggested that HCV might modify glucose uptake and glycolysis to increase fatty acids synthesis, but underlying mechanisms have not been completely elucidated. We thus investigated how HCV may modulate glycolysis. We observed that in infected Huh7.5 cells and in subgenomic replicon-positive Huh9.13 cells, glucose consumption as well as lactate secretion was increased. Using protein complementation assays and coimmunoprecipitation, we identified a direct interaction between the HCV NS5A protein and cellular hexokinase 2 (HK2), the first rate-limiting enzyme of glycolysis. NS5A expression was sufficient to enhance glucose consumption and lactate secretion in Huh7.5 cells. Moreover, determination of HK activity in cell homogenates revealed that addition of exogenous NS5A protein, either the full-length protein or its D2 or D3, but not D1, domain, was sufficient to increase enzyme activity. Finally, determination of recombinant HK2 catalytic parameters (Vmax and Km) in the presence of NS5A identified this viral protein as an activator of the enzyme. In summary, this study describes a direct interaction between HCV NS5A protein and cellular HK2 which is accompanied by an increase in HK2 activity that might contribute to an increased glycolysis rate during HCV infection.
IMPORTANCE Substantial evidence indicates that viruses reprogram the central carbon metabolism of the cell to support their replication. Nevertheless, precise underlying mechanisms are poorly described. Metabolic pathways are structured as connected enzymatic cascades providing elemental biomolecular blocks necessary for cell life and viral replication. In this study, we observed an increase in glucose consumption and lactate secretion in HCV-infected cells, revealing higher glycolytic activity. We also identified an interaction between the HCV NS5A nonstructural protein and cellular hexokinase 2, the first rate-limiting enzyme of glycolysis. This interaction results in an enhancement of catalytic parameters of the enzyme, which might explain, at least in part, the aerobic glycolysis shift observed in HCV-infected cells.
INTRODUCTION
Chronic infection with hepatitis C virus (HCV) is associated with abnormalities in glucidolipidic metabolism that have been shown to play important roles in various aspects of the virus life cycle, such as genome replication, viral assembly, and secretion (1, 2). In vitro, HCV replication is efficiently inhibited by cholesterol and fatty acid biosynthetic pathway inhibitors (3), and release of viral particles from infected cells can be inhibited by modulation of the very-low-density lipoprotein (VLDL) assembly process (4–6). Indeed, HCV interference with lipid metabolism has been extensively studied since the virus associates with lipoproteins and forms infectious hybrid particles, the lipo-viral particles (LVP) that are secreted from hepatocytes (5, 7–9). Association of the virus with lipids confers low density to LVP and is correlated to higher infectivity (10–12). Secreted LVP are particularly rich in neutral lipids, such as triglycerides, which require fatty acids to be synthesized. Indeed, within HCV-infected hepatocytes, lipid metabolism homeostasis is disturbed, leading in particular to accumulation of neutral lipids within cytoplasmic lipid droplets. These perturbations, illustrated by the frequent clinically patent steatosis in HCV-infected patients, are supposed to result from increased lipogenesis, reduced β-oxidation, and fatty acid export (13, 14). Nevertheless, the mechanisms underlying such cell metabolism modifications have not yet been completely elucidated, and, in spite of a growing body of literature documenting this intriguing interaction between the host glucidolipidic pathway and the HCV life cycle, little is known about the ability of the virus to modify global cell metabolism and how it proceeds.
Using a proteomic approach, in order to characterize the cellular metabolic modifications induced by HCV, Diamond et al. have recently described the early modifications of host protein expression in the permissive cell line Huh7.5 infected with an HCV cell culture (HCVcc)-adapted strain (15). They showed that several enzymes implicated in glycolysis, the pentose phosphate shunt, the tricarboxylic acid cycle (TCA) cycle, and central carbon metabolism (CCM) were overexpressed during infection. This study thus supports the idea that HCV might reprogram host metabolism.
HCV likely reprograms cell metabolism in response to higher energy expenditure and nutrient needs for virus synthesis following infection. An increase in glucose uptake by virus-infected cells was observed as early as 1957 (16, 17), but it was only recently that specific CCM modifications were observed during viral infection of cells by Herpesviridae members (herpes simplex virus [HSV], Kaposi's sarcoma-associated herpesvirus [KSHV], and human cytomegalovirus [HCMV]). In particular, HCMV induces in human fibroblasts an increase in metabolite fluxes through glycolysis and the citric acid cycle within infected cells (18–20). These modifications lead in particular to enhanced fatty acid synthesis, a pathway essential for HCMV replication since pharmacological inhibition of fatty acid synthesis inhibits viral propagation. Interestingly, the same inhibition was observed for influenza A virus. These observations address the important role of CCM modification during viral infection. However, underlying molecular mechanisms are still hypothetical and to date are poorly defined.
We wondered whether modifications of metabolite consumption or production could be altered in HCV-infected cells and, if so, which molecular mechanisms could explain these modifications. Here, we show that HCV infection of hepatoma cell lines reinforces aerobic glycolysis with an increase in glucose uptake and lactate production. We identified an interaction between the HCV NS5A protein and hexokinase 2 (HK2), which is the first and one of the three rate-limiting enzymes of glycolysis. This interaction results in modulation of enzymatic HK2 parameters, which may explain, at least partially, the enhancement of glycolysis observed during HCV cell infection.
MATERIALS AND METHODS
Materials.
Unless otherwise indicated, all chemicals were from Sigma-Aldrich (Saint-Quentin Fallavier, France), and cell culture reagents were from Life Technologies (Saint-Aubin, France). Purified NS5A full-length protein, NS5A domains AH-D1 (amino acids 1 to 213 containing the amino-terminal amphipathic α-helix [AH] and domain 1), D2 (amino acids 250 to 342), and D3 (amino acids 356 to 447), and core protein (amino acids 1 to 117) were produced with a wheat germ cell-free expression system (Cell-Free Science, Japan) and kindly provided by F. Penin's team (Institut de Biologie et Chimie des Protéines [IBCP], Lyon, France) (21–23). Empty pGluc1 and pGluc2 plasmids were kindly provided by Yves Jacob (Institut Pasteur, Paris, France) (24).
Cell culture.
Huh7.5, HepG2, or HEK293T cells were grown in Dulbecco's modified minimal essential medium (DMEM) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 10% fetal calf serum (FCS) in a 95% humidified incubator containing 5% CO2 in air at 37°C. For infection experiments, 24 h after seeding, the medium containing FCS was removed and replaced by serum-free medium in order to synchronize cells in G0, which reduces cell-to-cell variation in response to infection (19, 20). The Huh9.13 cell line, supporting replication of the HCV NS3-NS5B subgenomic replicon (genotype 1b, Con1 strain), was routinely cultured in the same medium as Huh7.5 cells, complemented with 1 mg/ml G418. An Huh9.13-cured cell line was obtained after a 1-month treatment of Huh9.13 cells with 500 U/ml alpha interferon 2a (IFN-α2a) (Intron-A; Schering-Plough).
HCV infection.
Jc1 virus stocks were generated as previously described (25). Huh7.5 cells were seeded in six-well plates and cultured in serum-free medium 24 h before infection at a multiplicity of infection (MOI) of 1 in a minimal volume. In parallel, control cells were mock infected under the same conditions. Four hours later, the infection medium was removed and replaced by complete serum-free medium. Cultures were then followed for 8, 24, 48, and 72 h and stopped. At each time point the cellular proteins were extracted, and cell supernatants were collected and immediately frozen at −80°C before further analysis.
Glucose, glutamine/glutamate, pyruvate, and lactate quantifications.
Metabolites were quantified from cell supernatants using a Glucose (GO) Assay Kit, a Glutamine and Glutamate Determination Kit (GLN1) (Sigma-Aldrich, Saint-Quentin Fallavier, France), and a pyruvate or lactate assay kit (BioVision, Nanterre, France). Assays were performed according to the manufacturer's instructions, and quantifications were normalized to cell protein contents determined by a Coomassie (Bradford) protein assay (Pierce Science, Brebières, France) using bovine serum albumin (BSA) as a standard protein.
Western blotting.
Monolayers of cells were washed twice in cold phosphate-buffered saline (PBS) before incubation for 15 min on ice in lysis buffer (1% Triton X-100 and 5 mM EDTA in PBS) containing 1% protease inhibitor cocktail (P8340; Sigma-Aldrich). Homogenates were scrapped and submitted to centrifugation at 12,000 rpm. Supernatants were diluted in Laemmli buffer and heated at 95°C for 5 min prior to fractionation by SDS-PAGE and transfer to a polyvinylidene difluoride (PVDF) membrane. After an overnight incubation at 4°C in PBS–0.1% Tween 20 supplemented with 5% (wt/vol) nonfat milk powder, blots were probed with monoclonal antibody (MAb) against HCV core protein (clone 7-50; ThermoScientific), diluted in PBS–0.1% Tween 20 (1:2,000), for 90 min and then by a peroxidase-conjugated goat anti-mouse antibody (Perbio Science) (1:5,000 dilution for 1 h in the same buffer). Blots were developed with enhanced chemiluminescence reagents according to the manufacturer's instructions (SuperSignal West Femto Maximum Sensitivity Substrate; Perbio Science).
PCA.
The methodology used for a protein complementation assay (PCA) has been recently described in detail (24) and is based on reconstitution of Gaussia luciferase activity from its two fragments fused, respectively, with viral and cellular potential interactors. Plasmids coding for the Con1 NS5A protein (pGluc1-NS5A) and plasmids coding for cellular partners (pGluc2-X) were obtained through Gateway technology (Invitrogen) (26). HEK293T cells were seeded at a density of 35,000 cells per well in 96-well white plates (Greiner Bio-One). Twenty-four hours later, cells were transfected with 100 ng of pGluc1-NS5A and 100 ng of pGluc2-X constructs and a plasmid coding for the firefly luciferase to normalize transfection efficiency. For each 96-well plate, the interaction between interferon regulatory factor-3 (IRF3) and the human papillomavirus 16 E6 oncoprotein (HPV16 E6) was assayed as a positive control (27). At 48 h posttransfection, 40 μl/well of luciferase assay lysis buffer (Promega) was added directly to the cells. After incubation under orbital agitation, firefly and Gaussia luciferase activities were measured on an Infinite M1000 PRO (Tecan) plate reader using luciferase and Gaussia assay reagents (Promega), respectively. Each PCA experiment was performed in triplicate. Results were normalized with firefly signal and are expressed as the normalized luminescence ratio (NLR), as previously described (24). The NLR for a given interacting protein pair A and B corresponds to luminescence activity in cells expressing pGluc1-A and pGluc2-B divided by the sum of the luminescence measured in cells transfected with pGluc1-A and pGluc2-empty and cells transfected with pGluc1-empty and pGluc2-X.
Reverse transcription-PCR (RT-PCR).
Total cellular mRNA was extracted using a NucleoSpin RNA kit (Macherey-Nagel). RT was carried out on 1 μg of RNA using a High Capacity RNA-to-cDNA kit (Life Technologies) according to the manufacturer's instructions. Then, 30 cycles of PCR were performed with 5 μl of RT product using GoTaq DNA polymerase (Promega, France) in the presence of 50 pmol of sense- and antisense-specific primers for HK1, HK2, HK3, HK4, or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and in accordance with the manufacturer's instructions.
Coimmunoprecipitation.
HEK293T cells were seeded in 10-cm dishes 24 h before transfection with plasmids indicated on the figure using jetPEI (Polyplus-Transfection, Illkirch, France). Anti-FLAG and anti-Myc precipitations were performed at 48 h posttransfection. Briefly, cells were harvested, washed once with PBS, and lysed in 2 ml of lysis buffer (20 mM Tris/HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM Na pyrophosphate, 1 mM Na orthovanadate, 10% glycerol, 1% Triton X-114, 1% anti-protease cocktail). Cells lysates were incubated for 10 min on ice and centrifuged at 14,000 × g at 4°C for 30 min. Supernatants were collected, mixed with 4 μg of either anti-FLAG (clone M2) or anti-Myc (clone 9E10) (28) antibody, and incubated for 4 h at 4°C. Then, 50 μl of Dynabeads-protein G (Invitrogen) was added. Following 1 h of incubation at 4°C, beads were washed three times with PBS. Bound proteins were eluted with Laemmli buffer, denatured for 5 min at 95°C, and subjected to immunoblotting detection.
Hexokinase activity assay.
The method used for extracting and assaying the HK activity from Huh7.5 cells was a modification of that described by Monakhov et al. (40) and Bergmeyer et al. (41). Thawed cells were homogenized with a Dounce homogenizer in precooled homogenization buffer (1 g/3 ml, wt/vol) containing 0.05 M Tris-HCl, 0.25 M sucrose, 0.005 M EDTA (pH 7.4), and 0.005 M 2-mercaptoethanol. Homogenates were centrifuged at 2,100 × g for 15 min at 4°C. Supernatants were centrifuged at 104,300 × g for 45 min at 4°C (TL100 ultracentrifuge with a TLA110 rotor; Beckman). Supernatants were then stored at −80°C before determination of HK activity. HK activity was measured spectrophotometrically through NADP+ reduction in a glucose-6-phosphate (Glc-6-P) dehydrogenase-coupled reaction. Incubation medium for HK activity measurements contained 100 μl of 0.05 M triethanolamine buffer (pH 7.6), 100 μl of 0 to 0.555 M d-glucose solution, 20 μl of 0.1 M MgCl, 20 μl of 0.014 M NADP+, and 2 μl of 125 U/ml glucose-6-phosphate dehydrogenase (Saccharomyces cerevisiae). Depending on the experiment, 20 μl of cell extract or 0.1 U of purified recombinant HK2 (Sigma-Aldrich), in the presence or absence of the amount of viral protein indicated on the figure, was mixed with the incubation medium and equilibrated to 37°C. The reaction was started with the addition of 10 μl of 0.019 M ATP, and absorbance was continuously recorded for 30 min at 340 nm (Infinite M200 plate reader; Tecan). Standard HK solution (Sigma-Aldrich) was used as a positive control, and results of triplicates were expressed as nmol of glucose consumed per min.
Statistical analysis.
Metabolite secretion and excretion were compared between infected and noninfected (control) cells for 72 h postinfection. Areas under the curves were calculated for each condition either by the linear method (trapezoids) or by natural spline interpolation and compared using the means for the two groups and a Fisher t test.
RESULTS
HCV infection enhances aerobic glycolysis.
Glucose, glutamine, and pyruvate are the three main carbon sources for cultured cells. Modifications of glycolysis enzymes suggest that HCV modulates the glycolytic pathway (15). We thus measured key metabolites within supernatants of Huh7.5 cells infected with HCV Jc1 (MOI of 1) at different times postinfection (i.e., 0, 8, 24, 48, and 72 h) Quantifications were normalized by cellular protein content in culture wells (Fig. 1). HCV infection was monitored by Western blot detection of core protein in cell homogenates. Increasing amounts of the viral protein between 24 h and 72 h postinfection indicated effective replication of the virus (Fig. 1A). Glucose, glutamine, and pyruvate concentrations decreased more rapidly in supernatants of HCV-infected cells than in control cells (Fig. 1B, C, and D). Interestingly, the increase in glutamine consumption occurred earlier following infection than that of glucose (24 h versus 48 h postinfection). Concomitant with these increased uptakes, lactate secretion in control cells was lower than in infected cells (Fig. 1E), whereas glutamate levels remained steady (Fig. 1F). These observations were confirmed in Huh9.13 cells that support replication of the subgenomic replicon (Con1 strain). Indeed, glucose consumption was increased in correlation with enhancement of lactate secretion in cells harboring the replicon in comparison to cells that were previously cured (Fig. 2). Thus, in two different HCV cell culture models with two different strains of the virus, we observed an increase in glucose consumption and lactate secretion, two features that suggest a higher flux of glucose through aerobic glycolysis.
FIG 1.
HCV infection modifies consumption and secretion of metabolites in Huh7.5 cells. Replicate cultures of Huh7.5 cells were serum starved for 24 h and subsequently mock infected (control) or HCV infected at an MOI of 1 (infected). Supernatants and cells were harvested at 0, 8, 24, 48, and 72 h postinfection and processed for metabolites and protein quantifications. (A) Protein extracts from cell homogenates at each time point were subjected to Western blotting and stained with anti-Core antibody (clone C7-50). Variations in glucose (B), glutamine (C), pyruvate (D), lactate (E), and glutamate (F) concentrations in cell culture medium were normalized with total protein amounts in the well and are expressed as nmol consumed or secreted (compared to time zero) per μg of protein (Δnmol/μg). Error bars show ± standard errors of the means (n = 3). *, P < 0.01, between infected and control cells (t test). MW, molecular weight (in thousands).
FIG 2.
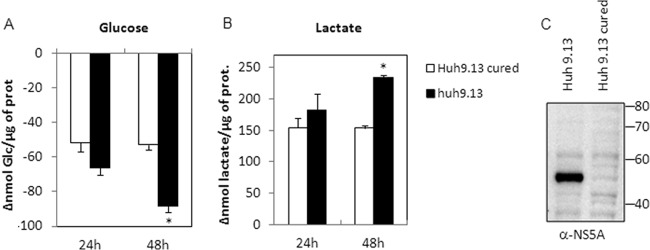
Metabolite consumption and secretion modifications in cells harboring subgenomic replicon. Replicate cultures of Huh9.13 or Huh9.13-cured cells were seeded and incubated for 24 h before the medium was changed (time zero). Supernatants and cells were harvested at 24 and 48 h postseeding, and metabolites and proteins were quantified as described in Materials and Methods. Variations in glucose (A) and lactate (B) concentrations in cell culture medium were normalized with total protein amounts in the well and are expressed as nmol consumed or secreted (compared to time zero) per μg of protein (Δnmol/μg). Error bars show ± standard errors of the means (n = 3). *, P < 0.01 (t test), for a comparison between Huh9.13 and cured Huh9.13 cells at 48 h. (C) Huh9.13 and Huh9.13-cured cells were harvested in homogenization buffer before protein denaturation and analyzed by Western blotting. NS5A protein was stained using a specific anti-NS5A (α-NS5A) polyclonal antibody.
HCV NS5A interacts with hexokinase 2.
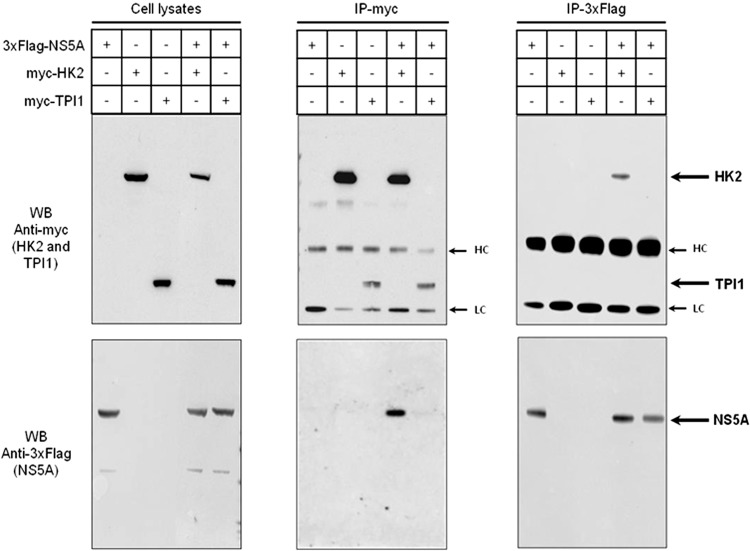
An increase in aerobic glycolysis may result from several factors. For example, in transformed cells, modification of HK2 activity through the relocalization of HK2 to the outer membrane of mitochondria contributes to the switch from oxidative phosphorylation to aerobic glycolysis (29, 30). Since the presence of the HCV Con1 subgenomic replicon was sufficient to enhance aerobic glycolysis in Huh9.13 cells, we investigated whether the viral proteins expressed by this replicon (i.e., NS3, NS4A-NS3, NS4A, NS4B, NS5A, and NS5B) were interactors of glycolytic enzymes. Each viral protein was coexpressed with each enzyme in HEK293T cells. Using PCA methodology, we determined for each pair of potential interactors the normalized luminescence ratio (NLR) that characterizes the interaction (24). Results of the PCA indicate that only NS5A potentially interacts with HK1 and HK2 (NLRs of 51.6 and 47.0, respectively) (Fig. 3) and, to a lesser extent, with GAPDH (NLR of 37.7). HK is the first of the rate-limiting enzymes of glycolysis and mediates phosphorylation of glucose to generate glucose-6-phosphate (Glc-6-P). Four isoforms of HK exist and are differentially expressed depending on cell types. Whereas HK1, HK2, and HK3 possess a high affinity for glucose, allowing a high activity rate, HK4 (i.e., glucokinase [GCK]) is less active under physiological concentrations of glucose. Because HK1 is not expressed in the hepatoma cell lines tested (Fig. 4), we wanted to confirm the interaction between HK2 and NS5A by coimmunoprecipitation following overexpression of both proteins in HEK293T cells. Triosephosphate isomerase 1 (TPI1), a glycolytic enzyme that did not interact with NS5A in a PCA assay, was used here as a negative control of interaction. Indeed, HK2 and NS5A were coimmunoprecipitated, whether the viral or the cellular protein was captured (Fig. 5). As expected, TPI1 was not precipitated with NS5A. Altogether, these results indicate an interaction between NS5A and HK2 proteins.
FIG 3.
Identification by PCA of protein interactions between viral and glycolytic enzymes. HEK293T cells were transfected with plasmids coding for viral and cellular protein expression as described in Materials and Methods. At 48 h posttransfection, luciferase activities were measured, and normalized luminescence ratios (NLRs) were calculated. Results for each pair of potential interactors were ranked from 0 to >30 and are presented in the figure according to the scale shown. In each reaction plate, the NLR for the two control proteins IRF3 and HPV16 E6 was >30. ORF, open reading frame.
FIG 4.

Expression of hexokinase isoforms in hepatoma cell lines. Total cellular mRNAs were extracted from 1 × 106 Huh7.5, HepG2, and Huh9.13 cells as described in Materials and Methods. Distinct pairs of primers were used to specifically amplify by PCR transcripts of HK1, HK2, HK3, and HK4 before separation on agarose gels. GAPDH transcripts were used for normalization.
FIG 5.
HCV NS5A protein interacts with HK2. Coimmunoprecipitations were performed from cell homogenates obtained 48 h posttransfection with the indicated expression plasmid for HK2, NS5A, or TPI1. Immunoprecipitations (IP) were realized using anti-3×Flag or anti-myc antibodies before analysis of captured complexes by Western blotting (WB). Expression controls of proteins in total cell lysates are presented in the left panel. Arrows indicate Ig heavy and light chains (HC and LC) or HK2, TPI1, or NS5A protein.
Apparent HK activity is enhanced by NS5A.
To determine the implications of the NS5A-HK2 interaction for the increase in glucose consumption observed in cells infected with HCVcc or in subgenomic replicon-positive cells (Fig. 1 and 2), Huh7.5 cells were transfected with an expression plasmid for NS5A or structural protein E2. Interestingly, overexpression of NS5A increased glucose consumption in cell culture whereas E2 had no effect (Fig. 6). Once again, lactate secretion mirrored glucose consumption. This suggested that NS5A expression within cells was sufficient to modulate glycolysis. To further evaluate the impact of NS5A on cellular HK activity, we measured Glc-6-P production in Huh7.5 cell homogenates in the presence or absence of purified NS5A produced in a wheat germ cell-free expression system. Glc-6-P was quantified using an in vitro enzymatic assay coupling HK activity to glucose-6-phosphate dehydrogenase activity and performing spectrophotometric detection of NADP+ reduction. Indeed, addition of purified full-length NS5A protein within cellular fractions during the assay increased the apparent observed HK activity by 2.5-fold, whereas addition of a control protein under the same conditions had no effect (Fig. 7A). Interestingly, purified D2 and D3 domains, but not D1, had the same effect in a dose-dependent manner (Fig. 7B). Therefore, addition of purified NS5A protein into the reaction mix was sufficient to increase HK activity, strengthening the idea that NS5A interaction with HK2 modulates its activity.
FIG 6.
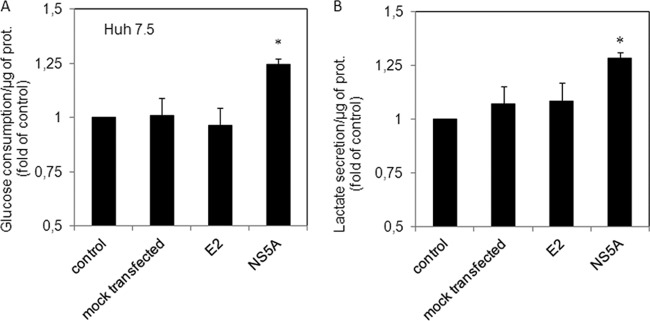
NS5A expression modifies glucose consumption and lactate secretion in Huh7.5 cells. Replicate cultures of Huh7.5 cells were seeded at 24 h before transfection with expression plasmids for HCV E2 and NS5A proteins. Supernatant and cells were harvested at 24 and 48 h posttransfection, and glucose, lactate, and proteins were quantified as described in Materials and Methods. Average changes in the glucose consumption (A) or lactate secretion (B) are presented as fold change relative to values of control cells. Error bars show ± standard errors of the means (n = 3). *, P < 0.05 (t test), for a comparison between control cells and NS5A-transfected cells.
FIG 7.
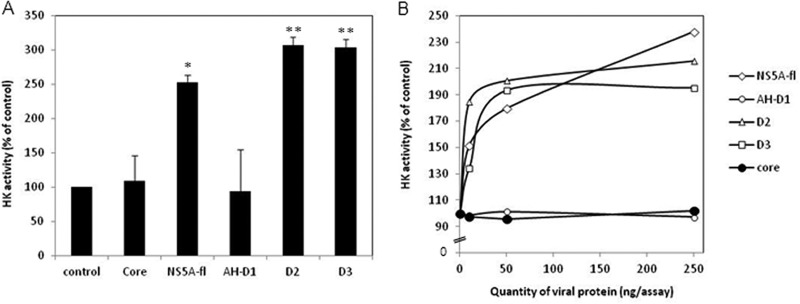
Full-length NS5A and its domains D2 and D3 enhance HK activity. (A) Hexokinase activity was measured in cytosolic fractions of Huh7.5 cells as described in Materials and Methods in the absence (control) or presence of 250 ng/assay of core protein (amino acids 1 to 117), full-length NS5A, or its domains AH-D1 (amino acids 1 to 213), D2 (amino acids 250 to 342), and D3 (amino acids 356 to 447). Averages of replicates are expressed as percentages of control values, and error bars show ± standard errors of the means (n = 3). (B) Hexokinase activity was measured as described for panel A in the presence of increasing concentrations of viral proteins (0, 10, 50, or 250 ng/assay). Results are presented as percentages of control values. *, P < 0.05; **, P < 0.01 (relative to the controls; t test).
NS5A is an activator of hexokinase 2.
To further understand how the NS5A interaction with HK2 could impact HK activity, we determined whether the viral protein had an influence on maximum velocity (Vmax) and Michaelis constant (Km) values, two catalytic parameters of the enzyme. For this, we used purified recombinant human HK2, produced in Escherichia coli, in the presence or absence of purified NS5A. Under these acellular conditions, we measured the HK2 velocity at increasing substrate concentrations. These measures allowed establishment of Michaelis-Menten saturation curves (Fig. 8A), which revealed an increase in Vmax in the presence of NS5A. The core protein, added under the same conditions, had no effect. Vmax and Km were calculated using the Lineweaver-Burk method and representation (Fig. 8B). The Km of HK2 decreased by 27%, which shows that HK2 affinity for glucose was increased in the presence of the viral protein. Moreover, Vmax increased by about 48%. Overall, these results indicate that interaction of NS5A with HK2 modifies the catalytic parameters of the enzyme and thus increases its activity.
FIG 8.
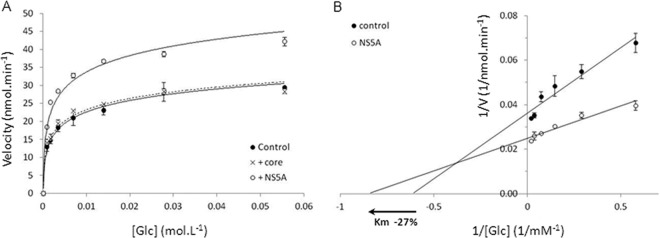
HCV NS5A protein modifies HK2 catalytic parameters. (A) Initial velocity of hexokinase activity was measured in vitro using purified recombinant human HK2 under different concentrations of substrate (i.e., glucose) in the presence (+NS5A) or absence (control) of purified full-length NS5A. Core protein was added under the same conditions as a control. (B) Double reversible plot of velocity versus glucose concentration (Lineweaver-Burk representation) in the presence (NS5A) or absence (control) of NS5A. Experiments were repeated twice, and means ± standard errors of triplicates are represented. Intersections of regression lanes with the y axis indicate 1/Vmax, and intersections with the x axis indicate −1/Km of the enzyme. Graphic determinations indicate an increase in Vmax of 45% and a decrease in Km of 27% in the presence of NS5A.
DISCUSSION
In the present study, we described an interaction between HCV-NS5A and hexokinase, which modifies the catalytic parameters of this enzyme. This interaction and its consequences on HK2 activity correlate with enhanced glucose consumption and higher lactate production in infected cells and in subgenomic replicon-positive or NS5A-overexpressing cells.
At the steady state, in the presence of oxygen, nonproliferating cells metabolize glucose via glycolysis to produce ATP through oxidative phosphorylation within mitochondria. In proliferating cells glucose is metabolized in a distinct manner from that in normal cells with production of ATP from aerobic glycolysis, a phenomenon referred as the “Warburg effect” (for a review, see reference 31). Indeed, under nonproliferating conditions, cells require energy (i.e., ATP or GTP) to ensure specific cell functions, but their needs for cellular building components are limited. During mitosis, proliferating cells must replicate all of their cellular components, thus increasing their need for biosynthesis of elementary molecules (i.e., nucleotides, amino acids, and lipids). The cellular events leading to such a metabolic shift have been intensively studied and are indeed very complex. A major trait of this metabolic modification is an increase in the glucose uptake by the cell and in the aerobic glycolysis rate.
It is tempting to speculate that viral infection may fulfill biosynthesis in a similar manner as proliferative dividing cells do to ensure the intracellular resources necessary for viral replication. The first rate-limiting enzyme of glycolysis is HK, which thus appears as an interesting target for viruses to modulate this cellular pathway. Indeed, HCV-NS5A interacts with HK2 and directly modifies the catalytic parameters of the enzyme (i.e., Vmax increase and Km reduction) at physiological glucose concentrations. The NS5A-induced increase in HK2 activity and avidity for glucose may contribute to enhanced glucose consumption and higher lactate production, thus inducing a Warburg-like phenomenon, as previously described for HCMV (18, 19). We thus confirmed in another viral context the previous observation of the viral impact on CCM and suggest a mechanism that could explain, at least partly, the effect on glycolysis. Of note, whereas studies of HCMV were performed on primary fibroblasts, our observations with HCV were performed on cell lines derived from carcinoma cells that have already shifted their metabolism to aerobic glycolysis. Nevertheless, the presence of NS5A further enhances this phenomenon.
HK4 (or GCK) is the predominant isozyme expressed in normal hepatocytes, whereas HK2 is the predominant form expressed in hepatoma cell lines. Whereas HK1, HK2, and HK3 possess a high affinity for glucose, GCK is less active under physiological concentrations of glucose. Moreover, GCK is not inhibited by its product (Glc-6-P); thus, the rate of reaction is driven by the supply of glucose, not by the demand for end products (32) and, indeed, GCK is effective only when glucose is abundant. In transformed cells, HK2 is overexpressed and associated to the outer mitochondrial membrane. This association confers high enzymatic activity since it helps couple the phosphorylation of glucose to ATP production by mitochondria and is accompanied by loss of product inhibition of the enzyme (33). In normal cells the bulk of HK is not associated to mitochondria, and HK2 is poorly expressed. The NS5A interaction with HK2 might be an efficient strategy for HCV to enhance glucose flux through glycolysis by further activating the most active HK isoform at the mitochondrion surface.
Recently, the question of how HCV could impair cell metabolism was addressed by Diamond et al., who identified several modifications of the cellular proteome, especially of the CCM enzymes in infected cells (15). Indeed, six of the glycolysis pathway enzymes were overexpressed; nevertheless, no observation was reported concerning the modulation of HK expression following HCV infection. The present observation of the catalytic modulation of HK by NS5A extends the work of Diamond et al. The increase in HK activity, the first rate-limiting enzyme of glycolysis, will promote the increase of glucose flux due to the upregulation of downstream glycolytic enzymes. Boosting HK activity may be an efficient strategy for HCV to allow the generation of intermediate metabolites for aerobic glycolysis and biosynthesis and to finely tune the balance between oxidative phosphorylation and aerobic glycolysis.
The importance of the glycolytic rate-limiting enzymes for viral replication was also revealed for human immunodeficiency virus (HIV) through several large-scale small interfering RNA (siRNA) screens. Indeed, extinction of HK4 expression in HeLa cells transfected with the IIIB strain of HIV-1 decreases viral translation by 48% and thus identifies the first rate-limiting enzyme of glycolysis as an HIV dependency factor (34). Transfection of 293T cells with siRNA targeting phosphofructokinase, the second rate-limiting enzyme of glycolysis, inhibits the first cycle of infection by vesicular stomatitis virus G protein (VSV-G)-pseudotyped HIV-1 vectors encoding luciferase (35). Finally, down-modulated expression of pyruvate kinase, the third and last rate-limiting enzyme of glycolysis, reduces replication of HIV-1 HXB2 in HeLa P4/R5 cells (36). Altogether, these observations indicate that perturbations of the glycolysis pathway impair HIV replication. The direct modulation of hexokinase activity by HCV suggests that viruses not only depend on the integrity of glycolysis for their replication but also may actively modify this pathway.
In addition to the metabolic effects that could result from the direct interaction of HK2 with HCV-NS5A, targeting of this enzyme by HCV may help the virus escape innate antiviral defenses. Indeed, recent reports suggest a link between glycolysis and innate immunity. Glucokinase regulatory protein (GCKR), which inhibits HK activity, has been shown to be an IFN-α effector gene required to suppress HCV replication in Huh7.5 cells (37). Indeed, inhibition of GCKR expression by siRNA rescues HCV replication from IFN-α-mediated suppression. Recently, microRNA 143 (miR-143) was identified as a regulator of cancer-associated aerobic glycolysis by targeting HK2 (38). In human lung cancer samples, miR-143 expression was inversely associated with the HK2 protein level, and miR-143 overexpression suppressed HK2 expression in CRL-5807 and CRL-5803 bronchoalveolar carcinoma cells. This microRNA was also identified as an IFN-α-induced gene and a potent inhibitor of HCVcc replication (39). HK2 activity enhancement by HCV-NS5A protein may thus be crucial for HCV replication to counteract the inhibition of glycolysis by IFN responses and therefore to ensure the biomolecular supply needed for viral replication.
ACKNOWLEDGMENTS
We are grateful to Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for providing the Jc1 construct and Huh9.13 cells, to Yves Jacob (Institut Pasteur, Paris, France) for providing pGluc1 and pGluc2 expression plasmids, to Charles M. Rice (Rockefeller University, New York, NY, USA) for providing Huh7.5 cells, and to Aurelie Badillo, Marie-Laure Fogeron, and François Penin (IBCP, Lyon, France) for providing purified viral proteins. We acknowledge the contribution of the AniRA level 3 security laboratory of SFR Biosciences Gerland-Lyon Sud (AMS3444/US8).
This research was supported by a grant from the Agence Nationale de Recherche sur le Sida et les Hépatites, by the Institut National pour la Santé et la Recherche Médicale, and by the European Union's 7th program (FP7/2007-2013) under grant agreement number 267429 (SysPatho).
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Scheel TKH, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 19:837–849. 10.1038/nm.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. 10.1371/journal.ppat.0030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566. 10.1073/pnas.0409834102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Popescu C-I, Dubuisson J. 2010. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol. Cell 102:63–74. 10.1042/BC20090125 [DOI] [PubMed] [Google Scholar]
- 5.Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramière C, Bartenschlager R, Penin F, Lotteau V, André P. 2009. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS One 4:e4233. 10.1371/journal.pone.0004233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 83:12680–12691. 10.1128/JVI.01476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103. 10.1016/j.tim.2010.11.005 [DOI] [PubMed] [Google Scholar]
- 8.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland J, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low-and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919–6928. 10.1128/JVI.76.14.6919-6928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz O, Delers F, Maynard M, Demignot S, Zoulim F, Chambaz J, Trépo C, Lotteau V, André P. 2006. Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins. J. Gen. Virol. 87:2983–2991. 10.1099/vir.0.82033-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz O, Cubero M, Trabaud MA, Quer J, Icard V, Esteban JI, Lotteau V, André P. 2008. Transmission of low-density hepatitis C viral particles during sexually transmitted acute resolving infection. J. Med. Virol. 80:242–246. 10.1002/jmv.21037 [DOI] [PubMed] [Google Scholar]
- 11.Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805–3809. 10.1073/pnas.0511218103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steenbergen RHG, Joyce MA, Thomas BS, Jones D, Law J, Russell R, Houghton M, Tyrrell DL. 2013. Human serum leads to differentiation of human hepatoma cells, restoration of VLDL secretion and a 1000-fold increase in HCV JFH-1 titers. Hepatology 58:1907–1917. 10.1002/hep.26566 [DOI] [PubMed] [Google Scholar]
- 13.Patel JH, Cobbold JFL, Thomas HC, Taylor-Robinson SD. 2010. Hepatitis C and hepatic steatosis. QJM 103:293–303. 10.1093/qjmed/hcp192 [DOI] [PubMed] [Google Scholar]
- 14.Roingeard P. 2013. Hepatitis C virus diversity and hepatic steatosis. J. Viral Hepat. 20:77–84. 10.1111/jvh.12035 [DOI] [PubMed] [Google Scholar]
- 15.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters K-A, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG, II, Waters KM, Smith RD, Rice CM, Katze MG. 2010. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 6:e1000719. 10.1371/journal.ppat.1000719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy HB, Baron S. 1957. The effect of animal viruses on host cell metabolism. II. Effect of poliomyelitis virus on glycolysis and uptake of glycine by monkey kidney tissue cultures. J. Infect. Dis. 100:109–118 [DOI] [PubMed] [Google Scholar]
- 17.Henle G, Deinhardt F, Bergs VV, Henle W. 1958. Studies on persistent infections of tissue cultures. I. General aspects of the system. J. Exp. Med. 108:537–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. 2011. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 7:e1002124. 10.1371/journal.ppat.1002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2:e132. 10.1371/journal.ppat.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munger J, Bennett BD, Parikh A, Feng X-J, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26:1179–1186. 10.1038/nbt.1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanoulle X, Badillo A, Wieruszeski J-M, Verdegem D, Landrieu I, Bartenschlager R, Penin F, Lippens G. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589–13601. 10.1074/jbc.M809244200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verdegem D, Badillo A, Wieruszeski J-M, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. 2011. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic α-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 286:20441–20454. 10.1074/jbc.M110.182436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne J-P. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353–11365. 10.1128/JVI.79.17.11353-11365.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, Jones L, Muller M, Demeret C, Gaud G, others 2011. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods 8:990–992. 10.1038/nmeth.1773 [DOI] [PubMed] [Google Scholar]
- 25.Merz A, Long G, Hiet M-S, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 286:3018–3032. 10.1074/jbc.M110.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, André P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4:230. 10.1038/msb.2008.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronco LV, Karpova AY, Vidal M, Howley PM. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061–2072. 10.1101/gad.12.13.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evan GI, Lewis GK, Ramsay G, Bishop JM. 1985. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol. 5:3610–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neary CL, Pastorino JG. 2013. Akt inhibition promotes hexokinase 2 redistribution and glucose uptake in cancer cells. J. Cell. Physiol. 228:1943–1948. 10.1002/jcp.24361 [DOI] [PubMed] [Google Scholar]
- 30.Saks V. 2007. Molecular system bioenergetics: energy for life. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 31.Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robey RB, Hay N. 2006. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 25:4683–4696. 10.1038/sj.onc.1209595 [DOI] [PubMed] [Google Scholar]
- 33.Mathupala SP, Ko YH, Pedersen PL. 2009. Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 19:17–24. 10.1016/j.semcancer.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- 35.König R, Zhou Y, Elleder D, Diamond TL, Bonamy GMC, Irelan JT, Chiang C-Y, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 37.Fusco DN, Brisac C, John SP, Huang Y-W, Chin CR, Xie T, Zhao H, Jilg N, Zhang L, Chevaliez S, Wambua D, Lin W, Peng L, Chung RT, Brass AL. 2013. A genetic screen identifies interferon-α effector genes required to suppress hepatitis C virus replication. Gastroenterology 144:1438–1449.e9. 10.1053/j.gastro.2013.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, Feng Y, Li L, Wang Y, Liu X, Chen H, Liu X-Y, Ji H. 2012. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 287:23227–23235. 10.1074/jbc.M112.373084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, Jin Q, Yang W. 2013. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J. Virol. 87:9707–9718. 10.1128/JVI.00802-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monakhov NK, Neistadt EL, Shavlovskil MM, Shvartsman AL, Neifakh SA. 1978. Physicochemical properties and isoenzyme composition of hexokinase from normal and malignant human tissues. J. Natl. Cancer Inst. 61:7–34 [DOI] [PubMed] [Google Scholar]
- 41.Bergmeyer HU, Grassl M, Walter HE. 1983. Methods of enzymatic analysis, 3rd ed, vol 2, p 222–223 Verlag Chemie, Deerfield Beach, FL [Google Scholar]