ABSTRACT
Elite controllers or suppressors (ES) are HIV-1-infected patients who maintain undetectable viral loads without antiretroviral therapy. The mechanism of control remains unclear, but the HLA-B*57 allele is overrepresented in cohorts of these patients. However, many HLA-B*57 patients develop progressive disease, and some studies have suggested that infection with defective viruses may be the cause of the lack of high levels of virus replication and disease progression in ES. We therefore performed a comprehensive comparative in vivo and in vitro characterization of viruses isolated from well-defined ES. For this purpose, we first performed full-genome sequence analysis and in vitro fitness assays on replication-competent isolates from HLA-B*57 ES and HLA-B*57 chronic progressors (CPs). Under our experimental conditions, we found that isolates from ES and CPs can replicate in vitro. However, since inherently these assays involve the use of unnaturally in vitro-activated cells, we also investigated the replication competence and pathogenic potential of these HIV isolates in vivo using humanized BLT mice. The results from these analyses demonstrate that virus isolates from ES are fully replication competent in vivo and can induce peripheral and systemic CD4 T cell depletion. These results provide the first direct in vivo evidence that viral fitness does not likely determine clinical outcome in HLA-B*57 patients and that elite suppressors can control replication-competent, fully pathogenic viruses. A better understanding of the immunological bases of viral suppression in ES will serve to inform novel approaches to preventive and therapeutic HIV vaccine design.
IMPORTANCE Elite suppressors are HIV-1-infected patients who have undetectable levels of viremia despite not being on antiviral drugs. One of the most fundamental questions about this phenomenon involves the mechanism of control. To address this question, we isolated virus from elite suppressors and from HIV-1-infected patients who have the usual progressive disease course. We compared how well the isolates from the two groups of patients replicated in culture and in humanized mice. Our results suggest that elite suppressors are capable of controlling HIV-1 due to the possession of unique host factors rather than infection with defective virus.
INTRODUCTION
Understanding the mechanisms involved in the natural control of human immunodeficiency virus type 1 (HIV-1) replication may lead to the design of an effective HIV-1 vaccine. Patients known as elite controllers or suppressors (ES), who maintain viral loads below the limit of detection of clinical assays without antiretroviral therapy (ART), represent fewer than 1% of all HIV-1 infected patients (1–3). Previous reports have suggested that some ES and long-term nonprogressors, who maintain stable CD4 counts for prolonged periods, are infected with attenuated or defective virus (4–12). However, in other studies, replication-competent HIV-1 isolates were cultured from some ES (13–15), and full-genome sequence analysis of these replication-competent isolates did not reveal any large deletions or signature mutations (13). It has been very challenging to isolate virus from ES, and full-length genotypic analyses have been performed on replication-competent isolates obtained from fewer than 10 ES (11, 13, 16–18) and just 3 HLA-B*57-positive ES (13, 16, 18). Furthermore, studies comparing the growth kinetics of replication-competent virus from ES to those of multiple isolates from patients with progressive disease have not been performed. Host factors clearly contribute to elite suppression of viral replication. The HLA-B*57 allele is overrepresented in cohorts of ES (19–24) and has been associated with HIV control in large genome-wide association studies (25, 26). HIV-1 epitopes that are presented by HLA-B*57 proteins are conserved and immunodominant, and robust HIV-1-specific T cell responses have been documented in HLA-B*57 ES (19, 20, 27–30). However, many HLA-B*57 patients are viremic and develop progressive disease (19). To determine the in vivo contribution of viral fitness to the clinical outcome in HLA-B*57 patients, we (i) isolated replication-competent virus from CD4+ T cells from 18 HLA-B*57 patients, (ii) performed full-genome sequence analysis, (iii) demonstrated their in vitro replication competence, and (iv) evaluated their ability to establish a de novo infection, deplete CD4+ T cells, and establish latency in vivo using BLT humanized mice. Our results strongly suggest that some ES are indeed capable of controlling replication of fully pathogenic HIV-1 isolates.
MATERIALS AND METHODS
Patient population.
We studied 24 HIV-1-seropositive individuals. Eleven were elite suppressors (ES), who maintained viral loads of <50 copies/ml without antiretroviral therapy; 3 were viremic controllers (VCs), who had viral loads of greater than 50 copies/ml but less than 2,000 copies/ml; and 10 were chronic progressors (CPs), who had pre-ART viral loads of >2,000 copies/ml. Replication-competent virus was obtained from 5 of the 11 ES. Table 1 lists the clinical characteristics of the patients used for the study. The protocol was approved by the Institutional Review Board of Johns Hopkins University School of Medicine. Informed consent was obtained before phlebotomy. The study was approved by the Johns Hopkins University Institutional Review Board. All study subjects were older than 21 years of age, and informed written consent was obtained from all subjects prior to enrollment into the study.
TABLE 1.
Clinical dataa
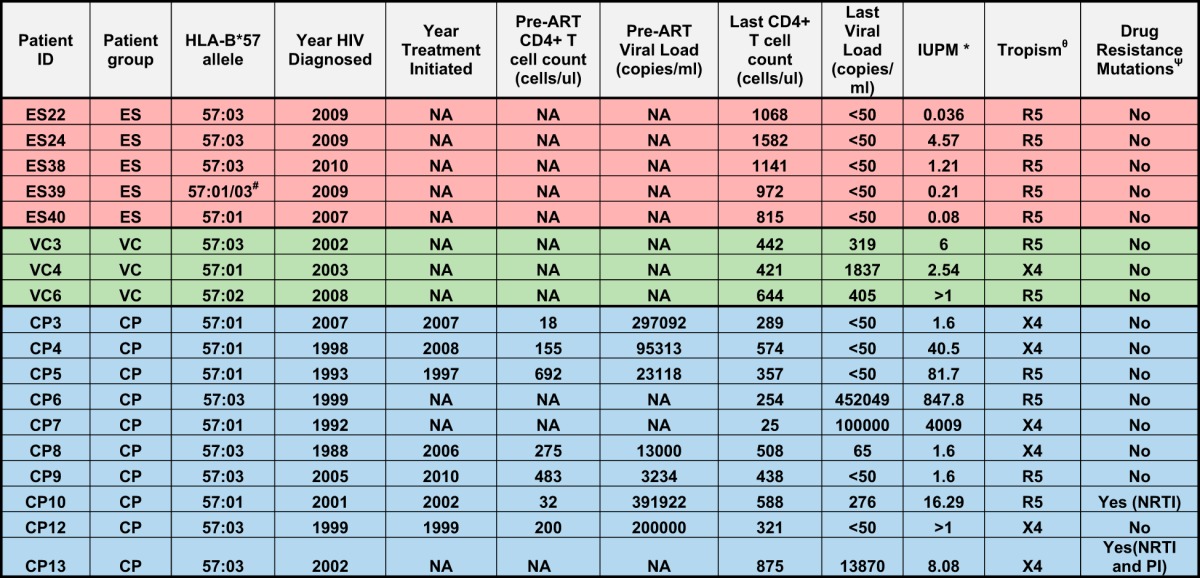
#, could not distinguish between HLA-B*57:01 and HLA-B*57:02; *, infectious units per million (IUPM) was determined by limiting-dilution culture analysis using purified CD4+ T cells; θ, tropism of isolates as determined by culture analysis in MT-2 cells; Ψ, drug resistance mutations were predicted using the Geno2Pheno database.
Virus isolation and sequence analysis.
Isolation of replication-competent virus from bulk CD4+ T cells was performed as previously described (13). Briefly, total CD4+ T cells were isolated by negative selection using the Miltenyi CD4+ T cell isolation kit II. The CD4+ T cells were cultured with irradiated donor cells and phytohemagglutinin (PHA) at a final concentration of 0.5 μg/ml in the presence of interleukin-2 (IL-2) and T cell growth factors. After 2 days, PHA was removed and PHA-activated donor blasts were added to the culture. HIV p24 Gag antigen was measured at days 14 and 21 (Perkin-Elmer). Replication-competent isolates were obtained from 10 CPs, 3 VCs, and 5 ES (Table 1). We were not able to isolate replication-competent virus from 6 other ES. Isolates from one ES (ES 38) have been previously described (18). RNA was isolated from positive-well supernatants. Full-genome sequence analysis of viral isolates was performed as previously described (13). nef clones from the replication-competent virus were compared to proviral nef clones amplified by PCR. Phylogenetic analysis suggested that the replication-competent isolates were representative of the virus archived in the latent reservoir of these patients (data not shown). Resistance mutation predictions were performed using Geno2Pheno database (http://www.geno2pheno.org/).
Sequence analysis of virus amplified from humanized mice.
Mouse spleen and thymus samples were collected, and proviral DNA was extracted using the QIAamp DNA Blood Minikit (Qiagen). Nested PCR was then performed to amplify proviral Gag and Nef, and sequence analysis was performed as previously described (13).
Generation of humanized BLT mice.
BLT mice were generated as previously described (31, 32). Briefly, 6- to 8-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory) were irradiated with 200 cGy and implanted with fetal thymus and liver tissue underneath the kidney capsule. CD34+ cells isolated from autologous fetal liver were used to transplant between 2 × 105 and 3 × 105 cells per mouse. Reconstitution of BLT mice with human immune cells was monitored in peripheral blood by flow cytometry every 3 to 4 weeks as previously described (31). Mice were maintained by the Division of Laboratory Animal Medicine under specific-pathogen-free conditions at the University of North Carolina at Chapel Hill in accordance with protocols approved by the Institutional Animal Care and Use Committee.
Flow cytometry analysis.
Mononuclear cells (MNCs) from BLT mice were isolated from the bone marrow, spleen, lymph nodes, lung, liver, and thymic organoid tissues as previously described (32). Live cells were identified based on their characteristic side scatter versus forward scatter. Subsequently, live human MNCs were identified with mouse anti-human CD45+ (clone HI30; BD Pharmingen) to determine the percentage of human reconstitution. Lymphocytes were gated through human CD45+ cells and CD3 (clone HIT3a; BD Pharmingen) cells for T cell subsets. T cells (CD45+ CD3+ gate) were further analyzed for CD4 (RPA-T4; BD Pharmingen) and CD8 (clone SK1; BD) subsets. For the identification of resting CD4+ T cells, analysis was performed as previously described (32). All flow cytometry data were collected and analyzed using BD FACSDiva software.
Exposure of BLT mice to HIV.
Humanized BLT mice were administered 200 μl of HIV at a concentration of approximately 325 ng/ml p24. All mice were exposed via tail vein injection. Mice were bled to determine the presence of viral RNA in the plasma beginning at 1 week postexposure.
Analysis of HIV infection.
RNA from the plasma was isolated using the RNeasy Minikit (Qiagen). Levels of viral RNA were quantified with a one-step real-time reverse transcriptase PCR (RT-PCR) assay using the following primers and probe: 5′-CATGTTTTCAGCATTATCAGAAGGA-3′, 5′-TGCTTGATGTCCCCCCACT-3′, and 5′-6-carboxyfluorescein (FAM)-CCACCCCACAAGATTTAAACACCATGCTAA-Q (nonfluorescent quencher)-3′ (Applied Biosystems).
Establishment and assessment of HIV latency in BLT mice.
HIV exposure was performed as described above. At 2 weeks postexposure, antiviral therapy consisting of tenofovir disoproxil fumarate, emtricitabine, and raltegravir was administered intraperitoneally (i.p.) as previously described (32). On day 20 of therapy, the dose of tenofovir disoproxil fumarate was lowered to 102 mg/kg body weight and maraviroc was added at 61.5 mg/kg body weight. After a period of suppression of viral replication was achieved, lymph nodes, spleen, liver, lung, and bone marrow were harvested from the animal, and cells were isolated from these tissues and from peripheral blood as described above. The cells from all tissues were combined, and the resting human CD4+ T cells were isolated via negative selection (Stemcell Technologies, Vancouver, Canada) (32). The enriched resting cells were cultured in the presence of 15 nM efavirenz and 1 μM raltegravir for 1 day as to limit the potential contribution of nonintegrated HIV DNA to the outgrowth assay results. The resting cells were then stimulated and cocultured with feeder cells as previously described (33). A maximum-likelihood method was used to calculate the frequency of resting cell infection (33). The results are expressed as infectious units per million resting CD4+ T cells (IUPM).
Viral tropism assay.
Positive supernatants from each patient were used to infect MT-2 cells (obtained from the NIH AIDS Research and Reference Program) as previously described (34). Tropism was determined by the degree of replication in these cells as determined by the p24 assay (Perkin-Elmer).
Viral fitness assay.
Viral fitness was analyzed as described previously (13). Peripheral blood mononuclear cells (PBMCs) from a healthy donor were activated for 2 days with IL-2 and PHA. CD4+ T cells were isolated (by magnetically activated cell sorting [MACS] with a CD4+ T cell isolation kit II) and infected by spinoculation (1,200 × g for 2 h) with equal quantities (200 ng/ml) of p24 from primary patient isolates or with Ba-L or IIIB laboratory HIV-1 strains as controls. Supernatant samples were taken over the course of 7 days. Viral replication was quantified using p24 enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer). The median p24 concentrations for each point were determined for each group of patients and were compared using the Mann-Whitney nonparametric test.
CD4 and HLA downregulation.
CD4+ T cells from healthy HLA-A2+ donors were obtained and infected as described above. On days 3, 5, and 7, the cells were stained with allophycocyanin (APC)-Cy7-conjugated anti-CD4 antibody, APC-conjugated anti-CD3 antibody, and phycoerythrin (PE)-conjugated HLA-A2 antibody (Becton Dickinson) and then fixed and permeabilized with Cytofix/Cytoperm solution (Becton Dickinson). Intracellular staining for Gag was then performed with phycoerythrin-conjugated KC57 antibody (Beckman Coulter) as previously described (35, 36). The HLA-A2 downregulation ratio was defined as the mean fluorescence intensity (MFI) of HLA-A2 on cells that were positive for intracellular Gag divided by the MFI of HLA-A2 on CD4+ T cells that were Gag negative. The CD4 downregulation ratio represents the fraction of all CD4+ T cells that were Gag positive and CD4 low. The Mann-Whitney nonparametric test was used to compare CD4 and HLA downregulation for each group of patients.
HLA typing.
Genomic DNA was isolated from peripheral blood mononuclear cells using the QIAamp DNA Blood Minikit (Qiagen). The HLA-B locus was amplified, followed by bidirectional sequencing of exons 2, 3, and 4 with AlleleSEQR HLA-B (Abbott). Sequences were then obtained on 3130 XL (Applied Biosystems) and assembled with Assign software (Conexio Genomics, Australia).
For HLA typing of the human tissue used to reconstitute the humanized BLT mice, the thymus/liver organoid was harvested at necropsy and cells were extracted. DNA was purified using the QIAamp DNA Blood Minikit (Qiagen).
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank (accession numbers KF384798 to KF384908).
RESULTS
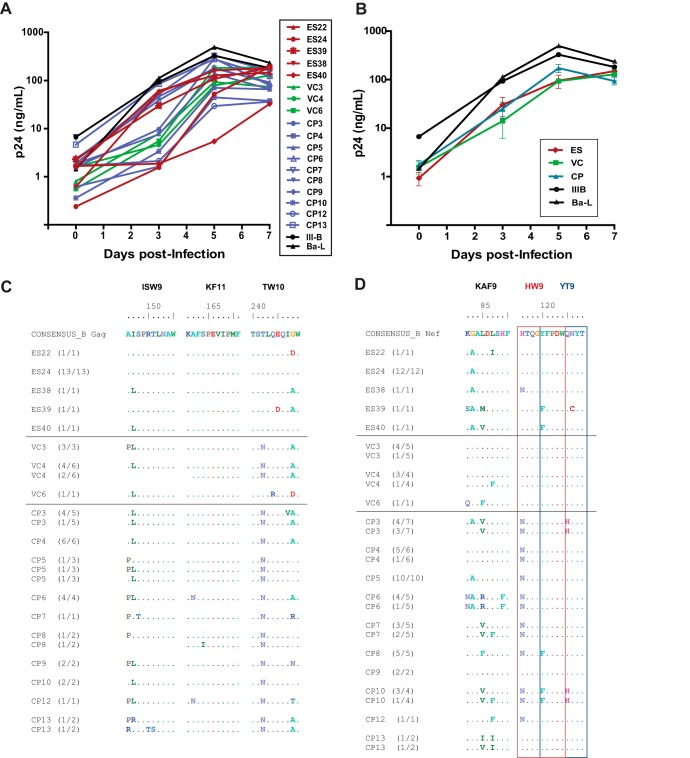
We studied virus isolated from 18 HLA-B*57 patients; 5 of these were ES, 10 were chronic progressors (CPs), and 3 were patients known as viremic controllers (VCs), who maintained plasma virus levels of between 50 and 2,000 copies of HIV-1 RNA/ml (Table 1). As shown in Fig. 1, all the isolates from the 3 patient groups replicated vigorously in vitro in IL-2/PHA-activated CD8-depleted CD4+ T cells, and the median growth curves from the 3 patient groups were not significantly different. Full HIV-1 genome sequencing performed for all 18 isolates revealed no large deletions in any of the genes, and drug resistance mutations, which could potentially affect viral fitness, were present in only 2 CPs (Table 1). However, viral isolates from CPs were more likely to be CXCR4-tropic than virus isolated from ES, consistent with higher levels of ongoing viral replication in CPs (Table 1).
FIG 1.
(A) Growth kinetics of 18 HIV-1 isolates cultured from CD4+ T cells from HLA-B*57+ patients. The patients were ES (red), VCs (green), or CPs (blue). Two laboratory strains (black) were included for comparison. The isolates were all cultured in activated CD4+ T cells from the same HIV-1 negative donor. (B) Median growth curve for each group of patients. (C and D) Sequence variation within the three HLA-B*57-03-restricted Gag (C) and Nef (D) epitopes for each isolate. The red and blue boxes denote two distinct but overlapping Nef epitopes, HW9 (116 to 124) and YT9 (120 to 128), respectively. Comparisons of the degree of sequence variation were made using the Mann-Whitney test.
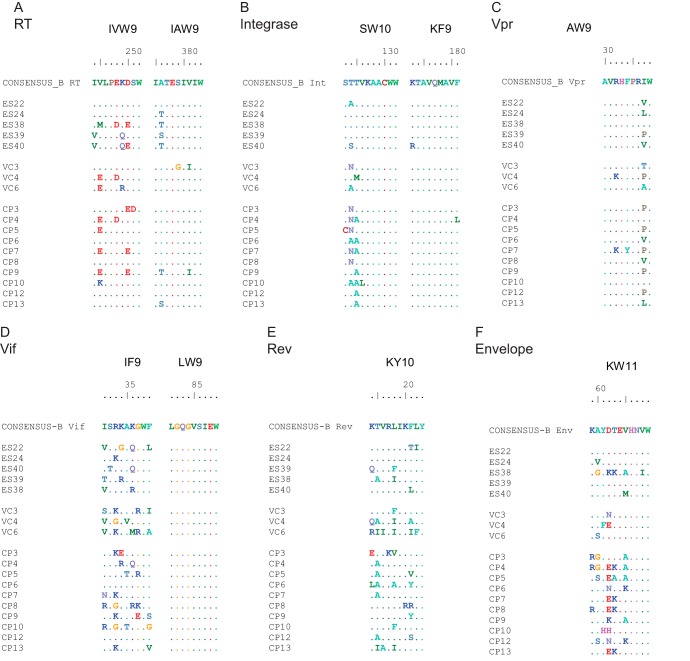
Escape mutations in HLA-B*57-restricted Gag epitopes have been well characterized and have been shown to have a fitness cost in vivo (37–39) and in vitro (40–42). We compared the frequency of mutations in HLA-B*57 epitopes in the different groups of patients and found that isolates from CPs and VCs were more likely to contain escape mutations in Gag, Nef, and integrase than isolates from ES (Fig. 1 and 2; see Table S1 in the supplemental material). In contrast, there was no significant difference in the frequency of escape mutants in HLA-B*57-restricted epitopes in other viral genes (Fig. 2). Multiple studies have highlighted the importance of Nef in viral pathogenesis in vivo (43, 44). Nef plays a key role in CD4 (45) and HLA-A and HLA-B molecule (46) downregulation. Sequence analysis demonstrated that all isolates had intact nef genes. We thus assessed these parameters in all 18 isolates. As shown in Fig. 3, there was no significant difference in the downregulation of CD4 or HLA-A2 by isolates from the different patient groups. Taken together, our findings suggested that isolates from HLA-B*57 ES are fully replication competent, with functional nef genes and fewer cytotoxic T lymphocyte (CTL) escape mutations in Gag and Nef than seen in CPs.
FIG 2.
Variations within HLA-B*57-restricted epitopes. Sequence analysis of epitopes in reverse transcriptase (A), integrase (B), Vpr (C), Vif (D), Rev (E), and Env (F) is shown.
FIG 3.
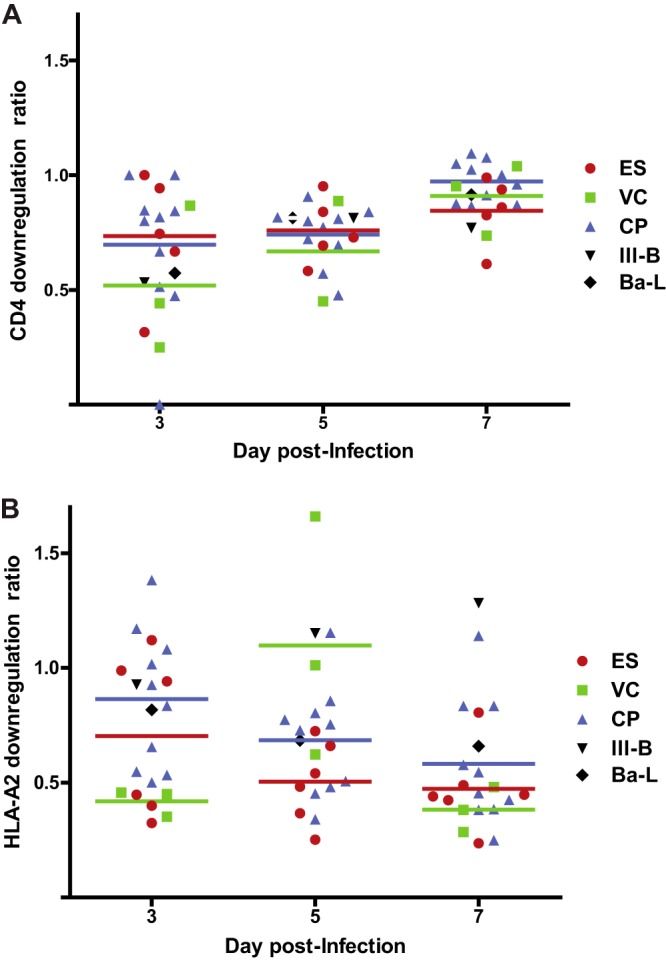
Downregulation of CD4 and HLA-A2 in primary CD4+ T cells infected with HIV-1 isolates. The isolates were laboratory isolates (black) or HIV-1 isolates cultured from ES (red), VCs (green), or CPs (blue). Each symbol represents an individual isolate. The HLA-A2 downregulation ratio (A) is the MFI of HLA-A2 on cells that were positive for intracellular Gag divided by the MFI of HLA-A2 on CD4+ T cells that were Gag negative. The CD4 downregulation ratio (B) represents the fraction of all CD4+ T cells that were Gag positive and CD4 low. The horizontal lines represent the median level of downregulation for each group of patients. The Mann-Whitney test was used to compare the degrees of CD4 and HLA-A2 downregulation in the different patient populations.
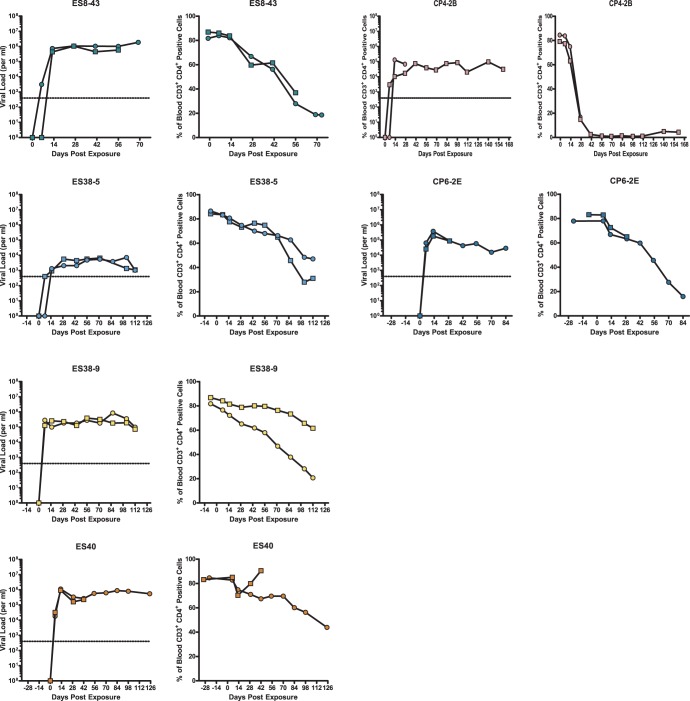
Having established the in vitro replication competence of these viruses, we proceeded to perform an in vivo analysis of their replication competence and their ability to induce CD4+ T cell depletion and to establish latency. To address these important issues, we used BLT humanized mice (47, 48). BLT mice are generated by transplantation of immunodeficient mice previously implanted with human fetal thymic and liver tissue with autologous CD34+ hematopoietic stem cells (31). The humanized BLT mice used for the experiments in this study were derived from 5 different tissue samples. Prior to infection, the presence of human cells in peripheral blood was confirmed by flow cytometry. The peripheral blood of the BLT mice used for experiments in this study had on average 68.5% (±6.6%) human (CD45+) cells, of which 47.9% (±12.45%) expressed human CD3. Of the CD3+ human T cells, 83.6% (±2.9%) also expressed human CD4. Once the presence of human cells was confirmed in all the animals, they were inoculated via tail vein injection with 4 isolates from 3 ES (including the previously described CCR5-tropic isolate ES8-43 [13]) and 2 isolates from 2 CPs. Infection was monitored longitudinally using plasma viral load analysis and by determining the levels of CD4+ T cells in peripheral blood. As shown in Fig. 4, mice infected with all ES and CP viral isolates developed persistent viremia, with steady-state viral loads ranging from 103 to 106 copies/ml. A significant decline (>20%) in peripheral CD4+ T cells was observed over the time course of the experiment in the majority of infected mice. Mice infected with the CXCR4-tropic CP4-2B isolate had the most dramatic decline in CD4+ T cells, consistent with data from prior studies in this model with R5 (68) and X4 (44) laboratory isolates. Interestingly, significant differences in viral loads were seen in 2 isolates cultured from the same ES (ES38-5 and ES38-9) that could not be attributed to sequence differences or to different donor tissue used for the generation of the humanized mice (see Table S2 in the supplemental material). These results demonstrate the in vivo replication capacity of these viruses and their intrinsic ability to induce CD4+ T cell depletion.
FIG 4.
In vivo replication and pathogenesis of HIV isolates from ES and CPs. BLT humanized mice were exposed to HIV-1 isolates that were cultured from either ES or CPs. Each isolate was used to infect two mice. The mice were bled periodically to obtain plasma for viral load analysis via real-time RT-PCR and blood mononuclear cells for flow cytometric analysis. For each isolate, the left panel shows viral load analysis (the dotted line represents the limit of detection for the viral load assay), and the right panel shows the percentage of peripheral blood CD4+ T cells. Different symbols represent different mice. One mouse infected with isolate CP4-2B and one mouse infected with isolate ES40 died shortly after day 28 and day 42 of infection, respectively.
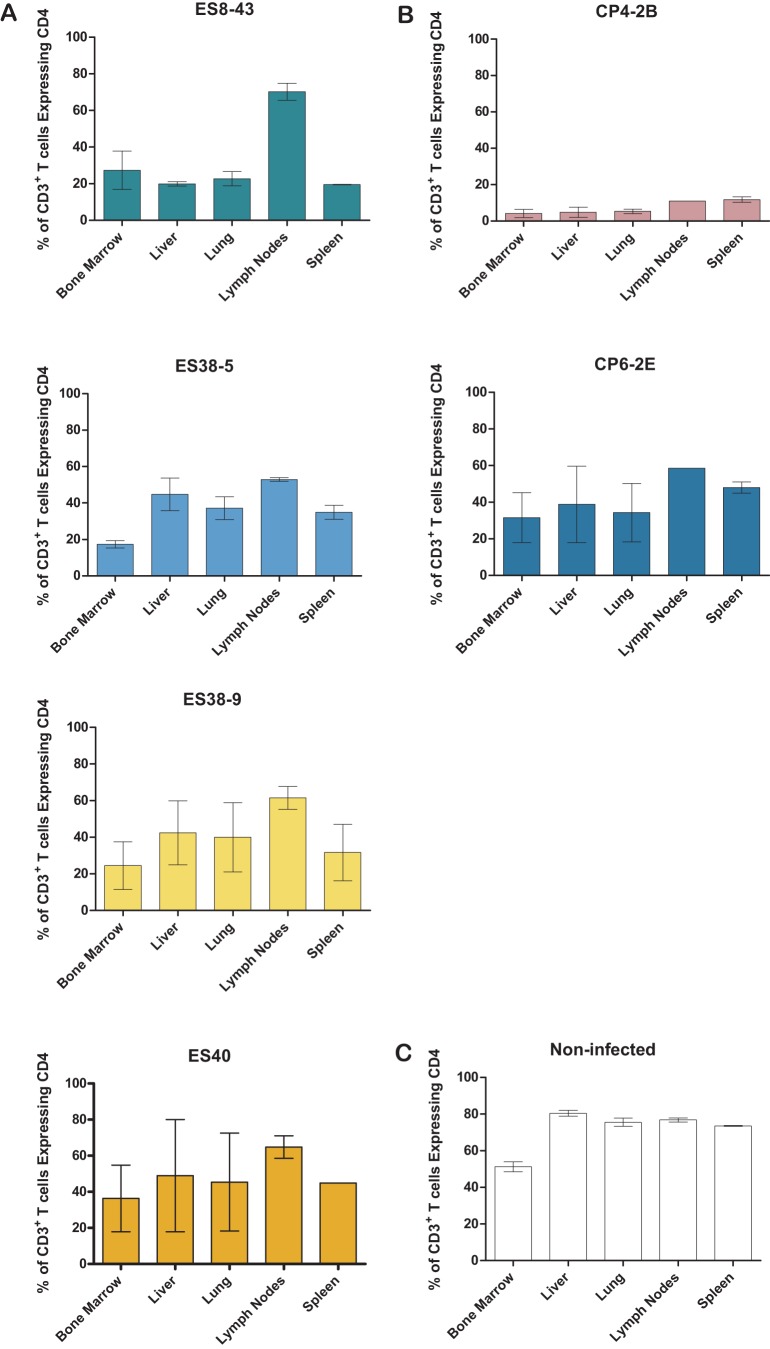
Having observed a decrease in the levels of peripheral blood CD4+ T cells, we investigated the effect of each of these viruses on the levels of CD4+ T cells in different tissues. For this purpose, tissues from each infected BLT mouse were collected and used to prepare single-cell suspensions for flow cytometry analysis. As shown in Fig. 5, there was a reduction in the levels of CD4+ T cells obtained from the tissues from infected animals that was similar to that observed in peripheral blood. Specifically, the mice infected with the CXCR4-tropic virus had the largest reduction in systemic CD4+ T cell levels (Fig. 5B). The rest of the mice infected with the ES viruses and the mice infected with the CP6 virus showed intermediate levels of CD4+ T cell depletion in all tissues analyzed. These results demonstrate that ES viruses are capable of replicating systemically and depleting CD4+ T cells in tissues in a manner that reflects peripheral blood levels.
FIG 5.
HIV isolates derived from elite suppressors are pathogenic in vivo and result in systemic CD4+ T cell depletion. Mononuclear cells were isolated from various tissues at necropsy. The percentage of T cells that express CD4 was calculated for each tissue. The bars represent the average for two animals. Results are from BLT mice infected with HLA*B57 elite suppressor-derived isolates (A), HLA*B57 chronic progressor-derived isolates (B), or noninfected BLT mice (C).
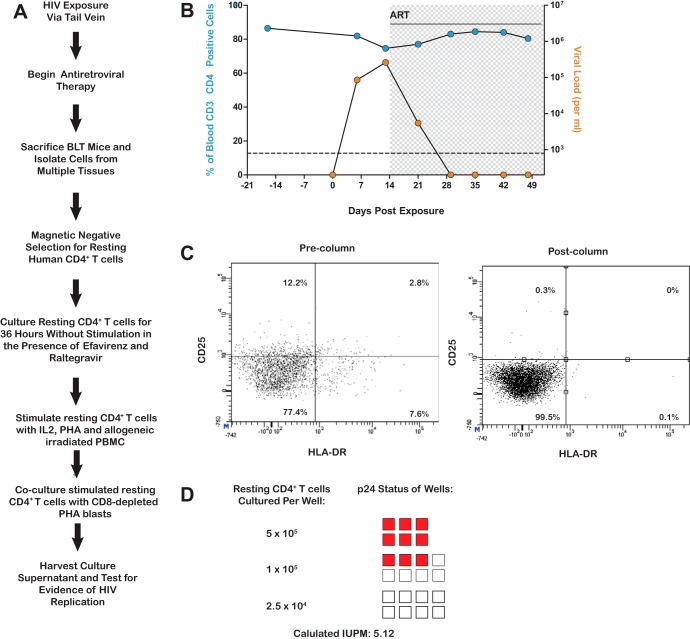
Having established that these viruses are capable of robust replication in vivo and systemic depletion of CD4+ T cells, we proceeded to investigate whether viruses from ES patients can establish latent infection in vivo. For this purpose we used an animal infected with the ES38-9 virus. We chose this virus because it demonstrated a typical profile of partial systemic CD4+ T cell depletion (Fig. 5). After confirming sustained HIV infection in peripheral blood at two different time points, ART consisting of raltegravir, tenofovir, and emtricitabine was initiated (Fig. 6). Upon therapy initiation, a dramatic drop in viral load was noted. The viral load remained suppressed for the duration of treatment. At 5 weeks after therapy initiation, lymphoid tissue was harvested and a mononuclear cell suspension from each tissue prepared. Resting cells were then isolated via negative antibody selection using magnetic beads. The resting state of the CD4+ T cells was confirmed by the lack of expression of HLA-DR and CD25 (Fig. 6C). HIV expression was induced by maximum stimulation of the cells via addition of medium containing PHA, IL-2, and allogeneic irradiated PBMCs. Induced HIV was then further propagated by the addition of allogeneic CD8 T cell-depleted, PHA-activated PBMCs. Under these conditions, HIV induction from latency was evident in 6/6 cultures containing 5 × 105 resting cells and in 3/8 cultures containing 105 resting cells. No outgrowth was observed in 8 wells containing 2.5 × 104 resting cells. Based on these data, the frequency of resting cell infection was determined, using a maximum-likelihood method, to be 5.2 infectious units per million resting CD4+ T cells (IUPM). These results are similar to what was observed previously in this system with the reference HIV-1 isolate JR-CSF and demonstrate the susceptibility of the ES virus to ART and its ability to establish latency in vivo (32).
FIG 6.
Establishment of a latent viral reservoir by an ES-derived HIV isolate. (A) Experimental overview for the ex vivo determination of latency. (B) Longitudinal analysis of plasma viral loads (orange circles) and the levels of CD4+ T cells in the blood (blue circles) of a BLT mouse exposed to the ES-derived isolate ES38-9. The shaded area indicates the duration that antiretroviral therapy was administered. The dashed line indicates the limit of quantitation of HIV RNA. (C) Enrichment of resting CD4+ T cells from the suppressed, ES38-9-infected BLT mouse. Mononuclear cells from all tissues were pooled and subjected to magnetic negative selection of CD4+ resting T cells. Samples were analyzed by flow cytometry for the expression of CD25 and HLA-DR before (left) and after (right) negative selection. (D) Results of a viral outgrowth assay to quantify the latent reservoir. Shaded squares indicate that the well was positive for p24 in the supernatant as determined by ELISA, while nonshaded squares represent wells that were negative for p24 in the supernatant.
Escape mutations in HLA-B*57 epitopes have been associated with diminished viral fitness, and reversion to the wild-type sequence has been observed after the virus is transmitted to HLA-B*5701-negative recipients (37, 39). Isolates ES38-5, ES38-9, and CP4-2B all contained multiple escape mutations and were inoculated into humanized mice that were negative for the B*57 allele (see Table S2 in the supplemental material). No reversion of these escape mutations was seen in bulk sequence of virus amplified from thymus and spleen, even after 4 months of infection in the case of mice infected with ES38 isolates (Fig. 7), consistent with a recent study that showed that no reversion of Gag escape mutations occurred until after more than a year after transmission of HIV-1 to HLA-B*57-negative donors (39). These data suggest that the virus can replicate efficiently in vivo and induce CD4+ T cell depletion even when potentially attenuating escape mutations are present.
FIG 7.
Limited variation occurs within three HLA-B*57-03-restricted Gag and Nef epitopes during HIV infection of BLT Mice. HIV-1 isolates were amplified from the thymuses and spleens of BLT humanized mice at week 16 (isolates ES38-5 and ES38-9), week 8 (isolate ES8-43), and week 4 (isolates CP4-2B and CP6-2E), and Gag and Nef epitopes were analyzed. The red and blue boxes denote two distinct but overlapping Nef epitopes, HW9 (116 to 124) and YT9 (120 to 128), respectively.
DISCUSSION
In the vast majority of cases, when untreated, HIV infection results in progressive loss of CD4+ lymphocytes, resulting in immunodeficiency, susceptibility to rare opportunistic infections and cancers, and ultimately death. The most notable exemptions are individuals who can naturally and completely control HIV infection. These rare individuals are designated elite suppressors. The mechanisms by which ES control viral replication and avoid disease progression are still not fully understood. Studies have shown that some macaques are capable of controlling pathogenic simian immunodeficiency virus (SIV) isolates (49, 50), but studies in human ES have yielded conflicting results. While some studies have suggested that some ES are infected with attenuated or defective virus, others have shown that some ES are infected with replication-competent virus. We have documented the transmission of replication-competent HIV-1 isolates from CPs to ES (16, 18), and studies have shown persistent viremia in ES (51–53), evolution of plasma virus over time (54–56), and a decrease in the frequency of latently infected CD4+ T cells in ES treated with highly active ART (HAART) (57). However, other studies comparing individual viral proteins from ES and CPs have reported reduced fitness of ES Gag (10, 58), Env (59), reverse transcriptase (60), and Nef (61) proteins. It seems unlikely that the majority of ES are infected with isolates that have four or more different attenuated genes, and a major caveat is that these studies have uniformly analyzed plasma isolates. There is strong evidence that ES plasma isolates have accumulated escape mutations which may have a negative effect on fitness (62–64). These attenuating mutations are largely absent from proviral clones and replication-competent isolates cultured from the latent reservoir of ES (62–64). Since isolates from the reservoir are more likely to be representative of the transmitted virus, it is important to study the fitness of virus from this compartment rather than virus that has subsequently evolved to evade the immune response. This is illustrated by a prior study where we demonstrated a significant reduction in fitness of a replication-competent isolate containing escape mutations in HLA-B*57-restricted epitopes in Gag compared to an isolate without escape mutations in these epitopes obtained from the same ES (65). In this study, we show that the growth kinetics of replication-competent virus isolated from CD4+ T cells of HLA-B*57-positive ES and CPs are comparable. We also demonstrate that the isolates have a similar ability to downregulate HLA and CD4 proteins in vitro.
Having established the in vitro fitness of the viruses obtained from the ES, we evaluated their replication capacity and ability to induce CD4+ T cell depletion in vivo. This analysis is particularly important because the in vivo substrate for replication represents a rich milieu of different components, all interacting in multiple ways that cannot be recapitulated ex vivo with cultured cells that represent only a single substrate entity in an artificial activation state. Furthermore, prior studies have shown that SIV isolates that appeared to be fully replication competent in vitro were attenuated when they were inoculated into nonhuman primates (43, 66). We therefore determined the replication capacity of a subset of these isolates in humanized mice. We chose BLT humanized mice because they represent the most advanced and complete system to investigate HIV replication in vivo. Specifically, BLT mice are fully reconstituted with all the types of human cells involved in HIV replication, including T cells, macrophages, and dendritic cells. BLT mice have been validated for the study of HIV transmission, pathogenesis, and HIV persistence (32, 68), and our recent studies have shown significant differences in the level of viremia in vivo when these mice are infected with attenuated versus wild-type viral isolates (44, 67). The presence of robust replication of all the isolates studied in BLT mice and their ability to induce CD4+ T cell depletion presented here represent the first evidence to date that isolates from ES are capable of establishing a pathogenic infection in vivo. Our data also show that viral fitness is not likely to determine whether an HLA-B*57-infected patient becomes an ES or a CP. The fact that viruses from ES patients replicate efficiently in vivo allowed us to show that replication of ES viruses in vivo is efficiently suppressed by ART. The ability to suppress the replication of ES viruses by ART made it possible to then demonstrate that these viruses can persist in vivo and establish a latent infection.
Our study has some limitations. We were not able to isolate virus from all ES, which is consistent with our prior work that showed that these patients have a very low frequency of latently infected CD4+ T cells. However, we cannot rule out the possibility that some HLA-B*57 ES are not infected with replication-competent virus. Furthermore, the in vitro evaluation of virus fitness is dependent on the use of primary cells activated in vitro in an artificial manner. This can mask important effects such as the role of Nef in HIV replication. However, the demonstration that isolates from HLA-B*57 ES have intact nef genes, can replicate efficiently in vitro, and are capable of effective downregulation of CD4 and HLA-A2 strongly suggests that Nef is functional in these viruses and not likely to contribute to the ES phenotype observed. Another limitation is the fact that the in vivo evaluation of these viruses was performed in a mouse model where human cells replace the endogenous immune system. However, it should be noted that this type of experiment cannot be performed in humans. Also, because of the limited species tropism of HIV, these experiments cannot be performed in nonhuman primates either. Therefore, BLT humanized mice represent a viable and useful alternative to perform these types of investigations.
In summary, this is the first study to show that these ES isolates replicate effectively in vivo and eventually cause CD4+ T cell depletion. This study also shows that ES isolates are pathogenic and capable of causing immunosuppression in vivo. Therefore, our results imply that infection with attenuated or defective viruses is not likely to be the cause of elite suppression in all patients. The finding that control of fully pathogenic HIV-1 is possible has major implications for the design of HIV-1 vaccines. In addition, in future experiments it will be important to determine whether this ES phenotype can be recapitulated in vivo by creating humanized mice with hematopoietic stem cells derived from these patients and challenging them with autologous and heterologous viruses.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the HHMI (R.F.S.), by NIH grants 5R01DA030156 (D.M.M.), AI096113, AI073146, and AI096138 (J.V.G.), AI080328 (J.N.B.), and AI100775 (M.D.S), and by the “Sara Borrell” grant from the Spanish Health Institute (M.S.).
Footnotes
Published ahead of print 3 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03380-13.
REFERENCES
- 1.Deeks SG, Walker BD. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27:406–416. 10.1016/j.immuni.2007.08.010 [DOI] [PubMed] [Google Scholar]
- 2.Migueles SA, Connors M. 2010. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA 304:194–201. 10.1001/jama.2010.925 [DOI] [PubMed] [Google Scholar]
- 3.Blankson JN. 2010. Control of HIV-1 replication in elite suppressors. Discov. Med. 9:261–266 [PubMed] [Google Scholar]
- 4.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988–991. 10.1126/science.270.5238.988 [DOI] [PubMed] [Google Scholar]
- 5.Calugi G, Montella F, Favalli C, Benedetto A. 2006. The entire genome of a nef-deleted strain of human immunodeficency virus type 1 recovered 20 years after primary infection: large pool of env-deleted proviruses. J. Virol. 80:11892–11896. 10.1128/JVI.00932-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariani R, Kirchhoff F, Greenough TC, Sullivan JL, Desrosiers RC, Skowronski J. 1996. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 70:7752–7764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. 1995. Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228–232. 10.1056/NEJM199501263320405 [DOI] [PubMed] [Google Scholar]
- 8.Iversen AK, Shpaer EG, Rodrigo AG, Hirsch MS, Walker BD, Sheppard HW, Merigan TC, Mullins JI. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J. Virol. 69:5743–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Zhang L, Ho DD. 1998. Characterization of gag and pol sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology 240:36–49. 10.1006/viro.1997.8913 [DOI] [PubMed] [Google Scholar]
- 10.Miura T, Brumme ZL, Brockman MA, Rosato P, Sela J, Brumme CJ, Pereyra F, Kaufmann DE, Trocha A, Block BL, Daar ES, Connick E, Jessen H, Kelleher AD, Rosenberg E, Markowitz M, Schafer K, Vaida F, Iwamoto A, Little S, Walker BD. 2010. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J. Virol. 84:7581–7591. 10.1128/JVI.00286-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander L, Weiskopf E, Greenough TC, Gaddis NC, Auerbach MR, Malim MH, O'Brien SJ, Walker BD, Sullivan JL, Desrosiers RC. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361–4376. 10.1128/JVI.74.9.4361-4376.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander L, Aquino-DeJesus MJ, Chan M, Andiman WA. 2002. Inhibition of human immunodeficiency virus type 1 (HIV-1) replication by a two-amino-acid insertion in HIV-1 vif from a nonprogressing mother and child. J. Virol. 76:10533–10539. 10.1128/JVI.76.20.10533-10539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, Gandhi SK, Siliciano JD, Williams TM, Siliciano RF. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J. Virol. 81:2508–2518. 10.1128/JVI.02165-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamine A, Caumont-Sarcos A, Chaix ML, Saez-Cirion A, Rouzioux C, et al. 2007. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS 21:1043–1045. 10.1097/QAD.0b013e3280d5a7ac [DOI] [PubMed] [Google Scholar]
- 15.Julg B, Pereyra F, Buzón MJ, Piechocka-Trocha A, Clark MJ, Baker BM, Lian J, Miura T, Martinez-Picado J, Addo MM, Walker BD. 2010. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin. Infect. Dis. 51:233–238. 10.1086/653677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey JR, O'Connell K, Yang HC, Han Y, Xu J, Jilek B, Williams TM, Ray SC, Siliciano RF, Blankson JN. 2008. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J. Virol. 82:7395–7410. 10.1128/JVI.00800-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaillard S, Dinoso JB, Marsh JA, DeZern AE, O'Connell KA, Spivak AM, Alwood K, Durand CM, Ambinder RF, Blankson JN. 2011. Sustained elite suppression of replication competent HIV-1 in a patient treated with rituximab based chemotherapy. J. Clin. Virol. 51:195–198. 10.1016/j.jcv.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckheit RW, III, Allen TG, Alme A, Salgado M, O'Connell KA, Huculak S, Falade-Nwulia O, Williams TM, Gallant JE, Siliciano RF, Blankson JN. 2012. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat. Commun. 3:716. 10.1038/ncomms1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, Hallahan CW, Selig SM, Schwartz D, Sullivan J, Connors M. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714. 10.1073/pnas.050567397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. 10.1016/j.immuni.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, Ueda P, Lu Z, Cohen D, Wrin T, Petropoulos CJ, Rosenberg ES, Walker BD. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563–571. 10.1086/526786 [DOI] [PubMed] [Google Scholar]
- 22.Han Y, Lai J, Barditch-Crovo P, Gallant JE, Williams TM, Siliciano RF, Blankson JN. 2008. The role of protective HCP5 and HLA-C associated polymorphisms in the control of HIV-1 replication in a subset of elite suppressors. AIDS 22:541–544. 10.1097/QAD.0b013e3282f470e4 [DOI] [PubMed] [Google Scholar]
- 23.Lambotte O, Boufassa F, Madec Y, Nguyen A, Goujard C, Meyer L, Rouzioux C, Venet A, Delfraissy JF, SEROCO-HEMOCO Study Group 2005. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin. Infect. Dis. 41:1053–1056. 10.1086/433188 [DOI] [PubMed] [Google Scholar]
- 24.Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, McCune JM, Deeks SG. 2008. HLA class I-restricted T cell responses may contribute to the control of HIV infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398–5407. 10.1128/JVI.02176-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947. 10.1126/science.1143767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. 10.1126/science.1195271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, Van Baarle D, Kostense S, Miedema F, McLaughlin M, Ehler L, Metcalf J, Liu S, Connors M. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061–1068. 10.1038/ni845 [DOI] [PubMed] [Google Scholar]
- 28.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, Lederman MM, Benito JM, Goepfert PA, Connors M, Roederer M, Koup RA. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781–4789. 10.1182/blood-2005-12-4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sáez-Cirión A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, Barré-Sinoussi F, Delfraissy JF, Sinet M, Pancino G, Venet A, Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group 2007. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. U. S. A. 104:6776–6781. 10.1073/pnas.0611244104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR. 2010. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 6:e1000917. 10.1371/journal.ppat.1000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melkus MW, Estes JD, Padgett-Thomas A, Gatlin J, Denton PW, Othieno FA, Wege AK, Haase AT, Garcia JV. 2006. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 12:1316–1322. 10.1038/nm1431 [DOI] [PubMed] [Google Scholar]
- 32.Denton PW, Olesen R, Choudhary SK, Archin NM, Wahl A, Swanson MD, Chateau M, Nochi T, Krisko JF, Spagnuolo RA, Margolis DM, Garcia JV. 2012. Generation of HIV latency in humanized BLT mice. J. Virol. 86:630–634. 10.1128/JVI.06120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, Bandarenko N, Schmitz JL, Bosch RJ, Landay AL, Coffin JM, Margolis DM. 2008. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS 22:1131–1135. 10.1097/QAD.0b013e3282fd6df4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salgado M, Rabi SA, O'Connell KA, Buckheit RW, III, Bailey JR, Chaudhry AA, Breaud AR, Marzinke MA, Clarke W, Margolick JB, Siliciano RF, Blankson JN. 2011. Prolonged control of replication-competent dual-tropic human immunodeficiency virus-1 following cessation of highly active antiretroviral therapy. Retrovirology 8:97. 10.1186/1742-4690-8-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nou E, Zhou Y, Nou DD, Blankson JN. 2009. Effective downregulation of HLA-A*2 and HLA-B*57 by primary human immunodeficiency virus type 1 isolates cultured from elite suppressors. J. Virol. 83:6941–6946. 10.1128/JVI.00306-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampah ME, Ceccato CM, Blankson JN. 2011. HIV type 1-mediated downregulation of HLA-B*57/B*5801 proteins on elite suppressor CD4+ T cells. AIDS Res. Hum. Retroviruses 27:183–186. 10.1089/aid.2010.0144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. 10.1038/nm992 [DOI] [PubMed] [Google Scholar]
- 38.Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, Derdeyn CA, Tang J, Kaslow RA, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder PJ, Hunter E. 2008. Transmission of HIV-1 gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 12;. 205:1009–1017. 10.1084/jem.20072457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung'u T, Lakhi S, Gilmour J, Goepfert P, Walker BD, Kaslow R, Mulenga J, Allen S, Goulder PJ, Hunter E. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623. 10.1128/JVI.80.7.3617-3623.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, Desouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. 2007. Escape and compensation from early HLA-B57-mediated CTL pressure on HIV-1 gag alters capsid interactions with cyclophilin A. J. Virol. 81:12608–12618. 10.1128/JVI.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boutwell CL, Rowley CF, Essex M. 2009. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol. 83:2460–2468. 10.1128/JVI.01970-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kestler HW, III, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651–662. 10.1016/0092-8674(91)90097-I [DOI] [PubMed] [Google Scholar]
- 44.Zou W, Denton PW, Watkins RL, Krisko JF, Nochi T, Foster JL, Garcia JV. 2012. Nef functions in BLT mice to enhance HIV-1 replication and deplete CD4+CD8+ thymocytes. Retrovirology 9:44. 10.1186/1742-4690-9-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. 10.1038/350508a0 [DOI] [PubMed] [Google Scholar]
- 46.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 nef protein. Nat. Med. 2:338–342. 10.1038/nm0396-338 [DOI] [PubMed] [Google Scholar]
- 47.Denton PW, Garcia JV. 2011. Humanized mouse models of HIV infection. AIDS Rev. 13:135–148 [PMC free article] [PubMed] [Google Scholar]
- 48.Marsden MD, Kovochich M, Suree N, Shimizu S, Mehta R, Cortado R, Bristol G, An DS, Zack JA. 2012. HIV latency in the humanized BLT mouse. J. Virol. 86:339–347. 10.1128/JVI.06366-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mudd PA, Watkins DI. 2011. Understanding animal models of elite control: windows on effective immune responses against immunodeficiency viruses. Curr. Opin. HIV AIDS 6:197–201. 10.1097/COH.0b013e3283453e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandrea I, Gaufin T, Gautam R, Kristoff J, Mandell D, Montefiori D, Keele BF, Ribeiro RM, Veazey RS, Apetrei C. 2011. Functional cure of SIVagm infection in rhesus macaques results in complete recovery of CD4+ T cells and is reverted by CD8+cell depletion. PLoS Pathog. 7:e1002170. 10.1371/journal.ppat.1002170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinoso JB, Kim SY, Siliciano RF, Blankson JN. 2008. A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy. Clin. Infect. Dis. 47:102–104. 10.1086/588791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatano H, Delwart EL, Norris PJ, Lee TH, Dunn-Williams J, Hunt PW, Hoh R, Stramer SL, Linnen JM, McCune JM, Martin JN, Busch MP, Deeks SG. 2009. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 83:329–335. 10.1128/JVI.01763-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, Baker B, Rosenberg R, Cutrell E, Seaman MS, Coffin JM, Walker BD. 2009. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J. Infect. Dis. 200:984–990. 10.1086/605446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J. Virol. 84:7018–7028. 10.1128/JVI.00548-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salgado M, Brennan TP, O'Connell KA, Bailey JR, Ray SC, Siliciano RF, Blankson JN. 2010. Evolution of the HIV-1 nef gene in HLA-B*57 positive elite suppressors. Retrovirology 7:94. 10.1186/1742-4690-7-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mens H, Kearney M, Wiegand A, Shao W, Schønning K, Gerstoft J, Obel N, Maldarelli F, Mellors JW, Benfield T, Coffin JM. 2010. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J. Virol. 84:12971–12981. 10.1128/JVI.00387-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chun TW, Shawn Justement J, Murray D, Kim CJ, Blazkova J, Hallahan CW, Benko E, Costiniuk CT, Kandel G, Ostrowski M, Kaul R, Moir S, Casazza JP, Koup RA, Kovacs C, Fauci AS. 2013. Effect of antiretroviral therapy on HIV reservoirs in elite controllers. J. Infect. Dis. 208:1443–1447. 10.1093/infdis/jit306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B, Rothchild AC, Li B, Trocha A, Cutrell E, Frahm N, Brander C, Toth I, Arts EJ, Allen TM, Walker BD. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphotye recognition. J. Virol. 83:2743–2755. 10.1128/JVI.02265-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lassen KG, Lobritz MA, Bailey JR, Johnston S, Nguyen S, Lee B, Chou T, Siliciano RF, Markowitz M, Arts EJ. 2009. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 5:e1000377. 10.1371/journal.ppat.1000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brumme ZL, Li C, Miura T, Sela J, Rosato PC, Brumme CJ, Markle TJ, Martin E, Block BL, Trocha A, Kadie CM, Allen TM, Pereyra F, Heckerman D, Walker BD, Brockman MA. 2011. Reduced replication capacity of NL4-3 recombinant viruses encoding reverse transcriptase-integrase sequences from HIV-1 elite controllers. J. Acquir. Immune Defic. Syndr. 56:100–108. 10.1097/QAI.0b013e3181fe9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mwimanzi P, Markle TJ, Martin E, Ogata Y, Kuang XT, Tokunaga M, Mahiti M, Pereyra F, Miura T, Walker BD, Brumme ZL, Brockman MA, Ueno T. 2013. Attenuation of multiple nef functions in HIV-1 elite controllers. Retrovirology 10:1. 10.1186/1742-4690-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey JR, Williams TM, Siliciano RF, Blankson JN. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357–1369. 10.1084/jem.20052319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bailey JR, Lassen KG, Yang HC, Quinn TC, Ray SC, Blankson JN, Siliciano RF. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 80:4758–4770. 10.1128/JVI.80.10.4758-4770.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey JR, Brennan TP, O'Connell KA, Siliciano RF, Blankson JN. 2009. Evidence of CD8+ T-cell-mediated selective pressure on human immunodeficiency virus type 1 nef in HLA-B*57+ elite suppressors. J. Virol. 83:88–97. 10.1128/JVI.01958-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Connell KA, Hegarty RW, Siliciano RF, Blankson JN. 2011. Viral suppression of multiple escape mutants by de novo CD8(+) T cell responses in a human immunodeficiency virus-1 infected elite suppressor. Retrovirology 8:63. 10.1186/1742-4690-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mudd PA, Ericsen AJ, Walsh AD, León EJ, Wilson NA, Maness NJ, Friedrich TC, Watkins DI. 2011. CD8+ T cell escape mutations in simian immunodeficiency virus SIVmac239 cause fitness defects in vivo, and many revert after transmission. J. Virol. 85:12804–12810. 10.1128/JVI.05841-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watkins RL, Zou W, Denton PW, Krisko JF, Foster JL, Garcia JV. 2013. In vivo analysis of highly conserved Nef activities in HIV-1 replication and pathogenesis. Retrovirology 10:125. 10.1186/1742-4690-10-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denton PW, Estes JD, Sun Z, Othieno FA, Wei BL, Wege AK, Powell DA, Payne D, Haase AT, Garcia JV. 2008. Antiretroviral pre-exposure prophylaxis prevents vaginal transmission of HIV-1 in humanized BLT mice. PLoS Med. 5:e16. 10.1371/journal.pmed.0050016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.