ABSTRACT
The nonstructural protein NSs is the main virulence factor of Rift Valley fever virus (RVFV; family Bunyaviridae, genus Phlebovirus), a serious pathogen of livestock and humans in Africa. RVFV NSs blocks transcriptional upregulation of antiviral type I interferons (IFN) and destroys the general transcription factor TFIIH subunit p62 via the ubiquitin/proteasome pathway. Here, we identified a subunit of E3 ubiquitin ligases, F-box protein FBXO3, as a host cell interactor of NSs. Small interfering RNA (siRNA)-mediated depletion of FBXO3 rescued p62 protein levels in RVFV-infected cells and elevated IFN transcription by 1 order of magnitude. NSs interacts with the full-length FBXO3 protein as well as with a truncated isoform that lacks the C-terminal acidic and poly(R)-rich domains. These isoforms are present in both the nucleus and the cytoplasm. NSs exclusively removes the nuclear pool of full-length FBXO3, likely due to consumption during the degradation process. F-box proteins form the variable substrate recognition subunit of the so-called SCF ubiquitin ligases, which also contain the constant components Skp1, cullin 1 (or cullin 7), and Rbx1. siRNA knockdown of Skp1 also protected p62 from degradation, suggesting involvement in NSs action. However, knockdown of cullin 1, cullin 7, or Rbx1 could not rescue p62 degradation by NSs. Our data show that the enzymatic removal of p62 via the host cell factor FBXO3 is a major mechanism of IFN suppression by RVFV.
IMPORTANCE Rift Valley fever virus is a serious emerging pathogen of animals and humans. Its main virulence factor, NSs, enables unhindered virus replication by suppressing the antiviral innate immune system. We identified the E3 ubiquitin ligase FBXO3 as a novel host cell interactor of NSs. NSs recruits FBXO3 to destroy the general host cell transcription factor TFIIH-p62, resulting in suppression of the transcriptional upregulation of innate immunity.
INTRODUCTION
Rift Valley fever virus (RVFV; family Bunyaviridae, genus Phlebovirus) is a serious, arthropod-borne pathogen affecting wild animals, livestock, and humans (1, 2). The RVFV outbreaks that regularly occur in Africa result in devastating losses of lives and wealth. In ruminants like cattle and sheep, the typical onset of an RVFV epidemic is characterized by so-called abortion storms and the deaths of about 100% of the newborns. In adult animals, the infection causes hepatitis and hemorrhage with a mortality rate of 10 to 20%. Human infections are mostly restricted to a self-limiting febrile illness, but in 1% to 2% of cases the symptoms can aggravate to fulminant hepatitis, encephalitis, retinitis, blindness, or a hemorrhagic syndrome (3). In hospitalized patients, the typical case fatality rate is 10% to 20%, but higher rates have been reported (4).
RVFV is transmitted by a multitude of mosquito and sandfly species (5, 6) but also through aerosols or contact with aborted fetuses (1, 2). It is capable of infecting a wide range of animals and has repeatedly proven its potential to spread into new areas and cause large epidemics (7). These features, together with the fact that preventive and therapeutic possibilities are limited, have led to the classification of RVF as a notifiable disease and a potential biological weapon (8).
The particles of RVFV consist of a lipid envelope with two integral glycoproteins, Gn and Gc, surrounding a core of three ribonucleoproteins (RNPs) (9). These RNPs are complexes of nucleoprotein (N), RNA-dependent RNA polymerase (L), and a viral genomic RNA (vRNA). RVFV has three different vRNA segments, of which two (designated L and M) are in negative sense and one (S) is in ambisense. The L segment encodes the L protein, the M segment encodes a polyprotein that is processed to the glycoproteins Gn and Gc along with the nonstructural proteins NSm1 and NSm2, and the S segment encodes the N protein and the nonstructural protein NSs.
NSs plays a central role in the pathogenesis of RVFV by downregulating the antiviral type I interferon (IFN-α/β) system (10, 11). IFNs are cytokines that are produced by infected cells to establish an antiviral state in the surrounding cells, mediated by products of the so-called IFN-stimulated genes (ISGs) (12). The induction of the IFN response occurs on the transcriptional level. RVFV NSs was shown to block both the specific upregulation of IFN genes and the general transcription of host cell mRNAs (reviewed in references 13 and 14). The molecular basis of the specific IFN inhibition by NSs was proposed to be the recruitment of the transcriptional repressor SAP30 to the IFN promoter (15), whereas the general effect was attributed to depletion of the general transcription factor TFIIH from its subunit p44 (16). These mechanisms are reliant on stoichiometric sequestration of target proteins, which is a relatively slow process requiring NSs to be present at least at a 1:1 ratio with the target protein. IFN gene transcription, however, is triggered by just a few viral signature structures detected by the host cell (17, 18) and occurs shortly after infection (19, 20). It remains therefore questionable whether the rapid and potent shutdown of IFN induction imposed by NSs (10) could be completely explained by a stoichiometric mechanism involving binding and sequestration of host cell targets, which at least in the initial phase of infection outnumber the viral proteins. In fact, a recent report shows that RVFV can act on host transcription through a different mechanism, namely, the proteasomal degradation of p62, a subunit of the general transcription factor TFIIH (21). It remained unclear, however, how this destructive activity is achieved and how it contributes to IFN antagonism. Therefore, in the present study, we investigated the connection between the RVFV NSs IFN antagonism, TFIIH-p62, and the proteasomal system. We demonstrate that p62 is degraded by NSs almost immediately after virus entry. Based on results from a large-scale proteomic screen (22), we identified the F-box protein FBXO3 as an interactor of RVFV NSs. FBXO3 and its cofactor Skp1 seem to form part of an E3 ubiquitin ligase responsible for the NSs-driven p62 degradation and IFN suppression. To our knowledge, this is the first report on a host cell interactor of RVFV NSs that is involved in IFN suppression via rapid degradation of host cell targets.
MATERIALS AND METHODS
Cells, viruses, and reagents.
HeLa, A549, Vero E6, 293T, and BHK-21 cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% (HeLa cells) or 10% (all other cells) fetal calf serum (FCS). RVFV strains rZH548 and Clone 13 and the newly generated recombinant RVFV strains (see below) were propagated on Vero E6 cells under biosafety level 3 (BSL-3) conditions. MG132 was purchased from Biomol.
Plasmid constructs.
The RVFV plasmids for rescuing recombinant RVFV, pI.18_RVFV_L, pI.18_RVFV_N (helper plasmids), pHH21_RVFV_vL, and pHH21_RVFV_vM (rescue plasmids) were described previously (23). The rescue plasmids used to express tandem affinity purification (TAP)-tagged fusion proteins via RVFV are based on an ambisense S segment rescue construct (termed pHH21_RVFV_vN_MCS-CTAP) containing a tandem AarI cloning site followed by a C-terminal TAP tag sequence instead of the NSs gene. The 5′ and 3′ AarI restriction sites are compatible with NcoI and HinDIII sites, respectively, and the C-terminal TAP sequence is in frame with the ATG start codon created by the NcoI-compatible 5′ AarI site (23). To express NSs fused with a C-terminal TAP tag via recombinant RVFV, the NSs gene of strain ZH548 was cloned into pHH21_RVFV_vN_MCS-CTAP, giving rise to the S segment rescue plasmid pHH21_RVFV_vN_NSsZH-CTAP. In a similar manner, the negative-control plasmid pHH21_RVFV_vN_GFP-CTAP was generated, expressing the enhanced green fluorescent protein (EGFP) gene fused with a C-terminal TAP tag, as well as the S segment rescue plasmids expressing a C-terminal Flag epitope-tagged RVFV NSs (pHH21_RVFV_vN_NSsZH_CFlag) or a 3×Flag-tagged ΔMx control protein (pHH21_RVFV_vN_3×Flag_ΔMx).
To express epitope tag-equipped FBXO3 isoforms, total RNA isolated from 293T cells was used for reverse transcription (RT) followed by amplification of the respective cDNAs with specific primers. The amplicons were subcloned, and the resulting constructs were used as the templates for generating amplicons with N-terminal 3× hemagglutinin (HA) tags. These amplicons were cloned to pcDNA3.1 by T/A cloning (Invitrogen). The constructs pcDNA3.1-3×HA-FBXO3/1 and pcDNA3.1-3×HA-FBXO3/2 were sequenced to verify that the inserts corresponded to GenBank entries NM_012175.3 and NM_033406.2, respectively. FBXO3 cDNAs were PCR amplified from these constructs and ligated to pDONR221 vector using the Gateway technology (Invitrogen). These constructs were used for cloning plasmids expressing tandem affinity tagged (StrepII-HA) proteins and transfected into 293 FLP-IN TREX cells in order to generate stable, doxycycline (DOX)-inducible cell lines. Detailed cloning strategies and primers are available upon request.
Western blot analyses.
Samples for Western blot analysis were separated by SDS-PAGE, blotted to polyvinylidene difluoride (PVDF) membranes, and blocked using 5% (wt/vol) milk powder in Tris-buffered saline with 0.1% Tween 20. The membranes were probed with antibodies against the following targets: HA tag, GTF2H1 (p62), Skp1 (monoclonal antibody [MAb] EPR3304), cullin 1 (MAb EPR3102Y), ROC1 (Rbx1) (all by Abcam), Flag tag (Sigma), cullin 7 (Thermo Scientific), cdk7 (MAb MO1) and β-actin (MAb 8H10D10) (both by Cell Signaling), PKR (MAb 71/10) (24), and RVFV N (25). Peroxidase-conjugated anti-mouse and anti-rabbit IgG polyclonals (Thermo Fisher) were used as secondary antibodies.
Generation of recombinant RVF viruses.
Recombinant Rift Valley fever virus strain ZH548 expressing TAP-tagged NSs (rZHΔNSs::NSsZH548-CTAP or TAP-tagged GFP (rZHΔNSs::GFP-CTAP), Flag-NSs (rZHΔNSs::CF-NSsZH548; rZH-CF-NSs in short), and 3×Flag-ΔMx (rZHΔNSs::F-ΔMx; rZH-NF-ΔMx in short) were rescued from cloned cDNA using the pol I/pol II system (23). Briefly, cocultures of 293T and BHK-21 cells grown in 6-well dishes were transfected with the helper constructs pI.18_RVFV_L and pI.18_RVFV_N (0.5 μg each) and the rescue constructs pHH21_RVFV_vL (L segment) and pHH21_RVFV_vM (M segment) combined with an S segment plasmid (1 μg each). After 5 days of incubation, cell supernatants were harvested and transferred onto Vero E6 cells to grow virus stocks. The genetic and biological properties of the recombinant viruses were confirmed by RT-PCR, sequencing of the NSs locus, and functional assays (data not shown).
Preparation of lysates for proteomics.
293T cells seeded in 15-cm dishes were infected with RVFVΔNSs::NSsZH548-CTAP or RVFVΔNSs::GFP-CTAP at a multiplicity of infection (MOI) of 5 for 16 h. Then, cells were washed once with cold phosphate-buffered saline (PBS), scraped off in 5 ml cold PBS, and centrifuged at 1,000 × g for 5 min at 4°C, and the pellet was snap-frozen in liquid nitrogen. After addition of 5 ml of TAP buffer (50 mM Tris-HCl [pH 7.5], 5% glycerol, 0.2% NP-40, 1.5 mM MgCl2, 100 mM NaCl, and protease and phosphatase inhibitors), pellets were again snap-frozen in liquid nitrogen and stored at −80°C.
Real-time RT-PCR.
Total cellular RNA was isolated with the RNeasy Minikit (Qiagen) and eluted in 40 μl of double-distilled water (ddH2O). An aliquot of 100 ng RNA was then used as a template for cDNA synthesis and PCR using the QuantiTect Reverse Transcription and QuantiTect SYBR green PCR kits (Qiagen) and a StepOne Real-Time PCR system (Applied Biosystems). mRNA levels of human IFN-β, IP-10, FBXO3, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were determined with QuantiTect primers (Qiagen) QT00203763, QT01003065, QT00061285, and QT01192646, respectively. All values obtained were normalized against the GAPDH mRNA signal using the ΔΔCT method (26). For the cytokine assays, mock samples that showed no PCR amplification were arbitrarily set to a threshold cycle (CT) value of 40. RVFV L RNA was measured by TaqMan quantitative RT-PCR (RT-qPCR) as described by Bird et al. (27).
Immunofluorescence assays.
HeLa cells were grown on coverslips to 30 to 50% confluence and infected and incubated for the indicated time. Then, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and washed with PBS containing 1% FCS. The following primary antibodies were used, diluted in PBS containing 1% FCS: Anti-Flag (Sigma), Anti HA.11 (Covance), and Anti-GTF2H1 (Abcam). After incubation at room temperature for 1 h, the coverslips were washed three times with 1% FCS in PBS and treated at room temperature for 45 min with anti-mouse and anti-rabbit IgG polyclonals coupled with Alexa Fluor 488 or Alexa Fluor 555 dyes. DAPI (4′,6-diamidino-2-phenylindole) was used at 0.1 μg/ml to counterstain nuclei. After washing three times with PBS and once with ddH2O, coverslips were mounted using Fluorsave solution (Calbiochem). Stained cell samples were examined using a Leica SP5 confocal microscope.
siRNA knockdown.
Knockdown of gene expression was achieved by 2-fold reverse transfection of small interfering RNAs (siRNAs). AllStar Negative Control siRNA as well as validated pools of four siRNAs (Qiagen) against mRNAs for FBXO3 (GeneSolution GS26273), Rbx1 (GeneSolution GS9978), Skp1 (GeneSolution GS6500), cullin 1 (GeneSolution GS8454), and cullin 7 (GeneSolution GS9820) were used. For reverse transfections, siRNAs were transfected with 50 nM total concentrations using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Four hours after transfection, the medium was changed. Three days after the first transfection, the cells were harvested and counted, and equal amounts of cells were again reverse transfected as described above. Experiments were performed 1 day after the second transfections.
Coimmunoprecipitation assays.
293 FLP-IN TREX cells stably expressing SII-HA-tagged FBXO3 isoforms 1 or 2 were used for coimmunoprecipitation experiments. Expression was induced by treating the cells with 2 μg/ml doxycycline hyclate for 2 days. The cells were then infected with rZH-CF-NSs or rZH-NF-ΔMx (MOI of 3). Doxycycline was included in the medium added after removing the inoculum. Three hours later, 20 μM MG132 was added, and the incubation was continued for 1 h. Cells were scraped into cold PBS and pelleted by centrifugation. The cells were lysed in cold TAP buffer. After centrifugation, lysis supernatant was incubated with Dynabeads (Invitrogen) coupled with either Anti HA.11 antibody (Covance) or Anti-Flag M2 antibody (Stratagene) for 1 h at +4°C. After three washes with TAP buffer, the complexes were eluted by boiling the beads in SDS-loading buffer and analyzed by Western blotting.
RESULTS
Rapid degradation of NSs targets.
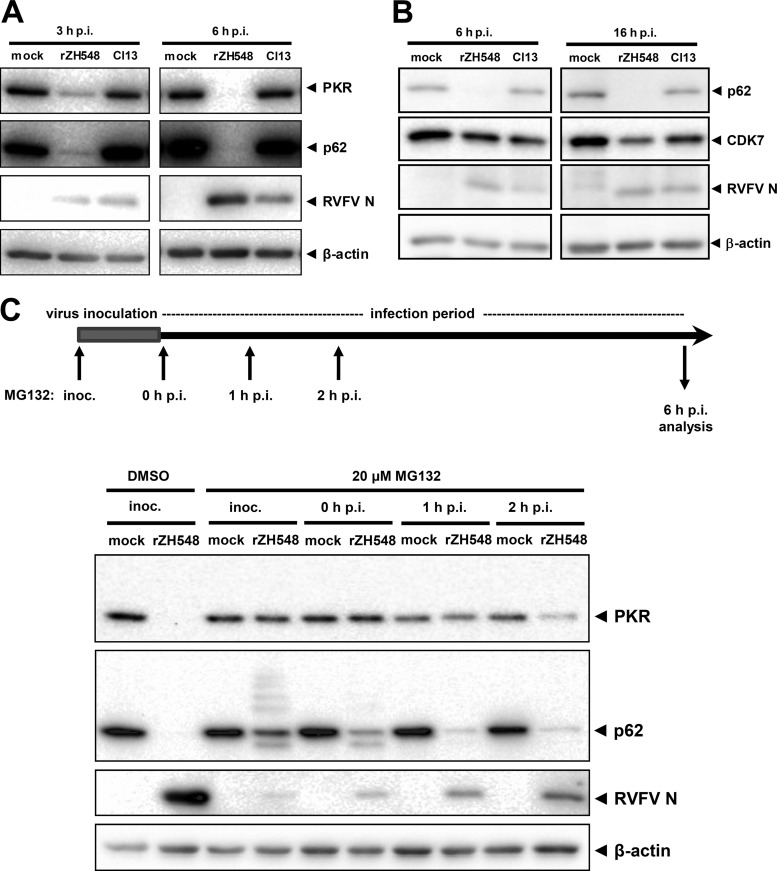
So far, two main host cell targets have been described to be degraded by RVFV NSs: p62, the TFIIH subunit mentioned above, and double-stranded RNA (dsRNA)-dependent protein kinase R (PKR), one of the main ISGs inhibiting RVFV replication (25, 28). We performed a time course experiment to obtain kinetics for p62 and PKR degradation after virus infection. In agreement with previous studies (16, 21, 25, 28), we observed rapid degradation of both PKR and p62 after infection with the recombinant wild-type (wt) strain ZH548, whereas the NSs-truncated strain Clone 13 had no such effect (Fig. 1A). A much weaker susceptibility to NSs-mediated degradation was observed for CDK7, a regulatory kinase of TFIIH (Fig. 1B). Similar results were obtained using human A549 cells (see below). Thus, we were able to confirm in our system that p62 and PKR are efficiently removed by RVFV NSs.
FIG 1.
Rapid proteasomal degradation of p62 by RVFV NSs. (A and B) Time course of target degradation. HeLa cells were infected with recombinant wt RVFV strain ZH548 (rZH548) or the NSs-truncated mutant Clone 13 (Cl13) at an MOI of 5 (A) or 10 (B) or were left uninfected (mock). Cells were lysed after the indicated time periods and subjected to Western blot analyses against PKR and p62 (A) or p62 and CDK7 (B). RVFV N and β-actin were detected as infection and loading control, respectively. (C) Time course of MG132 addition. HeLa cells were mock infected or infected with rZH548 at an MOI of 10 for 1 h in infection medium (inoculum). Then, cells were incubated either in 0.2% dimethyl sulfoxide (DMSO) alone or in 20 μM MG132 and 0.2% DMSO at different time points of infection. Inoc., DMSO or MG132 was present in the virus inoculum and later in the cell culture medium; 0 h p.i., virus inoculum was replaced by cell culture medium containing MG132; 1 h p.i., MG132 addition 1 h after taking off the inoculum and incubation in cell culture medium; 2 h p.i., MG132 addition 2 h after taking off the inoculum and incubation in cell culture medium. All cells were lysed at 6 h p.i. and analyzed by immunoblotting using the indicated antibodies.
To investigate the onset of p62 and PKR degradation in more detail, we added the proteasomal inhibitor MG132 at different time points of infection. As seen in Fig. 1C, addition of MG132 at 1 h after infection is required to preserve PKR levels, whereas later addition had no protective effect. To preserve p62 levels, the inhibitor had to be present even earlier. MG132 had to be added either from the first moment the virus contacted the cell (i.e., already in the inoculum) or directly after the virus inoculum was replaced by cell culture medium (Fig. 1C). Later addition of MG132 did not prevent p62 degradation, although the inhibitor largely slows down virus infection. Interestingly, immediate early addition of MG132 revealed the presence of a high-molecular-weight ladder of p62 as well as a short form of it, suggesting ubiquitin attachment and a degradation intermediate. Nonetheless, we were unable to directly demonstrate ubiquitination, possibly due to the fact that only a fraction of p62 is affected at any given time (data not shown). The data indicate that RVFV NSs induces the rapid proteasomal destruction of both PKR and p62, with the latter being even more quickly or more efficiently degraded than the former.
NSs interacts with the E3 ubiquitin ligase component FBXO3.
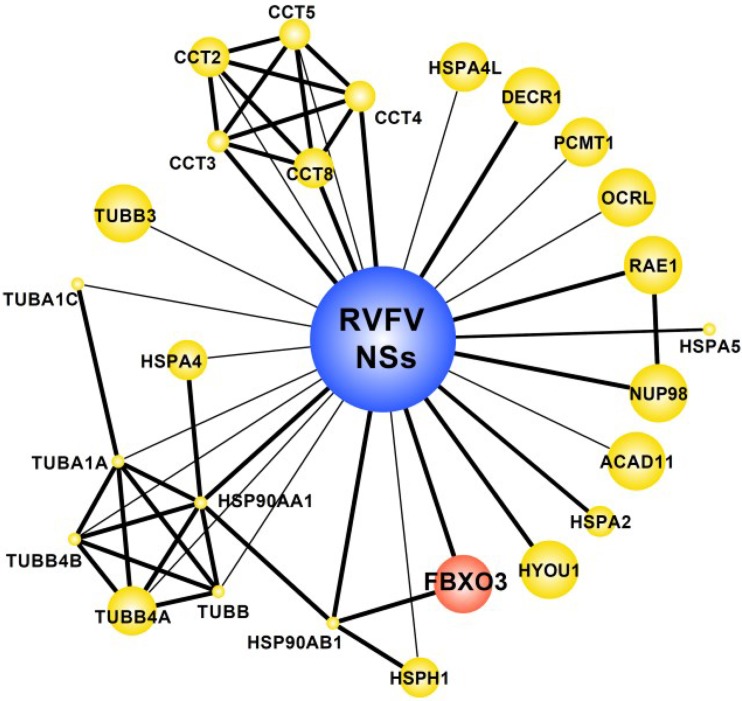
We have recently performed a proteomic screen for host cell interactors of 70 viral IFN antagonists, including the NSs of the virulent RVFV strain ZH548 (22). Since RVFV NSs inhibits RNA polymerase II-driven mRNA synthesis, it is difficult to express from eukaryotic plasmids. We had to construct a recombinant RVFV encoding an NSs with a C-terminal TAP tag. Approximately 2 × 108 293T cells were infected with the TAP-NSs virus at an MOI of 5 and lysed after overnight incubation. A recombinant RVFV encoding the EGFP gene fused with a C-terminal TAP tag served as a negative control. TAP tag-purified protein complexes were analyzed by one-dimensional (1D) gel liquid chromatography-mass spectrometry. The purified TAP-NSs complexes contained several significant NSs interactors, among them a poorly characterized F-box protein termed FBXO3 (Fig. 2). F-box proteins are components of the so-called SCF ubiquitin ligases, multisubunit complexes that are named after their main components, Skp1, cullin, and an F-box protein. Mammals have around 70 different F-box proteins (29). The F-box protein component constitutes the variable part of the SCF ubiquitin ligase and directs the complex to specific substrates. The signature F-box motif connects to the Skp1 subunit, and the ubiquitination substrate is recruited through another protein-protein interaction domain, which is situated at the C terminus. According to their type of C-terminal interaction domain, F-box proteins are classified as either FBXW (containing WD40 repeats), FBXL (containing leucine-rich repeats), and FBXO (for “other”). Since FBXO3 was the only host cell interactor with a connection to the ubiquitin-proteasome system (UPS), we decided to further investigate its potential link to the action of RVFV NSs.
FIG 2.
Host cell interactors of RVFV NSs. Interactors of RVFV NSs protein were identified by affinity purification followed by mass spectrometry (AP-MS) using RVFV NSs as bait and as published in reference 22. Network representation shows interactions between NSs and AP-MS identified proteins (preys). Known direct protein interactions between preys are also shown (22). The edge thickness represents the number of spectra identified in the RVFV NSs precipitates (1, thin edge; >1, thick edge). The size of the symbols (nodes) indirectly correlates with numbers of identifications in the published data set, i.e., big symbols show proteins uniquely identified in RVFV NSs precipitates, whereas smaller symbols indicate identification with RVFV NSs as well as other (non-NSs) viral open reading frames. Red, presence of domains suggesting involvement in ubiquitin-dependent degradation (i.e., RING or FBX domains) based on SMART domain annotations; yellow, all other proteins.
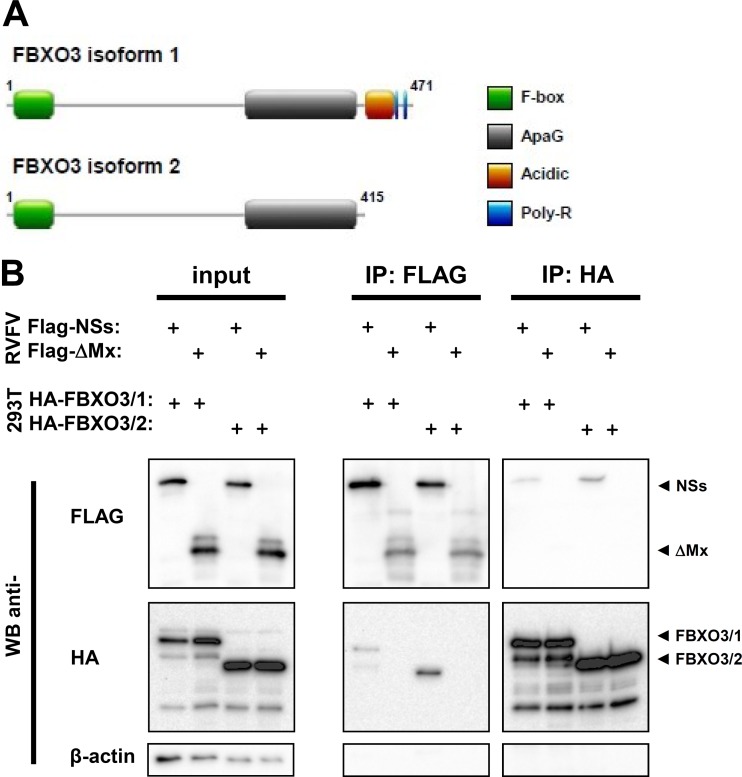
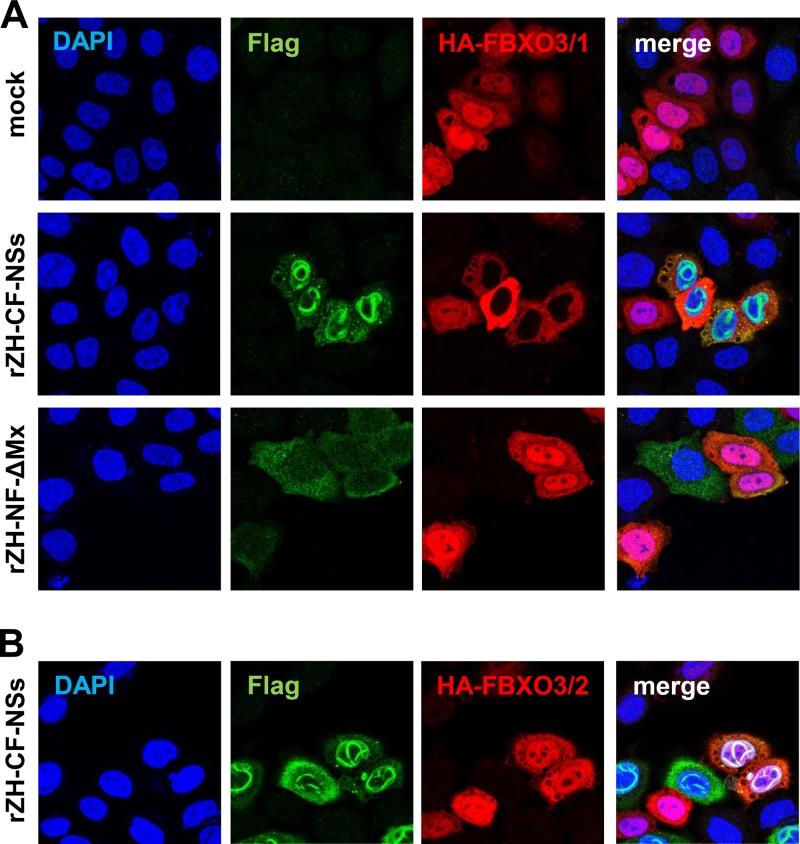
Human FBXO3 is 471 amino acids (aa) long, with several domains (Fig. 3A). Besides the full-length protein (NCBI sequence NM_012175), a shorter splice variant is predicted (NCBI sequence NM_033406), lacking the C-terminal acidic domain and poly(R) region. Due to a paucity of appropriate antisera to detect endogenous FBXO3, we cloned both the full-length protein (termed isoform 1; FBXO3/1) and the shorter isoform (isoform 2; FBXO3/2) and equipped them with an N-terminal HA epitope tag. In a first set of experiments, we confirmed the interaction with NSs. Stable 293 FLP-IN TREX cell lines were generated, containing an expression cassette for the HA-tagged FBXO3 isoforms under the control of a doxycycline-inducible promoter. The 293 FLP-IN TREX cells were treated with DOX to induce FBXO3 expression and then infected with a recombinant RVFV encoding the NSs of wt strain ZH548 equipped with a C-terminal Flag tag (Flag-NSs). As a control, we employed a recombinant RVFV encoding a Flag-tagged, biologically inactive fragment of the human MxA protein (Flag-ΔMx). Three hours after infection, cells were treated with MG132 and incubated for another hour. Cells were lysed and subjected to immunoprecipitations using antibodies against the epitope tags. Both the cell lysates and the immunoprecipitates were analyzed by immunoblotting, using epitope tag-specific antibodies. The Western blot of the cell lysates shows that all investigated proteins are present at similar, detectable levels (Fig. 3B, left panel). The Western blots of the immunoprecipitates show that NSs was able to bind both isoforms of FBXO3, whereas the ΔMx control was unable to do so (Fig. 3B, middle and right panels). These results thus confirm the findings of the proteomics study (22) and establish the F-box protein FBXO3 as a host cell interactor of RVFV NSs.
FIG 3.
Interaction of FBXO3 with RVFV NSs. (A) Cartoon depicting the domain structure of FBXO3 isoforms (UniProt accession number Q9UK99), drawn by using the image creator MyDomains of the ExPASy website (45). (B) Immunoprecipitation. 293 FLP-IN TREX cells expressing SII-HA-tagged FBXO3/1 or FBXO3/2 were induced with doxycycline for 2 days and then infected with rZH-CF-NSs or rZH-NF-ΔMx (MOI of 3). Three hours p.i., MG132 was added to the medium, cells were incubated for another hour, lysed, and subjected to immunoprecipitation using the indicated antibodies.
FBXO3 is important for p62 degradation and IFN antagonism by NSs.
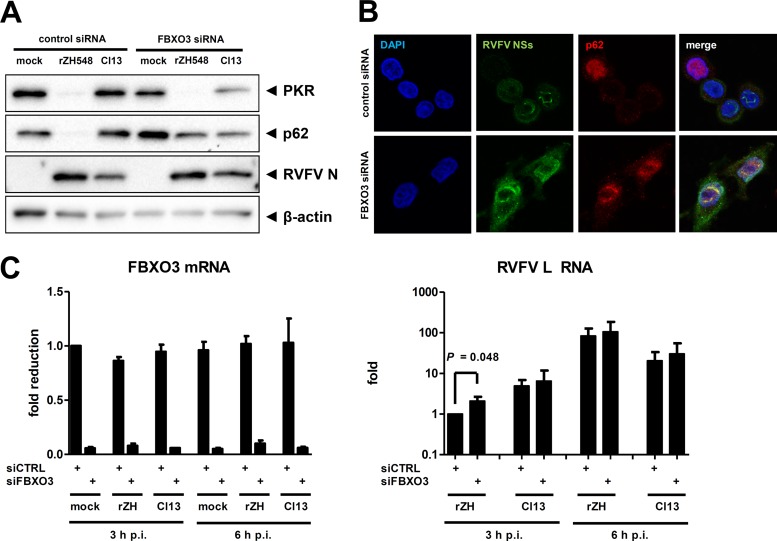
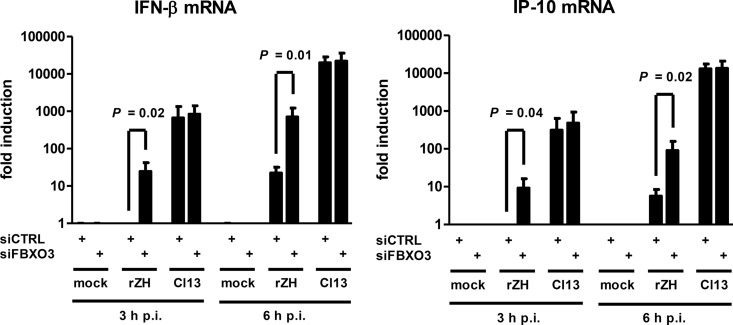
To clarify the contribution of FBXO3 to the UPS-dependent activities of NSs, we employed an siRNA knockdown approach. The Western blot analysis shown in Fig. 4A demonstrates that abrogation of FBXO3 expression had no influence on the NSs-driven destruction of PKR but prevented the degradation of p62. The degradation of p62 after infection with wt RVFV could also be seen in immunofluorescence experiments, and knockdown of FBXO3 rescued p62 levels (Fig. 4B). Moreover, in the absence of FBXO3 there is a colocalization of p62 with the typical filamentous structures formed by RVFV NSs, indicating a close proximity. Real-time RT-qPCR analyses demonstrated an FBXO3 mRNA knockdown efficiency of approximately 90% (Fig. 4C, left panel). FBXO3 itself does not have a major influence on RVFV RNA synthesis (Fig. 4C, right panel) or virus replication (data not shown). Importantly, however, the abrogation of FBXO3 expression (and hence preservation of p62) increased IFN-β induction in response to wt RVFV by a factor of 25 to 30 (Fig. 5, left panel). Similarly, wt RVFV-triggered expression of the antiviral chemokine IP-10 is elevated 9- to 16-fold under conditions of FBXO3 knockdown (Fig. 5, right panel). IFN and IP-10 induction in response to the NSs-truncated Clone 13, in contrast, was not influenced by FBXO3. This confirms that the increase of IFN induction under FBXO3-depleted conditions is not an unspecific effect but is connected to NSs. Further, reducing FBXO3 levels is sufficient to partially alleviate the anti-IFN activity of NSs. These data demonstrate that the E3 ubiquitin ligase component FBXO3 is necessary for the NSs-mediated destruction of p62, whereas PKR is degraded by another pathway. The NSs-driven elimination of p62 via FBXO3 contributes to the suppression of innate immunity by RVFV.
FIG 4.
Effect of FBXO3 on p62 degradation by RVFV NSs. (A) Western blot analysis of knockdown cells. Human A549 cells were treated with siRNA directed against the FBXO3 mRNA or with a control siRNA. Cells were then infected with wt RVFV (rZH548) or Clone 13 (Cl13) at an MOI of 10 and 6 h later analyzed for the presence of PKR, p62, RVFV N, and β-actin using Western blot analysis. (B) Immunofluorescence analysis. siRNA knockdown of FBXO3 mRNA was achieved in HeLa cells as described for panel A. Cells were infected for 6 h with a recombinant RVFV expressing Flag-tagged NSs (rZH-CF-NSs), and the presence of p62 and Flag-RVFV NSs was analyzed 6 h later using appropriate antibodies. DAPI was used for counterstaining the nuclei. (C) Real-time RT-PCR of the knockdown cells shown in panel A. Cells were treated and infected as described for panel A and analyzed for the presence of RNAs for FBXO3 (left panel) and RVFV L (right panel) at 3 and 6 h postinfection. Mean values and standard deviations of 3 independent experiments are shown. A two-tailed, paired t test using log-transformed data was used to compare the siRNA pairs.
FIG 5.
Effect of FBXO3 on IFN suppression by RVFV NSs. Real-time RT-PCR of knockdown cells shown in Fig. 4A and C, analyzed for the presence of IFN-β mRNA and IP-10 mRNA at 3 and 6 h postinfection. A two-tailed, paired t test using log-transformed data was used to compare the siRNA pairs.
We observed that the increased IFN induction in FBXO3-depleted cells does not impair RVFV replication (Fig. 4C, right panel). Most likely, this is because NSs still destroys PKR, one of the major IFN effectors against RVFV (Fig. 4A).
RVFV NSs removes the long isoform of FBXO3 from the nucleus.
We investigated the fate of FBXO3 in cells infected with RVFV. In uninfected cells, the full-length isoform 1 of FBXO3 is concentrated in the nucleus but also present in the cytoplasm (Fig. 6A, upper panels). However, in cells infected with wt RVFV, the nuclear form is no longer detectable, whereas the cytoplasmic pool is unchanged (Fig. 6A, middle panels). This phenomenon is caused by NSs expression, since the mutant RVFV strain with the control protein ΔMx in lieu of NSs maintained nuclear FBXO3 (Fig. 6A, lower panels). Employing the nuclear export inhibitor leptomycin B could not rescue nuclear FBXO3/1 in RVFV-infected cells (data not shown). Moreover, applying the host cell transcription inhibitor alpha-amanitin could not mimic the effect of NSs on nuclear FBXO3/1 (data not shown). This suggests that the selective disappearance of full-length FBXO3 from the nucleus is neither caused by nuclear expulsion nor an unspecific consequence of the massive transcriptional inhibition imposed by NSs. Interestingly, the disappearance of nuclear FBXO3 does not occur in the case of the short isoform FBXO3/2 (Fig. 6B). Thus, the interaction and functional importance of FBXO3 for RVFV NSs function are reflected by a disappearance of nuclear full-length FBXO3.
FIG 6.
Influence of RVFV NSs on FBXO3 isoforms. (A) Long isoform FBXO3/1. HeLa cells were transfected with 0.5 μg of cDNA expression construct pcDNA3.1-3xHA-FBXO3/1. After overnight incubation, cells were infected at an MOI of 10 with recombinant RVFV expressing either Flag-tagged NSs (rZH-CF-NSs) or the negative-control protein ΔMx (rZH-NF-ΔMx) or were left uninfected (mock). At 6 h p.i., cells were fixed, permeabilized, and double immunostained for the Flag (green) and the HA (red) epitopes. (B) Short isoform FBXO3/2. HeLa cells were transfected with the cDNA expression construct pcDNA3.1-3xHA-FBXO3/2 and infected and immunostained as described for panel A.
Degradation of p62 is dependent on Skp1.
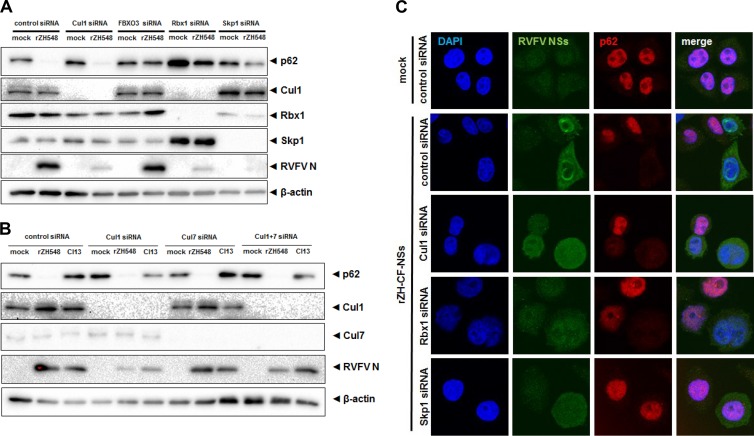
E3 ubiquitin ligases of the SCF type typically consist of an F-box protein (which defines the substrate specificity), the linker protein Skp1, the scaffold protein cullin 1, and the docking factor Rbx1, which connects the complex to an E2 ubiquitin conjugase (29). We wanted to determine whether any of the other SCF components is involved in FBXO3-related NSs action. For this purpose, we knocked down individual SCF components using siRNA and monitored levels of p62 in infected cells. A Western blot analysis is shown in Fig. 7A. Surprisingly, knockdown of cullin 1 expression could not protect p62 from degradation by NSs, although it reduced RVFV infection to some extent. Knockdown of cullin 7, which sometimes acts as an alternative to cullin 1 (29), also had no effect, neither alone nor in combination with a cullin 1 knockdown (Fig. 7B). In contrast, siRNA knockdown of FBXO3, Rbx1, and Skp1 seemed to rescue p62 levels in wt RVFV-infected cells (Fig. 7A). However, the abrogation of Rbx1 and Skp1 expression resulted in a diminished RVFV infection. Therefore, it remained difficult to distinguish whether Rbx1 and Skp1 are indeed required for p62 degradation or whether RVFV infection is simply too inefficient for a proper NSs effect. To obtain a clearer picture, we analyzed the degradation of p62 on the single-cell level (Fig. 7C). Knockdown of any of the three basic SCF components, but especially Rbx1, had a strong impact on cell morphology. Nuclei and cells were rounded and enlarged, and the increased presence of twin nuclei may hint to possible cell division problems. Only low levels of NSs were detectable. Nonetheless, in cells deficient in cullin 1 or Rbx1, expression of NSs resulted in a disappearance of the p62 signal. In cells deficient in Skp1, by contrast, NSs-positive cells also exhibited a p62 signal. Thus, only Skp1 (but neither cullin 1, nor cullin 7, nor Rbx1) appears to be essential for p62 degradation by NSs.
FIG 7.
NSs and components of canonical SCF ubiquitin ligases. (A and B) Western blot analysis of knockdown cells. HeLa cells were treated with a control siRNA or with siRNAs directed against mRNAs of cullin 1 (Cul1), FBXO3, Rbx1, or Skp1 (A) or cullin 1 and/or cullin 7 (B). Cells were then infected with recombinant wt RVFV (rZH548) or Clone 13 (Cl13) at an MOI of 10 and analyzed 6 h later for the presence of the indicated proteins. (C) Immunofluorescence analysis. HeLa cells with siRNA knockdown of cullin 1, Rbx1, or Skp1 mRNAs were infected for 6 h with rZH-CF-NSs at an MOI of 3, and the presence of nuclear DNA, p62, and Flag-RVFV NSs was analyzed 6 h later using DAPI stain and appropriate antibodies, respectively.
In summary, our collective data demonstrate that the NSs of RVFV drives the degradation of the general transcription factor p62 by means of the F-box protein FBXO3 and the linker protein Skp1. The rapid destruction of p62 substantially contributes to the block in IFN and cytokine induction observed in wt RVFV-infected host cells.
DISCUSSION
RVFV is an eminent pathogen for humans and animals alike. The major virulence factor of RVFV is the nonstructural protein NSs, a strong suppressor of the innate immune responses. NSs is a multifunctional protein that recruits the transcriptional repressor SAP30 to the IFN promoter (15), depletes the general transcription factor TFIIH from its subunit p44 (16), and degrades the TFIIH subunit p62 (16, 21) and the antiviral effector PKR (25, 28). The aim of our study was to identify the host cell factors that are engaged by NSs. Our data show that one aspect of destructive NSs action, the degradation of the TFIIH subunit p62, is mediated by the F-box protein FBXO3.
FBXO3 knockdown experiments demonstrated that the protection of p62 from NSs action partially relieved RVFV-mediated IFN suppression. To our knowledge, this is the first example of an NSs-interacting host cell partner that was positively demonstrated to be essential for NSs action. The study on the p44 host cell partner of NSs deduced its conclusions exclusively on the interaction data and on in vitro assays (16). The study on the SAP30 partner of NSs additionally demonstrated a correlation between the inability of the NSs Δ210-230 mutant to interact with SAP30 and the inability to inhibit IFN induction (15). Meanwhile, however, it is known that most mutations of NSs lead to a complete loss of function (30). In fact, Head et al. had demonstrated that NSs mutants deleted at aa 6 to 30, 31 to 55, 56 to 80, 81 to 105, 106 to 130, 131 to 155, 156 to 180, 181 to 205, 206 to 230, 231 to 248, and 249 to 265 all were unable to inhibit IFN induction or to degrade PKR. This indicates that the null phenotype of a certain NSs mutant alone is difficult to interpret, as it may affect stability or correct folding (30). In our study, in contrast, we show that the absence of the interactor FBXO3 partially neutralizes the strong block of IFN induction imposed by wt NSs whereas the PKR degradation function of NSs remains intact. We are therefore confident that FBXO3 is a crucial and specific interactor of NSs that is engaged to degrade p62 and block IFN transcription.
Compared to the deletion of NSs from the RVFV genome, siRNA depletion of FBXO3 was approximately 1 order of magnitude less efficient in rescuing IFN induction. This incomplete restoration of IFN induction, combined with the untouched PKR degradation activity, most likely is responsible for the absence of a measurable impact of FBXO3 depletion on RVFV growth. Several reasons are possible for the residual IFN suppression under conditions of FBXO3 depletion. First, it is possible that the siRNA treatment did not completely remove FBXO3 from the cells. Second, the binding of NSs to p62 (21) (Fig. 4B) to the TFIIH subunit p44 (16) and to SAP30 (15) also contributes to IFN inhibition. Third, we showed that FBXO3 depletion does not protect from PKR degradation by NSs. Since PKR is also involved in cytokine induction (31–33), the still-intact destruction of PKR by NSs may further contribute to an incomplete recovery of IFN induction in FBXO3-depleted cells. Most likely, it is the combination of all these effects that results in the remaining IFN suppression in RVFV-infected cells devoid of FBXO3.
Our study revealed the involvement of the SCF cofactor Skp1 in p62 degradation by NSs, whereas, somewhat surprisingly we failed to assign any role to the other cofactors, cullin 1, cullin 7, or Rbx1. Since siRNA depletion does not result in a complete removal of the target protein, we cannot exclude that the residual amounts of these cofactors are sufficient for the NSs mechanism. However, in the standard model for SCF E3 ligase F-box proteins, Skp1, cullins, and Rbx1 form an equimolar, stoichiometric complex (29). Moreover, both Skp1 and cullin 1 were previously identified as interactors of FBXO3 (34). It appears hence unusual that siRNA depletion of cullin 1/cullin 7 or Rbx1 does not preserve p62 from destruction by NSs. In fact, however, there are precedents of Skp1/F-box protein-only complexes that act on their own, i.e., independently of a canonical SCF-type ubiquitin ligase (35, 36). An alternative explanation therefore could be that NSs assembles a so-called “non-SCF F-Box/Skp1 complex” to degrade p62.
FBXO3 is an E3 ubiquitin ligase component that only recently gained wider attention. An earlier study suggested an involvement in transcription regulation by p53, the master regulator of apoptosis and DNA damage responses (34). TFIIH-p62, by contrast, seems to be a target of FBXO3 only in connection with RVFV NSs, as in uninfected cells neither depletion nor overexpression of FBXO3 had any effect on p62 levels. Strikingly, a recent study revealed that FBXO3 is indirectly stabilizing TRAF (tumor necrosis factor receptor-associated factor) proteins 1 to 6 (37). TRAF proteins are essential signaling adaptors for innate, inflammatory, and adaptive immune responses (38). FBXO3, through regulation of TRAF protein stability, turned out to be a key factor for production of inflammatory cytokines (37). Thus, the NSs interactor FBXO3 plays a key role in p53-mediated host cell regulation as well as in enabling TRAF-dependent inflammatory responses. Interestingly, RVFV NSs activates p53, apoptosis, and DNA damage responses (39, 40) and downregulates inflammatory cytokines (41). With respect to TRAF regulation, however, we observed that RVFV drives TRAF disappearance in an FBXO3-independent manner (data not shown), indicating an alternative, dominant pathway of counteracting inflammation.
The nuclear pool of the long isoform FBXO3/1 is removed by NSs, whereas the shorter isoform, which lacks the C-terminal acidic and poly(R)-rich domains, is not affected. It is known that F-box proteins can fall victim to their own ubiquitination activity (42) and become degraded along with their target protein (“unstable when active” phenomenon [35]). Thus, the disappearance of FBXO3/1 along with p62 could point at the C terminus of FBXO3/1 as the effector domain of NSs action, leading also to an autocatalytic degradation of FBXO3/1. In any case, the removal of FBXO3 from the nucleus may have implications for NSs regulation, liberating NSs to pursue another activity such as, e.g., PKR degradation. Interestingly, a recent chromatin immunoprecipitation-DNA sequencing (CHIP-Seq) study revealed that RVFV NSs also downregulates the promoter of the FBXO3 gene (43). Although in our experiments mRNA levels of FBXO3 were unaffected by NSs at 6 h postinfection (p.i.), it could be speculated that NSs blocks FBXO3 gene expression for the long term, for the same reason that it removes FBXO3 protein from the nucleus. Investigating the global consequences of FBXO3 removal could reveal novel insights into the functions of both FBXO3 and NSs.
In addition to the effects on IFN induction, host transcription, inflammatory cytokine production, p53 regulation, and PKR degradation, RVFV NSs induces chromosomal anomalies (44) and regulates many other genes involved in innate immunity, inflammation, cell adhesion, axonal guidance, development, and blood coagulation (43). Thus, NSs of RVFV is a multifunctional protein that dysregulates and damages the host at several levels. However, an NSs-truncated RVFV strain (Clone 13) kills mice even faster than the wt strain does, provided the animals are devoid of an IFN system (11). This argues for IFN antagonism (encompassing the blockade of IFN mRNA transcription and the degradation of the PKR) being the prime function of NSs, to which our newly identified host cell factor FBXO3 contributes substantially.
ACKNOWLEDGMENTS
We thank Christina Lückel for technical assistance and Eliane Meurs for kindly donating the anti-PKR antibody.
Work in our laboratories was supported by the Deutsche Forschungsgemeinschaft grants We 2616/5-2, SFB 593, and SFB 1021 to F.W., grant PI 1084/2-1 to A.P., an Alexander von Humboldt stipend to M.H., and an ERC starting grant (iViP) to A.P.
Footnotes
Published ahead of print 8 January 2014
REFERENCES
- 1.Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. 2009. Rift Valley fever virus. J. Am. Vet. Med. Assoc. 234:883–893. 10.2460/javma.234.7.883 [DOI] [PubMed] [Google Scholar]
- 2.Boshra H, Lorenzo G, Busquets N, Brun A. 2011. Rift valley fever: recent insights into pathogenesis and prevention. J. Virol. 85:6098–6105. 10.1128/JVI.02641-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikegami T, Makino S. 2011. The pathogenesis of Rift Valley fever. Viruses 3:493–519. 10.3390/v3050493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. 2007. Rift Valley fever outbreak—Kenya, November 2006-January 2007. MMWR Morb. Mortal. Wkly. Rep. 56:73–76 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5604a3.htm [PubMed] [Google Scholar]
- 5.Fontenille D, Traore-Lamizana M, Diallo M, Thonnon J, Digoutte JP, Zeller HG. 1998. New vectors of Rift Valley fever in West Africa. Emerg. Infect. Dis. 4:289–293. 10.3201/eid0402.980218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moutailler S, Krida G, Schaffner F, Vazeille M, Failloux AB. 2008. Potential vectors of Rift Valley fever virus in the Mediterranean region. Vector Borne Zoonotic Dis. 8:749–753. 10.1089/vbz.2008.0009 [DOI] [PubMed] [Google Scholar]
- 7.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61. 10.1051/vetres/2010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, Jahrling PB, Ksiazek T, Johnson KM, Meyerhoff A, O'Toole T, Ascher MS, Bartlett J, Breman JG, Eitzen EM, Jr, Hamburg M, Hauer J, Henderson DA, Johnson RT, Kwik G, Layton M, Lillibridge S, Nabel GJ, Osterholm MT, Perl TM, Russell P, Tonat K. 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391–2405. 10.1001/jama.287.18.2391 [DOI] [PubMed] [Google Scholar]
- 9.Bouloy M, Weber F. 2010. Molecular biology of rift valley Fever virus. Open Virol. J. 4:8–14. 10.2174/1874357901004020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806. 10.1128/JVI.78.18.9798-9806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371–1377. 10.1128/JVI.75.3.1371-1377.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568. 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott RM, Weber F. 2009. Bunyaviruses and the type I interferon system. Viruses 1:1003–1021. 10.3390/v1031003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollidge BS, Weiss SR, Soldan SS. 2011. The role of interferon antagonist, non-structural proteins in the pathogenesis and emergence of arboviruses. Viruses 3:629–658. 10.3390/v3060629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. 10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le May N, Dubaele S, De Santis LP, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. 10.1016/S0092-8674(04)00132-1 [DOI] [PubMed] [Google Scholar]
- 17.Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. 2011. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J. Biol. Chem. 286:27278–27287. 10.1074/jbc.M111.256974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815–819. 10.1038/266815a0 [DOI] [PubMed] [Google Scholar]
- 19.Overby AK, Popov VL, Niedrig M, Weber F. 2010. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84:8470–8483. 10.1128/JVI.00176-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, Garcia-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. 10.1016/j.chom.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of Rift Valley fever virus promotes posttranslational downregulation of the TFIIH subunit p62. J. Virol. 85:6234–6243. 10.1128/JVI.02255-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Burckstummer T, Muller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G. 2012. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487:486–490. 10.1038/nature11289 [DOI] [PubMed] [Google Scholar]
- 23.Habjan M, Penski N, Spiegel M, Weber F. 2008. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J. Gen. Virol. 89:2157–2166. 10.1099/vir.0.2008/002097-0 [DOI] [PubMed] [Google Scholar]
- 24.Laurent AG, Krust B, Galabru J, Svab J, Hovanessian AG. 1985. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc. Natl. Acad. Sci. U. S. A. 82:4341–4345. 10.1073/pnas.82.13.4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375. 10.1128/JVI.02148-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 27.Bird BH, Bawiec DA, Ksiazek TG, Shoemaker TR, Nichol ST. 2007. Highly sensitive and broadly reactive quantitative reverse transcription-PCR assay for high-throughput detection of Rift Valley fever virus. J. Clin. Microbiol. 45:3506–3513. 10.1128/JCM.00936-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287. 10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skaar JR, Pagan JK, Pagano M. 2013. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14:369–381. 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Head JA, Kalveram B, Ikegami T. 2012. Functional analysis of Rift Valley fever virus NSs encoding a partial truncation. PLoS One 7:e45730. 10.1371/journal.pone.0045730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811. 10.1016/j.biochi.2007.03.001 [DOI] [PubMed] [Google Scholar]
- 32.McAllister CS, Taghavi N, Samuel CE. 2012. Protein kinase PKR amplification of interferon beta induction occurs through initiation factor eIF-2alpha-mediated translational control. J. Biol. Chem. 287:36384–36392. 10.1074/jbc.M112.390039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. 10.1371/journal.pone.0043031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I. 2008. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell. Biol. 28:7126–7138. 10.1128/MCB.00897-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermand D. 2006. F-box proteins: more than baits for the SCF? Cell Division 1:30. 10.1186/1747-1028-1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skaar JR, D'Angiolella V, Pagan JK, Pagano M. 2009. SnapShot: F box proteins II. Cell 137:1358, 1358.e1. 10.1016/j.cell.2009.05.040 [DOI] [PubMed] [Google Scholar]
- 37.Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, Zou C, Ellis B, Sciurba FC, Zhang Y, Mallampalli RK. 2013. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 14:470–479. 10.1038/ni.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradley JR, Pober JS. 2001. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20:6482–6491. 10.1038/sj.onc.1204788 [DOI] [PubMed] [Google Scholar]
- 39.Austin D, Baer A, Lundberg L, Shafagati N, Schoonmaker A, Narayanan A, Popova T, Panthier JJ, Kashanchi F, Bailey C, Kehn-Hall K. 2012. p53 Activation following Rift Valley fever virus infection contributes to cell death and viral production. PLoS One 7:e36327. 10.1371/journal.pone.0036327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baer A, Austin D, Narayanan A, Popova T, Kainulainen M, Bailey C, Kashanchi F, Weber F, Kehn-Hall K. 2012. Induction of DNA damage signaling upon Rift Valley fever virus infection results in cell cycle arrest and increased viral replication. J. Biol. Chem. 287:7399–7410. 10.1074/jbc.M111.296608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McElroy AK, Nichol ST. 2012. Rift Valley fever virus inhibits a pro-inflammatory response in experimentally infected human monocyte derived macrophages and a pro-inflammatory cytokine response may be associated with patient survival during natural infection. Virology 422:6–12. 10.1016/j.virol.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galan JM, Peter M. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. U. S. A. 96:9124–9129. 10.1073/pnas.96.16.9124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benferhat R, Josse T, Albaud B, Gentien D, Mansuroglu Z, Marcato V, Soues S, Le Bonniec B, Bouloy M, Bonnefoy E. 2012. Large-scale chromatin immunoprecipitation with promoter sequence microarray analysis of the interaction of the NSs protein of Rift Valley fever virus with regulatory DNA regions of the host genome. J. Virol. 86:11333–11344. 10.1128/JVI.01549-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. 2010. Nonstructural NSs protein of Rift Valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J. Virol. 84:928–939. 10.1128/JVI.01165-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40:W597–W603. 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]