Abstract
Cells are equipped with pattern recognition receptors (PRRs) such as the Toll-like and RIG-I-like receptors that mount innate defenses against viruses. However, viruses have evolved multiple strategies to evade or thwart host antiviral responses. Viral inclusion bodies (IBs), which are accumulated aggregates of viral proteins, are commonly formed during the replication of some viruses in infected cells, but their role in viral immune evasion has rarely been explored. Severe fever with thrombocytopenia syndrome (SFTS) is an emerging febrile illness caused by a novel phlebovirus in the Bunyaviridae. The SFTS viral nonstructural protein NSs can suppress host beta interferon (IFN-β) responses. NSs can form IBs in infected and transfected cells. Through interaction with tank-binding kinase 1 (TBK1), viral NSs was able to sequester the IKK complex, including IKKε and IRF3, into IBs, although NSs did not interact with IKKε or IRF3 directly. When cells were infected with influenza A virus, IRF3 was phosphorylated and active phosphorylated IRF3 (p-IRF3) was translocated into the nucleus. In the presence of NSs, IRF3 could still be phosphorylated, but p-IRF3 was trapped in cytoplasmic IBs, resulting in reduced IFN-β induction and enhanced viral replication. Sequestration of the IKK complex and active IRF3 into viral IBs through the interaction of NSs and TBK1 is a novel mechanism for viral evasion of innate immunity.
INTRODUCTION
Viruses have adopted various mechanisms to evade or subvert host antiviral responses initiated by viral RNA or DNA through Toll-like receptor (TLR) (1, 2) and RIG-I-like receptor (RLR) (3–5) signaling pathways. Viral double-stranded RNA (dsRNA) can be sequestered by VP35 of Ebola virus (6) to avoid the activation of TLRs or RLRs. Viral proteins that contain the TIR domain can be interferon (IFN) antagonists such as A46R of vaccinia virus, which blocks upstream IFN signaling by directly targeting MyD88, MAL, TRIF, and TRAM, the TIR domain-containing adaptors (2). RLR sensors such as RIG-I and MDA5 and adaptor IPS1 are targeted by NS1 of influenza virus (7), V protein of paramyxoviruses (3), and the protease precursor protein 3ABC of hepatitis A virus (5). The adaptor protein TRIF of the TLR signaling pathway can be targeted for degradation by viral proteins containing protease activities, such as NS3/4A of flavivirus hepatitis C virus (HCV) (8) and 3C of picornavirus EV71 (9), which can also cleave transcription factor (TF) IRF7, compromising IFN induction. In addition, the N-terminal fragment of cleaved IRF7 interacts with and inhibits IRF3, another TF essential for IFN induction, thereby enhancing viral infection (10). Downstream TFs are also targeted by viral proteins, such as NS1 of West Nile virus, which inhibits TLR3 signaling by preventing IRF3 translocation into the nucleus (11). Between TLR/RLR sensors and downstream TFs, IKK complexes are also targeted by proteins encoded by HCV, vaccinia virus, and rabies virus (12–14), among others.
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging febrile illness caused by a novel phlebovirus, SFTS bunyavirus (SFTSV), in the family Bunyaviridae (15). SFTSV is a vector-borne zoonotic arbovirus transmitted to humans by tick bites, causing high fever, loss of white blood cells and platelets, and, in severe cases, multiorgan failure. It is a single-stranded negative-sense RNA virus with three genomic segments, L, M, and S (15). The L segment encodes viral RNA polymerase, while the M segment encodes the two viral envelope glycoproteins, Gc and Gn. The S segment is an ambisense RNA of 1,744 bases encoding a nucleoprotein (NP) and a nonstructural protein (NSs). Our recent study demonstrated that NSs inhibits host IFN-β and NF-κB responses, although the mechanism remains unclear (16). Here we report that NSs forms unique cytoplasmic inclusion bodies (IBs) in infected or transfected cells that appear to play a role in immune evasion through the interaction of NSs and the IKK complex. Our data indicate that SFTSV has adopted a novel host immune evasion strategy involving the sequestration of IKK complex components TBK1, IKKε, and IRF, which are critical for the activation of the IFN signaling pathway, into IBs.
MATERIALS AND METHODS
Cells, viruses, and reagents.
African green monkey kidney Vero, HeLa, human embryonic kidney 293T, and MDCK cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Thermo Scientific, Logan, UT), 1 mM sodium pyruvate (HyClone), and 1% antibiotic-antimycotic solution (Gibco). Cells were cultured at 37°C under 5% CO2. A previously described SFTSV strain, JS-2010-014 (16), and the influenza virus strain A/Nanjing/NJU-108/2009 (here referred to as H1N1pdm) (17) were used in this study. All viral aliquots were stored at −80°C. Antibodies for β-tubulin, fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse IgG, and rhodamine (tetramethyl rhodamine isocyanate [TRITC])-conjugated goat anti-rabbit IgG were purchased from Cell Signaling Technology (Danvers, MA). Anti-Flag M2 antibody and Nile Red were obtained from Sigma-Aldrich (St. Louis, MO). Anti-β-actin antibody and antihemagglutinin (anti-HA) antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Construction of plasmids.
Plasmid pRK5-NSs has been described previously (16). cDNAs of NSs, TBK1, and IRF3 were individually subcloned into pEGFP-N3, pRK5-HA, and pRK5-Flag for the expression of an enhanced green fluorescent protein (EGFP)-NSs fusion and other tagged proteins. Either HeLa cells or HEK293T cells were transfected with various plasmids in equal quantity with Lipofactamine 2000 reagents (Invitrogen) for protein expression. Empty plasmids were used as transfection controls. Truncated mutants of NSs, which included NSs1-160, NSs66-205, and NSs66-249, were generated through PCR with specific primers. The resultant cDNA was cloned into pRK5 for transfection.
Immunoprecipitation and immunoblot analysis.
Cell lysates of transfected or cotransfected cells were incubated with either specific or control antibodies at room temperature (RT) for 2 h and then precipitated with protein A/G beads. After incubation, beads were washed four times with lysis buffer and the immunoprecipitates were electrophoresed on SDS-polyacrylamide gels and then transferred onto Immuno-blot polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) for primary antibody incubation overnight. Alkaline phosphatase (AP)- or horseradish peroxidase (HRP)-conjugated secondary antibodies were used for further incubation with the membranes for 90 min and developed by BCIP (5-bromo-4-chloro-3-indolylphosphate)/nitroblue tetrazolium (NBT) or enhanced chemiluminescence (ECL) reagents (Invitrogen).
Immunofluorescence assay and virus titration.
HeLa cells transfected with plasmids containing NSs and/or TBK1 cDNA were washed twice with phosphate-buffered saline (PBS) at various time points, fixed with 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 for 10 min, followed by three washes with PBS. In some experiments, HeLa cells were transfected with plasmids containing cDNA of truncated mutants of NSs or of full-length NSs and grown on coverslips, which were blocked with 5% bovine serum albumin (BSA; Sigma) in PBS for 30 min at 37°C; the cells were then incubated with a primary antibody at 1:50 to 1:100 dilutions at 4°C overnight. After washes with PBS-Tween 20 (PBST), cells were incubated with FITC-conjugated donkey anti-mouse IgG(H+L) or TRITC-conjugated rabbit IgG(H+L) at a dilution of 1:200 at 37°C for 1 h, washed, and stained with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml). After washes, the coverslips were analyzed under an Olympus confocal microscope. In some cases, the cells were pretransfected with plasmids, then infected 12 h later with H1N1pdm (17) at a multiplicity of infection (MOI) of 1. The cells were then treated as described above for antibody staining and confocal microscopy.
Luciferase reporter assay for IFN-β promoter activity.
HEK293 cells were grown in 24-well tissue culture plates in DMEM with 10% fetal bovine serum (FBS) until 50 to 70% confluence was reached. The cells were transiently transfected with a total of 505 ng of plasmid DNA, consisting of 100 ng of the reporter plasmid pIFN-β-Luc, 5 ng of pRL-SV8 for normalization, and 400 ng of empty control pRK5-HA, pRK5-TBK1, or pRK5-NSs. IFN-β activation was assessed using an IFN-β–luciferase reporter system. Twenty-four hours posttransfection, the cells were further stimulated with 50 ug/ml poly(I·C) for another 8 h. Then, the cells were washed in PBS and lysed in 0.1 ml reporter lysis buffer (Promega, Madison, MI). Firefly luciferase activities were measured by luciferase assay (Promega). Results were expressed as fold changes over the nontransfected controls.
Quantitative RT-PCR.
Two hundred nanograms of total RNA extracted from noninfected or H1N1pdm virus-infected cells was used for reverse transcription (Primescript reagent kit; TaKaRa, Shiga, Japan). Quantitative real-time PCR (RT-PCR) was conducted with 1 μl of cDNA in a total volume of 10 μl with the SYBR premix Ex Taq II (TaKaRa) by following the manufacturer's instructions. Relative expression values were standardized by an internal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control. Fold changes of mRNA transcripts of IFN-β and the viral M gene segment were calculated with the formula 2(ΔCT of gene − ΔCT of GAPDH), where CT is threshold cycle.
Statistical analysis.
For statistical analysis, a two-tailed Student t test was used to evaluate the data (IBM SPSS, Armonk, NY). A χ2 analysis was used to determine the significance of differences between two or more groups, with a 0.05 level of probability (P < 0.05) considered statistically significant.
RESULTS
Cytoplasmic viral inclusion bodies formed in infected cells.
When Vero cells were infected with SFTSV, a unique structure of IBs in the cytoplasm was observed with fluorescence-conjugated rabbit anti-SFTSV immune sera: IBs first appeared as granules and later changed to patches. Specific antibodies to various viral proteins, including NP, NSs, Gc, and Gn, were used to characterize the composition of the unique structure (18–20). The IBs in SFTSV infection were detected only by antibodies to NSs, the nonstructural protein encoded by the S segment (Fig. 1A). A plasmid construct for expressing a fusion protein of NSs and EGFP was prepared and used to transfect HeLa cells. In cells expressing EGFP-NSs, characteristic IBs appeared, which intensified in number and size over time (Fig. 1B), confirming that the component of IBs formed in infected cells was NSs. NSs in IBs could be functional since EGFP-NSs fusion proteins fluoresced, the same as other proteins (21). IBs were also observed in SFTSV-infected cells with electron microscopy (data not shown).
FIG 1.
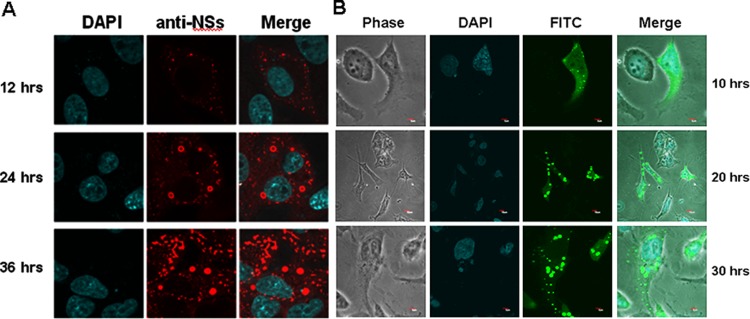
Inclusion bodies (IBs) formed by NSs in infected and transfected cells. (A) Vero cells were infected with SFTSV at an MOI of 1, and the cells were fixed and permeabilized at the indicated time points. The cells were stained with a rabbit anti-NSs antibody at 1:20 dilution in PBS, followed by staining with a TRITC-conjugated anti-rabbit IgG, and subjected to confocal microscopy (magnification, ×400). (B) A cDNA of NSs was cloned into pEGFP, and the plasmid was used to transfect HeLa cells. Transfected cells were subjected to confocal microscopy at the indicated time points.
Interaction of NSs, TBK1, and IKK complex.
We have demonstrated that NSs suppresses IFN-β and NF-κB signaling (16). We found that NSs could be coimmunoprecipitated (co-IP) with tank-binding kinase 1 (TBK1), a key kinase in the activation of IRF3/7 and IFN-β signaling, and that the interaction was specific, as NSs did not interact with IRF3 or IKKε when cells were cotransfected with NSs and IRF3 or IKKε (Fig. 2). We observed that various amounts of the cellular proteins (TBK1, IRF3, and IKKε) could be detected by immunoblotting in the transfected cells with or without NSs, despite the fact that identical plasmid quantities for the cellular proteins were used. We do not think that NSs affects the transcription or translation of the genes of these cellular proteins and would consider that the NSs interaction with these cellular proteins contributes to the amounts of the cellular proteins shown in the immunoblots, which will be further explained below.
FIG 2.
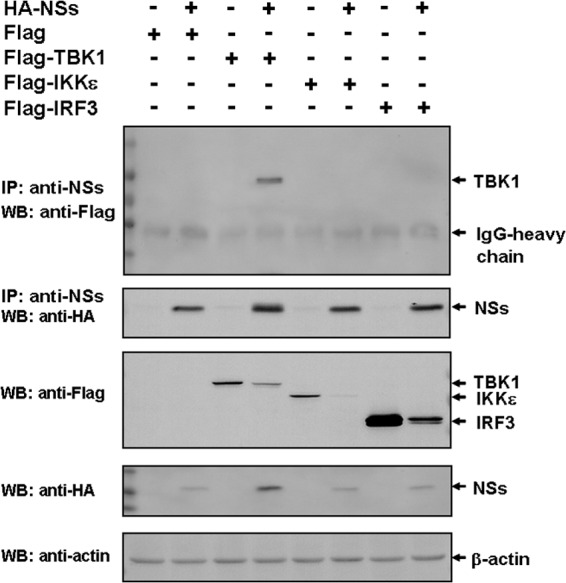
Interaction and coimmunoprecipitation of NSs and TBK1. HEK293 cells were transfected with HA-NSs alone or with Flag-TBK1, Flag-IKKe, or Flag-IRF3. The cell lysates were immunoprecipitated by anti-NSs antibodies, which were subjected to SDS-PAGE. Anti-Flag antibodies were used to blot the membrane (top) to show that TBK1 was the only protein interacting with NSs. The membrane was blotted with anti-NSs antibodies to show the precipitated NSs in the immune complexes (middle). The membrane was also blotted with anti-Flag and anti-HA antibodies to demonstrate the expression of TBK1, IKKε, and IRF3 as well as NSs (middle). IP, immunoprecipitation; WB, Western blotting.
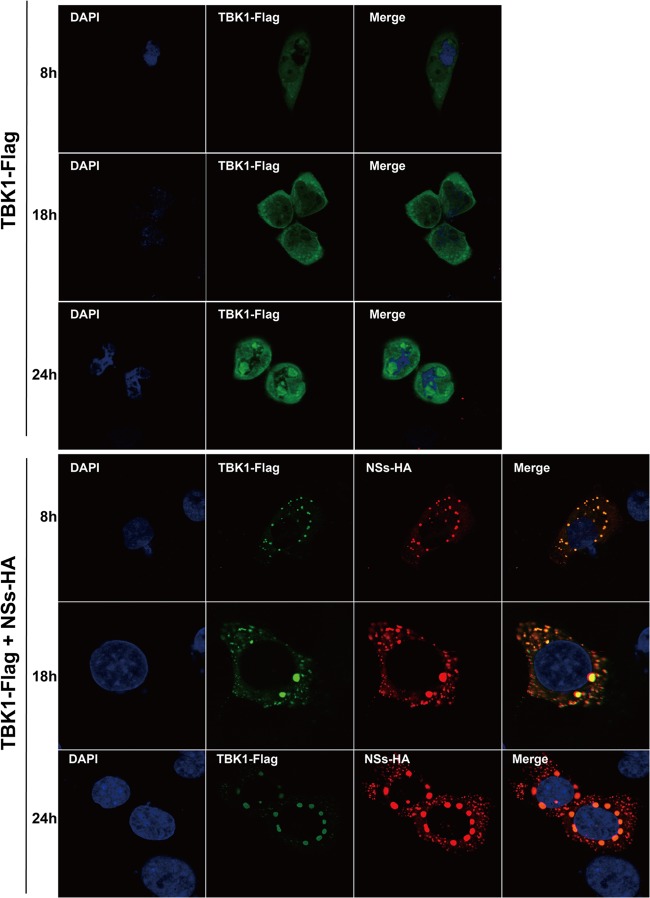
On the other hand, in HeLa cells transfected with Flag-tagged TBK1 (TBK1-Flag), TBK1 was dispersedly distributed in the cytoplasm (Fig. 3, top panels). When the cells were cotransfected with TBK1 and NSs, TBK1 colocalized with NSs and was predominantly found in IBs formed with NSs, as observed by confocal microscopy (Fig. 3, bottom panels). Translocation of TBK1 suggests that viral NSs interacted with and trapped TBK1 in the IBs.
FIG 3.
Colocalization of NSs with TBK1 through direct interaction in IBs. HeLa cells were cotransfected with HA-NSs and Flag-TBK1, and the cells were fixed at various time points and permeabilized before they were stained with anti-HA or anti-Flag antibodies. TBK1 proteins were dispersedly distributed in transfected cells (top), but they were all translocated and colocalized in IBs in the presence of NSs (bottom).
Colocalization of NSs and TBK1 was also observed in the infected cells. HeLa cells were mock infected or infected with SFTSV at an MOI of 1. The cells were stained at various time points postinfection with anti-NSs or anti-TBK1 antibodies. TBK1 was dispersedly distributed in the cytoplasm in noninfected cells (Fig. 4, top). However, TBK1 was relocated into the IBs predominantly in infected cells and colocalized with NSs, which formed the IBs (Fig. 4, middle and bottom), suggesting that endogenous TBK1 was translocated into the IBs probably through direct NSs-TBK1 interaction.
FIG 4.
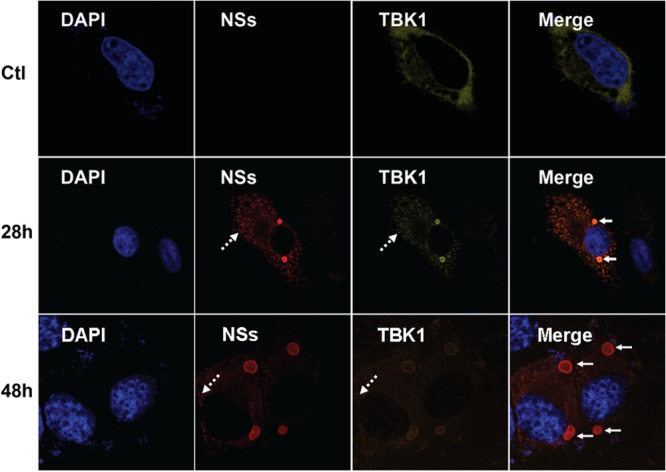
TBK1 colocalized with NSs in the IBs in SFTSV-infected cells. HeLa cells were either mock infected (Ctl) or infected with SFTSV at an MOI of 1 and incubated for 28 or 48 h before they were fixed and permeabilized. The cells were stained with anti-NSs and anti-TBK1 antibodies to detect NSs and TBK1 in infected cells by immunofluorescence. The cells were finally stained with DAPI for the nucleus and subjected to confocal fluorescence microscopy. The solid arrows indicate the IBs, while the dashed arrows show minute dots.
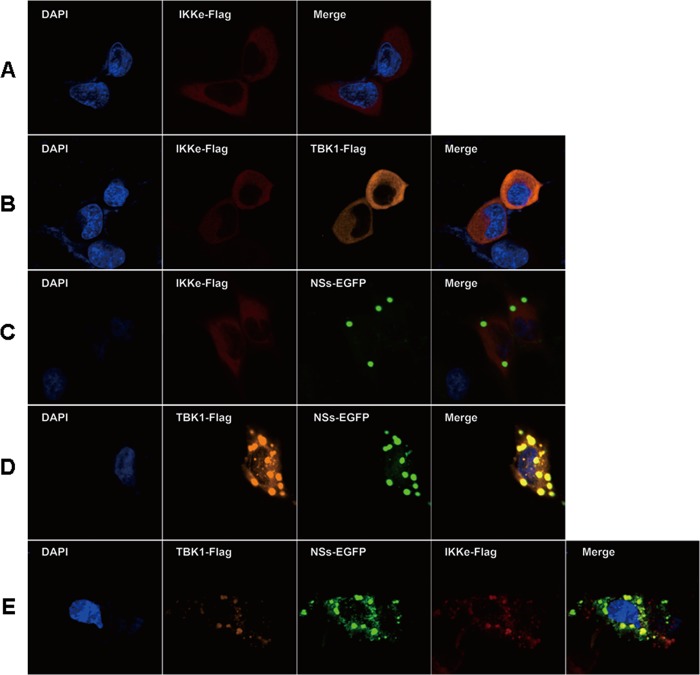
To examine whether IKKε, another component of the IKK complex, is also trapped in NSs-formed IBs, we transfected HeLa cells with IKKε alone or IKKε together with TBK1 and/or NSs (Fig. 5). IKKε was primarily located dispersedly in the cytosol with or without the expression of TBK1 (Fig. 5A and B). Cotransfection of IKKε and NSs did not bring IKKε into IBs (Fig. 5C), which corroborates the previous data indicating that IKKε and NSs did not co-IP or interact directly (Fig. 2). However, when the cells were transfected with IKKε, NSs, and TBK1, IKKε along with TBK1 was translocated into the IBs through their interaction (Fig. 5E), demonstrating that the components of the IKK complex could all be sequestered into the IBs in the presence of NSs.
FIG 5.
IKKε was translocated into IBs through interaction of TBK1 and NSs. HeLa cells were transfected with plasmids expressing Flag-IKKε alone (A), Flag-IKKε and Flag-TBK1 (B), Flag-IKKε and EGFP-NSs (C), Flag-TBK1 and EGFP-NSs (D), or Flag-IKKε, Flag-TBK1, and EGFP-NSs (E). Twenty-four hours posttransfection, the cells were fixed and stained with anti-IKKε (rabbit origin) and anti-TBK1 (goat origin) antibodies. The cells were stained subsequently with fluorescence-conjugated anti-goat or anti-rabbit secondary antibodies, followed by confocal microscopy.
Sequestration of IRF3 in NSs-formed IBs.
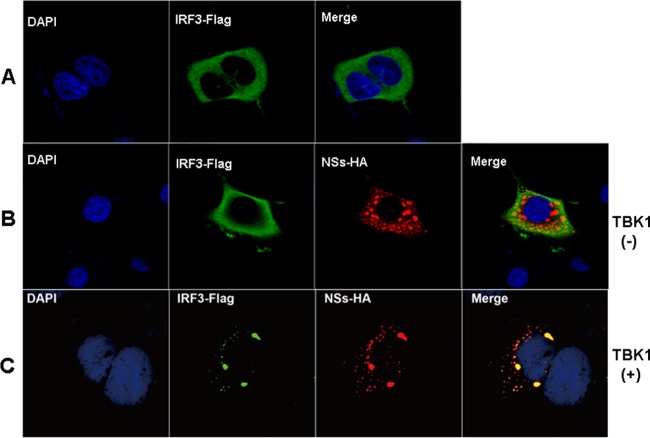
IRF3, as a TF, is dispersed in the cytoplasm in a nonphosphorylated form (Fig. 6A), which is phosphorylated upon activation by TBK1 in response to upstream stimulation (22, 23). In HeLa cells cotransfected with IRF3 and NSs, while NSs eventually migrated to IBs and remained there, IRF3 was diffusely distributed in the cytoplasm (Fig. 6B). However, in cells cotransfected with IRF3, NSs, and TBK1, IRF3 was located in IBs (Fig. 6C), apparently due to its interaction with TBK1. Therefore, NSs may have sequestered IRF3 in the IBs indirectly through the combination of an NSs-TBK1 interaction, as shown in this study, and an association between TBK1 and IRF3, as described previously (22, 23).
FIG 6.
Translocation of IRF3 into IBs in the presence of NSs and TBK1. (A to C) HeLa cells were transfected with Flag-IRF3 or with Flag-IRF3 and NSs with or without TBK1, and the cells were fixed and permeabilized, followed by staining with anti-Flag and/or anti-HA antibodies as well as FITC- or TRITC-conjugated secondary antibodies. The stained cells were subsequently subjected to confocal microscopy. (A) Cells were transfected with Flag-IRF3, which was found in the cytoplasm. (B) Cells were cotransfected with Flag-IRF3 and HA-NSs. While NSs was in IBs, IRF3 dispersed in the cytoplasm. (C) Cells were cotransfected with Flag-IRF3, HA-NSs, and nontagged TBK1. IRF3 was colocalized in IBs with NSs.
We noted that in the cells cotransfected with only IRF3 and NSs, IRF3 was not located in IBs, likely because endogenous TBK1 was not activated without an upstream stimulus and inactive TBK1 does not associate with IRF3 (Fig. 6C). In contrast, exogenous TBK1 could be self-activated due to overexpression, leading to transautophosphorylation (24, 25), and phosphorylated TBK1 bound to IRF3, subsequently directing IRF3 into IBs through its interaction with NSs.
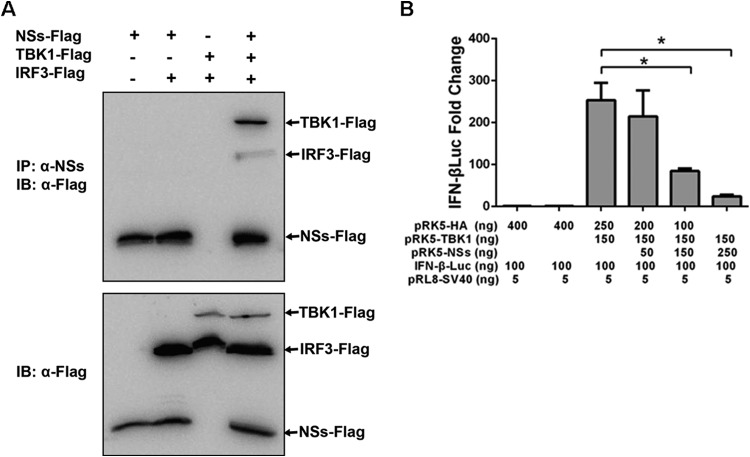
Coimmunoprecipitation confirmed that NSs and IRF3 do not interact with each other directly (Fig. 2), but in the presence of TBK1, NSs and IRF3 were associated, apparently due to the interaction of TBK1 with both NSs and IRF3 (Fig. 7A, top). Taken together, these data suggest that NSs interacted indirectly with IRF3 through an NSs-TBK1 interaction. At this point we have no data to indicate whether the TBK1-IRF3 interaction was or was not significantly disrupted by NSs.
FIG 7.
Indirect interaction of NSs and IRF3. (A) HEK293 cells were transfected with Flag-NSs alone or together with Flag-IRF3 and Flag-TBK1. Cell lysates were immunoprecipitated with anti-NSs (α-NSs) antibodies, which were subjected to SDS-PAGE. The membrane was blotted with anti-Flag antibodies (top). NSs was coimmunoprecipitated with IRF3 only in the presence of TBK1. The inputs of Flag-tagged NSs, IRF3, and TBK1 in transfected cells were examined by Western blot analysis of the cell lysates with anti-Flag antibodies (bottom). (B) Luciferase reporter assay to measure IFN-β promoter activity and quantitative RT-PCR to measure viral M gene transcripts in cells transfected with TBK1 and NSs. HEK293 cells were transfected with a luciferase reporter plasmid and plasmids expressing TBK1, with or without plasmids expressing NSs. The experiments were repeated at least three times, and the data from one representative experiment with two technical repeats were presented (*, P < 0.05).
These findings may also explain why cellular proteins TBK1, IRF3, and IKKε were detected at reduced levels in the presence of NSs (Fig. 2). Through the interaction of TBK1 with NSs, TBK1 was brought into the relatively insoluble IBs, leaving a reduced amount of TBK1 in the cytosol for detection by Western blotting in NSs-transfected cells; NSs may have similarly reduced the amounts of IRF3 and IKKε in the cytosol.
Using a luciferase reporter assay, we were able to determine that IFN-β reporter activities were increased over 250-fold when cells were transfected with TBK1. However, the increased IFN-β reporter activities were reduced significantly when the cells were cotransfected with TBK1 and NSs (Fig. 7B), which demonstrates that NSs suppressed IFN-β induction through an interaction with TBK1.
Blockage of active IRF3 from entry into the nucleus upon viral infection.
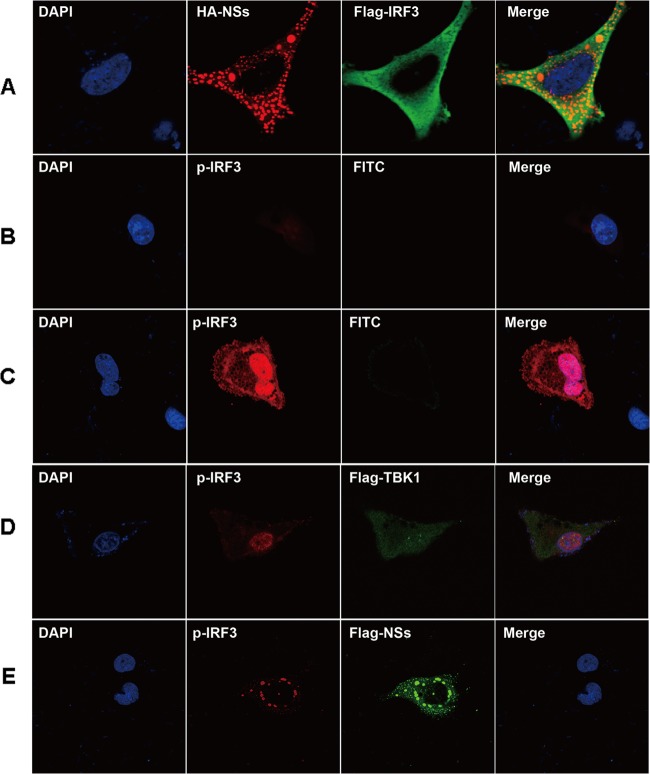
We next examined the functional effect of IBs in IFN-β signaling. While expressed IRF3 was dispersed in the cytoplasm of HeLa cells, NSs was mainly localized in IBs (Fig. 8A). Anti-phospho-IRF3 (anti-p-IRF3) antibody staining barely detected phosphorylated IRF3 in noninfected cells. When cells were infected with H1N1pdm, p-IRF3 increased dramatically and was subsequently translocated into the nucleus (Fig. 8C).
FIG 8.
Sequestration of active IRF3 into IBs and suppression of IFN-β induction in H1N1 influenza virus-infected cells. (A to E) HeLa cells were transfected with tagged plasmids, with or without subsequent infection with H1N1 influenza virus at an MOI of 1. The cells were fixed and permeabilized before staining with the indicated primary and conjugated secondary antibodies for confocal microscopy. (A) Cells were transfected with Flag-IRF3 and HA-NSs. No IRF3 moved into IBs formed by NSs. (B) IRF3- and NSs-cotransfected cells were stained with anti-phospho-IRF3, indicating that IRF3 was barely phosphorylated or activated. (C) Cells were infected with H1N1 influenza virus and later fixed and stained with anti-phospho-IRF3. IRF was phosphorylated and translocated into the nucleus. (D) Cells were transfected with Flag-TBK1, followed by H1N1 influenza virus infection. (E) Cells were transfected with Flag-TBK1 and HA-NSs, followed by H1N1 influenza virus infection. Phosphorylated IRF3 was trapped in IBs, colocalized with NSs.
When cells were pretransfected with Flag-tagged TBK1 followed by H1N1pdm infection, IRF3 was phosphorylated and p-IRF3 was translocated into the nucleus (Fig. 8D). However, when HeLa cells were cotransfected with TBK1 and NSs prior to viral infection, IRF3 was phosphorylated but p-IRF3 was sequestered into IBs with NSs, instead of being translocated into the nucleus (Fig. 8E).
Total RNA was prepared from transfected and infected cells for quantitative analyses of IFN-β induction and viral replication. As shown in Fig. 9, left, mRNA of IFN-β was upregulated over 340-fold 12 h after viral infection, but the induction was reduced to about 120-fold in the presence of NSs; the IFN-β levels increased up to 390-fold when TBK1 was expressed alone, but likewise the induction of IFN-β was reduced to less than 200-fold when TBK1 and NSs were coexpressed (Fig. 9, left). The transcript levels of the influenza virus M gene were affected as well in cells pretransfected with TBK1 or NSs prior to viral infection (Fig. 9, right). While the M gene transcripts were reduced significantly by TBK1 expression, the expression of NSs increased M gene transcripts. When cells were cotransfected with TBK1 and NSs, the reduction of the M gene transcripts by TBK1 was significantly attenuated in the presence of NSs. We concluded that NSs suppressed IFN-β induction and enhanced viral replication in influenza virus-infected cells through its interaction with TBK1.
FIG 9.
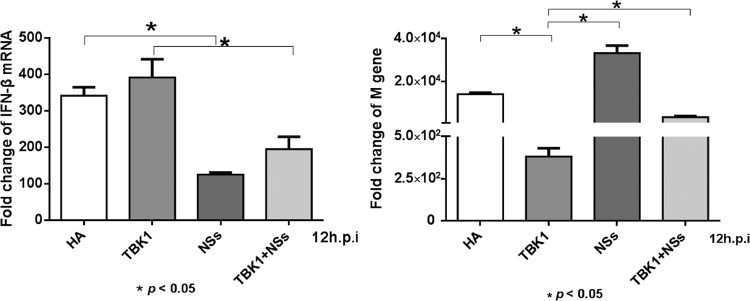
Reduced IFN-β induction in influenza virus-infected cells overexpressing NSs. Quantitative RT-PCR to measure mRNA transcripts of IFN-β (left) and the viral M gene (right). HEK293 cells were transfected with a blank plasmid or plasmids expressing TBK1, NSs, or both. Twelve hours later (p.i.), the cells were infected with H1N1 influenza virus. After another 12 h, total RNA was prepared from transfected and infected cells for RT-PCR. Fold changes were calculated from CT values for infected cells relative to that for noninfected cells for each transfection treatment. The experiments were repeated three times, and the data from one representative experiment with two technical repeats were presented.
The carboxyl-terminal region of NSs contributes to the formation of IBs and IFN suppression.
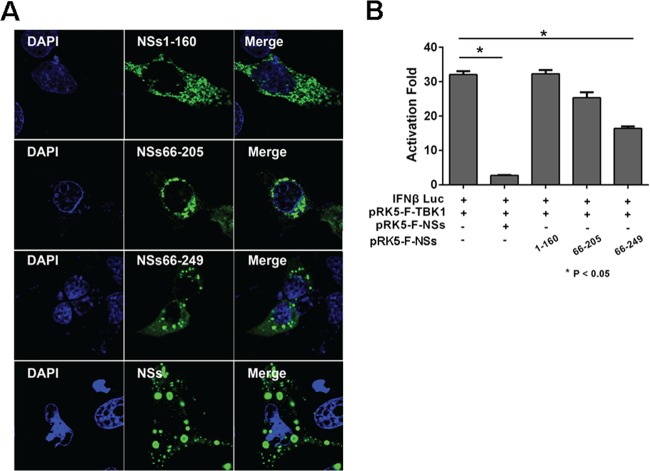
We generated NSs deletion mutants to determine which domains or regions of NSs are critical in the formation of IBs. HeLa cells were transfected with full-length NSs or its truncated fragments and the cells were stained at 28 h posttransfection. As shown in Fig. 10A, although the expression levels of both full-length and truncated NSs were comparable, distribution of the N-terminal region of NSs (NSs1-160) in transfected cells was evenly dispersed in the cytoplasm and no major IBs were observed. The middle portion of NSs (NSs66-205) had a distribution similar to that of the N-terminal region of NSs, with few IBs detectable. However, when the fragment included the C-terminal region, NSs66-249, characteristic IBs were observed (Fig. 10A). When HEK293 cells were cotransfected with TBK1 and IFN-β luciferase reporter genes, NSs1-160 and NSs66-205 did not suppress IFN-β reporter activities. In contrast, the C-terminal region significantly inhibited IFN-β activation (Fig. 10B). These data demonstrate that it is the C terminus of NSs that has immunomodulating capability through the formation of IBs.
FIG 10.
The C-terminal region of NSs contributed to IB formation and IFN suppression. (A) HeLa cells were transfected with pRK5-NSs1-160, pRK5-NSs66-205, pRK5-NSs66-249, or pRK5-F-NSs. The cells were fixed and permeabilized 28 h posttransfection. Expression of various lengths of NSs and the formation of the IBs were shown by staining with anti-NSs antibodies and detected by immunofluorescence. (B) Full-length and truncated NSs exhibited distinct abilities of inhibiting IFN-β promoter activity. HEK293 cells were cotransfected with plasmids expressing full-length and truncated NSs along with pGL3-IFN-β-Luc and pRL8 for 24 h, followed by stimulation with 50 μg/ml of poly(I · C) for another 8 h. Cell lysates were prepared and measured for luciferase activities. The experiments were repeated at least three times, and the data from one representative experiment with two technical repeats were presented.
DISCUSSION
IBs are cytoplasmic or nuclear aggregates of viral proteins that have been identified in various viral infections. Viral proteins in IBs are sometimes misfolded, but IBs in some viral infections are important functional sites for viral replication. These IBs are called virus factories or viroplasm (20, 26), in which viral proteins are functional. Since our EGFP-NSs fluoresced in IBs (Fig. 1B), the viral proteins appear to retain their native structures and are functional. In this report we present data to indicate that IBs formed by SFTSV NSs may play a role in viral modulation of host immune responses. IBs in SFTSV infection may sequester the IKK complex, including TBK1, IKKε, and IRF3, thus preventing p-IRF3 translocation into the nucleus, where it would activate the IFN-β response.
Various mechanisms are employed by viruses to evade host antiviral strategies. Viral proteins, accessory and nonstructural proteins in particular, function in many viruses to suppress host innate immunity (27, 28). By interacting with TIR domain-containing adaptors such as MyD88, TRIF, MAL, and TRAM, viral proteins can alter the functions of these adaptors, delink signaling pathways, and antagonize IFN induction upstream. Viral proteins can also target IKK complexes and IRF3/7 and block the activation and translocation of transcription factors into the nucleus to attenuate IFN responses downstream. The first IKK complex is composed of IKKα and IKKβ, which together with regulatory IKKγ phosphorylate IκB, leading to its degradation and subsequent translocation of activated NF-κB into the nucleus; the second IKK complex is composed of TBK1, IKKε, and DDX3 (29) and is critical for phosphorylating IRF3/7, which is activated and translocated into the nucleus to activate type I IFN gene transcription. The cytoplasmic tail of NY-1 hantavirus glycoprotein (Gn) interacts with TRAF3, disrupts TRAF3-TBK1 complex formation, and inhibits the activation of TBK1 (30). Activation of TBK1 is also inhibited by phosphoprotein (P) of rabies virus (12) and Borna disease virus (31). In both cases, these P proteins are phosphorylated by TBK1 as a viral decoy substrate and prevent activation of cellular target proteins of TBK1. The interaction of TBK1 with NS3 of HCV (14) or N1L of vaccinia virus (13) blocks subsequent association with IRF3, resulting in the failure of TBK1 to phosphorylate and activate IRF3. In addition, IRF3 can be directly inhibited by papain-like protease (PLpro) of severe acute respiratory syndrome coronavirus (SARS-CoV) for phosphorylation, dimerization, and translocation to the nucleus (32).
Our study suggests that NSs of SFTSV interacts with TBK1 but preserves TBK1's ability to phosphorylate IRF3 (Fig. 8). The C terminus of NSs has the ability to form IBs (Fig. 10), and this region of NSs may also be critical for binding to TBK1, which would consequently bring TBK1 and the IKK complex into the IBs. Interaction of TBK1 with NSs apparently did not interfere with its IRF3 binding and activation (Fig. 8E), although the detailed structural basis for this interaction remains unknown currently. Importantly, TBK1 interacted and phosphorylated IRF3, and the TBK1-IFR3 complex was subsequently directed to IBs, apparently due to the interaction of TBK1 and NSs, even though NSs did not interact with IRF3 directly (Fig. 2 and 7A). As a result, NSs succeeded in sequestering p-IRF3 into IBs and preventing active p-IRF3 from entry into the nucleus.
The mechanism by which IKK complex components are sequestered into viral IBs through virus-host protein interactions leading to suppressed IFN-β induction is obviously distinct from those shown by other viral proteins, which either interact with the TBK1/IKKε complex and inhibit TBK1 (12, 29–31), disrupt the TBK1-IRF3 interaction (14, 30), or target IRF3/7 directly (32), all resulting in the retention of inactive IRF3 in the cytoplasm. Therefore, sequestering TBK1/IKK complex components into the IB, a unique structure, through the interaction of viral NSs and TBK1 may provide a novel mechanism for viruses to evade antiviral immunity. Notably, a recent study found that the N protein of human respiratory syncytial virus interacts directly or indirectly with MDA5 and brings MDA5 into IBs, which are primarily formed by the N protein (33). This shows that IBs formed by a viral protein could sequester the viral RNA sensor, resulting in decreased downstream activation of TBK1/IKKε and IFN-β induction both in transfected and infected cells. Multiple functions of IBs in viral replication and evasion of innate immunity could be revealed with more studies on virus infections, and IBs may play critical roles in the virulence and pathogenesis of many viruses.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid from the Vice President Office of Research, University of Minnesota, Twin Cities, and by the Minnesota Rapid Agricultural Response Fund (FY13-14 to Z.X.). It was also supported by the State Key Laboratory of Pharmaceutical Biotechnology of Nanjing University (grant KFGW-200902 to Z.X.) and by the Jiangsu Province Key Medical Talent Foundation (RC2011084 to X.Q.). X.Q. also received support from the 333 Projects of Jiangsu Province.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O'Neill LA. 2000. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 97:10162–10167. 10.1073/pnas.160027697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. 2005. Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J. Exp. Med. 201:1007–1018. 10.1084/jem.20041442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269. 10.1073/pnas.0407639101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 5.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U. S. A. 104:7253–7258. 10.1073/pnas.0611506104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardenas WB, Loo YM, Gale M, Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178. 10.1128/JVI.02199-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr, Garcia-Sastre A. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514–524. 10.1128/JVI.01265-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992–2997. 10.1073/pnas.0408824102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei X, Sun Z, Liu X, Jin Q, He B, Wang J. 2011. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by Toll-like receptor 3. J. Virol. 85:8811–8818. 10.1128/JVI.00447-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. 2013. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 87:1690–1698. 10.1128/JVI.01855-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson JR, de Sessions PF, Leon MA, Scholle F. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262–8271. 10.1128/JVI.00226-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzozka K, Finke S, Conzelmann KK. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673–7681. 10.1128/JVI.79.12.7673-7681.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, Arvikar S, Latz E, Fitzgerald KA, Marshall WL. 2004. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J. Biol. Chem. 279:36570–36578. 10.1074/jbc.M400567200 [DOI] [PubMed] [Google Scholar]
- 14.Otsuka M, Kato N, Moriyama M, Taniguchi H, Wang Y, Dharel N, Kawabe T, Omata M. 2005. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibition of cellular antiviral responses. Hepatology 41:1004–1012. 10.1002/hep.20666 [DOI] [PubMed] [Google Scholar]
- 15.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364:1523–1532. 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu B, Qi X, Wu X, Liang M, Li C, Cardona CJ, Xu W, Tang F, Li Z, Wu B, Powell K, Wegner M, Li D, Xing Z. 2012. Suppression of the interferon and NF-kappaB responses by severe fever with thrombocytopenia syndrome virus. J. Virol. 86:8388–8401. 10.1128/JVI.00612-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao W, Sun W, Qu B, Cardona CJ, Powell K, Wegner M, Shi Y, Xing Z. 2012. Distinct regulation of host responses by ERK and JNK MAP kinases in swine macrophages infected with pandemic (H1N1) 2009 influenza virus. PLoS One 7:e30328. 10.1371/journal.pone.0030328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333–339 [DOI] [PubMed] [Google Scholar]
- 19.Netherton CL, Wileman T. 2011. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 1:381–387. 10.1016/j.coviro.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147–172. 10.1042/BC20040058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsumoto K, Umetsu M, Kumagai I, Ejima D, Arakawa T. 2003. Solubilization of active green fluorescent protein from insoluble particles by guanidine and arginine. Biochem. Biophys. Res. Commun. 312:1383–1386. 10.1016/j.bbrc.2003.11.055 [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- 23.Kopp E, Medzhitov R. 2003. Recognition of microbial infection by Toll-like receptors. Curr. Opin. Immunol. 15:396–401. 10.1016/S0952-7915(03)00080-3 [DOI] [PubMed] [Google Scholar]
- 24.Larabi A, Devos JM, Ng SL, Nanao MH, Round A, Maniatis T, Panne D. 2013. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 3:734–746. 10.1016/j.celrep.2013.01.034 [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Helgason E, Phung QT, Quan CL, Iyer RS, Lee MW, Bowman KK, Starovasnik MA, Dueber EC. 2012. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. U. S. A. 109:9378–9383. 10.1073/pnas.1121552109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Netherton C, Moffat K, Brooks E, Wileman T. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70:101–182. 10.1016/S0065-3527(07)70004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911–922. 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Negrate G. 2012. Viral interference with innate immunity by preventing NF-kappaB activity. Cell. Microbiol. 14:168–181. 10.1111/j.1462-5822.2011.01720.x [DOI] [PubMed] [Google Scholar]
- 29.Schroder M, Baran M, Bowie AG. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27:2147–2157. 10.1038/emboj.2008.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alff PJ, Sen N, Gorbunova E, Gavrilovskaya IN, Mackow ER. 2008. The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J. Virol. 82:9115–9122. 10.1128/JVI.00290-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unterstab G, Ludwig S, Anton A, Planz O, Dauber B, Krappmann D, Heins G, Ehrhardt C, Wolff T. 2005. Viral targeting of the interferon-{beta}-inducing Traf family member-associated NF-κB activator (TANK)-binding kinase-1. Proc. Natl. Acad. Sci. U. S. A. 102:13640–13645. 10.1073/pnas.0502883102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208–32221. 10.1074/jbc.M704870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE, Jr, Santangelo PJ. 2012. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 86:8245–8258. 10.1128/JVI.00215-12 [DOI] [PMC free article] [PubMed] [Google Scholar]