Abstract
In 2003, a review of the use of statistics in Infection and Immunity (IAI) found that more than half of articles had errors of statistical analysis or reporting of statistical results. This updated review of recent articles identifies and discusses the most common statistical methods reported in IAI and provides examples of both good reporting and common mistakes. Furthermore, it expands on the criteria for statistical analysis and reporting presented in the IAI “Instructions to Authors,” with the goal of helping both readers and authors better understand and apply the criteria.
TEXT
A decade ago, a review of 141 articles in Infection and Immunity (IAI) found errors of statistical analysis in 20% of articles, insufficient reporting of statistical methods in 22%, and both types of errors in 12% of articles (1). In 2011, IAI updated its instructions to authors with a more detailed set of criteria for statistics (2). To evaluate current statistical practice and reporting in IAI, 110 IAI research articles published in print or online in April and May 2013 were queried for the types of statistical methods used, and 10 articles were reviewed in-depth for errors.
Most articles in IAI report at least basic descriptive statistics, and two-thirds include some kind of hypothesis testing. Of the 110 articles searched, 83 articles included at least one of the words “mean,” “average,” “frequency,” or “percent,” and 73 articles included at least one of the words “standard deviation,” “standard error,” “standard error of the mean,” “variance,” or “range.” Seventy-four mentioned at least one of the following standard statistical tests: t test, analysis of variance (ANOVA), Wilcoxon/Mann-Whitney test, Kaplan-Meier test, log rank test, chi-square test, Fisher's exact test, or regression analysis.
Specifically, 51 articles used t tests to compare means between two groups. Forty-one articles used ANOVA to compare means across multiple groups. When more than two groups are being compared, ANOVA is better than using a series of t tests because it can adjust for the increased likelihood of finding significant differences when no true differences exist. The “Instructions to Authors” specify that multigroup comparisons should report both an overall P value (indicating whether there are any differences among the groups being compared) and P values for specific comparisons of interest between group means (post hoc tests). An example of this type of analysis is found in the work of Manivannan et al. (3), which reports both the significance of the overall ANOVA model and pairwise comparisons between all pairs of group means using the Newman-Keuls multiple-comparison test as shown in Fig. 1. Figure 2 is taken from the work of Navabi et al. (4) and shows an example where Dunnett's post hoc test was used to compare each group to the control group. Comparisons between all pairs of means (using the Tukey-Kramer or other tests) or between each mean and the mean from a control group (using Dunnett's test) are the most common applications of multiple-comparison procedures, but other approaches, such as the Dunn (for nonparametric analyses), Bonferroni, Holm-Šidák (less conservative than the Bonferroni test), or Scheffé test may be appropriate for different scenarios. The Bonferroni adjustment is particularly simple and can be applied in any multiple-comparison situation, such as when two groups are compared across multiple outcomes. It involves multiplying each P value by the total number of comparisons made. However, when the total number of comparisons is large, the Bonferroni adjustment is overly conservative.
FIG 1.
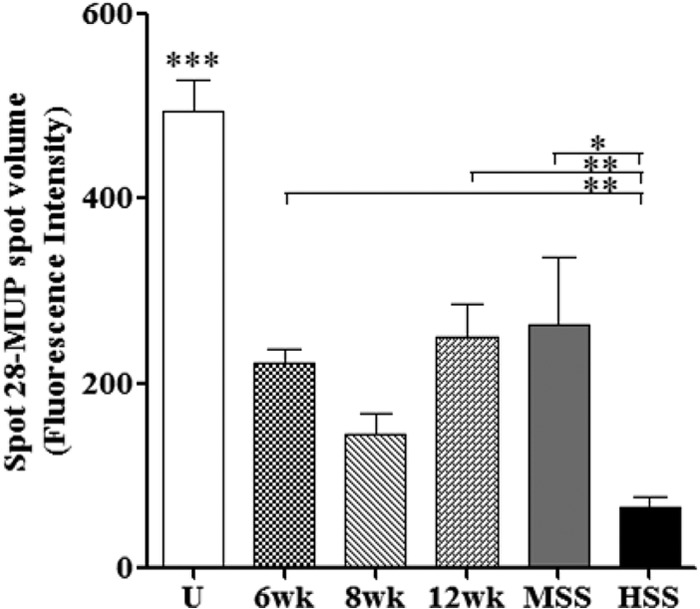
Liver candidate markers in different study groups in a mouse model, illustrating results of ANOVA followed by the Newman-Keuls pairwise multiple-comparison test (3). Shown are comparisons of spot volumes of mouse liver cytokeratin 18, interleukin 2 (IL-2), major urinary protein (MUP), Schistosoma mansoni phosphoenolpyruvate carboxykinase (PEPCK), peroxiredoxin 6, and liver hydroxyproline for uninfected (U), 6-week-infected (6wk), 8-week-infected (8wk), 12-week-infected (12wk), and 20-week-infected mice with moderate splenomegaly syndrome (MSS) and hypersplenomegaly syndrome (HSS). Data shown are means ± standard errors of the means (SEM). P ≤ 0.01 (overall ANOVA). Individual groups were compared using the Newman-Keuls multiple-comparison test. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05 (compared to all other study groups and as indicated). (This figure and its legend are reprinted or modified from reference 3 with permission.)
FIG 2.
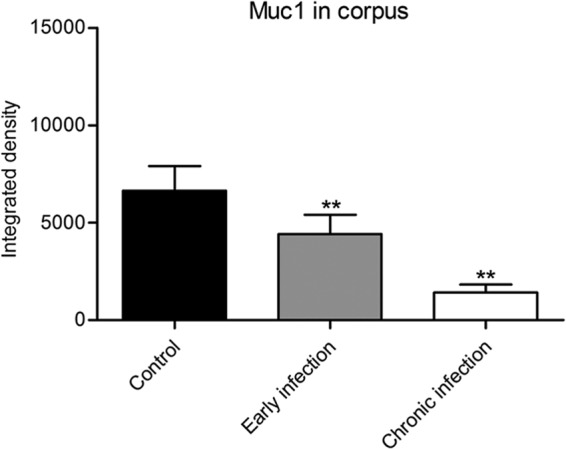
Quantification of Muc1 during early and chronic Helicobacter pylori infection, illustrating results of ANOVA followed by Dunnett's multiple-comparison test with a control (4). The integrated densities of fluorescence as a measure of Muc1 in the corpus during early and chronic infection are compared. Data were compared to control values (n = 6) by ANOVA with Dunnett's post hoc test (**, P < 0.01; NS, not significant). (This figure and its legend are reprinted or modified from reference 4 with permission.)
Twenty articles used nonparametric tests, such as the Wilcoxon signed-rank test or the Mann-Whitney U test. These tests are useful for comparing groups when data do not follow a normal distribution, because they are less sensitive to outliers and skewness than standard t tests and ANOVA. Note that the Wilcoxon rank sum and Mann-Whitney tests yield the same results and are analogous to t tests in that they are appropriate for comparing two unpaired groups. The standard nonparametric test for comparing more than two groups is the Kruskal-Wallis test.
The “Instructions to Authors” specify that “Data should be appropriately analyzed as parametric (normally distributed) or nonparametric data” but provide scant guidance on how to decide which method is appropriate. Common approaches are graphical examinations using histograms, stem-and-leaf plots, or normal probability plots or hypothesis tests, such as the Kolmogorov-Smirnov and Shapiro-Wilk tests. Histograms and stem-and-leaf plots should indicate the classic bell-shaped curve, normal probability plots should show all data points close to a 45° line, and hypothesis tests should indicate no significant departures from normality (P > 0.05). However, these approaches may be unreliable with a small number of samples; in particular, hypothesis tests are likely to provide a false sense of security with small numbers of samples, because they are unlikely to reject the hypothesis that the data are selected from a normally distributed population. In practice, it is important to consider prior information about the data along with graphs and hypothesis tests. For example, titers, counts, ratios, and proportions generally do not follow a normal distribution, while physical measurements, such as weight and length, are more likely to be at least approximately normal.
When data do not follow a normal distribution, the first approach is usually to transform the data and check whether the transformed data are approximately normal. Log transformation is the most commonly applied transformation in IAI articles, and it tends to work well for titers, cell counts, and other quantities that follow a skewed distribution with a long upper tail. In general, if it is natural to think of differences between groups as percent differences or fold changes, log transformation may be appropriate. Figure 3 illustrates data in which groups with higher mean values also have greater within-group variability, as indicated by a wider spread of data points. This data set, taken from the report of Dondji et al. (5), is a promising candidate for log transformation. A related strategy is to examine a bar chart with standard deviation bars. If the standard deviation bars are wider in groups that have higher means, a log transformation may be appropriate. Parametric tests may then be used to compare means of the log-transformed data. If desired, means can be exponentiated to display on the original scale; these reverse-transformed means are known as geometric means. If transformation does not result in standard deviations that are similar across groups, common approaches include t tests for unequal variances when two groups are compared and Welch's ANOVA for comparing three or more groups. Finally, nonparametric tests are often a good alternative if no suitable transformation can be found for nonnormal data. Keep in mind that nonparametric tests do not compare the means of the data. For this reason, data that are analyzed using nonparametric tests should be reported as medians and ranges, or medians and selected percentiles (typically 25th and 75th percentiles), instead of means and standard deviations.
FIG 3.
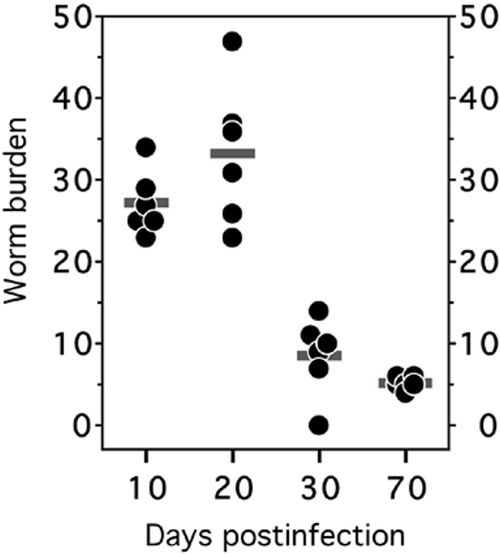
Intestinal worm burden of infected hamsters (5). At each time point postinfection, six infected hamsters were sacrificed and adult hookworms removed from the intestines. The horizontal bars represent the mean numbers of adult worms for each group. Days with higher mean worm burdens tend to have more-variable data, indicating that ANOVA on the raw data may be problematic and that ANOVA on log-transformed data may be preferable. (This figure and its legend are reprinted or modified from reference 5 with permission.)
Twelve articles used correlation or regression models to quantify associations among continuous variables. The most common measure of association is probably the Pearson (product moment) correlation, but the nonparametric Spearman rank correlation is an alternative when data are not normally distributed or the association is nonlinear. Eight articles compared proportions between groups using either the chi-square test or Fisher's exact test. Although the two methods give similar results for large samples, Fisher's exact test is better than the chi-square test when analyzing the small numbers of samples typical of studies published in Infection and Immunity. Tivendale et al. (6) provide an example of a comparison of proportions using Fisher's exact test, and their data are reproduced in Table 1. Of note, this table reports 22 distinct P values. Although this analysis compares proportions instead of means (as in ANOVA), the Infection and Immunity instruction to report both an overall P value and individual follow-up tests still applies. The Bonferroni or Holm-Šidák test could be used to adjust for multiple comparisons in this case.
TABLE 1.
Mortality rates among chick embryos inoculated with APEC and NMEC isolates compared using Fisher's exact testd
| Strain | Mortality ratec |
P value vsa: |
|
|---|---|---|---|
| DH5α | APEC O2 | ||
| Uninoculated | 0/6 | 1.000 | <0.001 |
| PBS | 3/10 | 0.300 | 0.001 |
| E. coli DH5α | 2/20 | <0.001 | |
| APEC 79 | 7/20 | 0.127 | <0.001 |
| APEC 353 | 12/20 | 0.002 | 0.064 |
| APEC 358 | 16/20 | <0.001 | 0.661 |
| APEC 380 | 16/20 | <0.001 | 0.661 |
| NMEC 15 | 14/20 | <0.001 | 0.235 |
| NMEC 18 | 11/20 | 0.005 | 0.031 |
| NMEC 38 | 19/20 | <0.001 | 1.000 |
| NMEC 58 | 14/20 | <0.001 | 0.235 |
| APEC O2b | 32/40 | <0.001 | |
P value determined by Fisher's exact test for comparing proportions.
Positive control for the chicken embryo lethality assay.
Data represent the number of embryos that died/total number of embryos tested.
This table is reprinted from reference 6 with permission. APEC, avian-pathogenic E. coli; NMEC, neonatal meningitis E. coli.
Eight articles used the Kaplan-Meier method to describe survival time and/or the log rank test to compare survival times between groups; a recent example is found in the paper of Goodyear et al. (Fig. 4) (7). These methods are more appropriate for comparing times to death than simple comparisons of means or median survival times because they can account for animals that survived to the end of the study or that were withdrawn from the study prior to the end for reasons other than death. Kaplan-Meier curves also provide a more complete summary of mortality data than survival percentages at the end of the study, because they consider the time of death, not just whether death occurred. Furthermore, they can be used to describe time to any event (such as infection clearance), not just mortality.
FIG 4.
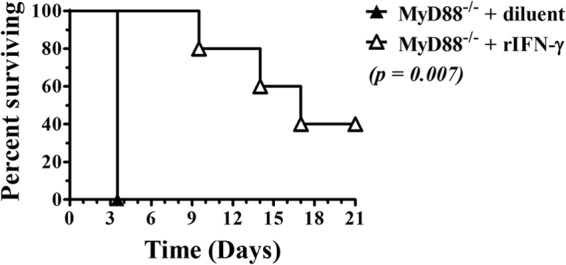
Example of Kaplan-Meier curves for describing survival data (7). Gamma interferon (IFN-γ) treatment protects MyD88−/− mice against lethal Burkholderia mallei infection. MyD88−/− mice were treated i.p. with recombinant IFN-γ (rIFN-γ) or a diluent (phosphate-buffered saline [PBS] plus 0.1% bovine serum albumin [BSA]) (5 mice per group) as described in reference 7. Mice were challenged with 500 CFU B. mallei intranasally. Survival was monitored, and mice were euthanized upon reaching a predetermined endpoint. Statistical differences were determined by using Kaplan-Meier curves and log rank analysis. (This figure and its legend are reprinted or modified from reference 7 with permission.)
A brief review of 10 IAI articles from 2013 found that four showed significant errors in reporting or analysis (although some showed more than one error). Two articles included figures with error bars that were not labeled, so it was unclear whether the bars represented standard deviation, standard error, or some other measure of variability or precision. The two measures should not be confused: standard deviation describes the variability of the data in the sample, and standard error describes the precision of the estimate of the mean. Standard error bars are popular in part because they are always narrower than standard deviation bars, but this is no excuse to use them when the goal is to describe the spread of the data. When the goal is to compare groups, a useful alternative to the standard error is the 95% confidence interval for the mean, described below, which has a direct connection to statistical significance. One article appeared to report 95% confidence intervals, but since the intervals were not described in Materials and Methods or the table caption, this was unclear.
Two articles used a series of t tests to compare several groups when ANOVA would be preferable. For example, one experiment involved three doses and three time periods, for a total of nine conditions being compared, so some adjustment for multiple comparisons should have been performed.
Two articles used t tests but were unclear about whether the t test for paired or unpaired data was used. Paired t tests should be used when comparing results from the same experimental units (e.g., following the same animals over time), and unpaired t tests should be used when comparing measurements from separate groups of experimental units (e.g., comparing two strains of animal). In one of these articles, the study design suggested that groups were unpaired, the figure captions indicated that paired t tests were used, and Materials and Methods stated that paired-sample analyses were performed using unpaired t tests.
This error highlights what is probably the most common major pitfall in statistical analysis: failure to account for correlated data. Most common statistical methods assume that observations are independent. As noted in the “Instructions to Authors,” when multiple samples are taken from one experimental unit (usually an animal), this assumption is violated. It is reasonable to expect that an animal with a higher infection load on day 1 will also have a higher infection load on day 2 or that two slices from the same brain will be more similar than two slices taken from different brains, violating the independence assumption. When multiple measures are taken over time for comparison, tests for paired or repeated data are appropriate. Examples include paired t tests or repeated-measures ANOVA for comparing means, the sign test or Wilcoxon signed-rank test for nonparametric comparisons, and McNemar's test for comparing proportions. When multiple measures are taken from the same experimental unit as technical replicates, they should be combined in some way (usually as averages or percentages), and statistical analysis can be performed on the summaries.
The editors of IAI have rightly emphasized the importance of good statistical practice in the “Instructions to Authors.” Statistical inference is important because it quantifies the role of chance in research results. A statistically significant finding is based on sufficient evidence that it is likely to be reliable and repeatable, whereas a nonsignificant finding could easily be a random occurrence. The role of chance in research results is often described by the P value. P values are common to all hypothesis tests, are generated by all major statistical analysis programs, and are reported in the majority of articles in IAI, so it is important to recognize some of the common fallacies in interpreting P values.
The P value represents the probability of observing a result at least as extreme as that observed in the experiment, if the null hypothesis is true. The most common null hypothesis is that two or more groups are equivalent, so the P value can often be interpreted as the probability of finding a difference between groups at least as large as the difference observed from chance alone if the groups are truly equivalent. In classical hypothesis testing, the researcher chooses to reject the null hypothesis and conclude that the groups are not equivalent if the P value is sufficiently small. Traditionally, the threshold for statistical significance is a P of <0.05, although this value is arbitrary.
Note that most P values correspond to two-tailed or two-sided hypothesis tests. They test whether the data deviate from a null hypothesis in either direction. For example, in studies of gene expression, a two-tailed test would yield a significant result if a gene is either overexpressed or underexpressed. In contrast, for a one-tailed or one-sided hypothesis test, the researcher would have to decide in advance whether to test for over- or underexpression. If the one-tailed test was designed to look for genes that are overexpressed and a particular gene is underexpressed instead, the result would not be statistically significant. Researchers are often tempted to report P values from one-tailed tests because they are always smaller than the corresponding P values from two-tailed tests. However, one-tailed tests are appropriate only if the researcher specifies the direction of the hypothesis before analyzing the data and should be accompanied by a strong justification for why a one-tailed test is appropriate.
The P value has several limitations as a summary of experimental results. First, a significant P value alone does not mean that a finding is important. The P value incorporates information about both the effect size (such as the difference between groups) and the precision of the estimates into a single summary. If the precision of the estimate is increased (e.g., by increasing the sample size), the P value will decrease even though the difference between groups is unchanged. If the sample size is large enough, even tiny differences between groups will be statistically significant. Therefore, P values should never be reported without some indication of the magnitude of the difference between groups. This does not seem to be a major problem of reporting in IAI, possibly because for many microbiologists, P values are (appropriately) viewed as adjunct to the main goal of presenting numerical results.
To illustrate the disconnect between P values and effect sizes, consider a series of experiments where proportions surviving are compared between two groups of animals. In each experiment, none of the animals in group A survive, and half of the animals in group B survive, so the effect size is the same in each experiment. When the sample size is six animals per group, the P value for this comparison is 0.18 (based on a two-sided Fisher exact test), a result that is not statistically significant. When the sample size is 12 animals per group, the P value for this comparison is 0.01, and when the sample size is 24 animals per group, the P value is <0.001. In this example, the smaller P values do not indicate a larger difference between groups, only that the difference between groups was measured with more precision because of the increased sample size.
A statistically significant P value does not prove that the results of the experiment are true. Using the standard probability criterion (P < 0.05) for statistical significance, 1 in 20 comparisons will be statistically significant by chance alone. Furthermore, the P value provides only information about chance. If the experiment is poorly designed and subject to bias, this will not be reflected in the P value. For example, a study might show that two groups differ significantly with a P value of 0.01, but if the two groups were tested in different labs or on different equipment, it is impossible to determine whether the difference is due to the treatment or to other differences in experimental conditions.
Conversely, lack of statistical significance does not prove that there is no difference between groups. It is also possible that a difference exists but that, due to bad luck, large variation in the data, and/or a small sample size, the difference did not reach statistical significance in this particular experiment. Interpreting nonsignificant P values as proof of equivalence can lead to abandonment of promising ideas when small initial studies do not yield statistical significance. When a result is not statistically significant, the observed difference and the variability in the data can provide an indication of whether the result was biologically meaningful, regardless of statistical significance, and further studies can be planned. Confidence intervals are especially informative in this situation.
Confidence intervals provide an alternate approach to quantifying the role of chance in research. Confidence intervals describe a range of values which is likely to include the true quantity that is being estimated. A 95% confidence interval, for example, is constructed so that if the experiment is repeated 100 times, 95 of the experiments will yield 95% confidence intervals that include the true value. Loosely, a confidence interval provides a range of plausible values for the truth, acknowledging that any individual experiment will deviate from the truth due to random variation. For example, the result stated earlier that 4 of 10 (or 40%) sampled articles in IAI showed statistical errors can be used to conclude, via confidence intervals, that the data support a proportion of statistically flawed IAI papers of between 14% and 71%. The “Instructions to Authors” require that authors report confidence intervals for data presented as endpoints, such as 50% lethal doses (LD50s) or 50% infectious doses (ID50s), but confidence intervals are useful in many other situations.
One common use of confidence intervals is to define the error bars on bar charts. Used in this way, 95% confidence interval bars that do not overlap generally imply a statistically significant difference between the two groups, with P being <0.05 (3). However, overlapping 95% confidence interval bars do not necessarily imply nonsignificance. van Belle (8) discusses this phenomenon in detail and suggests that as a rule of thumb, overlaps of 25% or less suggest statistical significance. Hypothesis tests should be conducted for confirmation, so confidence intervals do not entirely replace P values in this context.
Confidence intervals can also be constructed for the difference or ratio of summary measures between two groups. If the 95% confidence interval for the difference does not include 0, then the two groups are significantly different (P < 0.05). Similarly, if the 95% confidence interval for a ratio does not include 1, the P value is <0.05. Confidence intervals for differences and ratios are especially useful because most researchers have an intuitive idea of what sort of ratio or difference between groups is large enough to be scientifically important. Regardless of statistical significance, if a 95% confidence interval for the difference between groups includes effects that are scientifically important, then it would be shortsighted to rule out the possibility that such an effect might exist.
For example, suppose the ratio between two groups is 1.5, with a 95% confidence interval from 0.7 to 3.1. Because this confidence interval includes 1, the null hypothesis that the two groups are equivalent (their ratio equals 1) cannot be ruled out. However, it is also impossible to rule out a 3-fold-higher result in the first group. If this is a scientifically meaningful difference, it might be worth further exploration in a study that might compare the groups with greater precision.
In summary, when reading or reviewing statistical results in IAI, P values should be interpreted in proper perspective: they are useful for indicating that results are not likely due to chance, but they do not address bias, they do not provide information about the magnitude of differences or associations, and, if they are nonsignificant, they do not imply that differences do not exist. Confidence intervals provide a useful adjunct or alternative that incorporates information about both the magnitude and the precision or reliability of results.
Footnotes
Published ahead of print 16 December 2013
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Olsen CH. 2003. Review of the use of statistics in Infection and Immunity. Infect. Immun. 71:6689–6692. 10.1128/IAI.71.12.6689-6692.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Society for Microbiology 2011. 2011 instructions to authors. Infect. Immun. 79:1–20 [Google Scholar]
- 3.Manivannan B, Rawson P, Jordan TW, Karanja DM, Mwinzi PN, Secor WE, La Flamme AC. 2011. Identification of cytokeratin 18 as a biomarker of mouse and human hepatosplenic schistosomiasis. Infect. Immun. 79:2051–2058. 10.1128/IAI.01214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navabi N, Johansson ME, Raghavan S, Linden SK. 2013. Helicobacter pylori infection impairs the mucin production rate and turnover in the murine gastric mucosa. Infect. Immun. 81:829–837. 10.1128/IAI.01000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dondji B, Bungiro RD, Harrison LM, Vermeire JJ, Bifulco C, McMahon-Pratt D, Cappello M. 2008. Role for nitric oxide in hookworm-associated immune suppression. Infect. Immun. 76:2560–2567. 10.1128/IAI.00094-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect. Immun. 78:3412–3419. 10.1128/IAI.00347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodyear A, Troyer R, Bielefeldt-Ohmann H, Dow S. 2012. MyD88-dependent recruitment of monocytes and dendritic cells required for protection from pulmonary Burkholderia mallei infection. Infect. Immun. 80:110–120. 10.1128/IAI.05819-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Belle G. 2008. Statistical rules of thumb, 2nd ed. John Wiley and Sons, Inc., New York, NY [Google Scholar]


