Abstract
The virulence and fitness in vivo of the major human pathogen Staphylococcus aureus are associated with a cell-to-cell signaling mechanism known as quorum sensing (QS). QS coordinates the production of virulence factors via the production and sensing of autoinducing peptide (AIP) signal molecules by the agr locus. Here we show, in a wax moth larva virulence model, that (i) QS in S. aureus is a cooperative social trait that provides a benefit to the local population of cells, (ii) agr mutants, which do not produce or respond to QS signal, are able to exploit the benefits provided by the QS of others (“cheat”), allowing them to increase in frequency when in mixed populations with cooperators, (iii) these social interactions between cells determine virulence, with the host mortality rate being negatively correlated to the percentage of agr mutants (“cheats”) in a population, and (iv) a higher within-host relatedness (lower strain diversity) selects for QS and hence higher virulence. Our results provide an explanation for why agr mutants show reduced virulence in animal models but can be isolated from infections of humans. More generally, by providing the first evidence that QS is a cooperative social behavior in a Gram-positive bacterium, our results suggest convergent, and potentially widespread, evolution for signaling to coordinate cooperation in bacteria.
INTRODUCTION
Staphylococcus aureus is a major human pathogen, responsible for 1% of all hospital admissions and leading to an estimated cost of $9.5 billion per year in the United States alone (1–4). Infections caused by S. aureus range from relatively mild boils and infected skin wounds to those with high mortality rates, such as bacteremia, infective endocarditis, and toxic shock. S. aureus infections can be difficult to treat because of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), which are resistant to beta-lactam antibiotics. Globally, MRSA infection rates are increasing (2, 5, 6).
The virulence and growth in vivo of S. aureus depend upon quorum sensing (QS) via the accessory gene regulator (agr) locus (1, 7, 8). Cells release a small autoinducing peptide (AIP), the accumulation of which induces the production and secretion of both more AIP and a range of virulence factors. These virulence factors include hemolysins α, β, δ, and γ, toxic shock syndrome toxin (TSST), enterotoxins, Panton-Valentine leukocidin (PVL), and exfoliatins A and B (8–10). At high cell densities, or when cells are enclosed in small spaces, the agr system leads to positive feedback in the production of AIP and a significant increase in the production of virulence factors (11–15).
It is commonly assumed that QS is a cooperative social trait, in which the benefits of virulence factors are shared between the local population of cells and their production is coordinated across cells via QS (16, 17). However, there has been no empirical demonstration that QS in S. aureus, or any other Gram-positive bacteria, is a cooperative social trait (see Discussion). Evidence that QS is a social behavior has come primarily from work on the Gram-negative bacterium Pseudomonas aeruginosa (14, 18–27). Relative to P. aeruginosa, S. aureus represents a completely independent evolution of QS, based on different genetics and molecular biology, and so it is not clear that it is social in the same way. Furthermore, agr mutants appear to be better, not worse, at performing some potentially social traits, such as forming biofilms on abiotic surfaces, than the wild type (28, 29).
Whether or not QS via agr is a cooperative social trait has implications for virulence, epidemiology, and how strains will respond to medical intervention (11, 13, 30, 31). For example, if QS is a social trait, then this could explain the potentially contradictory results that agr mutants are less virulent in clonal infections of animal models (lower cooperation) but are still able to spread in infections of humans (“cheats” [see below] can exploit cooperators) (1–4). This is important because direct fitness, and not social, explanations have been invoked for the occurrence of agr mutants, based on the metabolic cost of agr expression or the fact that agr mutants are better adapted to certain environments (2, 5, 6).
Here, we examined whether QS in S. aureus is a cooperative social trait and determined the consequences for the evolution of virulence. We carried out all our work in vivo, using a greater wax moth (Galleria mellonella) larva virulence model (1, 7, 8). We first tested whether QS is a social trait that (i) provides a benefit at the group level and (ii) can be exploited by “cheats,” which avoid the cost of signaling and producing virulence factors while benefiting from those produced by others (8–10). We then tested the consequences of this social interaction for virulence, examining how virulence varies with the percentage of cheats in the population. Finally, we tested how the time spent within each host (transmission speed) and diversity within populations affect the evolution of virulence. These two effects are theorized to be influential on the evolution of virulence due to their effect on the pattern of transmission. However, they are often conflated, making them hard to disentangle (11–15), and so we designed an experiment to manipulate them independently.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
We used a fully QS-proficient wild-type strain (RN6390B) and a QS-deficient mutant RN6390B Δagr::ermB Ermr strain (ROJ37, with agr and RNAIII deleted) (16, 17). To determine the timing of QS induction in vivo, we used an RN6390B agr reporter strain, RN6390B(pSB2035), in which lux and green fluorescent protein (GFP) expression is under the control of the P3 promoter on a plasmid (14, 18–27). We grew overnight cultures of strains in 5 ml of lysogeny broth (LB) at 37°C with shaking. We centrifuged the required amounts of overnight culture and resuspended them in phosphate-buffered saline (PBS) to an optical density of approximately 0.2 at 600 nm. We then mixed the solutions as required.
Galleria virulence assay.
We purchased Galleria mellonella wax moth larvae from Wiggly Wigglers and modified an existing infection protocol (28, 29). We injected 20 μl of S. aureus culture (containing approximately 2.2 × 106 cells) into healthy 5th-instar larvae between the rear two sets of prolegs by using a microfine insulin syringe (U-100 [29 G]; Becton Dickinson) attached to a Tridak Stepper dose dispenser. We then incubated the wax moths at 37°C for different lengths depending upon the experiment. We determined the proportion of bacteria in the larvae by homogenizing the wax moths by using a Pro Scientific Pro200 handheld homogenizer. We serially diluted the homogenate and plated it out on Baird-Parker agar (Sigma-Aldrich). After 24 h, we picked a sample of the colonies and placed it onto Baird-Parker agar containing 5 μg/ml erythromycin, which selected for agr mutants. We determined the actual proportions of wild-type strains to Δagr mutant strains used when inoculating larvae in the same way.
Determining the virulences of wild-type, agr mutant, and mixed population infections.
We examined the virulences of groups with the following characteristics (20 wax moths per group): wild type alone, agr mutant alone, 1% Δagr mutant–99% wild type, 10% Δagr mutant–90% wild type, 50% Δagr mutant–50% wild type, 90% Δagr mutant–10% wild type, and PBS control. Periodically over 72 h, we assessed the proportion of Galleria spp. that had died. We determined death when wax moths no longer responded to stimulation with a pipette tip. In addition, in most cases, we noted that the larvae completely melanized.
Determining growth of S. aureus strains in Galleria.
To determine how RN6390B (wild type) and ROJ37 (agr mutant) grew in Galleria over time, we used 60 wax moths and sacrificed 10 larvae per group (preselected at the start of the experiment) at preselected time points and homogenized them separately. We serially diluted the homogenate of each larva and spread them onto separate Baird-Parker agar plates to count CFU.
Determining invasion and relative fitness of agr mutants in mixed populations in Galleria.
To determine how agr mutants grow in a mixed population with wild-type bacteria, we tested the 1% Δagr mutant–99% wild-type and 50% Δagr mutant–50% wild-type groups. We included 60 wax moths in each group. We sacrificed 10 larvae per group (preselected at the start of the experiment) at selected time points and homogenized them. We plated each larval homogenate onto separate Baird-Parker agar plates, and we picked 50 colonies from each plate and set them on erythromycin selection plates to determine the proportion of agr mutants.
We determined the relative fitness of the agr mutant in vivo when in competition with the wild type by inoculating wax moths with 3 different treatments (10 wax moths per treatment): 1% Δagr mutant–99% wild type, 10% Δagr mutant–90% wild type, and 50% Δagr mutant–50% wild type. We sacrificed the larvae after 48 h and homogenized them separately. We calculated the proportion of agr mutants subsequently by picking 100 colonies per larva and setting them on erythromycin selection plates. We calculated relative fitness (w) of agr mutants using the equation w = x2 (1 − x1)/x1 (1 − x2), where x1 is the starting mutant proportion of the population and x2 is the end mutant proportion (11, 13, 30, 31).
Varying transmission rate and relatedness.
We performed an experiment to determine the effect of transmission speed and relatedness on quorum sensing (the experimental design is shown in Fig. 4). Our experiment tested the effects of two variables, high/low relatedness and fast/slow transmission, on QS over time, and is an extension of previous experimental designs which looked at the effect of relatedness (18, 23, 32).
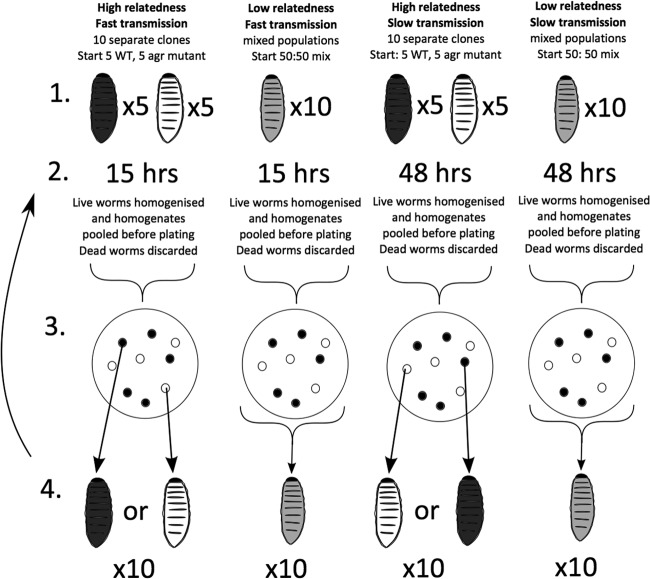
FIG 4.
Transmission selection experiment. (1) We varied relatedness between interacting individuals by initiating each subpopulation with either a single bacterial clone (relatively high r) or a large number of bacterial clones (>100; relatively low r). (2) We varied the transmission rate by altering the incubation period to either 15 h (relatively fast transmission) or 48 h (relatively slow transmission). (3) We pooled the samples from each larva by homogenizing all larvae together (excluding dead ones), and we plated them out onto Baird-Parker agar plates to isolate single colonies. (4) We injected 10 fresh larvae per group with 2.2 × 106 cells from overnight cultures. In the high-relatedness treatments, we injected each larva with culture grown from a separate, single clone, whereas for a low-relatedness line, we injected all the larvae with the same culture derived from >100 separate colonies. We used 10 worms per group per round and kept the transmission speed constant for each group. We ran the experiment for 3 rounds of selection. In the figure, black larvae symbolize individuals inoculated with QS wild-type (WT) cooperators, white larvae symbolize individuals inoculated with agr mutants (cheats), and gray larvae symbolize a mixture of cooperators and cheats (see Materials and Methods for full details of the selection method).
We manipulated relatedness by varying how the mix of colonies was used to initiate the next round of selection. In the relatively high-relatedness treatment, each wax moth larva was infected with a single clone, which could be either the wild type or the agr mutant, and so cells only had the potential to interact with genetically identical cells, corresponding to a relatedness (r) of 1. In contrast, in the relatively low-relatedness treatment, each larva was infected with multiple clones (>100), allowing the potential for both the wild type and the agr mutant to be in the same larva. In this case, cells had the potential to interact with both different and identical cells, corresponding to an r of <1. We manipulated transmission speed by varying the length of time that the bacteria spent in the host. For fast transmission, larvae were infected for 15 h, and for slow transmission, larvae were infected for 48 h. We therefore created 4 different treatment conditions (high relatedness/fast transmission, high relatedness/slow transmission, low relatedness/fast transmission, and low relatedness/slow transmission), and we replicated each treatment 4 times (16 selection lines in total).
We started our experiment by infecting 5 larvae with wild-type strains (all derived from a single clone) and 5 larvae with agr mutant strains (all derived from a single clone) (high relatedness) and infecting 10 larvae with a 50:50 mix of wild-type and agr mutant strains (low relatedness). Each larva received approximately 2.2 × 106 cells. We placed the larvae in a 37°C incubator for a given time before we sacrificed each larva. We manipulated transmission speed by allowing bacteria to grow in the host for either 15 h (fast transmission) or 48 h (slow transmission). We pooled the samples from each larva by homogenizing all larvae together (excluding dead ones), and we plated them out onto Baird-Parker agar plates to isolate single colonies. We excluded dead larvae in our experiment because during a natural infection cycle, bacteria in dead hosts would not necessarily be transmissible to new hosts and so they would be removed from the infection pool. Each colony on the plates represented either a wild type or an agr mutant. For high-relatedness treatment lines, we picked 10 individual colonies and grew them separately overnight in LB. For low relatedness, we took a sweep of >100 colonies to initiate the overnight culture in LB. We then injected 10 fresh larvae per group with 2.2 × 106 cells from the overnight cultures, which we had adjusted to an optical density of 0.2 in PBS. In the high-relatedness treatments, we injected each larva with culture grown from a separate, single clone, whereas in a low-relatedness line, we injected all the larvae with the same culture derived from >100 separate colonies. Again, we used 10 worms per group per round and kept the transmission speed constant for each group. We ran the experiment for 3 rounds of selection.
Analyses.
We analyzed the data using R version 3.0.1 and SPSS version 19 (33). We analyzed the survival data (see Fig. 3 and also Fig. S3 in the supplemental material) in two ways. First, we used a Cox proportional hazards model, fitting the percentage of agr mutants as a time-independent covariate, to test if host survival rate depended on the percentage of agr mutants. Such a model assumes that any effect of changing the percentage of agr mutants is not time dependent, i.e., affects on survival rates are not limited to the first 24 h. Regressing the Schoenfield residuals upon time can test this assumption. If there is no significant relationship, then the assumption holds. As an alternative, we analyzed the survival data by comparing the proportions of larvae still alive at the end of our experiment (72 h). We did this using a generalized linear model, GzLM, that allows a binary response (dead or alive) to vary as a proportion of the sample (e.g., 7 of 20 larvae still alive), depending on the percentage of agr mutants. However, this method provides only a snapshot of the effect (at the 72-h point) rather than comparing rates of survival over time as in the Cox proportional hazards model (34). In all other analyses, we used general linear models (GLM). Before data analysis of the population invasion data, we square root transformed the proportion of agr mutants to meet the assumptions of the GLM (data shown in Fig. 1B).
FIG 3.
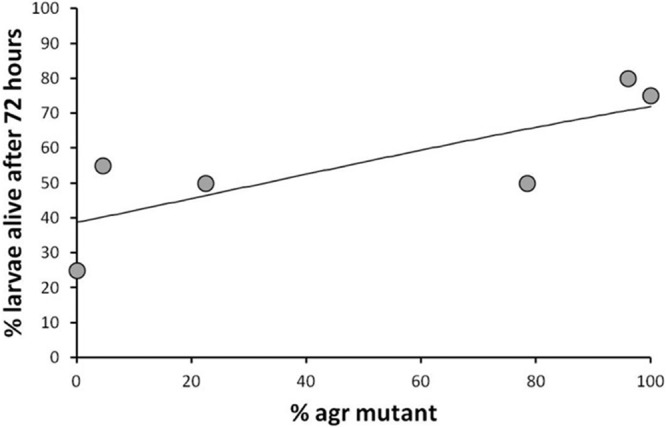
QS and virulence. The percentage of larvae alive after 72 h was positively correlated with the percentage of agr mutants in the original infection. The fitted line is a logistic regression curve, assuming binomial errors. Twenty larvae per treatment group were used. The fitted line is a logistic regression curve, assuming a binomial distribution of errors, resulting from a generalized linear model. The logistic regression was necessary because we were regressing a binary outcome (survival versus nonsurvival) (see Materials and Methods).
FIG 1.
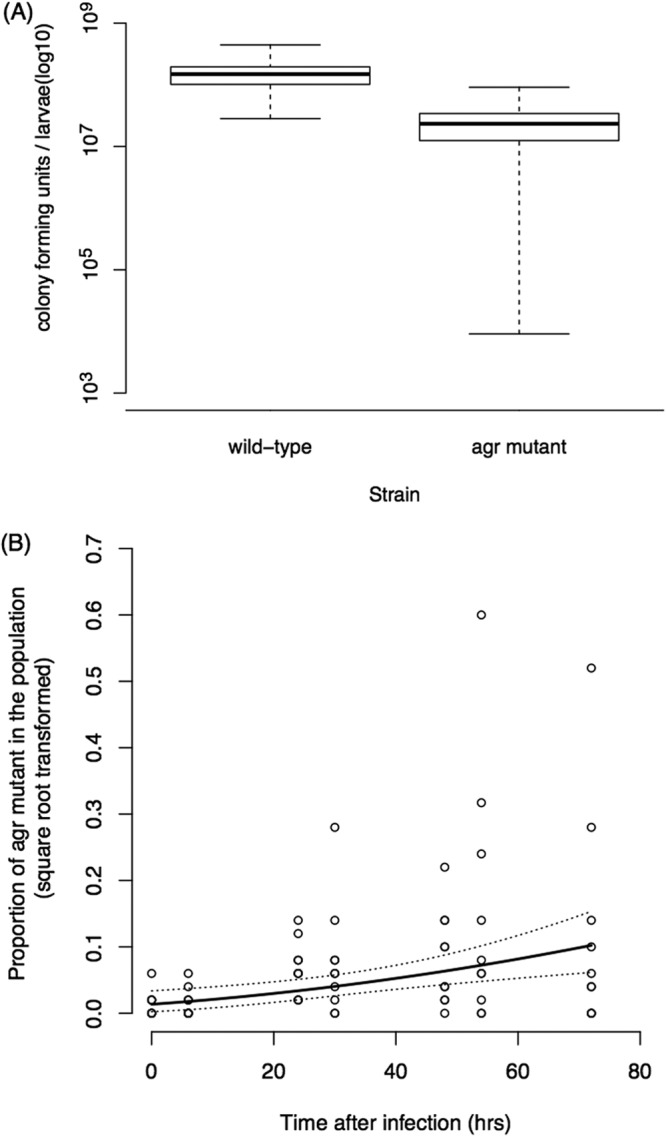
Social cost and benefit of QS. We grew the QS wild type (RN6390B) and an agr mutant that does not respond to QS (ROJ37) in single and mixed infections. (A) When only one strain was used to infect each host, the QS wild type grew to higher densities after 48 h than the agr mutant, suggesting a benefit of QS at the group level. The boxes represent the means ± the standard errors. The whiskers (error bars) show the minimum and maximum values. (B) When grown in mixed infections, the agr mutant increased in frequency, suggesting that the benefits of QS can exploited by individuals that do not respond to QS. The regression line is shown in solid black, and 95% confidence intervals are shown by dotted lines. Each circle represents one single larva. Overall, these results suggest that QS represents a cooperative social trait that can be exploited by cheats.
RESULTS
QS is a cooperative social trait in S. aureus infections.
We first tested whether agr QS is a cooperative social trait during infections of wax moth larvae. We predicted that, if it is, (i) populations of wild-type cells that exhibit QS should grow better than populations of agr mutants that do not produce or respond to signal and (ii) when grown in mixed populations, agr mutants should be able to exploit the signal and virulence factors produced by wild-type cells and hence increase in frequency (i.e., they act as social cheats) (9, 27). We tested these predictions using a fully QS proficient wild-type strain (RN6390B) and an agr-deficient QS mutant (ROJ37) (16).
We found that when inoculated into wax moths in single-strain infections, the wild-type strain grew to a higher density than the agr mutant after 48 h (analysis of variance [ANOVA]; F1,18 = 6.77, P = 0.018) (Fig. 1A; see also Fig. S1 in the supplemental material). In contrast, when we inoculated with a mixed infection containing 98.5% wild-type strains and 1.5% agr mutant strains, we found that the agr mutant increased in frequency over time (F1,66 = 11.29, P = 0.001) (Fig. 1B). Overall, these results suggest that QS via the agr locus is a cooperative social trait that provides a benefit at the group level (Fig. 1A) but which can be exploited by cheats (agr mutants) who do not perform the action (Fig. 1B).
Social evolution theory also predicts that the fitness of cheats should be frequency dependent, with their relative fitness decreasing as they become more common (30, 35–37). As cooperators become more common, there is greater population growth and more opportunity for cheats to exploit cooperators (30). As predicted, we found that the relative fitness of the agr mutant decreased as it became more common (R2 = 0.760; F1,27 = 85.62, P < 0.001) (Fig. 2). We found no increase in agr frequency when we initiated mixed infections containing high levels of agr mutants (P = 0.07) (see Fig. S2 in the supplemental material).
FIG 2.
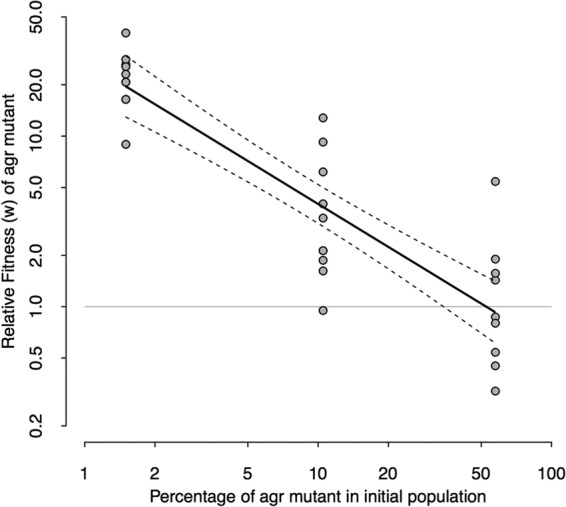
The fitness of cheats is frequency dependent. The relative fitness of the agr mutant, when coinfecting a host with the wild type, was negatively correlated with the percentage of agr mutants used to inoculate the infection. Each data point represents 48 h of growth in a wax moth larva. The regression line is shown in solid black, and 95% confidence intervals are shown by dotted lines. The gray horizontal line at 1.0 represents where the wild type and agr mutants have equal fitness levels.
QS and virulence.
We then examined the virulence consequences of QS, examining how virulence varies with the percentage of agr mutants in the population. We started infections with 100% wild-type strains, 100% agr mutant strains, or a mixture of the two. We found that as the percentage of agr mutants used to initiate infections increased, the proportion of hosts still alive after 72 h increased [GzLM; Wald X2(1) = 5.895, P = 0.015] (Fig. 3). The results of the Cox proportional hazards model showed that this was due to a significantly lower mortality rate over time [Cox proportional hazards model; Wald χ2(1) = 8.98, P = 0.003] (see Fig. S3 in the supplemental material). A regression of the Schoenfield residuals upon time showed no significant relationship, suggesting that this effect of agr mutants upon mortality rate did not vary over time (GLM; F1,51 = 0.025, P = 0.875). These results show that agr mutants are not only less virulent than the wild type but that the presence of agr mutants reduces the virulence of wild-type infections.
Transmission, relatedness, and the social evolution of virulence.
Our results described above showed that QS via the agr locus in S. aureus is a cooperative social trait and that the presence of agr mutants, which act as cheats, reduces virulence. Given this, anything that influences the relative fitness of cooperation and/or cheating will also influence the evolution of virulence. Theory predicts that the strain diversity within hosts can have two major influences, which act in opposite directions, through its influence on the genetic relatedness of the parasites infecting each host. First, low strain diversity within populations (high within-host genetic relatedness) can lead to increased virulence due to increased cooperation between highly related cells to exploit the host (11, 13). Secondly and in contrast, high strain diversity (low within-host genetic relatedness) can also lead to increased virulence due to incentives to outgrow nonrelated cells (38, 39).
In natural populations, the importance of these two influences of relatedness depends upon the transmission rate (see Fig. S4 in the supplemental material). If time spent in the host is long (slow transmission), reduced virulence may be favored to prevent host death before transmission to a new host occurs. In this case, a higher relatedness may lead to decreased competition for resources and hence select for decreased virulence, which could occur through the selection of agr cheats. In contrast, when time in the host is short (fast transmission), any influence of growth rate on host death will be relatively unimportant, because the host is unlikely to die before transmission. Here, a higher relatedness may favor cooperative traits such as QS and hence select for increased virulence. However, relatedness and transmission will usually be correlated, because faster transmission leads to higher strain diversity per host and hence lower relatedness, making it hard to disentangle these different effects.
We tested the influences of both relatedness and transmission by manipulating them independently with an experimental evolution study (Fig. 4). We found that there was a significant influence of relatedness but not the transmission rate. As predicted, we found that QS was favored by a relatively high relatedness (F1,12 = 5.576, P = 0.036) (Fig. 5A). At low relatedness, the agr mutant went to fixation, whereas at high relatedness, both the wild type and the agr mutant went to fixation or close to it, with approximately equal likelihoods (Fig. 5A). In contrast, the transmission rate had no influence on the relative success of the QS wild type versus the agr mutant (F1,12 = 0, P = 0.990) (Fig. 5A). There was no interaction between relatedness and transmission rate (F1,12 = 0.005, P = 0.948): the effect of relatedness did not depend on transmission rate or vice versa. Because the QS wild type led to greater growth (Fig. 1A) and virulence (Fig. 3), we then wanted to determine the consequences of relatedness on virulence. We did this by combining the results from the selection experiment with the data we collected on the mortality rate in infections. Consequently, we estimated that high relatedness would lead to a higher virulence (56.6% survival) than low relatedness (71.3% survival) after 3 rounds of selection (Fig. 5B).
FIG 5.
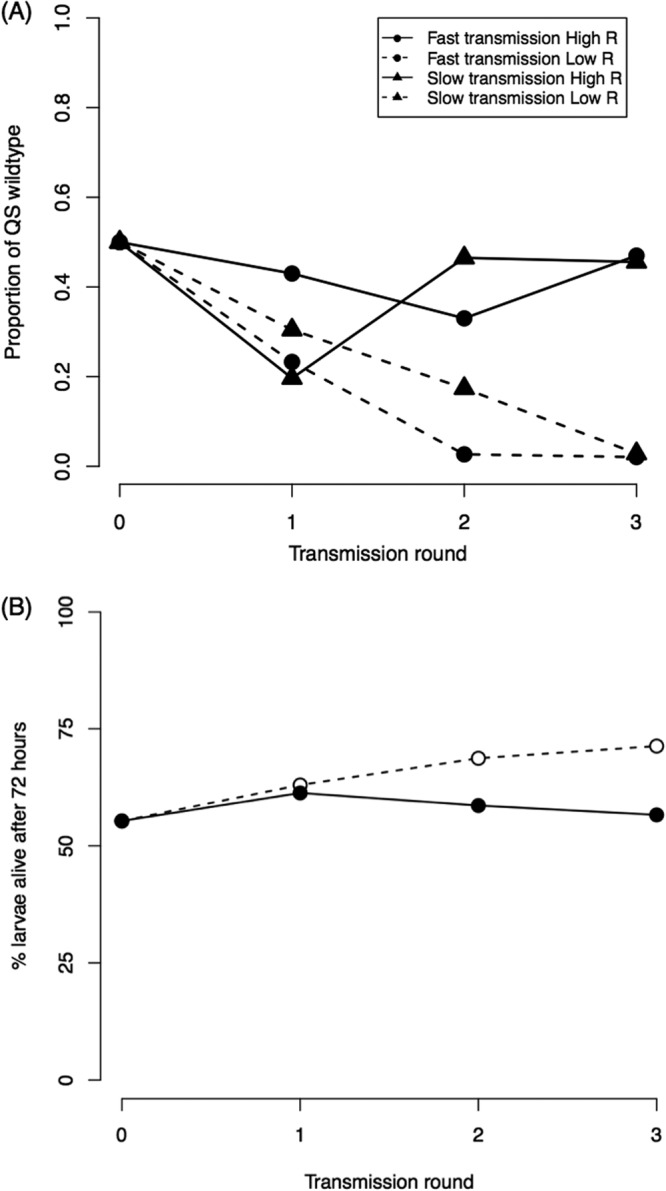
Transmission, relatedness, and virulence. (A) The mean proportion of QS wild-type strains is plotted against rounds of selection in the wax moth larvae. Each point represents the mean of 4 independent replicate selection lines, with solid lines representing high relatedness (R), dashed lines representing low relatedness, circles representing fast transmission (15 h), and triangles representing slow transmission (48 h). (B) Predicted virulence (given as larva survival at 72 h) for the mean of the high relatedness (●, solid line) and low relatedness (○, dashed line) treatments over the rounds of the experiment. Overall, a higher relatedness led to a higher proportion of the QS wild type and a higher predicted virulence. In contrast, transmission rate had no significant effect on the proportion of QS wild-type strains or virulence.
DISCUSSION
We have shown that QS in S. aureus is a cooperative social trait that can be exploited by “cheats” which do not respond to QS and that social interactions between cells determine virulence. More specifically, we found that (i) QS provides a benefit at the group level (Fig. 1A; Fig. S1) but agr mutants who do not perform QS can increase in frequency when in coinfections with a QS wild type (Fig. 1B), (ii) the social interaction between bacterial cells determines virulence (Fig. 3), which is negatively correlated with the proportion of cheats in the population (Fig. 2), and (iii) in an experimental evolution study, a relatively high strain diversity (low relatedness) allows cheats to spread (Fig. 5A), leading to an estimated decrease in virulence (Fig. 5B), but transmission rate had no significant influence on the relative success of cheats (Fig. 5A).
Cooperation and clinical epidemiology.
Our results provide the first demonstration that QS is a cooperative social trait in a Gram-positive bacterial species (Fig. 1 to 3). In order to demonstrate this, we showed that QS provides a fitness benefit at the group level (Fig. 1) but that this can be exploited in mixed cultures by mutants who not respond to signal (Fig. 1 and 2) (27). Although some previous studies are consistent with QS being a cooperative social trait, they have not shown the fitness benefit of responding to signal at the group level (5, 40). Consequently, they are also consistent with a fitness benefit due to a nonsocial trait. Nonetheless, QS via potential agr systems occurs across the firmicutes, including Staphylococcus spp. and Clostridium spp., suggesting possible similar social behaviors (41). Furthermore, despite its different underlying genetic architecture and molecular biology, our results are strikingly similar to those obtained in the Gram-negative opportunistic pathogen P. aeruginosa (18, 19, 21, 22). This suggests a convergent evolution for signaling to coordinate cooperation, using different signaling mechanisms and signaling molecules (42).
Our results also provide an explanation for the potentially contradictory results seen with agr mutants in clinical and in vivo studies. It has been observed that agr mutants arise in frequencies between 5 and 50% in a wide range of staphylococcal infections of humans (3, 4, 43, 44). On the basis of these results, it has been argued that either an agr mutation confers a selective advantage or agr mutants are effectively neutral during infections. In contrast, experimental studies in animal models have shown that agr mutants are significantly less virulent and that agr expression confers a survival advantage to S. aureus during infection (1, 8). Our observation that QS via agr is a cooperative social trait can explain both of these results, because while QS is beneficial, agr mutants can exploit the QS of others (cheat) and therefore increase in frequency. Furthermore, agr mutants found in clinical infections have tended to be agrC and agrA (signal-blind) mutants, which do not respond to signal (22, 23). The relative lack of agrB (signal-negative) mutants that respond to signal but do not produce signal is consistent with a role of social evolution: in mixed populations with the wild type, agrB mutants would still pay the cost of responding to the signal produced by the wild type, while agrC and agrA mutants would not.
Our results have at least two possible implications for medical intervention strategies. First, the ability of agr mutants to act as cheats and invade populations can be exploited. Specifically, such cheats could be used to reduce virulence or introduce medically useful genes, such as those for antibiotic susceptibility, into populations (31). Second, the social nature of QS can influence the relative usefulness of antivirulence strategies. For example, a clinical study on P. aeruginosa showed that the use of a QS-inhibiting drug stopped the spread of naturally arising QS mutants in infections and therefore diminished natural selection toward reduced virulence, thereby increasing the potential spread of more-virulent genotypes in a hospital environment (45).
Transmission, relatedness, and virulence.
Theoretical models have predicted that increased strain diversity within hosts can lead to either higher or lower virulence, through its influence on genetic relatedness. Greater strain diversity leads to lower genetic relatedness, which can lead to greater competition for host resources, which would select for higher virulence (38, 46). In contrast, lower relatedness between interacting bacteria can also select against the cooperation required for coordination of virulence factor production and increased fitness and hence lead to lower virulence (11, 13). We attempted to disentangle these two influences by varying both the level of competition and relatedness in an experimental evolution study (Fig. 4). We manipulated relatedness by varying the number of clones with which we inoculated each host, using either 1 (relatedness [r] = 1) or >100 (r ≈ 0.5, at which the wild type and mutant are equally abundant) (47). As predicted, we found a significant influence of relatedness, with higher relatedness favoring QS and higher virulence (Fig. 5), as has been found previously in P. aeruginosa (23). We found a large variance in the high-relatedness treatment, with the wild type and the agr mutant going to fixation with approximately equal probabilities. This variance may reflect the influence of drift, which we expect to be important in experimental evolution studies with population structuring, because this leads to population bottlenecks and relatively small effective population sizes (48).
We manipulated the risk of host death by varying how long we allowed growth to occur in the host (transmission speed), such that a relatively small (15 h; fast transmission) or large (48 h; slow transmission) proportion of hosts would have died before the chance for transmission. Theory predicts that fast transmission should select for higher virulence because there is a lower risk of death and there are more opportunities to exploit a host (38). Therefore, as QS regulates virulence, we predicted that lengthening the time of transmission would select against QS and favor the selection of agr mutants. We found, in contrast to these theoretical predictions, no significant effect of transmission time. One possible explanation for this is that our transmission times were not sufficiently different. However, we chose 15 and 48 h in order to maximize the potential influence of transmission: at 15 h, the onset and expression of agr had occurred (see Fig. S5 in the supplemental material), but there were still very few deaths, and at 48 h, there was substantial death but not enough to kill all the hosts in each group and therefore bring the experiment to an end.
An alternative explanation for why transmission speed had no influence is that the influence of host mortality is swamped by selection for/against cooperation. Consistent with this, a previous theoretical model that combined the potential for cooperation and host mortality found that that while mortality could influence virulence, the effect was relatively negligible (11). This suggests that the extent to which a pathogen is able to grow in a host can be far more important than any consequences of higher growth for host mortality. Given that many virulence factors in bacteria appear to be cooperative public goods (49), this could be a relatively general pattern in bacteria. Consequently, while theoretical work on virulence has focused on the trade-off between parasite growth and host mortality (38, 50), these models may be of little relevance to microorganisms such as bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Williams for the strains used in this study and Alan Cockayne, Owen Darch, Rasmus Jensen, Roman Popat, Freya Harrison, and two anonymous referees for technical support and comments on the manuscript.
We thank the European Research Council, the Royal Society, the Medical Research Council, the Leverhulme Trust, the Natural Environment Research Council (grant NE/J007064/1), and the Human Frontier Science Program for funding.
Footnotes
Published ahead of print 16 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01216-13.
REFERENCES
- 1.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 2.Mandell G, Bennett J, Dolin R. 2010. Mandell, Douglas and Bennett's principles and practice of infectious disease, 7th ed. Churchill Livingstone Elsevier, London, United Kingdom [Google Scholar]
- 3.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. 2008. agr function in clinical Staphylococcus aureus isolates. Microbiology 154:2265–2274. 10.1099/mic.0.2007/011874-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP. 2008. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J. Infect. Dis. 198:1171–1174. 10.1086/592051 [DOI] [PubMed] [Google Scholar]
- 5.Paulander W, Nissen Varming A, Bæk KT, Haaber J, Frees D, Ingmer H. 2013. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio 3:e00459–12. 10.1128/mBio.00459-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. 2007. Generation of virulence factor variants in Staphylococcus aureus biofilms. J. Bacteriol. 189:7961–7967. 10.1128/JB.00789-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemp M, Massey RC. 2007. The use of insect models to study human pathogens. Drug Discov. Today 4:105–110. 10.1016/j.ddmod.2007.06.007 [DOI] [Google Scholar]
- 8.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem. Rev. 111:117–151. 10.1021/cr100370n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghoul M, Griffin AS, West SA. Toward an evolutionary definition of cheating. Evolution, in press [DOI] [PubMed] [Google Scholar]
- 10.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449. 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- 11.West SA, Buckling A. 2003. Cooperation, virulence and siderophore production in bacterial parasites. Proc. Biol. Sci. 270:37–44. 10.1098/rspb.2002.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carnes EC, Lopez DM, Donegan NP, Cheung A, Gresham H, Timmins GS, Brinker CJ. 2010. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat. Chem. Biol. 6:41–45. 10.1038/nchembio.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown SP, Hochberg ME, Grenfell BT. 2002. Does multiple infection select for raised virulence? Trends Microbiol. 10:401–405. 10.1016/S0966-842X(02)02413-7 [DOI] [PubMed] [Google Scholar]
- 14.Qazi SN, Counil E, Morrissey J, Rees CE, Cockayne A, Winzer K, Chan WC, Williams P, Hill PJ. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074–7082. 10.1128/IAI.69.11.7074-7082.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckling A, Brockhurst MA. 2008. Kin selection and the evolution of virulence. Heredity 100:484–488. 10.1038/sj.hdy.6801093 [DOI] [PubMed] [Google Scholar]
- 16.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. 2008. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 381:300–309. 10.1016/j.jmb.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 17.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67:43–63. 10.1146/annurev-micro-092412-155635 [DOI] [PubMed] [Google Scholar]
- 18.Diggle SP, Griffin AS, Campbell GS, West SA. 2007. Cooperation and conflict in quorum-sensing bacterial populations. Nature 450:411–414. 10.1038/nature06279 [DOI] [PubMed] [Google Scholar]
- 19.Sandoz KM, Mitzimberg SM, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881. 10.1073/pnas.0705653104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilder CN, Diggle SP, Schuster M. 2011. Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5:1332–1343. 10.1038/ismej.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rumbaugh KP, Diggle SP, Watters CM, Ross-Gillespie A, Griffin AS, West SA. 2009. Quorum sensing and the social evolution of bacterial virulence. Curr. Biol. 19:341–345. 10.1016/j.cub.2009.01.050 [DOI] [PubMed] [Google Scholar]
- 22.Köhler T, Buckling A, van Delden C. 2009. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U. S. A. 106:6339–6344. 10.1073/pnas.0811741106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rumbaugh KP, Trivedi U, Watters C, Burton-Chellew MN, Diggle SP, West SA. 2012. Kin selection, quorum sensing and virulence in pathogenic bacteria. Proc. Biol. Sci. 279:3584–3588. 10.1098/rspb.2012.0843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popat R, Crusz SA, Messina M, Williams P, West SA, Diggle SP. 2012. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 279:4765–4771. 10.1098/rspb.2012.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darch SE, West SA, Winzer K, Diggle SP. 2012. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 109:8259–8263. 10.1073/pnas.1118131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dandekar AA, Chugani S, Greenberg EP. 2012. Bacterial quorum sensing and metabolic incentives to cooperate. Science 338:264–266. 10.1126/science.1227289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West SA, Winzer K, Gardner A, Diggle SP. 2012. Quorum sensing and the confusion about diffusion. Trends Microbiol. 20:586–594. 10.1016/j.tim.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 28.Peleg AY, Monga D, Pillai S, Mylonakis E, Moellering RC, Eliopoulos GM. 2009. Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J. Infect. Dis. 199:532–536. 10.1086/596511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross-Gillespie A, Gardner A, West SA, Griffin AS. 2007. Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170:331–342. 10.1086/519860 [DOI] [PubMed] [Google Scholar]
- 31.Brown SP, West SA, Diggle SP, Griffin AS. 2009. Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:3157–3168. 10.1098/rstb.2009.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffin AS, West SA, Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430:1024–1027. 10.1038/nature02744 [DOI] [PubMed] [Google Scholar]
- 33.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299–314. 10.2307/1390807 [DOI] [Google Scholar]
- 34.Crawley MJ. 2012. The R book. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 35.Velicer GJ. 2003. Social strife in the microbial world. Trends Microbiol. 11:330–337. 10.1016/S0966-842X(03)00152-5 [DOI] [PubMed] [Google Scholar]
- 36.MacLean RC, Gudelj I. 2006. Resource competition and social conflict in experimental populations of yeast. Nature 441:498–501. 10.1038/nature04624 [DOI] [PubMed] [Google Scholar]
- 37.Gore J, Youk H, van Oudenaarden A. 2009. Snowdrift game dynamics and facultative cheating in yeast. Nature 459:253–256. 10.1038/nature07921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71:37–78. 10.1086/419267 [DOI] [PubMed] [Google Scholar]
- 39.Anderson RM, May RM. 1982. Coevolution of hosts and parasites. Parasitology 85:411–426. 10.1017/S0031182000055360 [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Evans BA, Rozen DE. 2010. Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc. Biol. Sci. 277:2991–2999. 10.1098/rspb.2010.0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiner E, Scott J, Minton NP, Winzer K. 2012. An agr quorum sensing system that regulates granulose formation and sporulation in Clostridium acetobutylicum. Appl. Environ. Microbiol. 78:1113–1122. 10.1128/AEM.06376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams P, Winzer K, Chan WC, Cámara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1119–1134. 10.1098/rstb.2007.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. 2010. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202:1593–1599. 10.1086/656915 [DOI] [PubMed] [Google Scholar]
- 44.Sakoulas G, Moise PA, Rybak MJ. 2009. Accessory gene regulator dysfunction: an advantage for Staphylococcus aureus in health-care settings? J. Infect. Dis. 199:1558–1559. 10.1086/598607 [DOI] [PubMed] [Google Scholar]
- 45.Köhler T, Perron GG, Buckling A, van Delden C. 2010. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000883. 10.1371/journal.ppat.1000883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alizon S, Hurford A, Mideo N, Van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22:245–259. 10.1111/j.1420-9101.2008.01658.x [DOI] [PubMed] [Google Scholar]
- 47.Grafen A. 1985. A geometric view of relatedness. Oxf. Surv. Evol. Biol. 2:28–89 [Google Scholar]
- 48.Kümmerli R, Gardner A, West SA, Griffin AS. 2009. Limited dispersal, budding dispersal, and cooperation: an experimental study. Evolution 63:939–949. 10.1111/j.1558-5646.2008.00548.x [DOI] [PubMed] [Google Scholar]
- 49.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. 2007. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38:53–77. 10.1146/annurev.ecolsys.38.091206.095740 [DOI] [Google Scholar]
- 50.Alizon S, de Roode JC, Michalakis Y. 2013. Multiple infections and the evolution of virulence. Ecol. Lett. 16:556–567. 10.1111/ele.12076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.