Abstract
Bordetella bronchiseptica is pervasive in swine populations and plays multiple roles in respiratory disease. Most studies addressing virulence factors of B. bronchiseptica utilize isolates derived from hosts other than pigs in conjunction with rodent infection models. Based on previous in vivo mouse studies, we hypothesized that the B. bronchiseptica type III secretion system (T3SS) would be required for maximal disease severity and persistence in the swine lower respiratory tract. To examine the contribution of the T3SS to the pathogenesis of B. bronchiseptica in swine, we compared the abilities of a virulent swine isolate and an isogenic T3SS mutant to colonize, cause disease, and be transmitted from host to host. We found that the T3SS is required for maximal persistence throughout the lower swine respiratory tract and contributed significantly to the development of nasal lesions and pneumonia. However, the T3SS mutant and the wild-type parent are equally capable of transmission among swine by both direct and indirect routes, demonstrating that transmission can occur even with attenuated disease. Our data further suggest that the T3SS skews the adaptive immune response in swine by hindering the development of serum anti-Bordetella antibody levels and inducing an interleukin-10 (IL-10) cell-mediated response, likely contributing to the persistence of B. bronchiseptica in the respiratory tract. Overall, our results demonstrate that the Bordetella T3SS is required for maximal persistence and disease severity in pigs, but not for transmission.
INTRODUCTION
Respiratory disease in pigs is the most serious concern for swine producers today. The most recent National Animal Health Monitoring System (NAHMS) survey found that respiratory problems are a major cause of mortality in swine herds, with 53.7% of nursery pig deaths and 60.1% of grower-finisher pig deaths attributed to respiratory problems (1). Bordetella bronchiseptica is widespread in swine populations and is an important contributor to respiratory disease in pigs. In young pigs, it is a primary cause of bronchopneumonia, and in older pigs it contributes to secondary pneumonia. It is the primary etiologic agent of nonprogressive atrophic rhinitis, a mild to moderately severe reversible condition, and it promotes colonization by toxigenic strains of Pasteurella multocida, which leads to severe progressive atrophic rhinitis (2, 3). In pigs with pneumonia, B. bronchiseptica is often isolated in combination with other pathogens (4). Numerous studies have demonstrated that coinfection with B. bronchiseptica increases colonization and exacerbates the severity of disease caused by both viral and bacterial pathogens, including swine influenza virus, porcine reproductive and respiratory syndrome virus, porcine respiratory coronavirus, Haemophilus parasuis, P. multocida, and Streptococcus suis (5–12).
B. bronchiseptica expresses many virulence factors, including adhesins, secretion systems, autotransporters, and toxins, that are globally regulated by the BvgAS two-component signal transduction system (13–16). In response to environmental cues, such as temperature or MgSO4 or nicotinic acid concentrations, BvgAS controls expression of a spectrum of phenotypic phases, transitioning between a virulent (Bvg+) phase and a nonvirulent (Bvg−) phase, a process referred to as phenotypic modulation. During the virulent Bvg+ phase, the BvgAS system is fully active and many of the known virulence factors are expressed, including the type III secretion system (T3SS). Conversely, BvgAS is inactive during the Bvg− phase, resulting in the maximal expression of motility loci, virulence-repressed genes (vrg genes), and genes required for the production of urease (17–19). The Bvg+ phase is necessary and sufficient for colonization of the respiratory tract (14, 20).
Many Gram-negative bacterial pathogens utilize a T3SS to translocate or inject protein effectors directly into the cytosol of a eukaryotic cell. These type III-secreted effector proteins have been demonstrated to interact with a variety of eukaryotic signal transduction pathways, thereby altering the physiological function of the eukaryotic cell (21–24). B. bronchiseptica expresses a T3SS similar to those shown to directly translocate effector proteins through a needlelike injection apparatus directly into eukaryotic cells, which causes disruption of host cell signaling and necrosis-like cell death (25, 26). Under Bvg+ conditions, the btr regulatory locus (including btrS, btrU, btrW, and btrV) is transcribed (27). BtrS is an extracytoplasmic-function (ECF) sigma factor that is necessary and sufficient for activating the more than 20 T3SS-related genes (e.g., bsc, bop, bsp, and bte) (27). BscN is the putative ATPase that provides energy for the secretion of effector proteins and is required for the function of the T3SS apparatus (26).
The mouse has been the primary animal model used to investigate virulence mechanisms of B. bronchiseptica, including the contribution of the T3SS to pathogenesis of B. bronchiseptica. T3SS mutants induce higher levels of serum anti-Bordetella antibodies than mice infected with the wild type (28). Recent experiments have also demonstrated that the T3SS gene products, BopB, BopC and BteA, are secreted and required for cytotoxicity of mammalian cells (29–31). Additionally, it has been demonstrated that the T3SS mediates persistent bacterial colonization of the lower respiratory tract by altering dendritic cell maturation and enhancing the production of the anti-inflammatory cytokine interleukin-10 (IL-10) (26, 28, 32). The induction of IL-10 as a mechanism to evade a host inflammatory response has recently been shown to be specifically mediated by the T3SS gene product BopN (33).
Using a B. bronchiseptica swine isolate within a natural swine host infection, we have demonstrated a number of key phenotypic differences associated with several B. bronchiseptica mutants compared to those reported using rodent infection models (20, 34). These differences include clinical presentation of disease, pathology, and host immune responses (20, 34). While the biological roles of the Bordetella T3SS-secreted gene products may be similar in swine and rodents, no definitive data exist with respect to the role of the T3SS in the pathogenesis of disease in swine, either in vivo or with swine tissue or cells in vitro. In addition, pigs provide an excellent natural host system to identify bacterial genetic components required for transmission. In this report, we investigated the role of T3SS in Bordetella pathogenesis in swine by constructing an in-frame deletion of the bscN structural gene in KM22, a virulent B. bronchiseptica swine isolate, and compared this mutant to KM22 for its ability to colonize, cause disease, and be transmitted by direct and indirect routes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. bronchiseptica strain KM22, isolated from a swine herd with atrophic rhinitis, harbors a ribotype and pertactin type shared with the majority of strains isolated from swine and has been used in a number of studies by our laboratory in which we have demonstrated its ability to cause disease and be transmitted among swine (5, 7, 8, 10, 20, 34–37). RB50 and WD3 (RB50ΔbscN) have been previously described (26, 28, 32, 38, 39). All B. bronchiseptica strains were grown at 37°C on Bordet-Gengou (BG) agar (Difco, Sparks, MD) supplemented with 10% sheep's blood or, when specified, in Stainer-Scholte (SS) broth (40). Escherichia coli One Shot TOP10 (Invitrogen, Carlsbad, CA) was used for all cloning steps, and E. coli SM10λpir was used to mobilize plasmid pTN53 into B. bronchiseptica KM22. When appropriate, antibiotics were included at the following concentrations: carbenicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 100 μg/ml; and streptomycin, 40 μg/ml.
Cloning and construction of the B. bronchiseptica T3SS mutant (KM22ΔbscN).
For construction of KM22ΔbscN, a 1,080-bp DNA fragment covering the region immediately 5′ of bscN and extending to codon 7 was amplified by PCR from KM22 genomic DNA using primers bscNf1-for and bscNf1-rev (Table 1), which were designed such that EcoRI and XbaI sites would be generated at the 5′ and 3′ ends, respectively. The resulting PCR product was cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to create pTN35. The insert of pTN35, containing the upstream region of bscN, was cloned into pUC19 (New England BioLabs, Ipswich, MA) via EcoRI and XbaI sites to obtain pTN41. A 1,258-bp DNA fragment extending from codon 436 of bscN into the adjacent downstream region was amplified from KM22 genomic DNA by PCR using primers bscNf2-for and bscNf2-rev (Table 1), which were designed such that XbaI and HindIII sites would be generated at the 5′ and 3′ ends, respectively. The resulting PCR product was cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to obtain pTN37. The insert of pTN37, containing the downstream region of bscN, was excised by XbaI and HindIII digestion and cloned into XbaI-HindIII-digested pTN41 to create pTN43. The 2,335-bp DNA fragment, containing the upstream and downstream regions of bscN, was amplified from pTN43 by PCR using primers bscNf1-for and bscNf2B-rev (Table 1), which were designed such that EcoRI and AvrII sites would be generated at the 5′ and 3′ ends, respectively. The resulting PCR product was cloned into pCR2.1 (Invitrogen, Carlsbad, CA) to create pTN52. The insert of pTN52 was excised by EcoRI and AvrII digestion and cloned into EcoRI-AvrII-digested allelic exchange vector pSS4245 (39, 41) to create pTN53. pTN53 was introduced into E. coli SM10λpir (42) and transconjugated into KM22 as previously described (39, 41). A single colony selected for subsequent use, designated KM22ΔbscN, was confirmed to have a deletion of the bscN gene by DNA sequence analysis of a PCR amplicon encompassing regions upstream and downstream of the bscN gene.
TABLE 1.
Primers used for cloning and quantitative real-time PCR in this study
| Primer name | Sequence (5′–3′) |
|---|---|
| bscNf1-for | GAATTCCAGTTGAAGGTCGCGCTATTGT |
| bscNf1-rev | TCTAGAGATGTAGTGGTACTGACGCATGCCCCT |
| bscNf2-for | TCTAGAATCATCGGACCCGAATCCTAAT |
| bscNf2-rev | AAGCTTAAGGCGTCAGGGCGCAGTGCCAGCACA |
| bscNf2B-rev | CCTAGGCTTAAGGCGTCAGGGCGCAGTGCCAGCACA |
| bscNck-for | ACGGAAGGAGCCGGGCATGACTGAGAAG |
| bscNck-rev | AGTCCGGCCATGCCTCGACAGACATCCT |
Precipitation of extracellular proteins.
B. bronchiseptica strains were grown for 48 h at 37°C in SS broth supplemented with 40 μg/ml streptomycin. Bacterial cells were pelleted by centrifugation at 4,500 × g for 15 min at 4°C, and 5-ml culture supernatant samples were collected. Secreted proteins were then precipitated by addition of sodium deoxycholate to 0.1% and trichloroacetic acid to a final concentration of 20% (wt/vol). Following overnight incubation at 4°C, samples were centrifuged for 2 h at 20,000 × g. Pellets were washed with ice-cold acetone, centrifuged an additional 15 min at 20,000 × g, air dried, and resuspended in 0.25 ml SDS-PAGE loading buffer (Invitrogen, Carlsbad, CA). Next, proteins in 0.02 ml of each sample were separated by SDS-PAGE with a 4 to 12% bis-Tris acrylamide gel (Invitrogen, Carlsbad, CA), and the bands were visualized by silver staining.
Cytotoxicity assays.
Cytotoxicity assays were carried out as previously described (26, 27). Briefly, J774 mouse macrophages were cultured in Dulbecco's modified Eagle medium (DMEM) broth supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, 1% nonessential amino acids, and 1% sodium pyruvate to 85% confluence at 37°C with 5% CO2. Warmed RPMI medium lacking phenol red, with 5% FBS, 1% l-glutamine, 1% nonessential amino acids, and 1% sodium pyruvate, was then used to replace the DMEM. Porcine alveolar macrophages were isolated from bronchoalveolar lavage fluid (BALF) of healthy, noninfected pigs as previously described (43). Isolated alveolar macrophages were seeded at 105 cells per well in 0.1 ml in 96-well flat-bottom plates in RPMI 1640 lacking phenol red with 2% swine sera, 1 mM l-glutamine, and 5 mM HEPES. Bacterial infections using strains KM22, KM22ΔbscN, RB50, and RB50ΔbscN were prepared from cultures collected in mid-log phase and were carried out using a multiplicity of infection (MOI) of 10:1. Bacterial suspensions were centrifuged onto the macrophages at 250 × g for 5 min, and cell cultures were incubated at 37°C with 5% CO2 for 4 h. The cell culture supernatants were collected, and percent lactate dehydrogenase (LDH) release was analyzed using the Cytotox96 kit (Promega, Madison, WI) according to the manufacturer's instructions. Results were analyzed for significance using GraphPad (San Diego, CA) Prism 5 software. Means were compared using a one-way analysis of variance (ANOVA) with Tukey's posttest and a significance level (P) of <0.001.
Pathogenesis experiments in swine.
B. bronchiseptica strains KM22 and KM22ΔbscN were cultured on BG agar supplemented with 10% sheep's blood at 37°C for 40 h. Suspensions of these cultures were prepared in phosphate-buffered saline (PBS) to contain approximately 2 × 109 CFU/ml, and 106 CFU diluted in PBS were used for inoculation of pigs. Serial dilutions of the inoculum for each strain were plated on BG agar plates to determine CFU/ml and to confirm the expected colony morphology and hemolytic phenotype. All colonies from all inocula displayed the expected colony morphology and hemolytic characteristics. Each experimental group was housed in a separate isolation room in animal biosafety level 2 (ABSL2) biocontainment facilities. B. bronchiseptica was not isolated from nasal swabs collected from all piglets prior to the start of the experiment. Forty-eight naturally farrowed early-weaned piglets were divided into 2 experimental groups of 16 pigs each and 1 control group of 16. Pigs were inoculated intranasally at 1 week of age with 1 ml (0.5 ml/nostril) of a bacterial suspension containing 106 CFU of KM22 or KM22ΔbscN or with 1 ml of sterile PBS. Bacterial colonization of the nasal cavity was quantified by nasal swabs on days 1, 3, and 5 and by nasal washes at all other sample days postinoculation. On days 7, 14, 28, and 56 postinoculation, 4 pigs from each group were euthanized for sample collection to evaluate the bacterial burden in the respiratory tract, immune responses, and lesion severity. At necropsy, snouts were transected and removed at the level of the first premolar tooth. A 1-cm cross section was cut from the caudal portion of the snout and used for scoring turbinate atrophy. The trachea was then severed just below the larynx, and the trachea and lung were removed. All housing, husbandry, and experiments performed with pigs were in accordance with the law and approved by the National Animal Disease Center Institutional Animal Care and Use Committee.
Determination of colonization.
Nasal swabs were placed into tubes containing 500 μl PBS and vortexed. Nasal washes were performed by instilling 5 ml of PBS into the nasal cavity through one nostril and collecting the effluent into a beaker. Tracheal washes were performed by placing a segment of trachea approximately 8 cm in length in a 15-ml centrifuge tube with 5 ml of PBS and shaking vigorously. Lung lavage was performed by filling the lungs with 50 ml of sterile PBS, gently massaging, and aspirating; approximately 25 ml of the PBS was recovered. Serial 10-fold dilutions were made from nasal swab fluids, nasal and tracheal washes, and lung lavages, and the number of CFU of B. bronchiseptica per ml was determined by plating 100 μl of the dilutions on duplicate selective blood agar plates containing 20 μg/ml penicillin, 10 μg/ml amphotericin B, 10 μg/ml streptomycin, and 10 μg/ml spectinomycin. The limit of detection was 10 CFU/ml. B. bronchiseptica was identified on the basis of colony morphology. Statistical analyses of the nasal colonization data were performed using a mixed linear model (SAS 9.2 for Windows XP; SAS Institute Inc., Cary, NC) for repeated measures and a heterogeneous autoregressive covariance structure to best account for unequal study day intervals. Linear combinations of the least-squares mean estimates for log10 CFU were used in a priori contrasts after testing for either a significant (P < 0.05) effect of the bacterial challenge strains or a significant strain by time interaction. Comparisons were made between challenge groups for each isolate at each time point using a 5% level of significance (P < 0.05) to assess statistical differences. Endpoint data for tracheal and lung bacterial loads were analyzed by analysis of variance using a general linear model for unbalanced data that included treatment group and study day, with CFU as the dependent variable. A 5% level of significance (P < 0.05) was used to assess statistical differences.
Turbinate atrophy scores.
Each of the four scrolls of the ventral turbinate in snout cross sections was assigned an atrophy score that ranged from 0 to 4 (0 = normal, 1 = more than half of the scroll remaining, 2 = half or less of the scroll remaining, 3 = the scroll is straightened with only a small portion left, and 4 = total atrophy of the scroll). The atrophic rhinitis score is the sum of the four scroll scores and ranges from 0 to 16. Statistical analysis on the turbinate scores was performed using the GraphPad Prism 6 software program by means of a one-way ANOVA with the Brown-Forsythe test and Bartlett test to test for equal variance across groups and Tukey's multiple-comparison test to compare differences among groups. A 5% level of significance (P < 0.05) was considered significant.
Pathological evaluation of the lung.
At necropsy, an estimate of gross lung lesion involvement was assigned based on the percentage of each lung lobe affected and the percentage of total lung volume represented by each lobe. Percentage of total lung volume of each lobe was estimated as 10% for the left cranial, 10% for the left middle, 25% for the left caudal, 10% for the right cranial, 10% for the right middle, 25% for the right caudal, and 10% for the intermediate lung lobes. Sections of lung taken at necropsy for microscopic evaluation were fixed in 10% neutral buffered formalin for 24 h and then placed in 70% ethanol. All sections were routinely processed and embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Quantitative real-time PCR (qRT-PCR).
Tracheal epithelial cells were collected from infected and control pigs on days 7 and 28 postchallenge. A longitudinal cut along the dorsal membrane of the trachea was made, and a clean razor blade was used to scrape the epithelial lining. Cells were placed into TRIzol and stored at −80°C. RNA was subsequently isolated using MagMAX-96 for Microarrays total RNA isolation kit (Invitrogen, Carlsbad, CA), and cDNA was synthesized using High Capacity RNA-to-cDNA master mix (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. SYBR green-based real-time PCR was performed for various mRNA targets, and data were analyzed as previously described (8). Fold change in transcript level was determined using the 2-ΔΔCT method (CT, threshold cycle) (44). Results were log2 transformed and analyzed for significance using an unpaired Student t test for statistical analysis.
Humoral response.
Serum was collected from the same four pigs from each experimental group on days 21, 28, 35, 42, and 56 postchallenge. BALF was collected from infected and control pigs on days 28 and 56 postchallenge. Titers of anti-Bordetella antibodies were measured by enzyme-linked immunosorbent assay (ELISA) as previously described (34). Briefly, plates were coated with heat-killed B. bronchiseptica KM22 or KM22ΔbscN (grown to an optical density at 600 nm [OD600] of 0.8). Sera and BALF were serially diluted, and B. bronchiseptica-specific antibody was detected using secondary antibodies specific for swine IgG (Kirkegaard and Perry Laboratories, Gaithersburg, MD). Endpoint antibody titers were expressed as the reciprocal of the highest dilution giving an OD405 of ≥0.1 higher than the average OD measured for the negative control (serum from cesarean-derived colostrum-deprived piglets for serum ELISA and PBS for BALF ELISA) after a 45-min incubation with substrate. For BALF, endpoint antibody titers were expressed as the reciprocal of the highest dilution giving an OD405 at least 3 standard deviations above the OD for the negative control (PBS). Statistical analyses of the serum antibody response were performed using a mixed linear model (SAS 9.2 for Windows XP; SAS Institute Inc., Cary, NC) for repeated measures and a heterogeneous autoregressive covariance structure to best account for unequal study day intervals. Linear combinations of the least-squares mean estimates for the log2 mean relative titers were used in a priori contrasts after testing for either a significant (P < 0.05) effect of the bacterial challenge strains or a significant strain by time interaction. Comparisons were made between challenge groups for each challenge group at each time point using a 5% level of significance (P < 0.05) to assess statistical differences. Statistical analyses of the anti-Bordetella IgG titers in the BALF were performed using a one-way analysis of variance with Tukey's posttest.
Cell-mediated response.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood as previously described (45), and tracheal-bronchial lymph node (TBLN) cells were isolated by mechanical homogenization followed by successive passages through filters with 70-μm and 40-μm pores. PBMCs and TBLN cells were collected from infected and control pigs on days 28 and 56 postchallenge. Isolated cells were enumerated and plated at 106 cells per well in 0.2 ml in triplicate for each treatment of 0.05 ml medium or 107 CFU of heat-killed KM22 in 0.05 ml. Cells were incubated for 3 days at 37°C in 5% CO2, and supernatants from triplicate wells were pooled and stored at −80°C. A commercial ELISA specific for swine IL-10 (R&D Systems, Minneapolis, MN) or swine IFN-γ (Invitrogen, Carlsbad, CA) was performed according to the manufacturer's recommendations. A one-way analysis of variance with Tukey's posttest was used for statistical analysis.
Transmission experiments in swine.
Twenty-eight naturally farrowed early-weaned piglets were used in the transmission experiments, and B. bronchiseptica was not isolated from nasal swabs collected from all piglets prior to the start of the experiment. One of three isolation rooms in ABSL2 biocontainment facilities were used to house each experimental group. Two isolation rooms were identically set up with two pens placed 18 in. apart. Two pigs were intranasally inoculated with 106 CFU of KM22 and placed in a pen together in one isolation room, and two pigs were intranasally inoculated with 106 CFU of KM22ΔbscN and placed in a pen together in a second isolation room. These pigs served as the primary challenged pigs in the study. A control group of 8 pigs was intranasally inoculated with 1 ml sterile PBS and housed together in a third isolation room with no pens. Two days after inoculation of primary challenged pigs, 3 naive pigs, serving as a direct contacts, were placed in each of the pens containing the primary challenged pigs and 5 naive pigs, serving as indirect contacts, were placed in each of the empty pens. Nasal colonization of all pigs was monitored by nasal swabs collected 5, 12, 19, and 26 days postcontact. Nasal swabs were obtained from indirect contacts first, followed by direct contacts, and then nasal swabs were obtained from primary challenged pigs. Necropsies were performed on day 26 postcontact, and all pigs were euthanized with an overdose of barbiturate. The trachea was then severed just below the larynx, and the trachea and lung were removed for determination of colonization. All housing, husbandry, and experiments performed with pigs were in accordance with the law and approved by the Institutional Animal Care and Use Committee.
RESULTS
In-frame deletion of bscN in KM22 leads to decreased secretion of polypeptides and decreased in vitro cytotoxicity.
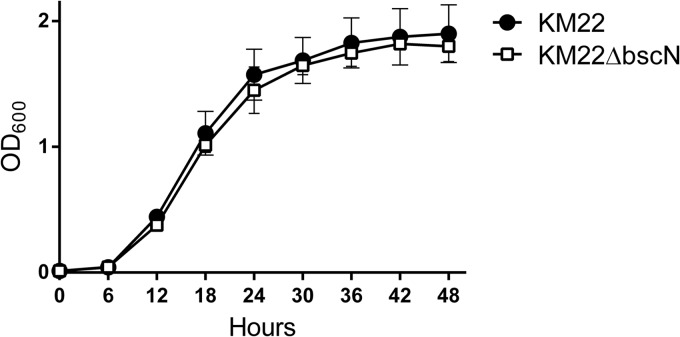
The virulent B. bronchiseptica swine isolate KM22 has been successfully used by our laboratory to develop a reproducible swine respiratory disease model reflective of clinical B. bronchiseptica infections within swine herds and host-to-host transmission by direct and indirect routes (5, 7, 8, 10, 20, 34–37). To examine the contribution of the T3SS to Bordetella pathogenesis in swine, we constructed an in-frame deletion in the bscN gene of KM22. Initial in vitro characterization indicated that strain KM22ΔbscN exhibited a growth rate (Fig. 1) and degree of hemolysis on BG blood agar similar to those of wild-type KM22 (data not shown), demonstrating that deletion of the bscN gene did not alter the growth rate or the secretion of the bifunctional adenylate cyclase toxin/hemolysin. To test if BscN is involved in protein secretion in KM22, similar to the function reported for the laboratory reference strain RB50 (26), we compared the protein profiles of culture supernatants from KM22 and KM22ΔbscN grown under Bvg+ conditions. As shown in Fig. 2, several proteins were present in the bacterial culture supernatant from KM22 and absent from the supernatant from KM22ΔbscN. Additionally, several proteins present in the bacterial culture supernatant from KM22 are present in greater amounts in the supernatant from KM22ΔbscN, some of which appear to be absent from the supernatant from KM22. These results are similar to the results reported for the corresponding isogenic mutant in RB50 and demonstrate that deletion of the bscN gene in KM22 results in an apparent decrease in a subset of Bvg+ phase-associated proteins in the culture supernatant, presumably caused by a defect in their secretion.
FIG 1.
Growth curve of wild-type B. bronchiseptica strain KM22 and isogenic mutant KM22ΔbscN. Optical densities at 600 nm (OD600) of KM22 and KM22ΔbscN versus time are shown. All data points represent averages obtained from triplicate cultures. Values are means ± standard deviations.
FIG 2.
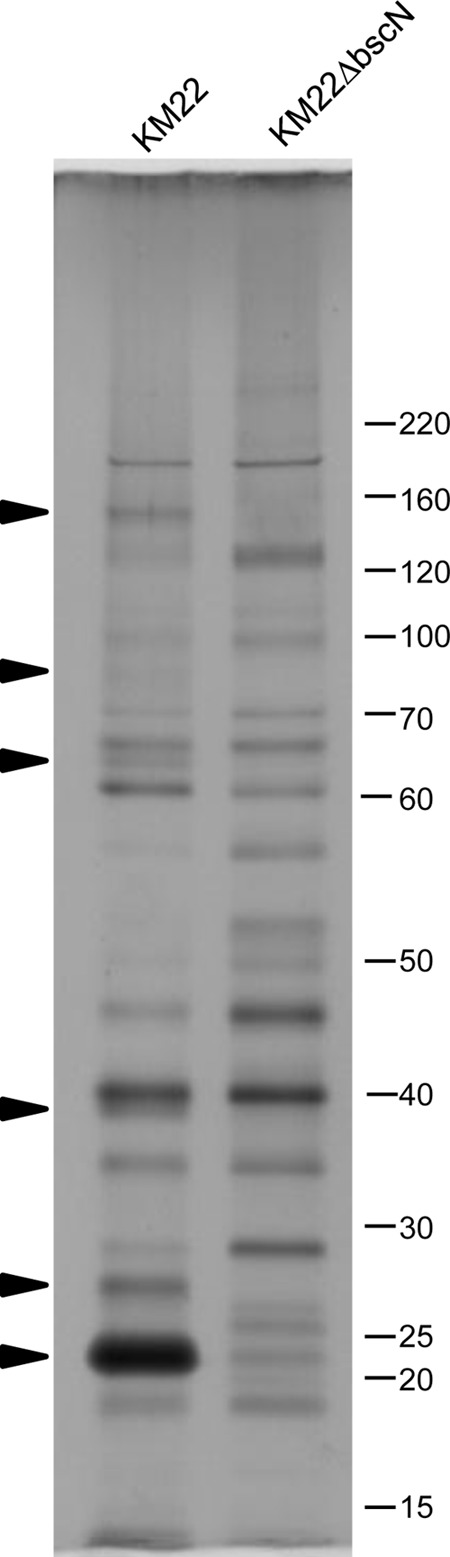
Culture supernatant protein profiles of wild-type B. bronchiseptica strain KM22 and isogenic mutant KM22ΔbscN. Secreted proteins were recovered from bacterial culture supernatants by precipitation with trichloroacetic acid, separated by SDS-PAGE, and visualized by silver staining. Arrowheads indicate proteins present in the supernatant from KM22 and not in supernatant from KM22ΔbscN. The positions of standard proteins and their molecular masses in kilodaltons are indicated on the right.
Perhaps the best-characterized phenotype of the Bordetella T3SS is killing of mammalian cells (26, 28–31, 39). Therefore, we assessed the ability of KM22 and KM22ΔbscN, along with RB50 and WD3 (RB50ΔbscN), to mediate cytotoxicity in J774 murine macrophages and primary porcine alveolar macrophages in vitro. Both KM22 and RB50 displayed a high level of cytotoxicity (Fig. 3A and B). In contrast, the cytotoxic activity of RB50ΔbscN and KM22ΔbscN was significantly lower than that of the corresponding wild-type strains RB50 and KM22, respectively (Fig. 3A and B). These data are in agreement with previous findings involving RB50 and RB50ΔbscN in J774 mouse macrophages and demonstrate that BscN is required for T3SS-mediated cytotoxicity by strain KM22.
FIG 3.
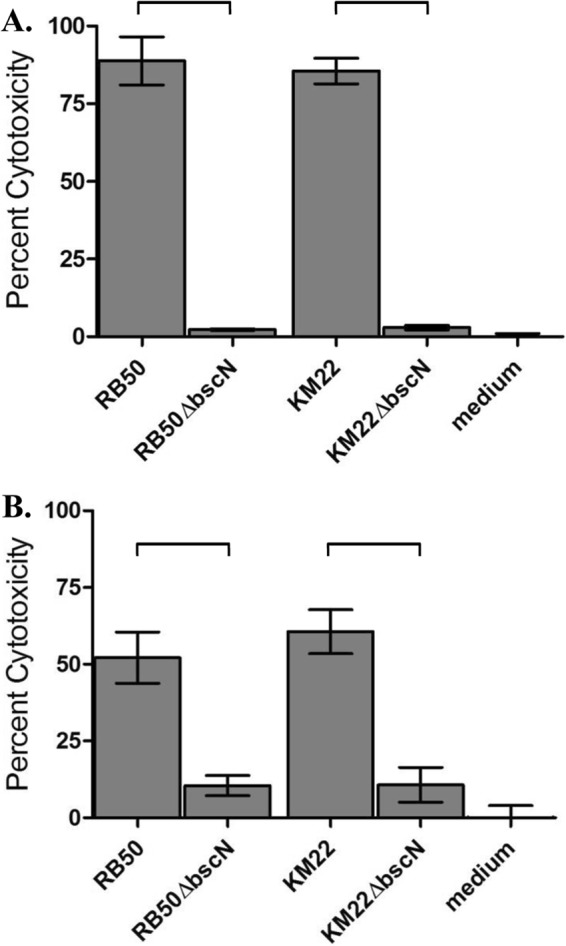
T3SS-mediated cytotoxicity of B. bronchiseptica KM22 and KM22ΔbscN. Cytotoxicity as measured by percent LDH release in J774 murine macrophages (A) and porcine alveolar macrophages (B) following 4 h of exposure to medium alone or to the indicated strain using an MOI of 10:1 is shown. Bars represent means of three independent experiments ± standard deviations. A one-way analysis of variance with Tukey's posttest was used for statistical analysis, and P values less than 0.001 are indicated with connecting bars.
The Bordetella T3SS is required for persistence in the swine respiratory tract.
To investigate the contribution of the Bordetella T3SS to the pathogenesis of B. bronchiseptica in swine, we intranasally inoculated groups of 1-week-old piglets with B. bronchiseptica strain KM22 or KM22ΔbscN or 1 ml of sterile PBS. Clinical signs were noted only in piglets from groups inoculated with KM22 or KM22ΔbscN, which mainly consisted of sneezing and coughing. No clinical signs were observed in piglets from groups inoculated with PBS.
Colonization of the nasal cavity was evaluated on days 1, 3, 5, and 7 and weekly thereafter until day 56 postinfection (Fig. 4A). B. bronchiseptica CFU were not recovered from any site in the respiratory tracts of piglets inoculated with PBS. In general, there was no significant overall difference in CFU levels recovered from the nasal cavities of pigs infected with either KM22 or KM22ΔbscN (Fig. 4A). However, focusing on specific days, there were statistically significant differences observed at some time points. Specifically, on day 7 postinfection, the CFU levels recovered from KM22ΔbscN-infected pigs were significantly (P < 0.0001) higher than the CFU levels recovered from KM22-infected pigs (Fig. 4A). In contrast, by day 21 postinfection the CFU levels recovered from KM22-infected pigs were significantly (P < 0.035) higher than the CFU levels recovered from KM22ΔbscN-infected pigs (Fig. 4A). Similar levels CFU were then recovered from KM22 and KM22ΔbscN-infected pigs until day 56 postinfection, when CFU levels recovered from KM22-infected pigs were significantly (P < 0.003) higher than the CFU levels recovered from KM22ΔbscN-infected pigs (Fig. 4A).
FIG 4.
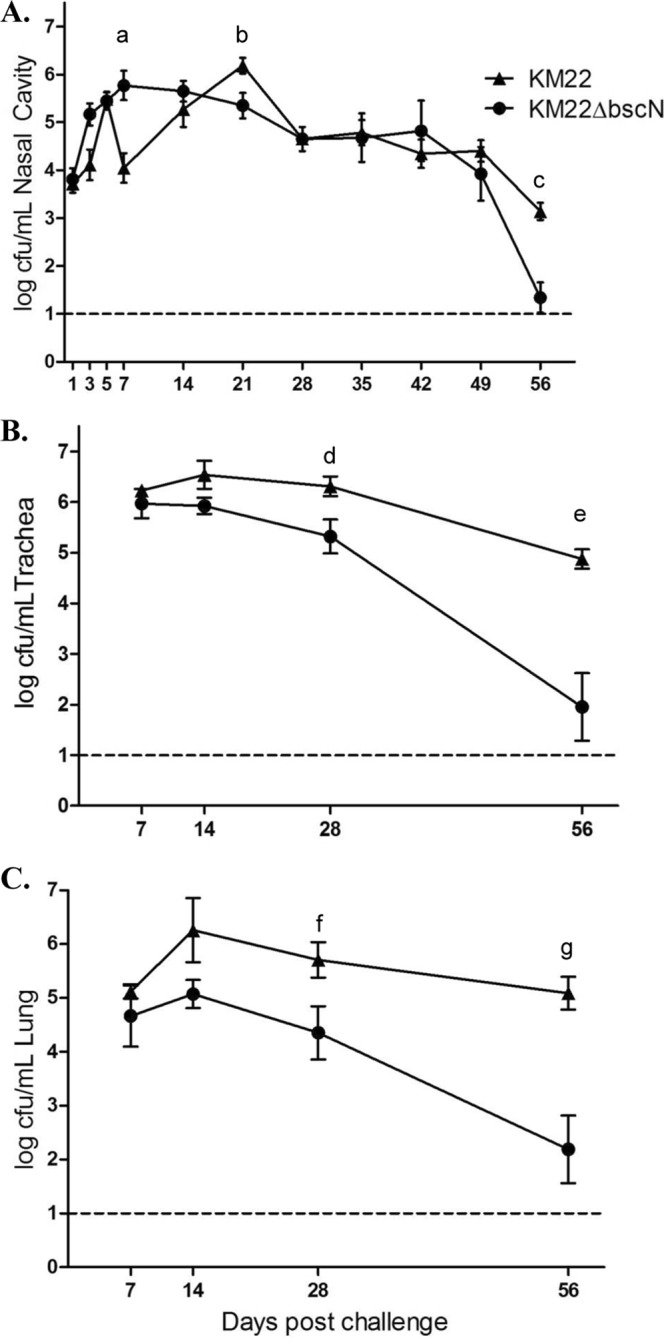
Colonization of the swine respiratory tract by wild-type B. bronchiseptica strain KM22 and KM22ΔbscN. Groups of 16 pigs were inoculated intranasally with KM22 or KM22ΔbscN. Bacterial load in the nasal cavity (A) was quantified 1, 3, 5, 7, 14, 21, 28, 35, 42, 49, and 56 days postinoculation. Bacterial load in the tracheae (B) and the lungs (C) was quantified 7, 14, 28, and 56 days postinoculation. The y axis indicates the mean log10 CFU ± standard errors. The dashed lines indicate the limit of detection (log10 of 1). Statistical difference is indicated for KM22ΔbscN compared to KM22 on day 7 postinoculation (P = 0.0001; a), for KM22 compared to KM22ΔbscN on day 21 postinoculation (P < 0.035; b), for KM22 compared to KM22ΔbscN on day 56 postinoculation (P < 0.003; c), for KM22 compared to KM22ΔbscN on day 28 postinoculation (P < 0.04; d), for KM22 compared to KM22ΔbscN on day 56 postinoculation (P < 0.0001; e), for KM22 compared to KM22ΔbscN on day 56 postinoculation (P < 0.05; f), and for KM22 compared to KM22ΔbscN on day 56 postinoculation (P = 0.0001; g). A P value less than 0.05 was considered significant.
Colonization of the trachea and lung was evaluated on days 7, 14, 28, and 56 postinfection (Fig. 4B and C). Starting on day 14 postinfection, lower CFU levels were recovered from both the tracheae and lungs of KM22ΔbscN-infected pigs than from those of pigs infected with the wild-type KM22. By day 28 postinfection, the difference between these groups for both tissues was statistically significant (P < 0.05), and significant differences were still apparent on day 56 postinfection (P < 0.0001). Taken together, the colonization data demonstrate that a functional Bordetella T3SS is required for persistence in the lower swine respiratory tract.
The Bordetella T3SS mediates increased pathology in the swine respiratory tract.
The degree of atrophic rhinitis was evaluated by determining the mean turbinate atrophy score for piglets inoculated with KM22, KM22ΔbscN, or PBS on days 7, 14, 28, and 56 postinfection. No substantial degree of turbinate atrophy (defined as an atrophic rhinitis score of greater than 2) was observed for pigs inoculated with PBS (Table 2). The group of piglets infected with KM22 had mean turbinate atrophy scores of greater than 2 at all time points examined (Table 2), and the mean scores were statistically higher than those for the pigs inoculated with PBS on days 14, 28, and 56 postinfection. Mean turbinate atrophy scores were greater than 2 for the group of piglets infected with KM22ΔbscN beginning on day 14 and continuing through day 56, and the mean scores were statistically higher than those for the pigs inoculated with PBS on days 14 and 56 postinfection; however the mean degree of turbinate atrophy was always lower for KM22ΔbscN-infected pigs than for KM22-infected pigs, and the difference was statistically significant at day 56 postinfection (Table 2). Combining the scores for each group across time points for the whole experiment showed that the mean turbinate scores for both the KM22-infected pigs (P < 0.001) and the KM22ΔbscN-infected pigs (P < 0.01) were statistically higher than the mean score for the PBS-inoculated pigs and that the mean score for the KM22-infected pigs was statistically higher than the mean score for the KM22ΔbscN-infected pigs (P < 0.05).
TABLE 2.
Mean turbinate atrophy scores and percentages of the lungs affected by pneumonia in pigs inoculated with B. bronchiseptica KM22 or KM22ΔbscN
| Day of necropsy | Group | Mean turbinate atrophy scorea (range) | Mean % pneumonia (range) | No. of pigs with pneumoniab |
|---|---|---|---|---|
| 7 | PBS control | 1.5 (0–2) | 0.875 (0–2) | 2/4c |
| KM22 | 3.75 (1–8) | 3.625 (0–10) | 3/4 | |
| KM22ΔbscN | 1.75 (0–4) | 5.0 (0–13) | 3/4d | |
| 14 | PBS control | 0.5 (0–1) | 0.75 (0–3) | 1/4c |
| KM22 | 4.5 (2–8)* | 7.5 (2–20) | 4/4 | |
| KM22ΔbscN | 4.25 (4–5)* | 7.38 (0–27) | 2/4e | |
| 28 | PBS control | 0.25 (0–1) | 0.25 (0–1) | 1/4c |
| KM22 | 5.75 (3–8)* | 9.75 (0–17) | 3/4 | |
| KM22ΔbscN | 3.67 (1–8) | 1.625 (0–6.5) | 1/4 | |
| 56 | PBS control | 1.25 (1–2) | 0 (0) | 0/4 |
| KM22 | 4.75 (4–6)** | 5.0 (2–13) | 4/4 | |
| KM22ΔbscN | 3.0 (2–4)* | 0.625 (0–2.5) | 1/4c |
*, mean turbinate scores were statistically different from those for pigs inoculated with PBS (P < 0.05); **, mean turbinate scores were statistically different from those for pigs inoculated with PBS (P < 0.001) or KM22ΔbscN-infected pigs (P < 0.05).
Data are reported as the number of pigs with pneumonia out of the total number possible for the respective strain of B. bronchiseptica.
Microscopically these lesions were not consistent with Bordetella infection.
Microscopically only 1 of the 3 pigs with pneumonia had lesions that were consistent with Bordetella infection, while 2 of the 3 pigs had only lesions similar to those seen in PBS control pigs.
Microscopically only 1 of the 2 pigs with pneumonia had lesions that were consistent with Bordetella infection, while the other had only lesions similar to those seen in PBS control pigs.
The degree of pneumonia and microscopic character of the lesions were also determined for piglets inoculated with KM22, KM22ΔbscN, or PBS on days 7, 14, 28, and 56 postinfection. We observed small areas of consolidation in the lungs of 4 of the pigs inoculated with PBS (Table 2). Microscopically, the areas of consolidation in these control pigs consisted of multifocal areas of interstitial pneumonia with septal infiltrates consisting mainly of macrophages and a few neutrophils, which is not typical of Bordetella infection. Fourteen of the 16 pigs infected with KM22 had areas of lung consolidation consistent with Bordetella infection. The gross lesions in KM22-infected pigs progressed from acute red to plum-colored lesions at days 7 and 14 to chronic gray to yellow firm and fibrotic lesions at days 28 and 56. Microscopic lesions in the lungs of the 14 pigs with pneumonia infected with KM22 were characterized by suppurative bronchopneumonia with areas of necrosis and hemorrhage and eventual fibroplastic replacement of lung parenchyma, which is typical of B. bronchiseptica pneumonia. There were microscopic lesions similar to those seen in the PBS control pigs in 5 of these pigs as well. In contrast, only 7 pigs out of 16 infected with KM22ΔbscN had areas of consolidation, and only 3 of the 7 exhibited evidence of suppurative bronchopneumonia. These 3 pigs had increased numbers of neutrophils in the airways but none of the necrosis, hemorrhage, or fibroplasia observed in the pigs infected with KM22. The other 4 pigs infected with KM22ΔbscN that had areas of lung consolidation had lesions only microscopically similar to those of the PBS control pigs. Combined, these data demonstrate that the Bordetella T3SS mediated increased pathology and significantly contributed to B. bronchiseptica-associated pneumonia.
The Bordetella T3SS modulates the swine immune response.
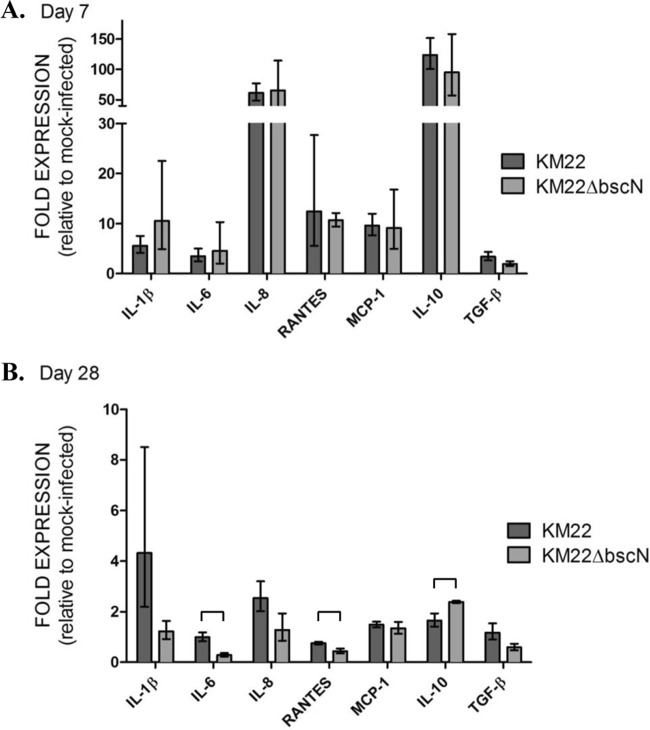
To determine if the Bordetella T3SS is capable of modulating local immune responses in the swine respiratory tract, changes in tracheal epithelial cell (TEC) mRNA levels of several cytokines and chemokines were evaluated on days 7 and 28 postinfection. On day 7, several proinflammatory mediators, including IL-1β, IL-8, and monocyte chemoattractant protein 1 (MCP-1), exhibited increased mRNA levels in TECs collected from both KM22- and KM22ΔbscN-infected pigs (Fig. 5A). In addition, mRNA levels of IL-10 were increased in both challenge groups over those for noninfected pigs (Fig. 5A). By day 28 postinfection, mRNA levels overall were lower than at day 7 postinfection for both challenge groups (Fig. 5A versus B). However, significant differences in mRNA levels for IL-6, RANTES, and IL-10 between challenge groups were observed on day 28 (Fig. 5B). Specifically, we detected higher mRNA levels for cytokines IL-6 and RANTES in TECs from KM22-infected pigs than in TECs from KM22ΔbscN-infected pigs, whereas IL-10 mRNA levels were greater in pigs infected with KM22ΔbscN (Fig. 5B). The levels of mRNA for the cytokine targets were also evaluated in alveolar macrophages collected from the same pigs; however, levels were minimally increased over those detected in noninfected pigs, and there were no significant differences between the challenge groups (data not shown).
FIG 5.
Gene expression changes in tracheal epithelial cells following infection. Tracheal epithelial cells were collected on days 7 (A) and 28 (B) postchallenge from pigs intranasally infected with KM22 or KM22ΔbscN or mock challenged with PBS, and gene expression changes were evaluated by qRT-PCR. The x axis indicates the cytokines whose genes were analyzed. The y axis indicates the mean log2 fold change in expression of each gene relative to mock challenge with PBS using the 2-ΔΔCT method. Error bars represent the standard deviations (SD) for 4 pigs per treatment group. An unpaired Student t test was used for statistical analysis, and P values less than 0.05 are indicated with connecting bars.
To investigate the effect of the Bordetella T3SS on the development of anti-Bordetella humoral immunity in swine, serum levels of anti-Bordetella IgG were quantified in pigs infected with KM22 or KM22ΔbscN or mock infected with PBS by ELISA using heat-killed KM22 or KM22ΔbscN whole cells as the antigen. The data represent samples collected from the same pigs throughout the experiment, with four pigs per group. When heat-killed KM22 whole cells were used as the antigen, similar serum IgG levels were detected in all groups at all time points examined except for day 56, where higher serum levels of anti-Bordetella IgG were observed in both KM22- and KM22ΔbscN-infected pigs (Fig. 6A). When heat-killed KM22ΔbscN whole cells were used as the antigen, higher serum anti-Bordetella IgG levels were detected in KM22- and KM22ΔbscN-infected pigs than in PBS-inoculated pigs beginning at 28 days postinfection and continuing through day 56 postinfection (Fig. 6B). Focusing on KM22ΔbscN-infected pigs, higher serum IgG levels were detected at days 35, 42, and 56 postinfection than were detected in KM22-infected pigs (Fig. 6B).
FIG 6.
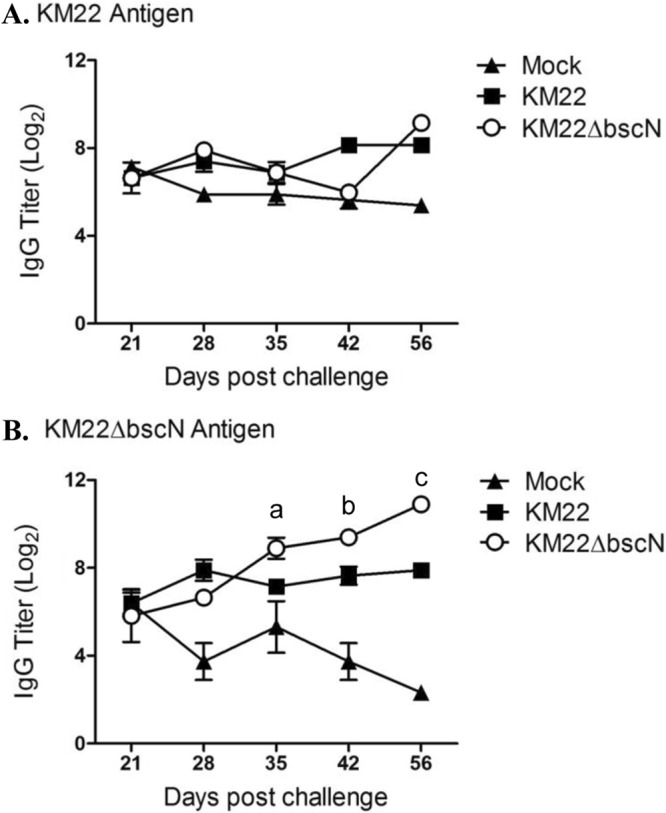
Serum anti-Bordetella IgG titers. Serum was collected from pigs intranasally inoculated with KM22 or KM22ΔbscN or mock challenged with PBS on days 21, 28, 35, 42, and 56 postchallenge. Titers of anti-Bordetella antibodies were measured by ELISA using B. bronchiseptica KM22 (A) or KM22ΔbscN (B) as the antigen. The y axis indicates the log2 mean relative titer ± standard errors for 4 pigs per treatment group. Statistical difference is indicated for KM22ΔbscN compared to KM22 on day 35 postinoculation (P = 0.08; a), for KM22 compared to KM22ΔbscN on day 42 postinoculation (P = 0.0306; b), and for KM22 compared to KM22ΔbscN on day 56 postinoculation (P < 0.001; c). A P value less than 0.05 was considered significant.
Next we quantified the levels of anti-Bordetella IgG in BALF collected from pigs infected with KM22 or KM22ΔbscN or mock infected with PBS by ELISA. Significantly (P < 0.05) higher IgG levels were detected in the BALF from KM22-infected pigs than in BALF from KM22ΔbscN-infected and uninfected control pigs when heat-killed KM22 whole cells were used as the antigen at day 28 postinfection (Fig. 7A). At day 56 postinfection, the IgG levels in the BALF of KM22ΔbscN-infected pigs increased so that both KM22- and KM22ΔbscN-infected pigs exhibited significantly (P < 0.05) higher IgG levels than PBS-inoculated pigs. A similar trend was observed when heat-killed KM22ΔbscN whole cells were used as the antigen (Fig. 7B).
FIG 7.
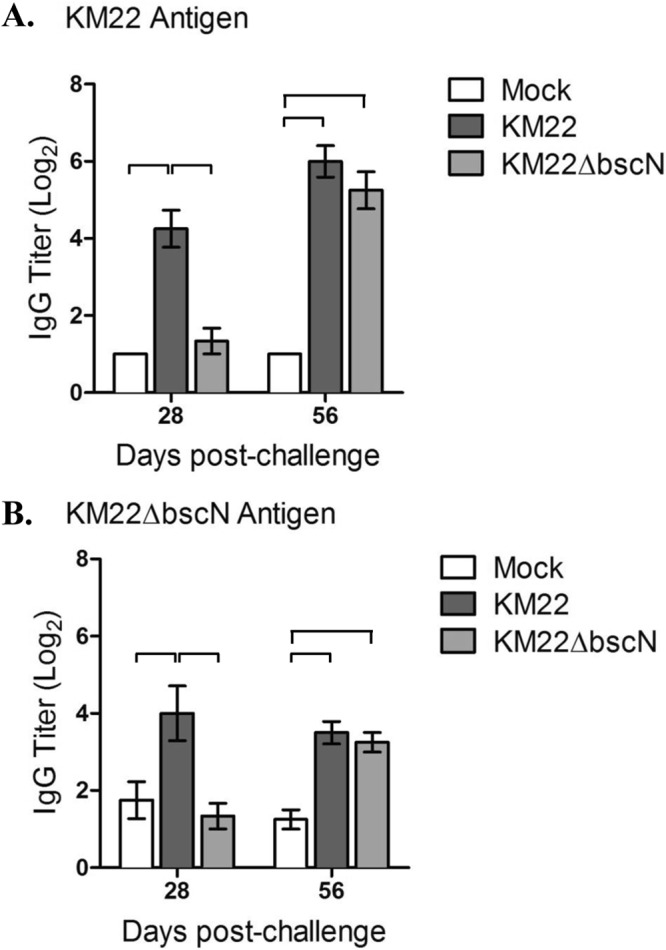
Anti-Bordetella IgG titers in BALF. Titers of anti-Bordetella antibodies were measured by ELISA using B. bronchiseptica KM22 (A) or KM22ΔbscN (B) as the antigen. The y axis indicates the log2 mean relative titer ± standard deviation (error bars) for 4 pigs per treatment group. A one-way analysis of variance with Tukey's posttest was used for statistical analysis, and P values less than 0.05 are indicated with connecting bars.
To evaluate cell-mediated immune responses elicited following infection with wild-type KM22 compared to the T3SS mutant, production of IL-10 and gamma interferon (IFN-γ) by PBMCs and TBLN cells following restimulation with heat-killed KM22 was measured. On day 28 postinfection significantly (P < 0.05) higher IL-10 levels were detected in the restimulated PBMCs from KM22-infected pigs than in PBMCs from pigs inoculated with KM22ΔbscN or PBS (Fig. 8A). At day 56 postinfection, IL-10 produced by PBMCs collected from KM22-infected pigs was reduced compared to the level at day 28 postinfection such that similar IL-10 levels were detected in the restimulated PBMCs from KM22- and KM22ΔbscN-infected pigs and PBS-inoculated pigs (Fig. 8A). At day 28 postinfection, higher IL-10 levels were produced by the restimulated TBLN cells from KM22-infected pigs than by those from KM22ΔbscN-infected pigs (Fig. 8B). On day 56 postinfection similar IL-10 levels were detected in the restimulated TBLN cells from KM22- and KM22ΔbscN-infected pigs; however these levels were not significantly increased over those for PBS-inoculated pigs (Fig. 8B). B. bronchiseptica-specific IFN-γ recall responses were also evaluated; however, very low, if any, levels of IFN-γ were detected in the supernatant of cells from any pig at any time point (data not shown).
FIG 8.
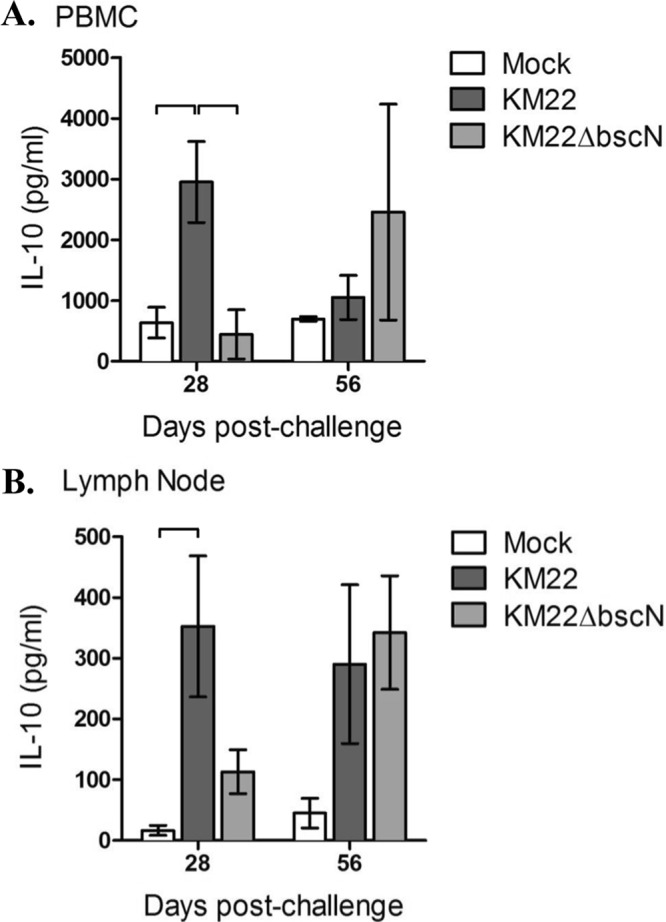
Anti-Bordetella IL-10 recall response following infection. PBMCs (A) and TBLN cells (B) were collected on days 28 and 56 postchallenge from pigs infected intranasally with KM22 or KM22ΔbscN or mock challenged with PBS. Cells were restimulated with heat-killed B. bronchiseptica KM22, and the supernatants were analyzed for IL-10 production by ELISA. IL-10 levels are presented as the means ± SD for 4 pigs per treatment group. A one-way analysis of variance with Tukey's posttest was used for statistical analysis, and P values less than 0.05 are indicated with connecting bars.
The Bordetella T3SS is not required for direct or indirect transmission of B. bronchiseptica in swine.
Despite numerous studies investigating the contribution of the Bordetella T3SS to general pathogenesis and specific mechanisms of immune modulation, a role for the Bordetella T3SS in transmission has never been investigated. Therefore, we directly tested whether the Bordetella T3SS is required for host-to-host transmission by direct or indirect routes. For this experimental design, two pigs serving as primary challenged pigs were intranasally inoculated with KM22 and placed in a pen within an isolation room. After 2 days, three naive pigs, serving as direct contacts, were added to the same pen, and five naive pigs were placed in a second pen placed approximately 18 in. away from the first pen. These five pigs served as indirect-contact pigs. The exact experimental procedure was replicated in a second isolation room except that the two pigs serving as primary challenged pigs were intranasally inoculated with KM22ΔbscN. In a third isolation room with no pens, pigs were intranasally inoculated with sterile PBS and allowed to comingle.
Direct transmission occurred for both KM22 and KM22ΔbscN, with all of the direct-contact pigs from each group becoming colonized by day 12 postcontact (Table 3). Transmission by indirect contact was detected on day 12 for only two pigs, both from the KM22ΔbscN group, but by day 19 postcontact B. bronchiseptica was recovered from the nasal cavities of all direct-contact and indirect-contact pigs from both the KM22 and KM22ΔbscN groups. Colonization of the trachea and lungs was also determined when pigs were necropsied 26 days postcontact. The B. bronchiseptica strain given to the primary challenged pigs from each group was isolated from the tracheae and lungs of all primary challenged, direct-contact, and indirect-contact pigs from both the wild-type KM22 and KM22ΔbscN groups (Table 4). B. bronchiseptica was not recovered from any site in piglets inoculated with PBS. These results show that both the wild-type KM22 and the T3SS mutant were equally capable of transmission in swine by both direct and indirect contact. Additionally, these data demonstrate that the T3SS of B. bronchiseptica is not required for direct or indirect transmission in swine.
TABLE 3.
Direct and indirect transmission to naive pigsa
| No. of days after contactb | No. of pigs infectedc with contact strain: |
|||||
|---|---|---|---|---|---|---|
| KM22 |
KM22ΔbscN |
|||||
| Primary | Direct | Indirect | Primary | Direct | Indirect | |
| 5 | 2/2 | 0/3 | 0/5 | 2/2 | 0/3 | 0/5 |
| 12 | 2/2 | 3/3 | 0/5 | 2/2 | 3/3 | 2/5 |
| 19 | 2/2 | 3/3 | 5/5 | 2/2 | 3/3 | 5/5 |
| 26 | 2/2 | 3/3 | 5/5 | 2/2 | 3/3 | 5/5 |
Two days after 2 primary pigs were challenged by the intranasal route with 106 CFU of the contact strain, three naive pigs serving as direct-contact pigs were added to the same pen. On the same day, five naive pigs serving as indirect-contact pigs were added to a separate pen. One isolation room was used per contact strain studied, and the experimental procedure was duplicated in the two isolation rooms used.
Days after initial contact with primary challenged pigs.
Data are reported as the number of pigs positive out of the total number possible.
TABLE 4.
Respiratory tract colonization in direct- and indirect-transmission pigs 26 days after initial contact with primary challenged pigsa
| Sample site | No. of pigs infectedb with contact strain: |
|||||
|---|---|---|---|---|---|---|
| KM22 |
KM22ΔbscN |
|||||
| Primary | Direct | Indirect | Primary | Direct | Indirect | |
| Nasal swab | 2/2 | 3/3 | 5/5 | 2/2 | 3/3 | 5/5 |
| Tracheal wash | 2/2 | 3/3 | 5/5 | 2/2 | 3/3 | 5/5 |
| Lung lavage | 2/2 | 3/3 | 5/5 | 2/2 | 3/3 | 5/5 |
Pigs were euthanized on day 28 of the experiment, 26 days after initial contact with primary challenged pigs.
Data are reported as the number of pigs positive out of the total possible number of pigs.
DISCUSSION
Despite widespread use of B. bronchiseptica vaccines by swine producers throughout the world, Bordetella-associated respiratory disease remains a significant problem for the industry (1, 46). The development of vaccines with improved efficacy is hindered by the absence of definitive data related to virulence mechanisms of B. bronchiseptica in swine. Evaluation of B. bronchiseptica-specific virulence factors involved in pathogenesis and transmission in swine provides data directly relevant and necessary for designing improved vaccines and therapeutic interventions. To examine the contribution of the T3SS to Bordetella pathogenesis and transmission in swine, we constructed an in-frame deletion in the bscN gene, which is required for a functional T3SS, in the virulent B. bronchiseptica swine isolate KM22. Altered in vitro phenotypes of the mutant compared to the parent strain include a decrease in a subset of proteins in the culture supernatant, presumably due to a defect in their secretion, and a significant decrease in T3SS-mediated cytotoxicity in both murine macrophages and primary porcine alveolar macrophages. These in vitro phenotypes of our T3SS deletion mutant, KM22ΔbscN, are similar to those reported for the corresponding RB50ΔbscN mutant (26, 28–31, 39).
To investigate the contribution of the Bordetella T3SS to pathogenesis of B. bronchiseptica in swine, we then compared the T3SS deletion mutant to the wild-type swine isolate for the ability to colonize and cause disease in pigs following intranasal inoculation. Colonization of the T3SS mutant in the nasal cavity was similar to that by KM22 except for three time points (days 7, 21, and 56). At day 7, the T3SS mutant was recovered at significantly higher levels than KM22. This finding was somewhat unexpected given that CFU levels in KM22-infected pigs on day 7 were sharply lower than those observed on days 5 and 14. On day 21 postinfection, levels of the T3SS mutant recovered from the nasal cavity were significantly lower than those of KM22, though this difference was less than 1 log, and no significant differences in CFU levels recovered between the challenge groups were observed at the subsequent time point, day 28. By day 56, levels of the T3SS mutant recovered from the nasal cavity were significantly lower than wild-type KM22 levels. The decreased colonization levels of the T3SS mutant at the last time point may imply a role for T3SS in persistence in the swine nasal cavity. A role for the Bordetella T3SS in persistence in the nasal cavity or upper respiratory tract has not been previously reported using rodent infection models (28). However, since the colonization data presented here did not continue past day 56 or until clearance of wild-type KM22, a definitive role for the Bordetella T3SS in persistence in the swine nasal cavity remains unclear. Even when nasal colonization levels were similar between pigs infected with wild-type KM22 or the T3SS mutant, the severity of turbinate atrophy was always lower in T3SS mutant-challenged pigs. This decrease suggests that the Bordetella T3SS contributes to the turbinate atrophy associated with B. bronchiseptica infections in swine.
When evaluating colonization of the lower respiratory tract, the T3SS mutant was recovered from both the tracheae and lungs at significantly lower levels than the wild-type on days 28 and 56 postinfection. The decreased bacterial burden of the T3SS mutant observed between days 28 and 56 postinfection suggests that the Bordetella T3SS may be required for persistence. While we observed a role for the T3SS in persistence within the nasal cavity, the impact of a functional T3SS was more profound in the tracheae and lungs. Prior studies utilizing rodent infection models have also reported a role for the T3SS in tracheal persistence, although significant differences have been reported depending on the mouse strain used (28). Coinciding with decreased bacterial burdens, disease severity was less in pigs infected with the T3SS mutant than in KM22-infected pigs. Overall, fewer lung lesions were observed in pigs infected with the T3SS mutant than in KM22-infected pigs, and, when present, the lesions were less extensive in terms of the necrosis, hemorrhage, or fibroplasia. Combined, these data demonstrate that the T3SS significantly contributes to B. bronchiseptica-associated pathology and disease severity in swine.
A previous study in rodents using passive transfer of immune sera demonstrated that antibody is sufficient for clearing B. bronchiseptica from the lower respiratory tract (45). Follow-up work indicated an important role for complement, in that antibody-mediated activation targeted the removal of B. bronchiseptica by Fc receptors on phagocytic cells (47). However, data have also demonstrated that B. bronchiseptica can persist in the lower respiratory tract despite the presence of B. bronchiseptica-specific antibody (32). This was particularly evident in the current study given the presence of circulating B. bronchiseptica-specific maternal antibody at the time of challenge, which continued to circulate for weeks following challenge. Our data indicate that maternal antibodies specific to B. bronchiseptica are directed toward T3SS proteins, as serum antibody levels specific to the T3SS mutant decreased in mock-infected pigs over time. In contrast, IgG levels specific to KM22 in noninfected pigs did not decrease over the same time period. B. bronchiseptica is ubiquitous in swine herds, and it is extremely difficult to find herds in which pigs have not been exposed to the bacterium and, thus, do not have circulating antibody. Sows are typically colonized at some point in their life, leading to the development of circulating B. bronchiseptica-specific IgG, which is transferred to piglets in colostrum. While our rearing protocol for piglets used in this study incorporates early weaning to eliminate vertical transmission of B. bronchiseptica from sow to piglets, it does not prevent colostrum ingestion and subsequent passive transfer of maternal antibody. Therefore, the immune status of piglets at challenge is significantly different from that of naive mice routinely used for identifying immune factors involved in B. bronchiseptica persistence and pathogenesis. While the disease caused by B. pertussis in humans is acute, carriage and transmission may not be acute. In fact using a nonhuman primate model, Warfel et al. demonstrated that baboons can remain colonized after 40 days postchallenge and transmit B. pertussis to naive hosts in the presence of robust levels of circulating antibodies (48). Therefore, B. bronchiseptica infection in sows and piglets may be more similar to the circumstances involved with B. pertussis in humans, and thus correlations between these two model systems may be more relevant to Bordetella pathogenesis in general.
Despite the presence of circulating B. bronchiseptica-specific IgG at the time of challenge, piglets were susceptible to B. bronchiseptica-mediated disease. T3SS proteins were able to modulate the B. bronchiseptica-specific immune response in challenged piglets given the significant differences observed in both humoral and cell-mediated immune responses. The ability of the T3SS to hinder development of the humoral immune response is shown in serum anti-Bordetella IgG titers, as titers increased steadily over time in pigs infected with the T3SS mutant when heat-killed KM22ΔbscN whole cells were used as the antigen. This antigen-specific response increased in piglets infected with the T3SS mutant as maternal antibody specific to B. bronchiseptica decreased over time. Pigs challenged with KM22 did not exhibit an increase in circulating B. bronchiseptica-specific IgG titers regardless of the B. bronchiseptica strain used as the antigen. In contrast to pigs infected with the T3SS mutant, KM22-infected pigs exhibited little evidence of an active humoral immune response greater than maternal antibody following KM22 challenge; however, it is difficult to distinguish between maternal antibody and newly developed antibody at these later time points.
Despite minimal differences in B. bronchiseptica-specific IgG levels in the sera, significantly higher IgG levels were detected in the in the BALF of KM22-infected pigs on days 28 and 56 following challenge. Immunoglobulin G circulating in the periphery is capable of transuding into the lower respiratory tract, and, as previously mentioned, antibody has been shown to play a role in clearance of B. bronchiseptica from the lower respiratory tract (45). At day 28 postinfection significantly higher mRNA expression levels for cytokines IL-6 and RANTES were detected in TECs from KM22-infected pigs than in TECs from pigs infected with the T3SS mutant, and lung lesions were more severe in KM22-infected pigs at day 28 as well. In addition, TEC mRNA levels of the anti-inflammatory cytokine IL-10 were increased in pigs infected with the T3SS mutant compared to those in KM22-infected pigs. Taken together, these results suggest that the presence of IgG in the lungs of KM22-challenged pigs was likely due to lung damage and subsequent transudation of immunoglobulin from circulation into the lung. Despite the increased levels of B. bronchiseptica-specific IgG in the lungs of KM22-challenged pigs, significantly higher CFU levels were recovered from the lungs of KM22 infected pigs than from lungs of pigs infected with the T3SS mutant on day 28 postchallenge, suggesting that the anti-Bordetella antibodies present in the lungs at this time point failed to contribute to clearance of wild-type B. bronchiseptica from the lower respiratory tract. This may be a result of the cytotoxicity of KM22 for alveolar macrophages, whose Fc receptors are required for antibody-mediated clearance (47). By day 56 postinfection, B. bronchiseptica-specific IgG levels were increased in pigs infected with the T3SS mutant and significantly fewer CFU were recovered from the lungs of pigs infected with the T3SS mutant. Given the minimal lung pathology exhibited in the pigs infected with the T3SS mutant at day 56 postinfection, it is likely that the IgG measured in the lungs at that time point was due to local production and not systemic IgG transuding into the lung. Future work aimed at identifying the specific B. bronchiseptica proteins that these IgG antibodies targeted may help distinguish between maternal antibody and antibody developed during the infection process, as well as specificity for clearance.
Various studies using mice have demonstrated that the T3SS activates the production of the cytokine IL-10 early in the infection process, thereby inhibiting the production of IFN-γ, which is also involved in antibody-mediated clearance of B. bronchiseptica (32, 33, 38, 49). To determine if B. bronchiseptica would employ a similar mechanism to skew the porcine immune response for persistent colonization, IL-10 production following restimulation of lymphocytes was evaluated. Consistent with previous studies, lymphocytes collected from peripheral blood and TBLNs of KM22-infected pigs on day 28 postinfection produced significantly more IL-10 following B. bronchiseptica restimulation than lymphocytes isolated from pigs infected with the T3SS mutant. On day 56 postinfection, no significant differences were observed for the IL-10 produced by PBMCs following restimulation, and the levels produced by PBMCs collected from KM22-infected pigs were less than those produced by PBMCs collected on day 28 postinfection. However, there was an increase in the amount of IL-10 produced by TBLN cells collected from pigs infected with the T3SS mutant on day 56 compared to day 28 postinfection. This IL-10 recall response in pigs challenged with the T3SS mutant parallels the levels of the anti-Bordetella antibodies present in the lungs, suggesting that IL-10 was involved in development of the B. bronchiseptica-specific immune response in these pigs. These data also provide evidence of the differential local versus systemic responses induced following infection with the T3SS mutant, as IL-10 recall responses were significantly elevated in TBLN cells, but not in the PBMCs at day 56. IL-10 was historically characterized as a Th2-polarizing cytokine, which supports the data currently presented. B. bronchiseptica-specific IFN-γ recall responses were also evaluated; however, very low, if any, levels of IFN-γ were detected in the supernatant of restimulated TBLN cells and PBMCs from any pig at any time point. Together, our results demonstrate that the Bordetella T3SS does alter the B. bronchiseptica adaptive immune response, likely allowing B. bronchiseptica to persist in the lower respiratory tract. Given that the local TEC responses at day 7 were not significantly different between groups, nor were colonization levels, our data support a role for the T3SS in altering the adaptive immune response, which significantly alters clearance.
Despite a high number of studies investigating the molecular mechanisms of immune modulation by the Bordetella T3SS, a potential role in transmission has never been investigated. We found that the T3SS mutant could be transmitted directly to naive pigs and indirectly to naive pigs when pigs were physically separated by pens. Our results demonstrate that the T3SS of B. bronchiseptica is not required for transmission in swine. Additionally, these data demonstrate that, even in animals exhibiting minimal disease, infection can still lead to transmission of B. bronchiseptica. These results highlight an important concern to consider when designing any vaccine or intervention strategy where the goal is to decrease transmission in addition to providing protection against clinically apparent disease.
ACKNOWLEDGMENTS
We thank Zahra Olson, Gwen Nordholm, and William Boatwright for excellent technical support and Brian Pottebaum, Jason Huegel, Jason Crabtree, and Dalene Whitney for animal care assistance.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print 23 December 2013
REFERENCES
- 1.U.S. Department of Agriculture 2008. Swine 2006, Part III: reference of swine health and health management in the United States. USDA/APHIS/VS CEAH N478.0308 USDA, Fort Collins, CO [Google Scholar]
- 2.Magyar T, Lax AJ. 2002. Atrophic rhinitis, p 169–197 In Brogden KA, Guthmiller J. (ed), Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 3.Chanter N, Magyar T, Rutter JM. 1989. Interactions between Bordetella bronchiseptica and toxigenic Pasteurella multocida in atrophic rhinitis of pigs. Res. Vet. Sci. 47:48–53 [PubMed] [Google Scholar]
- 4.Palzer A, Ritzmann M, Wolf G, Heinritzi K. 2008. Associations between pathogens in healthy pigs and pigs with pneumonia. Vet. Rec. 162:267–271. 10.1136/vr.162.9.267 [DOI] [PubMed] [Google Scholar]
- 5.Brockmeier SL, Palmer MV, Bolin SR. 2000. Effects of intranasal inoculation of porcine reproductive and respiratory syndrome virus, Bordetella bronchiseptica, or a combination of both organisms in pigs. Am. J. Vet. Res. 61:892–899. 10.2460/ajvr.2000.61.892 [DOI] [PubMed] [Google Scholar]
- 6.Loving CL, Brockmeier SL, Vincent AL, Palmer MV, Sacco RE, Nicholson TL. 2010. Influenza virus coinfection with Bordetella bronchiseptica enhances bacterial colonization and host responses exacerbating pulmonary lesions. Microb. Pathog. 49:237–245. 10.1016/j.micpath.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 7.Brockmeier SL, Palmer MV, Bolin SR, Rimler RB. 2001. Effects of intranasal inoculation with Bordetella bronchiseptica, porcine reproductive and respiratory syndrome virus, or a combination of both organisms on subsequent infection with Pasteurella multocida in pigs. Am. J. Vet. Res. 62:521–525. 10.2460/ajvr.2001.62.521 [DOI] [PubMed] [Google Scholar]
- 8.Brockmeier SL, Loving CL, Nicholson TL, Palmer MV. 2008. Coinfection of pigs with porcine respiratory coronavirus and Bordetella bronchiseptica. Vet. Microbiol. 128:36–47. 10.1016/j.vetmic.2007.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockmeier SL. 2004. Prior infection with Bordetella bronchiseptica increases nasal colonization by Haemophilus parasuis in swine. Vet. Microbiol. 99:75–78. 10.1016/j.vetmic.2003.08.013 [DOI] [PubMed] [Google Scholar]
- 10.Brockmeier SL, Register KB. 2007. Expression of the dermonecrotic toxin by Bordetella bronchiseptica is not necessary for predisposing to infection with toxigenic Pasteurella multocida. Vet. Microbiol. 125:284–289. 10.1016/j.vetmic.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 11.Vecht U, Arends JP, van der Molen EJ, van Leengoed LA. 1989. Differences in virulence between two strains of Streptococcus suis type II after experimentally induced infection of newborn germ-free pigs. Am. J. Vet. Res. 50:1037–1043 [PubMed] [Google Scholar]
- 12.Vecht U, Wisselink HJ, van Dijk JE, Smith HE. 1992. Virulence of Streptococcus suis type 2 strains in newborn germ-free pigs depends on phenotype. Infect. Immun. 60:550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter PA, Jones AM. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367–373. 10.1016/S0966-842X(03)00156-2 [DOI] [PubMed] [Google Scholar]
- 14.Cotter PA, Miller JF. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings CA, Bootsma HJ, Relman DA, Miller JF. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188:1775–1785. 10.1128/JB.188.5.1775-1785.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson TL. 2007. Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8:220. 10.1186/1471-2164-8-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerley BJ, Cotter PA, Miller JF. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611–620. 10.1016/0092-8674(95)90515-4 [DOI] [PubMed] [Google Scholar]
- 18.Akerley BJ, Monack DM, Falkow S, Miller JF. 1992. The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J. Bacteriol. 174:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMillan DJ, Shojaei M, Chhatwal GS, Guzman CA, Walker MJ. 1996. Molecular analysis of the bvg-repressed urease of Bordetella bronchiseptica. Microb. Pathog. 21:379–394. 10.1006/mpat.1996.0069 [DOI] [PubMed] [Google Scholar]
- 20.Nicholson TL, Brockmeier SL, Loving CL, Register KB, Kehrli ME, Jr, Stibitz SE, Shore SM. 2012. Phenotypic modulation of the virulent Bvg phase is not required for pathogenesis and transmission of Bordetella bronchiseptica in swine. Infect. Immun. 80:1025–1036. 10.1128/IAI.06016-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelis GR. 2010. The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin' delivery. Biol. Chem. 391:745–751. 10.1515/BC.2010.079 [DOI] [PubMed] [Google Scholar]
- 22.Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finlay BB, Cossart P. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276:718–725. 10.1126/science.276.5313.718 [DOI] [PubMed] [Google Scholar]
- 24.Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573. 10.1038/nature05272 [DOI] [PubMed] [Google Scholar]
- 25.Kubori T, Sukhan A, Aizawa SI, Galan JE. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U. S. A. 97:10225–10230. 10.1073/pnas.170128997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuk MH, Harvill ET, Miller JF. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945–959. 10.1046/j.1365-2958.1998.00850.x [DOI] [PubMed] [Google Scholar]
- 27.Mattoo S, Yuk MH, Huang LL, Miller JF. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 52:1201–1214. 10.1111/j.1365-2958.2004.04053.x [DOI] [PubMed] [Google Scholar]
- 28.Yuk MH, Harvill ET, Cotter PA, Miller JF. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 35:991–1004. 10.1046/j.1365-2958.2000.01785.x [DOI] [PubMed] [Google Scholar]
- 29.Kuwae A, Matsuzawa T, Ishikawa N, Abe H, Nonaka T, Fukuda H, Imajoh-Ohmi S, Abe A. 2006. BopC is a novel type III effector secreted by Bordetella bronchiseptica and has a critical role in type III-dependent necrotic cell death. J. Biol. Chem. 281:6589–6600. 10.1074/jbc.M512711200 [DOI] [PubMed] [Google Scholar]
- 30.Kuwae A, Ohishi M, Watanabe M, Nagai M, Abe A. 2003. BopB is a type III secreted protein in Bordetella bronchiseptica and is required for cytotoxicity against cultured mammalian cells. Cell. Microbiol. 5:973–983. 10.1046/j.1462-5822.2003.00341.x [DOI] [PubMed] [Google Scholar]
- 31.Panina EM, Mattoo S, Griffith N, Kozak NA, Yuk MH, Miller JF. 2005. A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58:267–279. 10.1111/j.1365-2958.2005.04823.x [DOI] [PubMed] [Google Scholar]
- 32.Pilione MR, Harvill ET. 2006. The Bordetella bronchiseptica type III secretion system inhibits gamma interferon production that is required for efficient antibody-mediated bacterial clearance. Infect. Immun. 74:1043–1049. 10.1128/IAI.74.2.1043-1049.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamatsu K, Kuwae A, Konaka T, Nagai S, Yoshida S, Eguchi M, Watanabe M, Mimuro H, Koyasu S, Abe A. 2009. Bordetella evades the host immune system by inducing IL-10 through a type III effector, BopN. J. Exp. Med. 206:3073–3088. 10.1084/jem.20090494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson TL, Brockmeier SL, Loving CL. 2009. Contribution of Bordetella bronchiseptica filamentous hemagglutinin and pertactin to respiratory disease in swine. Infect. Immun. 77:2136–2146. 10.1128/IAI.01379-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brockmeier SL, Halbur PG, Thacker EL. 2002. Porcine respiratory disease complex, p 231–258 In Brogden KA, Guthmiller J. (ed), Polymicrobial diseases. ASM Press, Washington, DC: [PubMed] [Google Scholar]
- 36.Brockmeier SL, Lager KM. 2002. Experimental airborne transmission of porcine reproductive and respiratory syndrome virus and Bordetella bronchiseptica. Vet. Microbiol. 89:267–275. 10.1016/S0378-1135(02)00204-3 [DOI] [PubMed] [Google Scholar]
- 37.Register KB, Ducey TF, Brockmeier SL, Dyer DW. 2001. Reduced virulence of a Bordetella bronchiseptica siderophore mutant in neonatal swine. Infect. Immun. 69:2137–2143. 10.1128/IAI.69.4.2137-2143.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinner JA, Pilione MR, Shen H, Harvill ET, Yuk MH. 2005. Bordetella type III secretion modulates dendritic cell migration resulting in immunosuppression and bacterial persistence. J. Immunol. 175:4647–4652 [DOI] [PubMed] [Google Scholar]
- 39.Buboltz AM, Nicholson TL, Weyrich LS, Harvill ET. 2009. Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica. Infect. Immun. 77:3969–3977. 10.1128/IAI.01362-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stainer DW, Scholte MJ. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211–220. 10.1099/00221287-63-2-211 [DOI] [PubMed] [Google Scholar]
- 41.Inatsuka CS, Xu Q, Vujkovic-Cvijin I, Wong S, Stibitz S, Miller JF, Cotter PA. 2010. Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect. Immun. 78:2901–2909. 10.1128/IAI.00188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (NY) 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 43.Loving CL, Brockmeier SL, Ma W, Richt JA, Sacco RE. 2006. Innate cytokine responses in porcine macrophage populations: evidence for differential recognition of double-stranded RNA. J. Immunol. 177:8432–8439 [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔ CT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 45.Kirimanjeswara GS, Mann PB, Harvill ET. 2003. Role of antibodies in immunity to Bordetella infections. Infect. Immun. 71:1719–1724. 10.1128/IAI.71.4.1719-1724.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerrero RJ. 1990. Respiratory disease: an important global problem in the swine industry, p 98 Proc. 11th Int. Pig Vet. Soc., Lausanne, Switzerland [Google Scholar]
- 47.Pishko EJ, Kirimanjeswara GS, Pilione MR, Gopinathan L, Kennett MJ, Harvill ET. 2004. Antibody-mediated bacterial clearance from the lower respiratory tract of mice requires complement component C3. Eur. J. Immunol. 34:184–193. 10.1002/eji.200324234 [DOI] [PubMed] [Google Scholar]
- 48.Warfel JM, Zimmerman LI, Merkel TJ. 25 November 2013. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc. Natl. Acad. Sci. U. S. A. 10.1073/pnas.1314688110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siciliano NA, Skinner JA, Yuk MH. 2006. Bordetella bronchiseptica modulates macrophage phenotype leading to the inhibition of CD4+ T cell proliferation and the initiation of a Th17 immune response. J. Immunol. 177:7131–7138 [DOI] [PubMed] [Google Scholar]