Abstract
Obtaining essential nutrients, such as carbohydrates, is an important process for bacterial pathogens to successfully colonize host tissues. The phosphoenolpyruvate phosphotransferase system (PTS) is the primary mechanism by which bacteria transport sugars and sense the carbon state of the cell. The group A streptococcus (GAS) is a fastidious microorganism that has adapted to a variety of niches in the human body to elicit a wide array of diseases. A ΔptsI mutant (enzyme I [EI] deficient) generated in three different strains of M1T1 GAS was unable to grow on multiple carbon sources (PTS and non-PTS). Complementation with ptsI expressed under its native promoter in single copy was able to rescue the growth defect of the mutant. In a mouse model of GAS soft tissue infection, all ΔptsI mutants exhibited a significantly larger and more severe ulcerative lesion than mice infected with the wild type. Increased transcript levels of sagA and streptolysin S (SLS) activity during exponential-phase growth was observed. We hypothesized that early onset of SLS activity would correlate with the severity of the lesions induced by the ΔptsI mutant. In fact, infection of mice with a ΔptsI sagB double mutant resulted in a lesion comparable to that of either the wild type or a sagB mutant alone. Therefore, a functional PTS is not required for subcutaneous skin infection in mice; however, it does play a role in coordinating virulence factor expression and disease progression.
INTRODUCTION
The ability to obtain essential nutrients during an infection is critical for bacterial pathogens to successfully colonize and proliferate within host tissues. One such key process involves the ability to import and catabolize optimal carbon sources such as carbohydrates. Bacteria have evolved elegant regulatory pathways that detect the presence of preferred carbohydrates, repress the utilization of nonpreferred sugars, and regulate their metabolism based on this information flow (1, 2). In fact, many pathogens tightly control the genes involved in carbohydrate utilization and regulation in response to in vivo growth, and these same genes have been shown to be important to the disease process (3–8). Therefore, it is apparent that bacterial pathogens have closely linked their sugar metabolic sensing networks to virulence gene expression during infection.
The primary bacterial system coupling the transport of carbohydrates across the cytoplasmic membrane with their phosphorylation is a multiprotein phosphorelay called the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) (9). The PTS is composed of at least three distinct proteins: the cytosolic proteins enzyme I (EI) and Hpr, encoded by ptsI and ptsH, respectively, and the membrane-bound sugar-specific enzyme II (EII) proteins. Each EII consists of one or two integral membrane-bound domains (EIIC/EIID) that are necessary for sugar translocation, and these domains form complexes with two hydrophilic components (EIIA and EIIB) that are required for substrate phosphorylation (1, 10). EI initiates the phosphorelay after autophosphorylation by PEP, followed by transfer of the phosphoryl group to the histidine at position 15 of HPr. P∼HPr–His then donates the phosphoryl group to one of several sugar-specific EIIA proteins, which in turn transfers it to its cognate EIIB, and finally onto the incoming cognate sugar transported by EIIC/D.
In addition to carbohydrate transport, the PTS in Gram-positive bacteria also plays an important role in modulating the activity of transcriptional regulators (e.g., antiterminators, activators) that control expression of sugar utilization genes. The PTS intermediates P∼HPr–His and P∼EIIsugar can directly phosphorylate transcriptional regulators that contain either PTS regulatory domains (PRD) and/or EII-like domains to affect their activity. In Bacillus subtilis, the phosphorylation state of EI and Hpr regulates the activity of PRD-containing antiterminators (e.g., LicT) and activators (e.g., LevR) in response to glucose availability, thus controlling their ability to regulate the expression of alternative sugar operons (1, 11). PTS-mediated signaling can also influence virulence gene expression in Gram-positive pathogens. The cellobiose PTS of Streptococcus pneumoniae is important for virulence in a murine pneumonia/sepsis model (12). Inactivation of ptsI (EI) and ptsH (Hpr) in Clostridium difficile led to induction of toxin gene expression in the presence of glucose (13). Activity of PrfA, the major virulence regulator in Listeria monocytogenes, is controlled by the phosphorylation state of PTS components (14). The Bacillus anthracis AtxA master virulence regulator contains both PRD and EIIB domains and requires an intact PTS pathway for full activity (15).
The PTS is also involved in carbon catabolite repression (CCR) via Hpr kinase (HprK)-mediated phosphorylation at Ser46 of Hpr in Gram-positive bacteria. In the presence of glucose, P∼Hpr-Ser interacts with the catabolite control protein CcpA, which is then able to bind to cre sites found in promoters of sugar utilization and metabolism operons to mediate CCR (1, 16). In addition to its central role in CCR, CcpA has been shown be important for the virulence of a number of important Gram-positive pathogens (17–22). CcpA-independent pathways for CCR also exist in Streptococcus mutans, which is exerted through a network of PTS permeases (23). Thus, the sugar status of the bacterial cell can have a profound impact on virulence and expression of important virulence phenotypes in Gram-positive pathogens.
The group A streptococcus (GAS) (Streptococcus pyogenes) is a Gram-positive human-adapted pathogen with the ability to cause benign and life-threatening diseases in various host niches, including the respiratory tract (pharyngitis), skin (impetigo), deep tissues (necrotizing fasciitis), and bloodstream (streptococcal toxic shock syndrome). A conservative estimate indicates that more than 745 million cases of GAS infection occur worldwide each year, leading to over half a million deaths (24). The clinical impact of GAS is based largely on its ability to produce a large array of surface-exposed (e.g., M protein, capsule) and secreted (e.g., streptolysins [SLS {streptolysin S} and SLO], SpeB protease) virulence factors that contribute significantly to its pathogenesis. GAS exhibits coordinate regulation of its virulence genes in response to changing host environments through global regulatory networks such as the stand-alone regulator Mga and the two-component system (TCS) CovRS. Mutations in the histidine kinase CovS are associated with an invasive GAS disease phenotype and the altered regulation of virulence genes, including sagA (SLS), speB (cysteine protease), and hasA (capsule).
GAS is a fermentative lactic acid bacterium that relies heavily on sugar metabolism for its energy production, since it lacks the necessary enzymes for a functional tricarboxylic acid (TCA) cycle and oxidative cytochromes for electron transport. Early studies established that glucose, sucrose, maltose, galactose, and mannose could support growth of an M3 GAS strain (25). The addition of PTS-transported sugars such as glucose, sucrose, or trehalose to a semisynthetic growth medium stimulates the synthesis of M protein, an important GAS virulence factor involved in resistance to phagocytosis and adherence to host tissues (26). Furthermore, genes involved in carbohydrate uptake and utilization are upregulated in vivo and contribute to virulence in GAS models of infection (19, 21, 27, 28). CcpA-mediated CCR via Hpr is also important for GAS virulence, regulating over 6% of the genome, including indirectly repressing transcription of the sag operon encoding SLS (18, 19, 21). Finally, we have recently shown that the Mga virulence regulator contains PRD domains that are phosphorylated by the PTS to control Mga-regulated gene expression and virulence (3). Despite the increasing evidence that carbohydrate utilization influences GAS pathogenesis, the roles of PTS transport and signaling on disease progression have not been investigated.
In this study, we characterize the PTS pathway in multiple strains of GAS by creating a deletion mutant of ptsI (EI), the first protein of the phosphorelay. We show that a functional PTS is not necessary for eliciting a GAS subcutaneous soft tissue infection. However, it does limit lesion severity at the site of subcutaneous (s.c.) infection in mice by regulating the temporal expression level of streptolysin S. Thus, PTS transport and signaling contributes to GAS pathogenesis.
MATERIALS AND METHODS
Bacterial strains and media.
Streptococcus pyogenes MGAS5005 (covS) is an invasive M1T1 strain with an available genome sequence (29). GAS strain 5448 is also an M1T1 strain isolated from an invasive infection, and 5448AP is an isogenic animal-passaged covS variant. GAS bacteria were either cultured in complete Todd-Hewitt medium (Alpha Biosciences) supplemented with 0.2% yeast extract (THY) or in 2× chemically defined medium (CDM) prepared by MP Biomedical as previously described (30, 31). Prior to use, freshly prepared sodium bicarbonate (59.51 μM [final concentration]) and l-cysteine (11.68 μM [final concentration]) were added along with a carbohydrate source at a final concentration of either 0.5% (d-glucose) or 1% (all other sources). Growth of GAS was assayed by measuring absorbance using a Klett-Summerson colorimeter (A filter). Alternatively, overnight cultures of GAS (10 ml) were adjusted to an optical density at 600 nm (OD600) of 0.2 in saline, the appropriate carbohydrate was added, and 50-μl aliquots were transferred to the wells on a 24-well microplate (Corning) containing CDM with various sugars and covered with sealing tape (Bio-Rad). Growth was monitored for 12 to 24 h at 37°C using a FLUOstar Omega microplate spectrophotometer (BMG), with measurements taken at 30-min intervals after shaking (10 s). Escherichia coli strain DH5α and C41(DE3) were used as the host for plasmid constructions (Table 1). All E. coli strains were grown in Luria-Bertani (LB) broth (EMD Chemicals).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5 | hsdR17 recA1 gyrA endA1 relA1 | 56 |
| C41(DE3) | F− ompT gal dcm hsdSB(rB− mB−) (DE3) | 57 |
| S. pyogenes strains | ||
| MGAS5005 | M1T1 covS | 29 |
| 5448 | M1T1 | 58 |
| 5448AP | M1T1 covS | 58 |
| MGAS5005.ΔptsI | ΔptsI mutant in strain MGAS5005 | This study |
| MGAS5005.ΔptsIc | ΔptsI complemented with pKSM456 insertion | This study |
| 5448.ΔptsI | ΔptsI mutant in strain 5448 | This study |
| 5448AP.ΔptsI | ΔptsI mutant in strain 5448AP | This study |
| Plasmids | ||
| pCRK | Temperature-sensitive conditional vector; Kmr | 59 |
| pBluescript II KS(−) | ColE1 ori Ampr lacZa | Stratagene |
| pSL60-1 | Vector containing nonpolar aad9 gene | 33 |
| pKSM645 | ΔptsI mutagenic plasmid; nonpolar aad9 | 3 |
| pBlue.ΔptsI | ΔptsI in pBluescript II KS(−); bla (Apr) | 3 |
| pKSM456 | Promoterless ptsI integration plasmid; Kmr | This study |
Antibiotics were used at the following concentrations: erythromycin at 500 μg/ml for E. coli and 1.0 μg/ml for GAS, spectinomycin at 100 μg/ml for both E. coli and GAS, and kanamycin at 50 μg/ml for E. coli and 300 μg/ml for GAS.
DNA manipulations.
Plasmid DNA was isolated from E. coli using the Wizard Plus SV miniprep system (Promega). DNA fragments were gel purified from agarose using the QIAquick gel extraction kit (Qiagen) or the Wizard SV gel and PCR clean-up system (Promega). PCR for cloning and generating probes was performed using Phusion (Finnzymes) according to the manufacturer's protocol. All DNA sequencing was done by Genewiz, Inc. Genomic DNA was extracted from GAS using the MasterPure complete DNA and RNA purification kit for Gram-positive bacteria (Epicentre).
Construction of a ΔptsI mutant in GAS.
The plasmid to generate the ΔptsI allele was created as previously described (3). Briefly, the primers ptsI-1a and ptsI-1b (Table 2) were used to amplify from S. pyogenes MGAS5005 genomic DNA (gDNA) an 819-bp upstream region containing 578 bp of ptsI and a BglII restriction site. Primers ptsI-2a and ptsI-2b (Table 2) were used to amplify an 812-bp region containing the 3′ end of ptsI with BglII ends and a 10-bp overlap with the first fragment at the 5′ end. These fragments were combined as the template DNA for PCR splicing by overlap extension (SOEing) (32) with primers ptsI-3a and ptsI-3b (Table 2) to generate the ptsI deletion. The resulting product was blunt end ligated into pBluescript II KS(−) to create pBlueΔptsI (Table 1). The nonpolar aad9 spectinomycin resistance cassette was amplified from pSL60-1 using primers aad9L2-bglII and aad9R2-bglII (Table 2), digested with BglII, and ligated into BglII-digested pBlueΔptsI (Table 1). The resulting XbaI/XhoI ΔptsI::aad9 fragment from pBlueΔptsI was ligated into XbaI/XhoI-digested pCRK to yield pKSM645, which was used to construct the ΔptsI mutant MGAS5005.ΔptsI (Table 1). Additional mutants were constructed from strain 5448 (5448.ΔptsI and 5448AP [5448AP.ΔptsI]) using the same protocol (Table 1). GAS ΔptsI mutants were screened for sensitivity to kanamycin and verified by PCR on genomic DNA for specific junction regions.
TABLE 2.
Primers used in this study
| Target gene | PCR primer | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| ptsI | ptsI-1a | agatctGCTGAGTGACTTGTACGA | This study |
| ptsI-1b | GGTATTCATGCGCGTCCA | This study | |
| ptsI-2a | gcgGCCTTGTGTTGGTGGTTTAAGAGCAAC | This study | |
| ptsI-2b | GTCACTCAGCagatctCGTGCGCTTACAGAATGT | This study | |
| ptsI-3a | tctagaGCGGCCTTGTGTTGGTGGTTT | This study | |
| ptsI-3b | ctcgagGGTATTCATGCGCGTCCA | This study | |
| ptsl-internal sense | CAAATTGGTCTGCAAGC | This study | |
| ptsl-internal antisense | CAGATACTGCTCAACTTAACA | This study | |
| ptsl-external sense | GCTAGCAAAAAAGAGCTGGTTTA | This study | |
| ptsl-external antisense | CTCTTGACTACAAAGGTAAAGCAGTAAA | This study | |
| 5005.645jxn-sense | GCGGGTTATTTTTTAAATGTTTCCGAAG | This study | |
| 5005.645jxn-antisense | GGGCCATCTGCAAAATACAAAGCAT | This study | |
| ptsIcomplementL | cccggatccCATCACTCTTGACTACAA | This study | |
| ptsIcomplementR | cccggatccTTAATCTTCAGAAACGTA | ||
| ptsI M1 RT L | CGGAAACCAAGGAATGGAT | This study | |
| ptsI M1 RT R | TGGCAAACCTGTTGTGGTT | ||
| aad9 | aad9L2-bglII | gcgcagatctGGGTGACTAAATAGTGAGGAG | 19 |
| aad9R2-bglII | gcgcagatctGGCATGTGATTTTCC | ||
| gyrA | gyrA M1 RT L | CGACTTGTCTGAACGCCAAAGT | 60 |
| gyrA M1 RT R | ATCACGTTCCAAACCAGTCAAAC | ||
| sagA | sagA M1 RT L | GCTACTAGTGTAGCTGAAACAACTCAA | 19 |
| sagA M1 RT R | AGCAACAAGTAGTACAGCAGCAA |
Lowercase letters indicate nucleotides not complementary to the target DNA; underlining indicates restriction sites. Italic nucleotides are in the overlap region for SOEing.
Single-copy complementation of strain MGAS5005.ΔptsI.
A thermosensitive integration plasmid for single-copy complementation was constructed using primers ptsIcomplementL and ptsIcomplementR (Table 2) to amplify a 1,919-bp fragment containing a promoterless wild-type copy of ptsI and BamHI restriction site on both ends. The resulting PCR fragment was digested with BamHI, ligated into BamHI-digested pCRK to yield pKSM456 (Table 1), and confirmed by sequence analysis. pKSM456 was electroporated into the mutant MGAS5005.ΔptsI at the permissive temperature (30°C), and integrants were isolated following growth at the nonpermissive temperature (37°C) to allow for complementation of the ΔptsI allele in single copy. The resulting strain MGAS5005.ΔptsIc was verified by PCR and sequence analysis.
Murine infections.
An overnight culture (10 ml) was used to inoculate 75 ml of THY and incubated statically at 37°C until late logarithmic phase. Approximately 3 × 109 CFU/ml, as determined by microscope counts and verified by plating for viable colonies, were used to infect 5- to 6-week-old female CD1 mice (Charles River Laboratories). The mice were anesthetized with ketamine, hair was removed from an ∼3-cm2 area of the haunch with Nair (Carter Products), and 100 μl of a cell suspension in saline (3 × 108 CFU/mouse) was injected subcutaneously (s.c.). Mice were monitored twice daily for 7 days and were euthanized by CO2 asphyxiation upon signs of morbidity. Lesion sizes (L × W) were measured at 36 h postinfection with length (L) determined at the longest point of the lesion and width (W) as the widest point. Lesion size data were analyzed using GraphPad Prism (GraphPad Software) and tested for significance using an unpaired two-tailed t test with 99% confidence.
RNA isolation.
GAS from an overnight culture was inoculated 1:20 into 10 ml THY supplemented with appropriate antibiotics and grown to the appropriate Klett unit (ca. 80 to 100). The cells were then pelleted by centrifugation at 8,000 × g for 20 min at 4°C. The cells were resuspended in 1 ml of TE buffer (10 mM Tris [pH 7.4] and 1 mM EDTA) with 0.2% (vol/vol) Triton X-100 and boiled for 10 min. The lysate was extracted three times using chloroform-isoamyl alcohol, ethanol (EtOH) precipitated for 20 min at −80°C, and pelleted at 13,000 × g for 15 min at 4°C, and the RNA pellet was resuspended in diethyl pyrocarbonate (DEPC)-treated H2O. RNA quality was assessed on a formaldehyde gel. The formaldehyde gel consisted of 18% (vol/vol) formaldehyde, 1% (wt/vol) agarose, 72% (vol/vol) DEPC-treated H2O, and 10% (vol/vol) 10× MOPS buffer [10× MOPS buffer is 0.4 M 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0), 0.1 M sodium acetate, 0.01 M EDTA]). To quantify RNA, absorbance at 260/280 nm was determined using a spectrophotometer. RNA (5 μg) was treated with DNase using the Turbo DNA Free kit (Ambion) per the manufacturer's protocol. Treated RNA was assessed for quality as mentioned above and for gDNA contamination by PCR analysis.
Real-time RT-PCR.
Twenty-five nanograms of DNase-treated total RNA was added to SYBR green master mix (Applied Biosystems) along with 6.5 μl of each gene-specific real-time primer from a stock of 20 nM (Table 2) using the one-step protocol. The real-time reverse transcription-PCR (RT-PCR) experiments were completed using a Light Cycler 480 (Roche), and the levels presented represent ratios of the wild type/experimental levels relative to the level of gyrA transcript as the internal control. The real-time primers were designed using Primer3, the WWW primer tool (biotools.umassmed.edu/bioapps/primer3_www.cgi).
Biolog phenotype microarrays.
Carbohydrate metabolic profiles were determined using the Biolog Omnilog system (Biolog Inc.) per the manufacturer's protocol. Tests were performed utilizing the PM1 and PM2 carbon panels, each as standard 96-well microplates containing 95 different carbon sources and one negative control. Each well contained a redox dye for colorimetric determination in response to metabolizing the carbon source. Strains were first cultured on tryptic soy agar (TSA) 5% blood agar plates for 24 h at 37°C. The cells were swabbed, picked off the plate, and resuspended in inoculating fluid (Biolog) to adjust to an absorbance at 600 nm of 0.14. An aliquot of 100 μl of the suspension was immediately dispensed into each well of the PM microplate with a multichannel pipette. The plate was incubated at 37°C for 48 h, and the data were analyzed using the Omnilog software (Biolog).
Hemolysis assay.
GAS bacteria were grown in THY supplemented with 10% heat-inactivated horse serum (Sigma). Samples were taken every hour for a total of 8 to 14 h and immediately frozen at −80°C, bacterial cells were pelleted, and a 1:10 dilution was made of the supernatant in saline. Five hundred microliters of the diluted supernatant was added to an equal volume of 2.5% (vol/vol) difibrinated sheep red blood cells (RBC) (Sigma) and washed three times with sterile phosphate-buffered saline (PBS), pH 7.5. This mixture was incubated at 37°C for 1 h and cleared by centrifugation at 3,000 × g. Supernatants were measured at 541 nm by using a spectrophotometer (Molecular Dynamics) to determine release of hemoglobin by lysed RBCs. Percent hemolysis was defined as follows: [(sample A − blank A)/(100% lysis A)] × 100, where A is absorbance. To assay for streptolysin O-specific hemolytic activity, the streptolysin S inhibitor trypan blue (13 μg/μl) was added to samples prior to incubation.
RESULTS
GAS EI mutant (ΔptsI) is defective for growth in several carbohydrates.
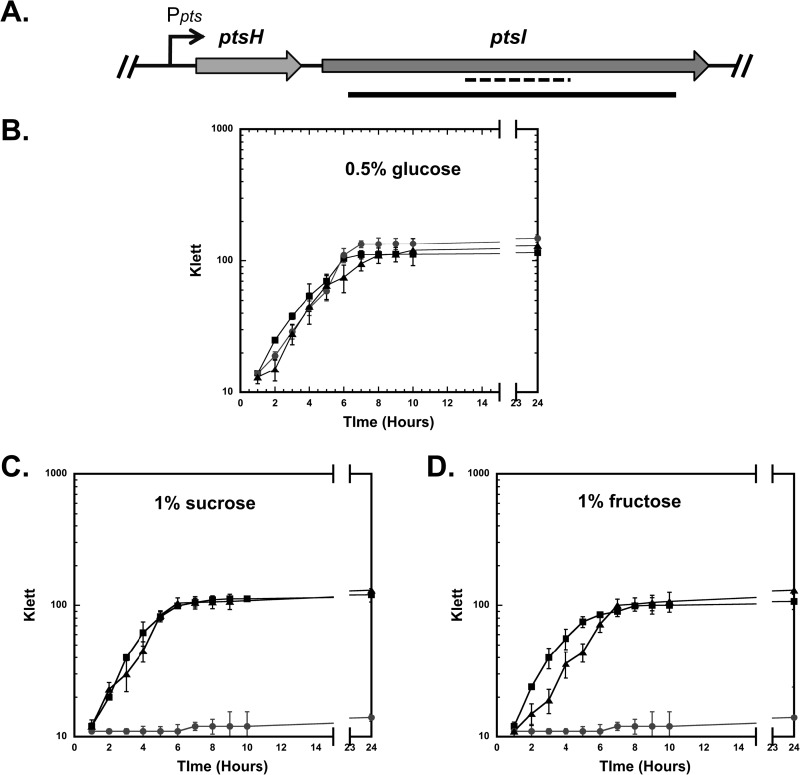
To assess the role of the PEP phosphotransferase system (PTS) in GAS, an EI mutant (ΔptsI) was constructed in the M1T1 strain MGAS5005 (Table 1), a representative strain of the most common GAS serotype associated with severe invasive disease worldwide (29). The genetic composition of the PTS in GAS is comparable to that found in many bacterial species (1). The “general” PTS proteins Hpr (encoded by ptsH) and EI (encoded by ptsI) are highly conserved among low-G+C Gram-positive bacteria, including multiple species of Streptococcus. The ptsHI (M5005_Spy1121 and M5005_Spy1120, respectively) genes are typically found in an operon (Fig. 1A). We replaced wild-type ptsI with an in-frame deletion (ΔptsI) containing a nonpolar aad9 spectinomycin resistance cassette (33) in the MGAS5005 genome (Fig. 1A). The resulting MGAS5005.ΔptsI mutant (Table 1) was verified by PCR (data not shown) and qRT-PCR (see Fig. 4A). The MGAS5005.ΔptsI strain exhibited a small-colony phenotype compared to wild-type MGAS5005 when grown on TSA agar supplemented with 5% sheep blood, but not on TSA or THY agar alone (data not shown). Although the MGAS5005.ΔptsI strain had a slightly increased lag phase compared to the lag phase of the wild type in rich THY medium, the growth kinetics were comparable (data not shown).
FIG 1.
Construction of a ΔptsI mutant from the MGAS5005 strain. (A) Schematic of the ptsHI genomic region from GAS MGAS5005 with the putative Ppts promoter (arrow) driving transcription of ptsH (Hpr) and ptsI (EI). The ΔptsI region (black bar) was replaced with a nonpolar aad9 cassette (spectinomycin resistance). The region amplified for qRT-PCR analysis is shown (dashed line). (B to D) Growth curves of wild-type MGAS5005 (black squares), ΔptsI mutant (gray circles), and complemented ΔptsI (black triangles) in CDM supplemented with 0.5% (wt/vol) glucose (B), 1% (wt/vol) sucrose (C), or 1% (wt/vol) fructose (D) are shown. Bacterial growth is measured by absorbance in Klett units on the y axes. Data are representative of three independent experiments.
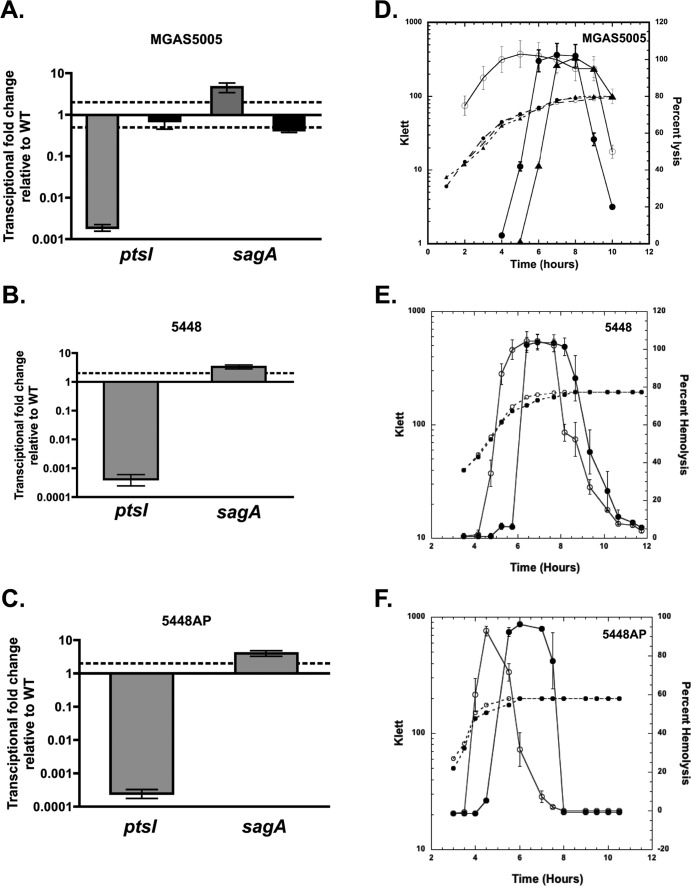
FIG 4.
Influence of ΔptsI on streptolysin S (SLS) production. (A to C) Transcript levels of sagA were measured by qRT-PCR at late log phase in THY for the wild type (WT) and the complemented MGAS5005.ΔptsIc strain and isogenic ptsI mutants of strains MGAS5005 (A), 5448 (B), and 5448AP (C). Twofold differences in expression for mutant compared to the wild type (dashed line) were considered significant. (D to F) SLS hemolytic activity was measured in culture supernatants for the wild type (closed circles) and isogenic ptsI mutants (open circles) of strains MGAS5005 (D), 5448 (E), and 5448AP (F) and the complemented ΔptsI mutant (closed triangles). Strains were grown in THY supplemented with 10% heat-inactivated horse serum and supernatants isolated throughout growth for analysis of SLS activity. Data are presented as percent hemolysis (solid lines) for three biological replicates. Representative growth curves for the strains are shown as dashed lines.
Growth of the parental MGAS5005 strain and the MGAS5005.ΔptsI mutant was further analyzed by phenotypic microarray (Biolog) using the PM1 and PM2 carbon panels, which contain a total of 190 different carbon sources. Each well was inoculated with either strain, and the plates were incubated at 37°C in an Omnilog plate incubator for 48 h to allow generation of independent metabolic curves (data not shown). Wild-type MGAS5005 showed reasonable metabolism on multiple carbon sources (Table 3). Of the 45 carbon sources able to support MGAS5005 metabolism, the ΔptsI mutant showed poor or no utilization of 19 carbon sources (Table 3), including both PTS-transported (boldface) and non-PTS-transported sugars. Carbohydrate-specific phenotypes observed by phenotype microarray were confirmed by growth assays in chemically defined medium (CDM) supplemented with 1% (wt/vol) of certain carbohydrate sources as the sole carbon source. MGAS5005.ΔptsI showed comparable growth to MGAS5005 (wild type) when grown in CDM supplemented with 0.5% glucose (Fig. 1B). However, MGAS5005.ΔptsI was unable to grow when the PTS-transported sugars sucrose and fructose (Fig. 1C and D and Table 3) or lactose, galactose, trehalose, and mannose were tested using CDM (Table 3 and data not shown). These results strongly support the idea that MGAS5005.ΔptsI lacks a functional PTS. The inability of the ΔptsI mutant to grow on some non-PTS sugars (Table 3) indicates the importance of a functional PTS for uptake and utilization of non-PTS sugars.
TABLE 3.
Growth of the wild-type MGAS5005 strain and MGAS5005.ΔptsI mutant strain on various carbon sources by Biolog
| Carbon sourcea | Metabolismb by the following strain on the indicated carbon source: |
|
|---|---|---|
| MGAS5005 | MGAS5005.ΔptsI | |
| d-Galactose | + | − |
| d-Trehalose | + | − |
| d-Fructose | + | − |
| d-Mannose | + | − |
| α-d-Lactose | + | − |
| Sucrose | + | − |
| Maltotriose | + | − |
| Salicin | + | − |
| β-Methyl-d-glucoside | + | − |
| l-Lactic acid | +/− | − |
| d-Glucose-6-phosphate | +/− | − |
| Thymidine | + | − |
| Lactulose | +/− | − |
| d-Fructose-6-phosphate | + | − |
| N-Acetyl-β-d-mannosamine | + | − |
| α-Cyclodextrin | + | − |
| β-Cyclodextrin | +/− | − |
| 5-Keto-d-gluconic acid | + | − |
| Glycerol | + | − |
| l-Arabinose | + | + |
| N-Acetyl-d-glucosamine | + | + |
| d-Xylose | + | + |
| d-Ribose | + | + |
| α-d-Glucose | + | + |
| Maltose | + | + |
| Uridine | + | + |
| Adenosine | + | + |
| Inosine | + | + |
| l-Lyxose | + | + |
| Pyruvic acid | + | + |
| Dextrin | + | + |
| Glycogen | + | + |
| β-d-Allose | + | + |
| d-Arabinose | + | + |
| 2-Deoxy-d-ribose | + | + |
| d-Fucose | + | + |
| 3-O-β-d-Galactopyranosyl-d-arabinose | + | + |
| 3-methyl-Glucose | + | + |
| Palatinose | + | + |
| l-Sorbose | + | + |
| d-Tagatose | + | + |
| Turanose | + | + |
| d-Glucosamine | + | + |
| Oxalomalic acid | + | + |
| Dihydroxyacetone | + | + |
PTS sugars are indicated in boldface type.
Symbols: +, metabolism of more than twofold the background level; −, metabolism not over twofold the background level; +/−, metabolism twofold the background level.
To confirm that the growth defect phenotype was specific to the ΔptsI allele, we generated a complemented ΔptsI strain by introducing a wild-type ptsI allele into the chromosome of the MGAS5005.ΔptsI strain (MGAS5005.ΔptsIc [Table 1]). The MGAS5005.ΔptsIc complemented strain showed a growth profile in all PTS-specific sugars comparable to wild-type MGAS5005 (Fig. 1C and D and data not shown).
The ΔptsI mutant shows increased soft tissue damage in a mouse model of GAS skin infection.
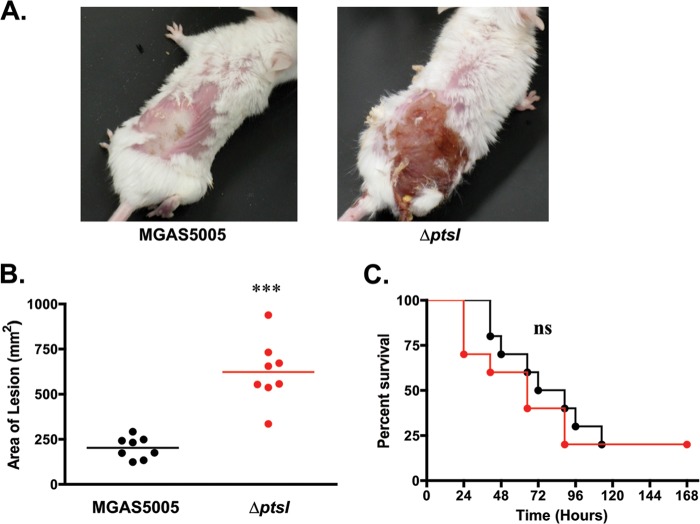
The roles of EI (ptsI) and PTS in GAS virulence was assessed using a murine model of streptococcal soft tissue infection. The MGAS5005 and MGAS5005.ΔptsI strains were grown to late log phase and then injected subcutaneously (∼3 × 108 CFU) into the haunches of 5- to 6-week-old, female CD1 mice. For 7 days postinfection, the progression of disease was monitored for both lesion size (at 38 h postinfection) and survival. Mice infected with wild-type MGAS5005 developed purulent abscessed lesions with minimal ulceration after 38 h (Fig. 2A). In contrast, mice infected with a comparable dose of MGAS5005.ΔptsI developed lesions at 16 h equivalent in size to the lesions of mice infected with the wild type at 38 h, and these lesions became fully ulcerative by 24 h postinfection (data not shown). The visual differences in both the size and severity of lesions between the wild type and the ΔptsI mutant at 38 h postinfection were quite striking (Fig. 2A). This coincided with a significant increase in the mean lesion size for mice infected with MGAS5005.ΔptsI compared to MGAS5005 measured at the same time postinfection (Fig. 2B). Interestingly, a corresponding increase in lethality over 7 days was not observed in mice infected with MGAS5005.ΔptsI, suggesting that there was no enhancement of dissemination from the skin or in systemic lethality (Fig. 2C). Thus, the ΔptsI mutant exhibits a hypervirulent phenotype at the site of infection compared to the wild type; however, systemic progression and lethality are not altered.
FIG 2.
Effect of M1T1 MGAS5005.ΔptsI mutant in mouse model of GAS infection. Mice were inoculated s.c. with ∼3 × 108 CFU and monitored for 7 days. (A) Representative images of mice infected with wild-type MGAS5005 or the ΔptsI mutant at 38 h postinfection. (B) Lesion sizes measured in mice infected with the MGAS5005 strain (black) and ΔptsI mutant (red) at 38 h postinfection. One of two independent experiments is shown (the total number of mice was 40). Each symbol represents the value for a single animal, and the horizontal bars indicate the mean value for the group. Statistical significance was determined by unpaired two-tailed t test of the animals shown. (C) Survival plot of mice infected with MGAS5005 or its ΔptsI mutant over the course of 7 days. Significance was determined by Kaplan-Meier survival analysis and log rank test of the animals shown (***, P ≤ 0.001; ns, not significant).
The ΔptsI mutant exhibits a comparable phenotype in multiple M1T1 GAS strains.
To explore the role of EI in different genetic backgrounds, we constructed independent ΔptsI mutants in two additional M1T1 strains, strains 5448 and 5448AP (covS), with the same mutagenic plasmid used for MGAS5005.ΔptsI (Table 1). Similar to MGAS5005.ΔptsI, a small-colony phenotype on blood agar plates was observed for 5448.ΔptsI and 5448AP.ΔptsI (data not shown). Furthermore, these ΔptsI mutants exhibited growth defects when grown in CDM supplemented with PTS sugars identical to that seen for MGAS5005.ΔptsI (see Fig. S1 in the supplemental material; also data not shown). Finally, similar growth phenotypes were observed in at least two independently isolated MGAS5005.ΔptsI strains, the 5448.ΔptsI and 5448AP.ΔptsI mutant strains. Thus, biologically independent ΔptsI mutants exhibit comparable phenotypes in multiple M1T1 backgrounds.
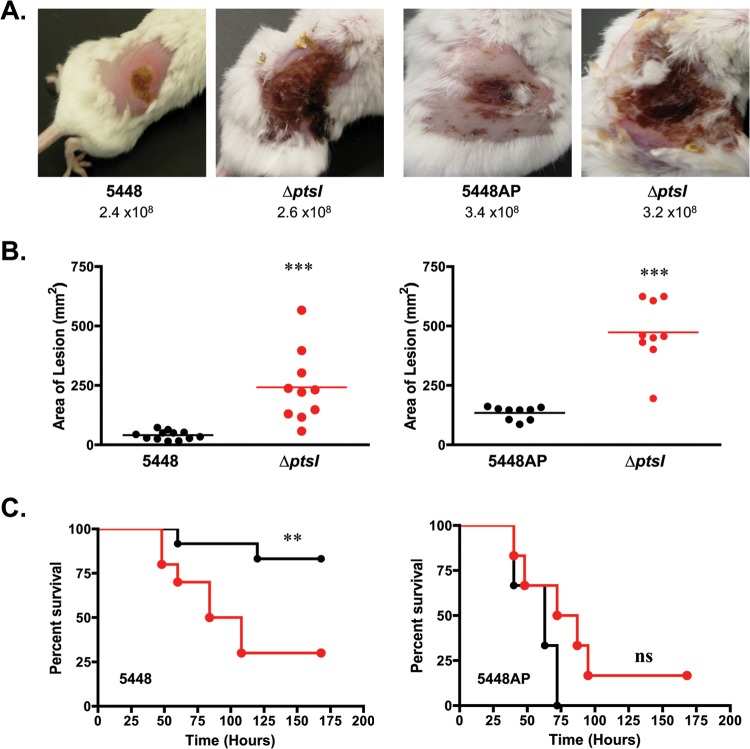
We also tested the 5448.ΔptsI and 5448AP.ΔptsI mutants and their respective parental strains in the murine model of streptococcal soft tissue infection (Fig. 3). As seen with MGAS5005.ΔptsI, mice infected by either ΔptsI mutant exhibited a more severe (Fig. 3A) and significantly larger (Fig. 3B) ulcerative lesion at 38 h postinfection compared to infection with its parental strain. In contrast to MGAS5005.ΔptsI and 5448AP.ΔptsI (covS mutants), only mice infected with 5448.ΔptsI (intact covS) showed a significant increase in systemic lethality compared to the wild type (Fig. 3C). These data suggest that EI is important in controlling virulence at the site of localized skin infections, as well as potentially serving a CovS-dependent role in limiting dissemination to sterile sites of the body.
FIG 3.
ΔptsI mutants of additional M1T1 GAS strains. (A) Representative images of mice infected s.c. with strain 5448 (CovS+) and its ΔptsI mutant (left) or 5448AP (CovS−) and its ΔptsI mutant (right) at 38 h postinfection. The numbers of CFU used in infection are indicated. (B) Lesion sizes of the same strains measured at 38 h postinfection from a representative experiment (the total number of mice is 60). Each symbol represents the value for a single animal, with the horizontal bars indicating the mean values for groups. Statistical significance was determined by unpaired two-tailed t test. (C) Survival plot of mice infected with either wild-type 5448 and 5448AP in black and their corresponding ΔptsI mutants in red. Significance was determined by Kaplan-Meier survival analysis and log rank test (**, P ≤ 0.01; ***, P ≤ 0.001; NS, not significant).
The ΔptsI mutant increases secretion of SpeB in a strain-specific manner.
The cysteine protease SpeB is a well-characterized secreted virulence factor that has been shown to be a key contributor in various models of GAS skin infection, including ulcerative lesion formation (34–36). To investigate whether the M1T1 ΔptsI mutants resulted in altered speB transcript levels, quantitative RT-PCR (qRT-PCR) was performed on mRNA isolated from the wild type and the corresponding isogenic ΔptsI mutant from all three M1T1 strains at the transition phase of growth. No significant differences (greater than 2-fold) in speB transcript levels were observed for any of the three ΔptsI mutants compared to their respective parental strain (data not shown), suggesting that transcription of speB is not altered in any of the mutants.
Increased secretion of active SpeB by GAS has been shown to negatively affect the formation of biofilms (36–38) and can be used as an indirect reporter for SpeB activity. A biofilm formation assay (see Supplemental Methods in the supplemental material) was performed on each M1T1 wild type and its respective ΔptsI mutant. Cells were grown in a 6-well plate and allowed to grow for 24 h at 37°C, and biofilm formation was quantified by crystal violet staining. The MGAS5005.ΔptsI and 5448AP.ΔptsI strains, both with covS mutant backgrounds, showed significantly less biofilm than their wild-type parental strains (Fig. S2A to S2C, left). Importantly, the reduction seen in these ΔptsI mutants could be reversed with the addition of the cysteine protease inhibitor E64 (Fig. S2A to S2C, left). In contrast, 5448.ΔptsI showed no significant difference compared to the wild-type 5448. It should be noted that 5448 showed reduced ability to form biofilm compared to MGAS5005 and 5448AP, and an MGAS5005.ΔspeB control exhibited no significance difference in biofilm formation compared to MGAS5005 (Fig. S2A).
Secretion of SpeB into the supernatants from all three ΔptsI mutants and their wild-type strains was assayed by Western blotting using a polyclonal SpeB antibody. No differences were observed in the amount of cell-bound SpeB of ΔptsI mutants compared with their corresponding wild-type strain (data not shown). However, dramatically increased levels of secreted SpeB (both 40-kDa zymogen and 28-kDa mature forms) were detected in the culture supernatants from the MGAS5005.ΔptsI and 5448AP.ΔptsI mutant strains compared to their isogenic covS parental strains (see Fig. S2A to S2C, right, in the supplemental material). Interestingly, 5448.ΔptsI had decreased levels of secreted SpeB compared to wild-type 5448, which correlates with the ability to form biofilm. Although this strongly suggests that EI (PTS) may reduce SpeB secretion in a CovS-dependent manner (MGAS5005ΔptsI and 5448APΔptsI), this does not provide a mechanism for how increased lesion severity occurs in 5448.ΔptsI.
ΔptsI mutants exhibit expression of SLS during log-phase growth.
Streptolysin S (SLS) is a secreted virulence factor that contributes to the formation of lesions in soft tissue models of mouse infection and GAS virulence in general (34, 39, 40). Biosynthesis and secretion of SLS in GAS require the 9-gene sag operon, with sagA encoding the toxin precursor and the first position in the locus (41). To determine whether the ΔptsI mutation affects sag operon expression, qRT-PCR was performed on the isolated mRNA of the MGAS5005 strain, MGAS5005.ΔptsI mutant, and the complemented MGAS5005.ΔptsIc to quantify sagA transcript levels in late-log-phase cells in THY. Changes in transcript levels greater than 2-fold were considered significant. A modest yet significant increase in sagA transcript levels was observed in the ΔptsI mutants compared to their corresponding wild-type strains (Fig. 4A to C) at this time point.
SLS-specific hemolytic activity was assayed using 2.5% defibrinated sheep RBCs incubated with culture supernatants from the wild-type MGAS5005 and mutant MGAS5005.ΔptsI GAS taken at 1-h intervals across growth. Addition of the SLS inhibitor trypan blue blocked all RBC lysis in each experiment, indicating that the observed hemolytic activity was due to SLS (data not shown). SLS hemolytic activity in the MGAS5005.ΔptsI mutant supernatants showed a dramatic increase early in logarithmic phase and remained elevated during stationary phase (Fig. 4D). In comparison, SLS hemolytic activity from the supernatants of the wild type and the complemented ΔptsI strain showed little activity during logarithmic phase and increased to maximum levels at the transition to stationary phase (Fig. 4D), as previously observed for SLS (19). Similarly, an early onset of SLS hemolytic activity was observed for the 5448.ΔptsI and 5448AP.ΔptsI mutants in comparison to their respective wild-type strains (Fig. 4E and F). These data suggest that the early onset of SLS activity during exponential-phase growth in all three M1T1 GAS strains lacking a functional PTS could lead to the increased severity of localized lesions observed in mice (Fig. 2 and 3).
SLS is required for the lesion severity observed in M1T1 ΔptsI mutants.
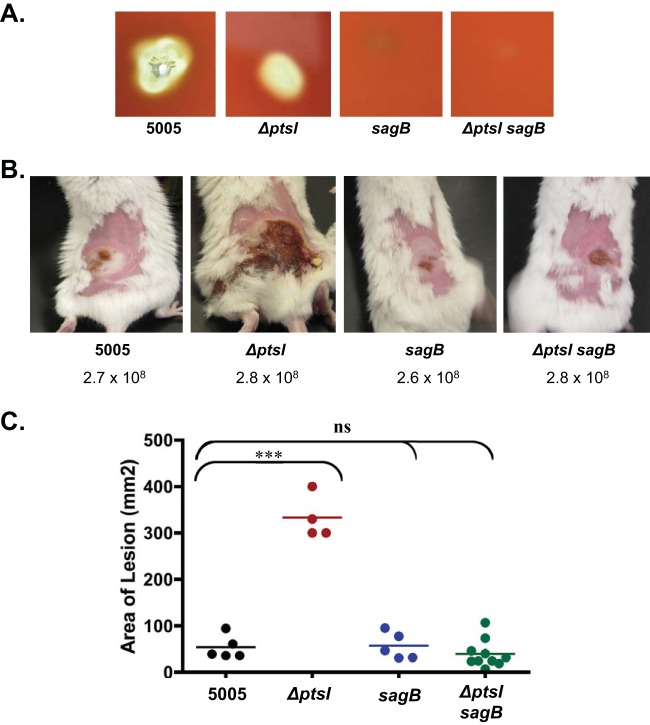
The role of SLS in the hypervirulence of lesions of mice infected with ΔptsI mutants was investigated by creating a ΔptsI sagB double mutant from the MGAS5005 strain. Inactivation of sagB has been shown to block SLS production in M1T1 GAS (19, 41). As expected, a ΔptsI sagB double mutant resulted in the absence of hemolytic activity both on blood agar plates (Fig. 5A) and also in RBC hemolysis assays (data not shown) comparable to an established MGAS5005.sagB mutant (Fig. 5) (19). To investigate the role of SLS in vivo, MGAS5005, MGAS5005.ΔptsI, MGAS5005.ΔsagB, and the MGAS5005.ΔptsI sagB double mutant were tested in the murine model of streptococcal soft tissue infection (Fig. 5B and C). The nonhemolytic ΔsagB mutant (19) showed lesion sizes comparable to those of the wild-type MGAS5005 (Fig. 5C), indicating that SLS does not normally contribute significantly to ulcer formation in this M1T1 background at the time point observed. Despite the absence of a functional PTS, the ΔptsI sagB double mutant lacking expression of SLS produced purulent lesions with a size and severity comparable to those of the wild type and ΔsagB strains, and not the ΔptsI mutant (Fig. 5B and C). These results strongly suggest that altered temporal expression of SLS during growth is the primary factor responsible for the increased lesion size and severity observed upon subcutaneous infection of mice with the ΔptsI mutant.
FIG 5.
Role of streptolysin S in ΔptsI increased lesion formation. (A) Zones of hemolysis for wild-type (WT) MGAS5005, ΔptsI single mutant, ΔsagB single mutant, and ΔsagB ΔptsI double mutant strains on 5% sheep blood agar plates after growth at 37°C. (B) Representative images of mice infected s.c. with WT MGAS5005, MGAS5005.ΔptsI mutant, ΔsagB single mutant, and ΔptsI sagB double mutant strains. The numbers of CFU used in infection are indicated. (C) Lesion size of the same experiment measured at 38 h after s.c. infection of MGAS5005 (black), ΔptsI mutant (red), ΔsagB mutant (blue), and ΔptsI sagB double mutant (green). Data represent two independent experiments, and significance was determined using the unpaired two-tailed t test (***, P ≤ 0.001; ns, not significant).
DISCUSSION
Although the connections between carbon metabolism and pathogenesis in GAS have been recognized for many years, the contribution of the PTS pathway to GAS virulence has yet to be investigated. In this study, we generated PTS mutants from multiple strains of GAS by deleting ptsI (EI) and characterized their growth profiles to identify the PTS-dependent carbon sources that support the growth of M1T1 GAS in vitro. Importantly, a PTS mutant was still able to colonize and elicit disease in a murine model of disseminating soft tissue infection. However, the PTS mutant resulted in a significant increase in localized lesion severity and size due to an increase in sag operon expression and an altered temporal expression of streptolysin S (SLS). Thus, a functional PTS appears to limit the pathogenesis of GAS during invasive skin infection.
PTS pathway and carbon utilization in GAS.
The PTS carbohydrate transport system is widespread in bacteria, where it shares the same basic composition in all genera; two “general” cytoplasmic components, EI and HPr, and sugar-specific EII transporters located in the cytoplasmic membrane (10). Typically, the genes encoding Hpr (ptsH) and EI (ptsI) are organized as a tightly regulated ptsHI operon (42), and this appears to be the case for GAS as well. Several attempts to construct either a nonpolar deletion or a polar insertion mutation of ptsH were unsuccessful (data not shown), suggesting that Hpr may be critical for GAS fitness under the conditions used here. This observation has been further supported by Tn-Seq essentiality studies ongoing in our lab (Y. Le Breton and K. S. McIver, unpublished results). Our results also confirm work done in closely related streptococci, such as S. pneumoniae (43) and S. mutans (23), as well as Staphylococcus aureus (44, 45) and Mycoplasma species (46, 47), where ptsH (Hpr) has also been reported to be essential for growth. In contrast to ptsH, we were able to generate a nonpolar ptsI (EI) deletion mutant in 3 different strains of M1T1 GAS and in an M4 GAS strain (3), indicating that the PTS itself is not required and that Hpr appears to possess a separate essential function in GAS. Despite being able to make ptsI null mutations in GAS, attempts to clone ptsI on a multicopy plasmid for complementation proved to be difficult, and we were successful only when the construct lacked a functional promoter. This suggests that overexpression of ptsI can be deleterious in E. coli and that the ptsHI operon is likely under tight regulation in GAS.
By screening with the Biolog phenotypic microarray platform, we established that 45 of 190 carbon sources tested were able to support the metabolism (defined as 2-fold over background) of the M1T1 GAS invasive throat strain MGAS5005 (Table 3). In contrast, Siemens et al. identified only 21 carbon sources with Biolog that supported the metabolism (defined as ≥10% of metabolism on glucose) of the M49 GAS “generalist” strain 591, a strain that is able to colonize both the throat and skin (48). Out of these 21 carbon sources, all but 5 (Tween 40, Tween 80, pectin, mannan, and gelatin) were in common with our studies in M1T1 (Table 3). The different metrics used to determine functional growth in the two studies could explain some of the disparity in carbon sources. However, it could also indicate that metabolic requirements differ between strains of GAS, likely due to their varied tissue sites of infection. S. pneumoniae, which is found primarily in the nasopharynx, can utilize 29 different carbon sources in Biolog, 16 of which overlapped with our M1T1 GAS findings (Table 3). Thus, many of the carbon sources are similar for streptococci; however, they appear to possess niche- and species-specific requirements.
The M1T1 MGAS5005 GAS genome possesses 14 putative EII loci (49) predicted for the transport of specific PTS sugars, although the exact substrates transported by most of these systems have not been experimentally determined. The isogenic ΔptsI M1T1 MGAS5005 mutant was still able to utilize 26 of the 45 carbon sources required for wild-type metabolism (Table 3), indicating that GAS does not need a functional PTS to utilize these carbon sources. However, the ΔptsI mutant did show metabolic defects with 19 carbon sources (Table 3), including 8 predicted PTS-specific carbohydrates (galactose, trehalose, fructose, mannose, lactose, sucrose, salicin, and maltotriose). Growth defects on maltose and glucose were not found in our study likely due to the presence of non-PTS uptake systems. Transport of maltose in GAS has been shown to occur through both the MalE-dependent ABC transport system as well as the MalT-specific PTS pathway (49, 50). In Lactococcus lactis, a non-PTS glucose uptake system (GlcU) has been reported; however, genes encoding a homologous system have not yet been identified in the genomes of S. pyogenes or S. pneumoniae (51). Interestingly, there is a putative glucokinase gene present in the MGAS5005 genome that could act to phosphorylate glucose uptake via a non-PTS transport system. The identity of the non-PTS glucose transporter in GAS remains to be determined. In addition to the six carbon compounds, ΔptsI blocked the utilization of α-glucosides (trehalose, sucrose, and maltotriose), β-galactosides (lactose and lactulose), β-glucosides (β-methyl-d-glucoside and salicin), cyclodextrins, lactic acid, and the three-carbon compound glycerol (Table 3). The defect in the use of non-PTS glycerol can be explained by the fact that glycerol kinase of Firmicutes is phosphorylated by PEP, EI, and HPr and that this modification is necessary for the activation of the enzyme (52). The results suggest that the PTS plays a broader role in regulating PTS-dependent and non-PTS-dependent transport, as well as the subsequent utilization of these substrates in central carbon metabolism.
PTS limits lesion severity during invasive skin infection in mice.
Despite the inability to utilize 19 PTS and non-PTS sugars (Table 3), all three independent M1T1 ΔptsI mutants were able to colonize mice following subcutaneous inoculation and elicit both localized and systemic disease to at least the levels of the parental wild-type strains. This provides strong evidence that GAS does not require a functional PTS or these specific sugars to infect at this tissue site and likely utilizes a carbon source that is not altered in the ΔptsI mutant (Table 3). In fact, lesion formation was significantly more rapid with increased necrosis and size in mice infected with mutant strains (Fig. 2, 3, and 5). Furthermore, mice infected by ptsI mutants exhibited highly ulcerative and spreading lesions that resulted in severe hemorrhaging and tissue damage. This is in sharp contrast to the phenotype of a ΔptsI mutant in Staphylococcus aureus, where virulence was attenuated compared to the wild type in an intraperitoneal (i.p.) model of systemic infection in BALB/c mice (53). The difference may merely reflect the different site of infections. Regardless, a functional PTS acts to limit localized tissue damage in GAS during invasive skin infection, and this appears to be linked to the metabolism of sugars that require PTS transport (Table 3).
Dissemination of GAS from the subcutaneous site of infection to the bloodstream and organs leading to lethality was not significantly altered in the MGAS5005.ΔptsI and 5448AP.ΔptsI mutant strains compared to their parental strains (Fig. 2C and 3C). Both of these M1T1 strains possess a mutation in the covS histidine kinase gene that is associated with a molecular switch to a highly invasive phenotype for GAS. In contrast, deletion of ptsI in the M1T1 strain 5448 harboring a wild-type covS gene exhibited a significant increase in systemic lethality due to dissemination (Fig. 3C) that correlated with the increase in lesion severity observed in all of the mutants. Thus, there might be a Cov-dependent influence on the ability of a functional PTS to limit systemic spread as well as localized lesion formation, although other genetic differences unlinked to cov could be involved. At this point, we do not know which Cov-regulated factor(s) may be involved in this dissemination phenotype. The cysteine protease SpeB is regulated by Cov and has been directly associated with the formation of severe lesions in skin models of GAS infection (34–36). We observed a PTS-dependent regulation on secretion of SpeB, but it was not common to all three M1T1 backgrounds regardless of Cov status (see Fig. S2 in the supplemental material).
PTS represses sag expression and early SLS production.
As mentioned above, the cytolysin SLS has been shown to contribute to the severity of skin lesions during GAS infection in mice (34, 39, 40). A modest upregulation of sagA at late log phase and increased SLS activity in the supernatants of the M1T1 ΔptsI mutants early in growth correlates nicely with the rapid onset of ulcerative lesions observed in the mice infected with the mutants. Importantly, inactivating sagB (an SLS-defective mutant) in the MGAS5005.ΔptsI background reversed this phenotype and resulted in localized lesions comparable to the lesions caused by the wild-type parental strain alone (Fig. 5). These data strongly suggest that early expression of SLS during growth is likely the primary factor responsible for the increased lesion size and severity observed upon subcutaneous infection of mice with the MGAS5005.ΔptsI mutant and other M1T1 strains. Despite the absence of SLS hemolytic activity in the ΔptsI sagB double mutant, lesion formation comparable to that of the wild type was still observed (Fig. 5). These results suggest that other virulence factors besides SLS are contributing to the lesion development seen in the parental M1T1 MGAS5005 strain.
A similar hypervirulent lesion phenotype, albeit not as significant, was previously observed by our group when a ΔccpA mutant derived from the MGAS5005 strain was tested in the murine skin infection model. Importantly, this phenotype was attributed to CcpA-dependent overproduction of sagA and SLS (19). Other studies have actually found attenuation in virulence upon infection with a ΔccpA mutant in M1T1 MGAS5005 and other GAS strains (18, 21). However, all studies observed an upregulation in sag operon transcription leading to increased SLS production in the mutants in vitro. Kietzman and Caparon showed that CcpA regulation of the sag operon in the M14 GAS strain HSC5 was indirect and that the repression did not appear to occur in infected skin tissue (18). The mechanism of indirect regulation of sag by CcpA has not been determined. Overall, this indicated that the absence of CCR in GAS modulates the regulation of SLS, but this appears to vary in strains. Since the CCR pathway remains intact in our ΔptsI mutants (Hpr kinase, Hpr, and CcpA), our findings may reflect a novel pathway for influencing sag expression and SLS production based on carbohydrate availability. However, we cannot rule out the possibility that both CcpA and the PTS pathways are impinging indirectly on the same regulatory pathway and leading to repression of SLS production.
In addition to CcpA, the expression of sagA is under the transcriptional control of GAS global regulators such as CovRS, Mga, RofA, FasBCA, and Nra (54). Our study suggests the effect of ptsI on SLS activity is CovS independent; yet, there is published evidence that CovR regulates the sag operon by direct binding to the PsagA promoter (55). Thus, although we think that CovS is not involved, we cannot rule out a role for CovR. Other known regulators of the sag operon include the newly described PTS regulatory domain (PRD)-containing virulence regulators (PCVR) such as Mga and RofA (3). Thus, it is possible that signaling through PTS might influence the expression of global regulatory networks to influence the expression of SLS and other virulence factors in GAS.
In conclusion, we have used PTS-defective strains of M1T1 GAS to identify the PTS and non-PTS carbon sources that allow these invasive isolates to grow in vitro. The ΔptsI mutants exhibited more severe and larger ulcerative lesions at the site of infection in a subcutaneous model of mouse infection. This phenotype was linked to upregulation of sagA and early onset of streptolysin S activity during exponential-phase growth in the mutant. Infection of mice with a ΔptsI sagB double mutant returned lesions to wild-type levels, implicating SLS in the observed phenotype. Therefore, a functional PTS is not required for subcutaneous skin infection in mice; however, it does limit early expression of SLS and thus the overall severity of lesions in vivo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoann Le Breton, Surya Sundar, and Kayla Valdes for critical review of the manuscript.
This work was supported by a grant from the NIH National Institute of Allergy and Infectious Diseases (AI047928) to K.S.M.
Footnotes
Published ahead of print 30 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01271-13.
REFERENCES
- 1.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031. 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorke B, Stulke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 3.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol. Microbiol. 88:1176–1193. 10.1111/mmi.12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho KH, Caparon MG. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545–1556. 10.1111/j.1365-2958.2005.04786.x [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Elias EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638–644. 10.1038/nm1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rollenhagen C, Bumann D. 2006. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 74:1649–1660. 10.1128/IAI.74.3.1649-1660.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer R, Camilli A. 2007. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 66:1–13. 10.1111/j.1365-2958.2007.05878.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313–5324. 10.1128/IAI.01807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87. 10.1159/000219373 [DOI] [PubMed] [Google Scholar]
- 10.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruger S, Gertz S, Hecker M. 1996. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J. Bacteriol. 178:2637–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAllister LJ, Ogunniyi AD, Stroeher UH, Paton JC. 2012. Contribution of a genomic accessory region encoding a putative cellobiose phosphotransferase system to virulence of Streptococcus pneumoniae. PLoS One 7:e32385. 10.1371/journal.pone.0032385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79:882–899. 10.1111/j.1365-2958.2010.07495.x [DOI] [PubMed] [Google Scholar]
- 14.Stoll R, Mertins S, Joseph B, Muller-Altrock S, Goebel W. 2008. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology 154:3856–3876. 10.1099/mic.0.2008/018283-0 [DOI] [PubMed] [Google Scholar]
- 15.Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. 2007. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol. Microbiol. 63:644–655 [DOI] [PubMed] [Google Scholar]
- 16.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245–259. 10.1271/bbb.80479 [DOI] [PubMed] [Google Scholar]
- 17.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349. 10.1128/JB.187.24.8340-8349.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kietzman CC, Caparon MG. 2010. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241–252. 10.1128/IAI.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinkel TL, McIver KS. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451–3463. 10.1128/IAI.00343-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seidl K, Stucki M, Ruegg M, Goerke C, Wolz C, Harris L, Berger-Bachi B, Bischoff M. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183–1194. 10.1128/AAC.50.4.1183-1194.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shelburne SA, III, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 105:1698–1703. 10.1073/pnas.0711767105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga J, Stirewalt VL, Melville SB. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186:5221–5229. 10.1128/JB.186.16.5221-5229.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng L, Burne RA. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol. Microbiol. 75:1145–1158. 10.1111/j.1365-2958.2009.07029.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. J. Gen. Microbiol. 129:643–651. 10.1099/00221287-129-3-643 [DOI] [PubMed] [Google Scholar]
- 25.Dassy B, Alouf JE. 1983. Growth of Streptococcus pyogenes and streptolysin O production in complex and synthetic media. J. Gen. Microbiol. 129:643–651. 10.1099/00221287-129-3-643 [DOI] [PubMed] [Google Scholar]
- 26.Pine L, Reeves MW. 1978. Regulation of the synthesis of M protein by sugars, Todd Hewitt broth, and horse serum, in growing cells of Streptococcus pyogenes. Microbios 21:185–212 [PubMed] [Google Scholar]
- 27.Musser JM, Shelburne SA., III 2009. A decade of molecular pathogenomic analysis of group A Streptococcus. J. Clin. Invest. 119:2455–2463. 10.1172/JCI38095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loughman JA, Caparon MG. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25:5414–5422. 10.1038/sj.emboj.7601393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771–782. 10.1086/432514 [DOI] [PubMed] [Google Scholar]
- 30.Podbielski A, Woischnik M, Kreikemeyer B, Bettenbrock K, Buttaro BA. 1999. Cysteine protease SpeB expression in group A streptococci is influenced by the nutritional environment but SpeB does not contribute to obtaining essential nutrients. Med. Microbiol. Immunol. 188:99–109. 10.1007/s004300050111 [DOI] [PubMed] [Google Scholar]
- 31.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton RM. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93–99 [DOI] [PubMed] [Google Scholar]
- 33.Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535–542. 10.1128/IAI.68.2.535-542.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engleberg NC, Heath A, Vardaman K, DiRita VJ. 2004. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect. Immun. 72:623–628. 10.1128/IAI.72.2.623-628.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukomski S, Montgomery CA, Rurangirwa J, Geske RS, Barrish JP, Adams GJ, Musser JM. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connolly KL, Roberts AL, Holder RC, Reid SD. 2011. Dispersal of group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS One 6:e18984. 10.1371/journal.pone.0018984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doern CD, Roberts AL, Hong W, Nelson J, Lukomski S, Swords WE, Reid SD. 2009. Biofilm formation by group A Streptococcus: a role for the streptococcal regulator of virulence (Srv) and streptococcal cysteine protease (SpeB). Microbiology 155:46–52. 10.1099/mic.0.021048-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts AL, Holder RC, Reid SD. 2010. Allelic replacement of the streptococcal cysteine protease SpeB in a Δsrv mutant background restores biofilm formation. BMC Res. Notes 3:281. 10.1186/1756-0500-3-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontaine MC, Lee JJ, Kehoe MA. 2003. Combined contributions of streptolysin O and streptolysin S to virulence of serotype M5 Streptococcus pyogenes strain Manfredo. Infect. Immun. 71:3857–3865. 10.1128/IAI.71.7.3857-3865.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heath A, DiRita VJ, Barg NL, Engleberg NC. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245–4254. 10.1128/IAI.68.7.4245-4254.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vadeboncoeur C, Frenette M, Lortie LA. 2000. Regulation of the pts operon in low G+C Gram-positive bacteria. J. Mol. Microbiol. Biotechnol. 2:483–490 [PubMed] [Google Scholar]
- 43.Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365–374 [PubMed] [Google Scholar]
- 44.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, Kedar GC, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387–1400. 10.1046/j.1365-2958.2002.02832.x [DOI] [PubMed] [Google Scholar]
- 45.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 10:291. 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, III, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430. 10.1073/pnas.0510013103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 69:67–76. 10.1111/j.1365-2958.2008.06262.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siemens N, Fiedler T, Normann J, Klein J, Munch R, Patenge N, Kreikemeyer B. 2012. Effects of the ERES pathogenicity region regulator Ralp3 on Streptococcus pyogenes serotype M49 virulence factor expression. J. Bacteriol. 194:3618–3626. 10.1128/JB.00227-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelburne SA, III, Keith DB, Davenport MT, Horstmann N, Brennan RG, Musser JM. 2008. Molecular characterization of group A Streptococcus maltodextrin catabolism and its role in pharyngitis. Mol. Microbiol. 69:436–452. 10.1111/j.1365-2958.2008.06290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelburne SA, Fang H, Okorafor N, Sumby P, Stikiewicz I, Keith D, Patel P, Austin C, Graviss EA, Musser JM, Chow D. 2007. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J. Bacteriol. 189:2610–2617. 10.1128/JB.01539-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. 2012. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7:e33320. 10.1371/journal.pone.0033320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darbon E, Servant P, Poncet S, Deutscher J. 2002. Antitermination by GlpP, catabolite repression via CcpA and inducer exclusion triggered by P-GlpK dephosphorylation control Bacillus subtilis glpFK expression. Mol. Microbiol. 43:1039–1052. 10.1046/j.1365-2958.2002.02800.x [DOI] [PubMed] [Google Scholar]
- 53.Kok M, Bron G, Erni B, Mukhija S. 2003. Effect of enzyme I of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology 149:2645–2652. 10.1099/mic.0.26406-0 [DOI] [PubMed] [Google Scholar]
- 54.Molloy EM, Cotter PD, Hill C, Mitchell DA, Ross RP. 2011. Streptolysin S-like virulence factors: the continuing sagA. Nat. Rev. Microbiol. 9:670–681. 10.1038/nrmicro2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, Gusa AA, Scott JR, Churchward G. 2005. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J. Biol. Chem. 280:38948–38956. 10.1074/jbc.M506121200 [DOI] [PubMed] [Google Scholar]
- 56.Hanahan D, Meselson M. 1983. Plasmid screening at high colony density. Methods Enzymol. 100:333–342 [DOI] [PubMed] [Google Scholar]
- 57.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 290:289–298. 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 58.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123–134 [DOI] [PubMed] [Google Scholar]
- 59.Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, McIver KS. 2013. Genome-wide identification of genes required for fitness of group A Streptococcus in human blood. Infect. Immun. 81:862–875. 10.1128/IAI.00837-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ribardo DA, McIver KS. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491–508. 10.1111/j.1365-2958.2006.05381.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.