Abstract
Bacillus anthracis, the causative agent of anthrax, relies on multiple virulence factors to subvert the host immune defense. Using Caenorhabditis elegans as an infection model, we screened approximately 5,000 transposon mutants of B. anthracis Sterne for decreased virulence. One of the attenuated mutants resulted in loss of expression of yceG and yceH, the last two genes in a six-gene cluster of tellurite resistance genes. We generated an analogous insertional mutant to confirm the phenotype and characterize the role of yceGH in resistance to host defenses. Loss of yceGH rendered the mutants more sensitive to tellurite toxicity as well as to host defenses such as reactive oxygen species and the cathelicidin family of antimicrobial peptides. Additionally, we see decreased survival in mammalian models of infection, including human whole blood and in mice. We identify a novel role for the yceGH genes in B. anthracis Sterne virulence and suggest that C. elegans is a useful infection model to study anthrax pathogenesis.
INTRODUCTION
Bacillus anthracis is a Gram-positive, spore-forming bacterium and the etiological agent of the deadly disease anthrax. The most lethal form of the disease, inhalational anthrax, occurs when inhaled spores are phagocytosed by resident macrophages or dendritic cells and are carried to regional lymph nodes (1). En route, the bacteria germinate, escape the phagolysosome, and break out of the cell to replicate extracellularly, eventually leading to massive septicemia, shock, and death (2). The hardy nature of the spores, their relative ease of dissemination, and the high mortality rate make B. anthracis a paramount bioterrorism concern.
B. anthracis must avoid an array of host bacterial defenses during the course of infection. Major plasmid-encoded virulence factors include the lethal and edema toxins and a poly-d-glutamate capsule (2). Although the anthrax toxins and capsule play central roles in disease pathogenesis, increasing evidence indicates that chromosomal genes also contribute to virulence. Sequencing of the B. anthracis genome identified a number of genes that share homologies to known virulence factors of other pathogens (3). Subsequently, studies have coupled targeted mutagenesis with rodent infection models to identify chromosomal virulence genes, including the dltABCD operon for lipoteichoic acid modification (4), the asbAB siderophore (5), the ABC transporter mntA (6), the nitric oxide synthase nos (7), the ClpXP protease (8), the purine biosynthesis gene purH (9), and the stress resistance gene hrtA (10).
Forward genetic screens using transposon-based mutagenesis systems can be a powerful method to identify novel bacterial virulence genes. We successfully employed this system in our identification of the clpX gene, encoding part of the intracellular protease ClpXP, as necessary for B. anthracis virulence (8). One challenge of this approach is designing a productive screen. Due to ethical and logistical constraints, it is not possible to perform large-scale virulence screens in vertebrate models. Instead, surrogate in vitro phenotypes that directly or indirectly correlate with virulence are often examined, for example, loss of hemolysis in identification of clpX (8).
A more amenable, whole-animal model to screen for virulence factors emerged with the finding that B. anthracis can infect and kill the nematode Caenorhabditis elegans (11). C. elegans has gained attention as a whole-animal infection model in recent years (12) because it possesses an evolutionarily conserved innate immune system with host defenses such as antimicrobial peptides (AMPs) (13) and reactive oxygen species (ROS) (14), as well as amenable traits, including small size, rapid generation time, and production of large numbers of genetically identical offspring. C. elegans has served as a host model system for several bacterial pathogens, including Pseudomonas aeruginosa and Staphylococcus aureus, with mutants subsequently confirmed to have decreased virulence in murine challenge experiments (12).
In the present study, we employ a transposon mutant library that we developed previously (8) to screen for B. anthracis Sterne mutants unable to infect C. elegans. Our most highly attenuated mutant had a transposon insertion in the second-to-last gene of a six-gene cluster of bacterial tellurite resistance genes. We find that these last two genes are important not only for tellurite resistance but also for resistance to specific reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), the cathelicidin family of antimicrobial peptides, and human whole-blood killing. This increased susceptibility correlates with reduced virulence of our transposon mutant in C. elegans and murine models of infection.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. anthracis Sterne (pXO1+ pXO2−) was grown in brain heart infusion (BHI) medium (Hardy Diagnostics) at 37°C with continuous shaking. We used antibiotic selection at the following concentrations: 50 μg/ml kanamycin (Kan) and 5 μg/ml of erythromycin (Em). Construction of the transposon mutant library was described previously (8).
Construction of targeted insertional mutant and complementation plasmid.
Approximately 600 bp of yceG sequence was amplified using the forward primer 5′-ATGCAAGCTTATGTCGAATCCTTGGCGTT-3′ and the reverse primer 5′-GTCAGAATTCCGACGATCTGGAAATGGCT-3′, which contained 5′ extensions with restriction sites for HindIII and EcoRI, respectively. This amplicon was then cloned into the temperature-sensitive plasmid pHY304 (15), a derivative of pVE6007 (16), which replicates in a temperature-sensitive manner. Transformants were grown initially at 30°C under Em selection and then shifted to the nonpermissive temperature of 37°C to force plasmid integration into the bacterial chromosome. Integration was confirmed by PCR using the pHY304-specific primer 5′-ACGACTCACTATAGGGCGAATTGG-3′ and the primer 5′-CCTTCTGGTAATCGCTTATGTGGC-3′, which is located downstream of the original amplicon.
The yceG and yceH genes were amplified as one amplicon using the forward primer 5′-ATGGTACCGGACAAGCTAAAGAAGAAGTTGCAGCT-3′ and the reverse primer 5′-ATGAGCTCTTAGTCTTCTCCCTTCTTATCGGTGTTC-3′ and cloned into the pDCerm expression vector (17) at the KpnI and SacI restriction sites. This plasmid was designated pGH and transformed into the TN1 strain.
C. elegans infection assay.
The C. elegans glp-4(bn2) strain, which is sterile at 25°C, was maintained using standard techniques (18). The C. elegans infection assay previously described (11) was modified to accommodate a high-throughput 96-well assay. Two days prior to the infection assay, a synchronous population of L1 worms was seeded onto nematode growth plates and fed Escherichia coli strain OP50. The worms were incubated at 25°C for 48 h to sterilize them and allow them to reach the young adult stage. The worms were then collected from plates, washed twice, and resuspended in 1 ml of S medium (19). Five microliters of this medium (containing approximately 5 to 10 worms) was then transferred to 190 μl of S medium containing Kan and 10 μg/ml Cry5B. Five microliters of an overnight B. anthracis culture grown in 96-well plates was then added for a final volume of 200 μl. The plates were wrapped in Parafilm and incubated at 28°C. Wells were assessed at 48 and 72 h. Nematodes were counted as infected only if they were filled with bacteria, which can be detected as worms being dark, rigid, and immobile. Wells with more than 60% of worms surviving were noted for further confirmation in a larger volume (24-well plates) in an independent assay.
Identification of transposon insertions.
Transposon insertions were identified using two different methods. DNA flanking the transposon insertion site was identified by single-primer PCR as described previously (8). Alternatively, the insertion site was identified using a Y-linker method (20). The transposon insertion site was confirmed using the transposon-specific primer 5′-TATGCATTTAATACTAGCGACG-3′ and one of the following primers located downstream of the insertion site: TN1, 5′-GTGAAGCGAAAGGAATCGAAGT-3′; TN2, 5′-TGTACTTGAACATCAGTCGATTTCTCCCA-3′; TN3, 5′-TAACTGCCAACTTATGAATAAATCCA-3′; TN4, 5′-CATGCGTGGCAAGTTA-3′; TN5, 5′-TCTTAGTGCAGGGAAACATAACTGA-3′; TN6, 5′-TCCCAACCATAATTCCTATGACACC-3′; TN7, 5′-CAGAGATTACAATAGGTTCGGCT-3′; TN8, 5′-TTCGTCTTATCAGGATTACTTCAGTTAG-3′; TN9, 5′-TGTCATCGGAAGCATAAATCCT-3′; TN10, 5′-CAAGAAATGCTCGTTTGGCTATC-3′; and TN11, 5′-TCTTTGCATCGGAACATTGCT-3′.
RT-PCR.
Sixteen-hour bacterial cultures were lysed using Lysing Matrix B beads (MP Biomedical) by being pulsed two times for 45 s at 6 m/s in a bead beater (MP Biomedical). RNA was purified using the RNeasy kit (Qiagen) followed by DNase treatment using the Turbo DNA-free kit (Life Technologies). For reverse transcription-PCR (RT-PCR) shown in Fig. 2B, RNA was reverse transcribed using Superscript III reverse transcriptase (Life Technologies) at 55°C with one of the following gene-specific primers: RT 1, 5′-CTCGCTACTTCCACACTTTGC-3′; RT 2, 5′-GAATACGCCAGCGATACCGC-3′; or RT 3, 5′-CCTCAACTGCTTGCGTAACTT-3′. PCR was performed at 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min for 30 cycles with the following primer combinations: A (fwd, 5′-ACTATCGTCCTGAGAGCCAC-3′, and rev, 5′-GCTCCTCCAATACCAGTTCCA-3′), B (fwd, 5′-TAGAGGGCATTCATATAGTGCTGG-3′, and rev, 5′-GATCCCAACCTAAGCCAACCTG-3′), C (fwd, 5′-GTGAAGGCGCTGGTGATGAC-3′, and rev, 5′-GAAATCCTCTGCACCTGAAACT-3′), D (fwd, 5′-TACGATGGAGAAGGACGCAG-3′, and rev, 5′-CCTGTGCGGTTATCTCCTGT-3′), E (fwd, 5′-AGGTGGTTTAGGTGCGCTTG-3′, and rev, 5′-CATGAGCTTCCTCTTCTTCTGC-3′), F (fwd, 5′-GCGGTATCGCTGGCGTATTC-3′, and rev, 5′-CATTAGGAACGCCAAGGATTCG-3′), and G (fwd, 5′-TTACAACATGGGATTGCAGAGG-3′, and rev, 5′-CAATTTCACGGCCCATCGTT-3′). For the RT-PCR shown in Fig. 2C, RNA was reverse transcribed using a high-capacity cDNA reverse transcription kit (Life Technologies). PCR was performed at 95°C for 30 s, 54°C for 30 s, and 72°C for 1 min and limited to 26 cycles to maintain semiquantitative conditions. The following primers were used: yceC Fwd, 5′-CAATCAGGTGGGTTTCTGT-3′; yceC Rev, 5′-TGATGCCATATACAATTCCTCCTCTGT-3′; yceG Fwd, 5′-CGTCCACAATATGACCTGACAG-3′; yceG Rev, 5′-GAGCTACAGGTGTATAGGAACG-3′; yceH fwd, 5′-TGGACCGATTCGATAGCAA-3′; yceH Rev, 5′-CGACTTTCTTCACGCTCACGC-3′; fusA Fwd, 5′-AAGCTGGTGGTGCTGAAGCAC-3′; fusA Rev, 5′-TTCCCAATCAGCTTCTCCTTGAAG-3′. All RT-PCRs were repeated with at least 3 different RNA preparations; a representative image is shown.
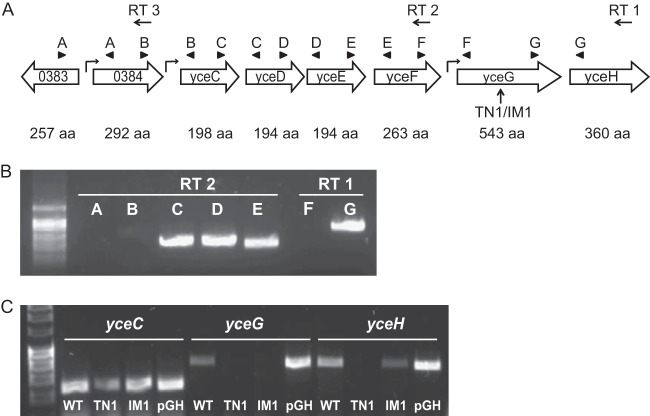
FIG 2.
Organization of the yce tellurite resistance gene cluster in B. anthracis. (A) Organization of the tellurite resistance gene cluster in B. anthracis with number of amino acids (aa) listed below each gene. The upward arrow indicates the site of transposon and plasmid insertion for the transposon mutant 1 and the insertional mutant 1 (TN1/IM1). Leftward arrows indicate primers used for reverse transcription (RT 1, RT 2, or RT 3). Arrowheads show location of PCR pairs A to G. The bent arrow indicates the predicted promoter region. (B) PCR using cDNA from indicated RT primers. Letters correlate with primer combinations shown in panel A. (C) Semiquantitative RT-PCR expression of the yceC, yceG, and yceH genes from cDNA prepared from WT, TN1, IM1, or TN1 complemented with an expression plasmid containing yceG and yceH (pGH).
Growth curves.
Growth curves were started using equal amounts of early-log-phase parental and mutant bacterial strains, and bacteria were grown at 37°C with shaking in BHI with the indicated amount of potassium tellurite (Sigma). Absorbance was recorded at 600 nm. Growth curves were repeated at least three times; a representative graph is shown.
Disk diffusion assays.
Parental or mutant B. anthracis stationary-phase cultures (16 h) were swabbed onto BHI agar plates. A 6-mm sterile paper disk (Becton, Dickinson) containing a specific antimicrobial was placed on the agar, the plate was incubated overnight at 37°C, the zone of inhibition was measured, and the area was calculated. Tellurite disks were prepared by soaking the disk in 10 mg/ml of potassium tellurite. Antibiotic disks containing 30 μg chloramphenicol, 30 μg tetracycline, and 10 U penicillin were obtained from Becton, Dickinson. For antibiotic disk diffusion performed with tellurite, the agar plates contained 30 μg/ml of potassium tellurite.
Oxidant assays.
For the growth assays, B. anthracis parental and mutant strains were grown to early log phase; diluted 1:2 (H2O2), 1:10 (hypochlorite), or 1:100 (methyl viologen); and incubated with the indicated amounts of each specific oxidant (Sigma). The final optical density (OD) after overnight incubation (≈16 h) was used as the measure of growth. A MIC assay was performed with 0, 10, and 40 mM methyl viologen (Sigma). For the H2O2 killing assay, bacteria were grown to an optical density of 0.4 at 600 nm and concentrated 10× in phosphate-buffered saline (PBS) and the concentrated bacteria were then diluted 1:2 into 0.1% H2O2 in PBS for a final concentration of 0.05% H2O2. Bacteria were incubated at 37°C. At the indicated time, an aliquot was removed and diluted 1:10 in PBS containing 3,000 U catalase (Sigma) to quench residual H2O2 and surviving bacteria were enumerated by serial dilution plating.
Antimicrobial peptide assays.
B. anthracis parental and mutant strains were grown to early log phase, diluted 1:10 in RPMI plus 5% Luria-Bertani broth, and incubated with 1.6 μM LL-37 (Anaspec) overnight at 37°C under static conditions. MIC assays were also performed using RPMI plus 5% Luria-Bertani broth and log-phase bacteria diluted to 1:20 with either 0, 125, and 250 μg/ml nisin; 0, 250, 500, and 1,000 μg/ml bacitracin; or 0, 25, 50, and 100 μg/ml polymyxin (all from Sigma) or log-phase bacteria diluted to 1:100 and 0, 0.4, and 0.8 μM HNP-2 (Anaspec).
Whole-blood assay.
Blood collected from three different healthy donors (use and procedures approved by the University of California, San Diego, Human Research Protections Program) was incubated with 105 CFU B. anthracis in a total volume of 500 μl and rotated at 37°C. After 15 min, aliquots were removed, blood was lysed in water, and surviving bacteria were enumerated by serial dilution plating.
In vivo infection.
Bacteria were grown to an optical density (OD) of 0.4 (600 nm), washed in PBS, and resuspended at a 1:30 dilution in PBS (approximately 7 × 105 CFU/ml). Parental (wild-type [WT]) and mutant (TN1) B. anthracis Sterne strains were mixed at a 1:1 ratio, and 0.1 ml was injected intravenously via the lateral tail vein into 8- to 10-week-old female CD1 mice. After 2 days, animals were euthanized and the kidneys were isolated and weighed. Kidneys were homogenized (twice for 1 min each at 6,000 rpm) using 1-mm zirconia-silica beads (BioSpec Products) in 2-ml screw-cap tubes containing sterile PBS using a MagNA lyser (Roche). Surviving bacteria were enumerated by serial dilution plating on BHI (total CFU) and BHI Kan50 plates (TN1 CFU). WT CFU was determined by subtracting the Kan-resistant CFU from the total CFU, and bacterial counts were calculated as CFU/g kidney.
Bioinformatics and statistics.
Protein sequences were aligned using the ClustalW program found at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html. The following sequences were used: the B. anthracis yceCDEFGH operon gene symbol BAS0385 through BAS0390, the R478 Ter operon with GenBank accession number U59239.2, and the Listeria monocytogenes TelA gene symbol lmo1967. Statistical analysis was performed using GraphPad Prism software.
RESULTS
Identification of an attenuated B. anthracis transposon mutant.
We have recently demonstrated that under the proper assay conditions, including supplementation with an exogenous pore-forming toxin, B. anthracis Sterne can infect C. elegans (11). Distinguishing between B. anthracis-infected and uninfected worms is based on visual appearance. Infected nematodes are rigid and dark and lack internal structures as they are filled with B. anthracis spores (11). Conversely, healthy animals are motile and curvilinear, displaying typical discernible internal structures. We screened approximately 5,000 B. anthracis transposon mutants created previously (8) for their ability to establish an infection in C. elegans using a high-throughput 96-well screening format. This is a nonsaturating screen; however, in our previous use of this library, we found that 5,000 mutants is a sufficient number to identify promising targets (8). Under our infection conditions, the mortality rate of C. elegans with wild-type (WT) B. anthracis Sterne was approximately 75%. We identified 11 transposon mutants that yielded a C. elegans mortality rate of 40% or less (Fig. 1) and determined the site of disruption (Table 1).
FIG 1.
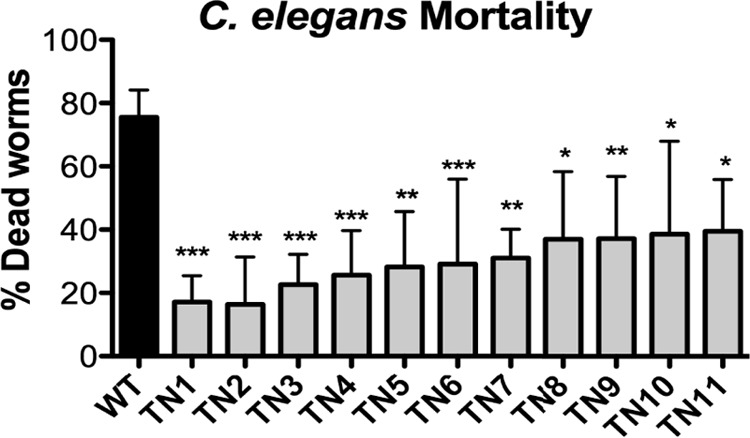
Transposon mutants have attenuated virulence in a C. elegans model of infection. C. elegans mortality rate after 72 h of infection with WT B. anthracis Sterne or transposon mutants (TN1 to -11). Data from at least three independent experiments are combined and presented as means ± standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001, from WT by one-way analysis of variance followed by Tukey's post hoc analysis.
TABLE 1.
Summary of attenuated mutants identified in C. elegans infection screen
| Mutant | Disrupted genea | Gene symbol | Sequence surrounding insertion siteb |
|---|---|---|---|
| TN1 | Putative tellurite resistance gene | BAS0389 | ACGTATCATTTGGTATGGTGATGCACAGGAAAGTCGTATATATTTCTTATATTTTCTTATTATGCTCGGTTGTGATGTGCTTTATTATCACCCAGAAGGA |
| TN2 | BNR repeat domain protein | BAS2814 | AACCAAATCTATCAGTTCGCATTTGTAAAGAAGTAAATTGAATATCATTATTTTCCAATATATCACCTCATTCTTATCATATATATATTCGTATGCTATT |
| TN3 | Conserved hypothetical protein | BAS0642 | GCACTTGTAAAAGATAAAACACAAGTGAAAATTCTTGTTTCACCTAAATAATATAGAATAGTAAAAAGTAGCTCTTTTCTAGAAATAGAAGAGGGCTACT |
| TN4 | Protease production regulatory protein | BAS0976 | TGCATAAAAACACGGCATATCTATGTGATATGCCGTGTCAAGGTCAATTTATATAAAAAATATTTGTTATATGCAAAAAACTAAGCAGGAATTTGTTTAC |
| TN5 | Hypothetical protein | BAS0432 | ATTGTAAAAATTGCGACCATTCAACTGATCCAGGACATTATATGTAAGTAAAAAAATAAGGGGATGGAAATCATGAGAGAACGTGAAAATTGGGATGTAT |
| TN6 | Spore germination protein | BAS3369 | GTATTAATAGGACCTATGACCTTTTCATCCTTTTTTCAAACGATAGATGATTACAGCTTCAGACCAATGATTCCATCTTTTATACGCTTACTTCGTTTCA |
| TN7 | Conserved hypothetical protein | BAS0645 | TAGAGATAGGAGATTCTATCTTGTTTCGCCATAGTAAAGCTGGAGAATTATGTGAGCGGTTTCCTTTTTTATATCGTGTGAAAGAAGGAGAGATTGTTGG |
| TN8 | Sensor histidine kinase | BAS0869 | GAGAGGAAAAGAATGTTTCCTTCTTTGTGACGGATATTGGGCACGATATTGTATTTGAAGTAATAGATAGAGGGATAGGAATACCGGCAGAAAAAATAAC |
| TN9 | PTS, IIB component | BAS2290 | GTGGTATCATTCCGCCGCAAGCATATACGCCACTTGGCGGACCGACTTTATTAAAGACTGTAAATGATTTAATTCGTTAGGGGGAACCGTTATGAATAAG |
| TN10 | Acetyl-CoA hydrolase | BAS1726 | ATTATTGAGAATTGTGCACATCTAATGTATCGTGATCAACTACGAGCTTATTATGAGGAAGCGAAAACAAGAGGTGGACAAACTCCTCATATTTTAGAGA |
| TN11 | Oxidoreductase | BAS0712 | ATATTTTAGTAAATAATGTCGCGCAGCAATATCCGCAGCAAGGACTAGAGTATATTACAGCAGAACAGTTAGAAAAGACTTTTCGTATTAATATTTTTTC |
Abbreviations: PTS, phosphotransferase system; CoA, coenzyme A.
Insertion site is shown in bold.
The most consistently attenuated mutant, designated TN1, induced a C. elegans mortality rate of less than 20% and contains a disruption in a putative tellurite resistance gene (BAS0389). Tellurite (TeO32−) is a water-soluble form of tellurium and is highly toxic to bacteria. Tellurite resistance genes are found in a number of bacterial species, although they have not been linked to pathogenesis (21). TN2 has a disruption in a gene containing BNR repeats (also known as ASP-box motifs) that are commonly found in glycosyl hydrolases such as sialidases, which have been linked to bacterial pathogenesis (22). TN3, TN4, and TN5 are all inserted in intergenic regions, and the disrupted gene is assumed to be the gene downstream of the insertion site. TN4 disrupts a predicted protease production regulatory protein, which could have implications in virulence. TN5 is in the middle of a gene cluster of conserved hypothetical proteins with little information on potential function. TN3 and TN7 also disrupt conserved hypothetical proteins of unknown function. These conserved hypothetical proteins are on either side of phospholipase C and sphingomyelinase, which have been implicated in B. anthracis virulence (23). While it is unlikely that either TN3 or TN7 directly disrupts these virulence genes, it is possible that the conserved hypothetical proteins have a functional cooperation with phospholipase C and/or sphingomyelinase that would account for this coincidence. TN6 inserts in an apparent operon containing spore germination proteins. TN9, TN10, and TN11 all disrupt metabolic genes and may impair general bacterial fitness. TN8 is found in a sensor histidine kinase (BAS0869) and likely also disrupts the response regulator (BAS0870) and lipoprotein (BAS0871) located directly downstream in a putative operon. Notably, after a short intergenic region of 220 bp, the next gene in this region is another putative tellurite resistance protein (BAS0872). We tried to assess whether TN1 and TN8 had similar phenotypes; however, this was complicated by a growth defect in TN8 (data not shown) that may itself account for the decreased virulence seen in the C. elegans model.
Although several of the disrupted genes are potentially of interest and would be interesting to pursue in future studies, we have focused this study on TN1. Not only is TN1 highly attenuated in our C. elegans model, it also disrupts an intriguing gene family that has been speculated, although to our knowledge never proven, to have a role in bacterial virulence (21).
Tellurite resistance genes in B. anthracis.
The B. anthracis genome contains a six-gene cluster of putative tellurite resistance genes (BAS3085 to BAS3090) in which the second-to-last gene is disrupted by the transposon insertion in TN1 (Fig. 2A, arrow). Homology with the corresponding proteins in Bacillus subtilis is high (Table 2), and we have maintained the gene nomenclature, yceCDEFGH, originally designated for B. subtilis (24). We used an RT-PCR approach for characterization of multigene operons (25) to test whether all 6 putative tellurite resistance genes were regulated within the same operon. Reverse transcription was performed using primer RT 1, RT 2, or RT 3 followed by PCR with primer sets A to G, whose forward and reverse primers are located on neighboring genes (Fig. 2A). PCR amplification of the cDNA should occur only with genes expressed on the same transcript, although all primers amplified using genomic DNA (data not shown). As a negative control, we looked at expression between BAS0383 and BA3084, which are orientated in opposite directions and cannot be transcribed together. As expected, no amplification was seen using primer set A with cDNA from RT 2 (Fig. 2B) or cDNA from RT 3 (data not shown). We find that yceCDEF are transcribed together on the same operon; however, there is little to no expression using a primer set amplifying between yceF and yceG (Fig. 2B). Primers amplifying between yceG and yceH have strong expression, indicating that these genes are transcribed together. A very faint band is consistently seen between the upstream gene BAS0384 and yceC, although it is difficult to visualize. Therefore, while there may be some transcription of yceCDEF from the promoter upstream of BAS0384, the majority of expression is likely driven by a regulatory element between BAS0384 and yceC. We conclude that the putative tellurite resistance genes contained in yceCDEF and yceGH are regulated independently of one another.
TABLE 2.
Homology of B. anthracis Yce proteins to other tellurite resistance proteins
| B. anthracis protein | B. subtilis protein(s) (% homology) | Ter/Tel protein(s) (% homology) |
|---|---|---|
| YceC | YceC (62.3) | TerE (40.1), TerD (37.0), TerZ (37.0) |
| YceD | YceD (68.6), YceE (67.0) | TerD (54.2), TerE (53.6) |
| YceE | YceE (71.1), YceD (63.6) | TerD (55.9), TerE (52.6) |
| YceF | YceF (64.3) | TerC (22.9) |
| YceG | YceG (41.4) | |
| YceH | YceH (58.1) | TelA (24.1) |
At least five unique bacterial tellurite resistance (Ter) determinants have been identified (21). The B. anthracis YceCDEF and YceGH proteins have homology with two of them, TerZABCDEF and TelA (Table 2). The terZABCDEF operon was originally identified on the R478 plasmid. and homologues have been found in the chromosomes of numerous species, including Yersinia pestis, E. coli, Bacillus subtilis, and Proteus mirabilis (21, 24, 26, 27). The telA gene has been recently characterized in the Gram-positive pathogen L. monocytogenes (28). At the amino acid level, B. anthracis YceCDEF has homology to several Ter proteins (Table 2). YceG, the site of the transposon insertion, has little identity to any other tellurite resistance proteins, while YceH has 24.1% identity to the TelA protein found in L. monocytogenes.
To confirm the involvement of the yceG gene in the observed phenotype of the transposon mutant, we constructed an independent mutation (IM1) in approximately the same location using targeted insertional plasmid mutagenesis. RT-PCR confirmed that yceG gene expression was absent in both the TN1 and IM1 mutants relative to the wild type (Fig. 2C), whereas the first gene, yceC, is expressed in all three strains as expected. The last gene, yceH, is absent or substantially reduced in TN1 but present in IM1 (Fig. 2C). This differential regulation of yceH by TN1 and IM1 was also confirmed by quantitative PCR (qPCR) (data not shown). Therefore, IM1 functions as a genetic knockout of yceG whereas the transposon mutant functions as a double knockout of yceG and yceH. For complementation analysis, yceG and yceH were cloned on an expression plasmid (designated pGH) and transformed into the TN1 strain. Although transcription of the yceG and yceH genes was confirmed by RT-PCR in our complemented strain (Fig. 2C, lanes marked pGH), there was no change in phenotype from TN1 lacking the complementation plasmid in any subsequent assays (data not shown). This lack of complementation may result from improper stoichiometry of the proteins involved. No difference in expression of an unrelated control gene, fusA, was seen in any strains (data not shown).
Since yceG and yceH are homologous to putative tellurite resistance genes, we first tested susceptibility to potassium tellurite. We observed that both WT and mutant strains of B. anthracis Sterne form the characteristic black precipitate caused by reduction of tellurite (TeO32−) to tellurium (Te0) (Fig. 3A). However, there is a small but statistically significant increase in the zone of growth inhibition surrounding a potassium tellurite-impregnated disk in the TN1 and IM1 strains in comparison to WT (Fig. 3A). Exposure to potassium tellurite in liquid culture also results in slower growth for both TN1 and IM1 relative to the WT bacteria (Fig. 3B), although no difference in growth is seen in BHI alone (data not shown). Although other genes likely contribute, yceGH appears to play a minor role in tellurite resistance in B. anthracis Sterne.
FIG 3.

Loss of yceG leads to decreased tellurite resistance. Growth inhibition of WT B. anthracis Sterne (black bars), transposon mutant (TN1, white bars), or insertional mutant (IM1, gray bars) spread on plates and exposed to a tellurite-impregnated disk. The area of growth inhibition was calculated and presented as mean ± standard deviation (right). *, P < 0.05, or **, P < 0.01, from WT by one-way analysis of variance followed by Tukey's post hoc analysis. The experiment was repeated at least three times, and representative pictures are shown (left). (B) Representative graph from a 24-hour growth curve in liquid BHI containing 30 μg/ml of potassium tellurite.
yceG and yceH genes contribute to innate immune defense mechanisms.
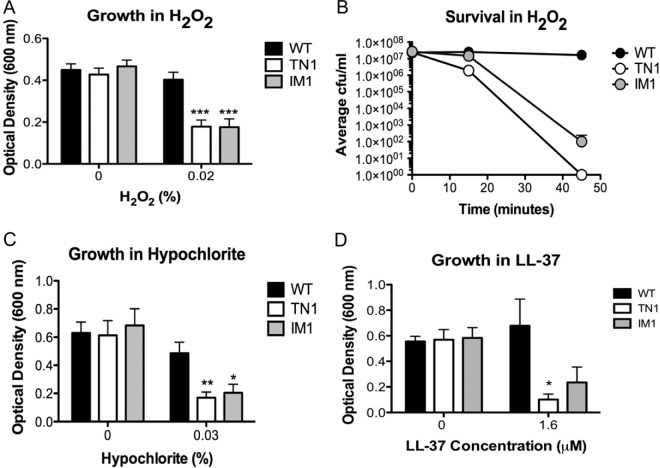
Because increased susceptibility to tellurite toxicity cannot by itself explain the attenuated virulence of TN1 in C. elegans, we next investigated whether yceGH could play a role in defense against aspects of the host innate immune system. A critical innate immune defense of C. elegans is the production of ROS in response to pathogens (14). In mammalian systems, professional phagocytes such as macrophages and neutrophils generate ROS (29). Tellurite is also a strong oxidant (30) and increases intracellular ROS production in exposed bacteria (31, 32). Therefore, we determined whether yceG and yceH contribute to ROS resistance. Early-log-phase bacteria were incubated overnight in BHI medium containing 0% and 0.02% H2O2. Although no difference in growth as measured by the final optical density was observed in BHI alone, addition of H2O2 significantly attenuated growth for both the TN1 and IM1 mutants (Fig. 4A). This killing occurred relatively rapidly, as exposure to 0.05% H2O2 killed all TN1 mutants by 45 min and reduced survival of IM1 by 100,000-fold in comparison to WT (Fig. 4B). While loss of yceG alone (IM1) appears sufficient to increase ROS sensitivity, yceH also likely contributes, since the phenotype seen in the TN1 mutant (loss of both yceG and yceH) is often stronger (compare survivals of TN1 and IM1 mutants in Fig. 4B). Additionally, we tested susceptibility for two additional ROS, sodium hypochlorite, the active ingredient in bleach, and superoxide generated by methyl viologen. As with H2O2, growth of TN1 and IM1 was significantly decreased compared with WT when bacteria were exposed to hypochlorite (Fig. 4C). However, no difference in survival was seen with superoxide (data not shown).
FIG 4.
YceG is necessary for resistance to reactive oxygen species and human cathelicidin. (A, C, and D) Growth of WT B. anthracis Sterne (black bars), transposon mutant (TN1, white bars), or insertional mutant (IM1, gray bars) for 16 h in H2O2 (A), hypochlorite (C), or cathelicidin (LL-37) (D). *, P < 0.05, **, P < 0.01, or ***, P < 0.001, from WT by one-way analysis of variance followed by Tukey's post hoc analysis. Data from at least three independent experiments are combined and presented as means ± standard errors of the means. (B) Survival of WT B. anthracis Sterne (black circles), the transposon mutant (TN1, white circles), or the insertional mutant (IM1, gray circles) after incubation with 0.05% H2O2 for 45 min. The experiment was repeated at least three times, and data from one representative experiment are presented as means ± standard deviations.
Antimicrobial peptides (AMPs) are a second innate immune defense mechanism employed by C. elegans (13). In L. monocytogenes, the tellurite resistance gene telA was identified in a transposon-based screen for susceptibility to the AMP nisin (28). Since yceH has homology to telA and is absent in our TN1 mutant, we tested whether TN1 and IM1 were more susceptible to several antimicrobial peptides, including the human cathelicidin LL-37, the α-defensin HNP-2, nisin, bacitracin, and polymyxin. A significant difference in survival was seen between WT and TN1 in LL-37 susceptibility (Fig. 4D); however, IM1 exhibited an intermediate phenotype (Fig. 4D). This is likely due to the fact that the TN1 insertion disrupts both yceG and yceH whereas IM1 disrupts only yceG. No difference in growth between WT B. anthracis Sterne, TN1, and IM1 exposed to HNP-2, nisin, bacitracin, or polymyxin was observed (data not shown). Thus, it appears that both yceG and yceH contribute to resistance to human cathelicidin; however, this did not represent a global change in membrane integrity as no difference in susceptibility was seen with any other AMP tested.
Tellurite enhances susceptibility to antibiotics independently of yceG and yceH.
It has recently been reported that exposure to tellurite enhances susceptibility of E. coli to a broad range of antibiotics, including chloramphenicol, tetracycline, and ampicillin (33). Similarly, we observed a significantly increased zone of inhibition for B. anthracis Sterne in antibiotic disk diffusion assays for chloramphenicol, tetracycline, and penicillin when 30 μg/ml of potassium tellurite was added to the agar plates in comparison to plates without potassium tellurite (Fig. 5A). We next asked whether mutations in the tellurite resistance genes would further enhance this sensitivity but found no difference in growth inhibition between the WT and TN1 mutant bacteria with any antibiotic in the absence (data not shown) or presence (Fig. 5B) of potassium tellurite. Therefore, while tellurite does increase B. anthracis susceptibility to antibiotics, this effect appears to be independent of yceG and yceH.
FIG 5.
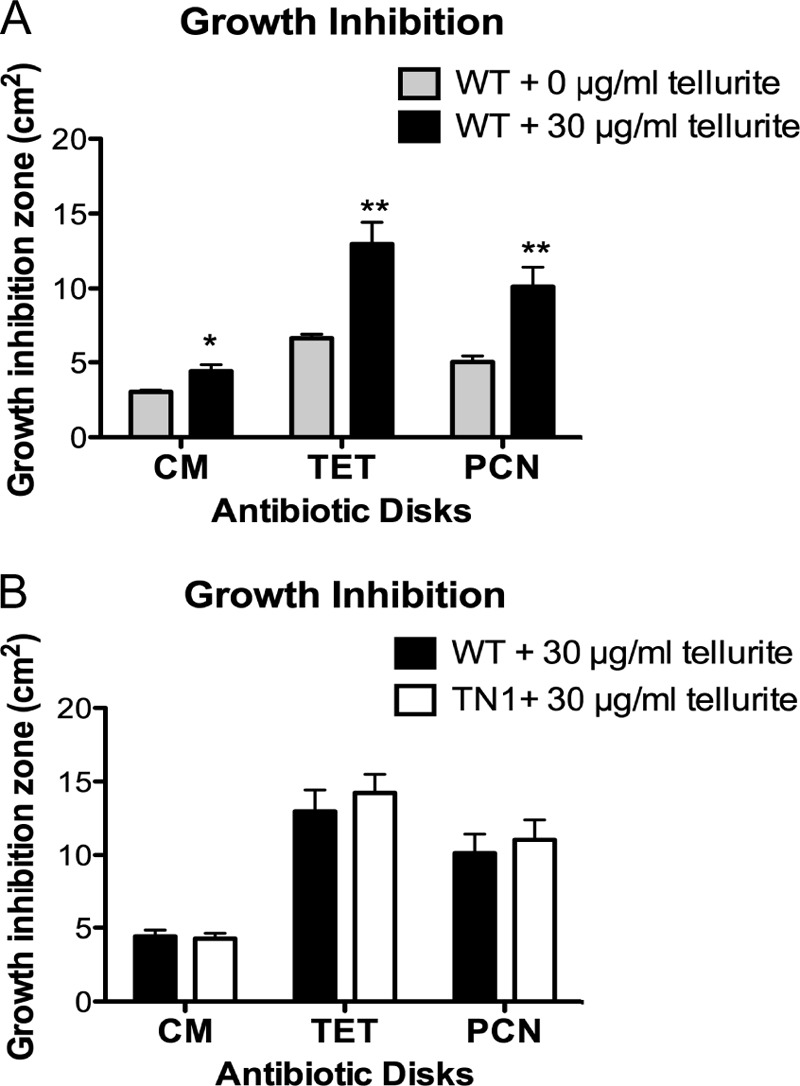
Tellurite enhances susceptibility to antibiotics independently of yceG and yceH. (A) Growth inhibition of WT B. anthracis Sterne spread on plates containing either 0 μg/ml tellurite (gray bars) or 30 μg/ml tellurite (black bars) and exposed to chloramphenicol (CM)-, tetracycline (TET)-, or penicillin (PCN)-impregnated disks. (B) Growth inhibition of WT B. anthracis Sterne (black bars) or TN1 mutant (white bars) spread on plates containing 30 μg/ml tellurite and exposed to chloramphenicol (CM)-, tetracycline (TET)-, or penicillin (PCN)-impregnated disks. Results from three independent experiments were combined and presented as means ± standard errors of the means. *, P < 0.05, or **, P < 0.01, by unpaired t test.
yceG and yceH contribute to B. anthracis pathogenicity in mammalian models of infection.
Since our transposon mutant is more susceptible to specific host defenses, we next assessed whether it was attenuated in mammalian models of infection. We found decreased survival of TN1 in human whole blood ex vivo (Fig. 6A), which contains a multitude of host defenses, including phagocytic cells, complement, and AMPs. We next tested whether TN1 was attenuated in an in vivo infection model. WT B. anthracis Sterne and the TN1 mutant were mixed at a 1:1 ratio and injected intravenously into mice. Bacterial survival in the kidneys was assessed when mice were moribund. Although equivalent numbers of WT and TN1 bacteria were injected (data not shown), survival of the transposon mutant was decreased about 10-fold in comparison to the WT bacteria (Fig. 6B), indicating that loss of yceG and yceH leads to attenuated virulence in a murine model of infection. Thus, yceG and yceH contribute to the overall virulence of B. anthracis in mammalian infection models. Furthermore, these data corroborate our results in the C. elegans screen (Fig. 1) and validate C. elegans as a screening model to identify B. anthracis Sterne mutants attenuated for virulence.
FIG 6.
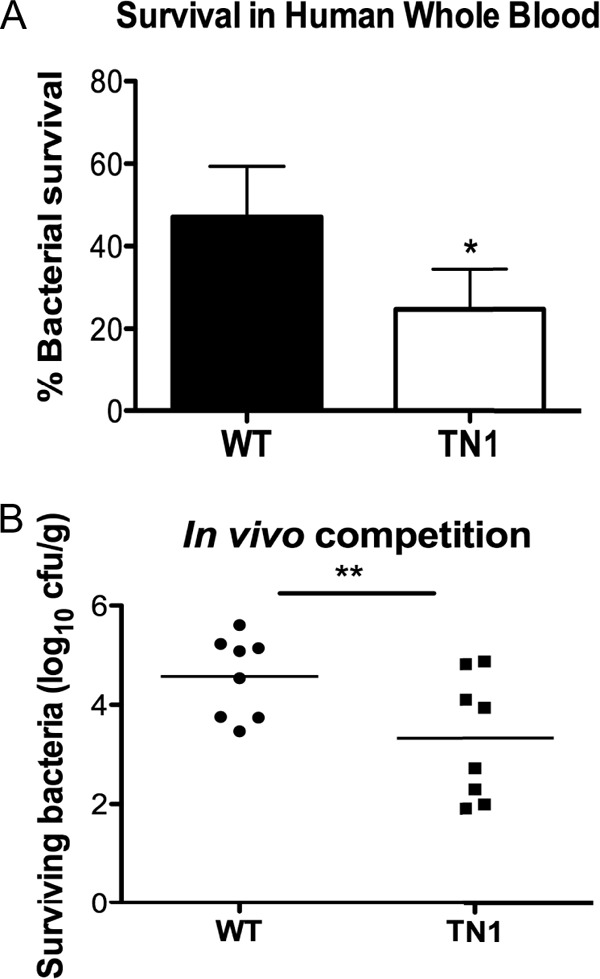
Loss of yceG and yceH increases susceptibility to mammalian defenses. (A) Survival of WT B. anthracis Sterne (black bars) and the TN1 mutant (white bars) after a 15-min incubation with human whole blood. Experiments were performed using blood from three individual donors, and results are combined and presented as means ± standard errors of the means. *, P < 0.05 by paired t test. (B) Enumeration of surviving CFU from kidneys of mice infected intravenously with a 1:1 ratio of WT B. anthracis Sterne to TN1. **, P < 0.01 by paired t test.
DISCUSSION
Novel virulence genes in several pathogens have been identified through screens utilizing C. elegans, including, as we demonstrate here, B. anthracis. Our ability to successfully use C. elegans to identify virulence factors was verified by the identification of the transposon mutant TN1, which is more susceptible to ROS and the human cathelicidin LL-37. TN1 has a disruption in yceG, the second-to-last gene in a six-gene cluster of tellurite resistance genes, that results in loss of expression of both yceG and yceH. Although originally identified as a putative operon (24), our results indicate independent regulation of yceCDEF and yceGH. There are at least five unique bacterial Ter determinants that have been identified, including the Ter proteins (similar to YceCDEF) and TelA (similar to YceH) (21). YceG remains poorly characterized with little sequence homology to other known tellurite resistance proteins. YceG has strong hydrophobic regions that may allow for interaction with the membrane, and across bacterial species, it is often found in association with TelA, indicating that these two proteins might be functionally linked (34). Despite their being widespread, surprisingly little is known regarding the actual function of any of the Ter genes.
As their name implies, the Ter genes are important for tellurite resistance in a number of bacterial species, including E. coli, P. mirabilis, and Y. pestis (21, 26, 27). While highly toxic to bacteria, tellurite is not necessarily commonly encountered, which raises the question of why these genes exist in such a wide range of species. It has been argued that the Ter genes are part of a general stress response system rather than solely tellurite resistance (21, 30, 34). In support of this, the Ter genes in E. coli are also implicated in resistance to bacteriophage and colicin (35). In Y. pestis, TerE and TerD proteins are upregulated along with other general stress response proteins during intracellular replication in macrophages (26).
Our data show that loss of yceG and yceH in B. anthracis results in a small growth impairment in the presence of tellurite. However, the more pronounced phenotype is the increased susceptibility to host defenses such as ROS. Both our transposon and insertional mutants demonstrated increased sensitivity to H2O2 and hypochlorite, the active ingredient in bleach. In a separate study in B. anthracis, yceCDEF were identified in an analysis of genes upregulated in response to H2O2 (36). The Ter operon of Proteus mirabilis is also induced by H2O2 and superoxide (27). Tellurite is also a strong oxidant (30), and tellurite toxicity has been linked to ROS production (31, 32). A recent comparative genomics study has found that the Ter genes are functionally linked to many enzymes involved in DNA processing and repair (34). It is therefore possible that these genes may play a role in repairing the DNA damage that would occur after exposure to oxidative stressors such as ROS or tellurite.
B. anthracis yceG and/or yceH is also important for resistance to the human cathelicidin LL-37. YceH has homology to TelA, which has been linked to resistance to nisin and some cell wall-targeting antibiotics in L. monocytogenes (28). Surprisingly, no difference in susceptibility was observed in the TN1 or IM1 mutants with any other AMP tested, including nisin, HNP-2, bacitracin, or polymyxin. This could be due to differences in mechanisms of action of the AMPs and/or in the responses of YceG/H. Although most AMPs form transmembrane pores, peptide insertion into the membrane can differ and some AMPs may have additional bactericidal mechanisms that could alter the bacterial response (37). In L. monocytogenes, loss of TelA increases susceptibility to some cell wall-acting antibiotics (the cephalosporins, cefotaxime, and cefuroxime) but not all (no difference was seen with methicillin or oxacillin) (28). We also saw no contribution of the yceG or yceH gene to resistance to several common antibiotics, even when antibiotic susceptibility was magnified by coincubation with tellurite. Therefore, although yceGH may function as part of a general stress response system, there is still specificity in their mechanism of action.
A potential role of Ter genes in host defense has long been speculated (21), but to our knowledge, this is the first report directly linking any of them to increased susceptibility in mammalian infection models. We find that loss of yceG and yceH results in decreased survival of B. anthracis Sterne in human whole blood and a murine in vivo competition assay, both of which expose the bacteria to a synergistic array of host defenses. Taken together, our results and those of others support the argument that tellurite resistance genes are likely part of a more general chemical stress response system. Loss of these genes could render bacteria more susceptible to a variety of potential environmental stresses, including, but not limited to, exposure to strong oxidants. This would help explain their widespread and ubiquitous nature in a wide variety of bacterial species, including several human pathogens. Further study will clarify the roles and mechanism of action of this intriguing and mysterious family of genes.
ACKNOWLEDGMENTS
This research was supported by NIH grant AI065993 (R.V.A. and V.N.). S.M.M. and C.Y.O. were supported by an IRCADA postdoctoral fellowship from NIGMS (GM068524), and S.M.M. was also supported by The Hartwell Foundation and through the Texas Christian University Research and Creative Activities Fund (60670).
Footnotes
Published ahead of print 23 December 2013
REFERENCES
- 1.Tournier JN, Paccani SR, Quesnel-Hellmann A, Baldari CT. 2009. Anthrax toxins: a weapon to systematically dismantle the host immune defenses. Mol. Aspects Med. 30:456–466. 10.1016/j.mam.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 2.Dixon TC. 1999. Anthrax. N. Engl. J. Med. 341:815–826. 10.1056/NEJM199909093411107 [DOI] [PubMed] [Google Scholar]
- 3.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, Kolstø AB, Fraser CM. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81–86. 10.1038/nature01586 [DOI] [PubMed] [Google Scholar]
- 4.Fisher N, Shetron-Rama L, Herring-Palmer A, Heffernan B, Bergman N, Hanna P. 2006. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 188:1301–1309. 10.1128/JB.188.4.1301-1309.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cendrowski S, MacArthur W, Hanna P. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 51:407–417. 10.1046/j.1365-2958.2003.03861.x [DOI] [PubMed] [Google Scholar]
- 6.Gat O, Mendelson I, Chitlaru T, Ariel N, Altboum Z, Levy H, Weiss S, Grosfeld H, Cohen S, Shafferman A. 2005. The solute-binding component of a putative Mn(II) ABC transporter (MntA) is a novel Bacillus anthracis virulence determinant. Mol. Microbiol. 58:533–551. 10.1111/j.1365-2958.2005.04848.x [DOI] [PubMed] [Google Scholar]
- 7.Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. 2008. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 105:1009–1013. 10.1073/pnas.0710950105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGillivray SM, Ebrahimi CM, Fisher N, Sabet M, Zhang DX, Chen Y, Haste NM, Aroian RV, Gallo RL, Guiney DG, Friedlander AM, Koehler TM, Nizet V. 2009. ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J. Innate Immun. 1:494–506. 10.1159/000225955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins A, Cote C, Twenhafel N, Merkel T, Bozue J, Welkos S. 2011. Role of purine biosynthesis in Bacillus anthracis pathogenesis and virulence. Infect. Immun. 79:153–166. 10.1128/IAI.00925-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chitlaru T, Zaide G, Ehrlich S, Inbar I, Cohen O, Shafferman A. 2011. HtrA is a major virulence determinant of Bacillus anthracis. Mol. Microbiol. 81:1542–1559. 10.1111/j.1365-2958.2011.07790.x [DOI] [PubMed] [Google Scholar]
- 11.Kho MF, Bellier A, Balasubramani V, Hu Y, Hsu W, Nielsen-LeRoux C, McGillivray SM, Nizet V, Aroian RV. 2011. The pore-forming protein Cry5B elicits the pathogenicity of Bacillus sp. against Caenorhabditis elegans. PLoS One 6:e29122. 10.1371/journal.pone.0029122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sifri CD, Begun J, Ausubel FM. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119–127. 10.1016/j.tim.2005.01.003 [DOI] [PubMed] [Google Scholar]
- 13.Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K, Zhang H. 2002. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361:221–230. 10.1042/0264-6021:3610221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chávez V, Mohri-Shiomi A, Garsin DA. 2009. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect. Immun. 77:4983–4989. 10.1128/IAI.00627-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaffin DO, Beres SB, Yim HH, Rubens CE. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466–4477. 10.1128/JB.182.16.4466-4477.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeng A, Sakota V, Li Z, Datta V, Beall B, Nizet V. 2003. Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208–1217. 10.1128/JB.185.4.1208-1217.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulston JE, Brenner S. 1974. The DNA of Caenorhabditis elegans. Genetics 77:95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon YM, Ricke SC. 2000. Efficient amplification of multiple transposon-flanking sequences. J. Microbiol. Methods 41:195–199. 10.1016/S0167-7012(00)00159-7 [DOI] [PubMed] [Google Scholar]
- 21.Taylor DE. 1999. Bacterial tellurite resistance. Trends Microbiol. 7:111–115. 10.1016/S0966-842X(99)01454-7 [DOI] [PubMed] [Google Scholar]
- 22.Quistgaard EM, Thirup SS. 2009. Sequence and structural analysis of the Asp-box motif and Asp-box beta-propellers; a widespread propeller-type characteristic of the Vps10 domain family and several glycoside hydrolase families. BMC Struct. Biol. 9:46. 10.1186/1472-6807-9-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heffernan BJ, Thomason B, Herring-Palmer A, Shaughnessy L, McDonald R, Fisher N, Huffnagle GB, Hanna P. 2006. Bacillus anthracis phospholipases C facilitate macrophage-associated growth and contribute to virulence in a murine model of inhalation anthrax. Infect. Immun. 74:3756–3764. 10.1128/IAI.00307-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumano M, Tamakoshi A, Yamane K. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22 degrees-25 degrees) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology 143:2775–2782. 10.1099/00221287-143-8-2775 [DOI] [PubMed] [Google Scholar]
- 25.Gupta A. 1999. RT-PCR: characterization of long multi-gene operons and multiple transcript gene clusters in bacteria. Biotechniques 27:960–972 [DOI] [PubMed] [Google Scholar]
- 26.Ponnusamy D, Hartson SD, Clinkenbeard KD. 2011. Intracellular Yersinia pestis expresses general stress response and tellurite resistance proteins in mouse macrophages. Vet. Microbiol. 150:146–151. 10.1016/j.vetmic.2010.12.025 [DOI] [PubMed] [Google Scholar]
- 27.Toptchieva A, Sisson G, Bryden LJ, Taylor DE, Hoffman PS. 2003. An inducible tellurite-resistance operon in Proteus mirabilis. Microbiology 149:1285–1295. 10.1099/mic.0.25981-0 [DOI] [PubMed] [Google Scholar]
- 28.Collins B, Joyce S, Hill C, Cotter PD, Ross RP. 2010. TelA contributes to the innate resistance of Listeria monocytogenes to nisin and other cell wall-acting antibiotics. Antimicrob. Agents Chemother. 54:4658–4663. 10.1128/AAC.00290-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambeth JD. 2004. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4:181–189. 10.1038/nri1312 [DOI] [PubMed] [Google Scholar]
- 30.Chasteen TG, Fuentes DE, Tantaleán JC, Vásquez CC. 2009. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33:820–832. 10.1111/j.1574-6976.2009.00177.x [DOI] [PubMed] [Google Scholar]
- 31.Pérez JM, Calderón IL, Arenas FA, Fuentes DE, Pradenas GA, Fuentes EL, Sandoval JM, Castro ME, Elías AO, Vásquez CC. 2007. Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. 10.1371/journal.pone.0000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tremaroli V, Fedi S, Zannoni D. 2007. Evidence for a tellurite-dependent generation of reactive oxygen species and absence of a tellurite-mediated adaptive response to oxidative stress in cells of Pseudomonas pseudoalcaligenes KF707. Arch. Microbiol. 187:127–135. 10.1007/s00203-006-0179-4 [DOI] [PubMed] [Google Scholar]
- 33.Molina-Quiroz RC, Muñoz-Villagrán CM, de la Torre E, Tantaleán JC, Vásquez CC, Pérez-Donoso JM. 2012. Enhancing the antibiotic antibacterial effect by sub lethal tellurite concentrations: tellurite and cefotaxime act synergistically in Escherichia coli. PLoS One 7:e35452. 10.1371/journal.pone.0035452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anantharaman V, Iyer LM, Aravind L. 2012. Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. Biosyst. 8:3142–3165. 10.1039/c2mb25239b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whelan KF, Colleran E, Taylor DE. 1995. Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 177:5016–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pohl S, Tu WY, Aldridge PD, Gillespie C, Hahne H, Mäder U, Read TD, Harwood CR. 2011. Combined proteomic and transcriptomic analysis of the response of Bacillus anthracis to oxidative stress. Proteomics 11:3036–3055. 10.1002/pmic.201100085 [DOI] [PubMed] [Google Scholar]
- 37.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]