Abstract
Exposure to the mold Aspergillus fumigatus may result in allergic bronchopulmonary aspergillosis, chronic necrotizing pulmonary aspergillosis, or invasive aspergillosis (IA), depending on the host's immune status. Neutrophil deficiency is the predominant risk factor for the development of IA, the most life-threatening condition associated with A. fumigatus exposure. Here we demonstrate that in addition to neutrophils, eosinophils are an important contributor to the clearance of A. fumigatus from the lung. Acute A. fumigatus challenge in normal mice induced the recruitment of CD11b+ Siglec F+ Ly-6Glo Ly-6Cneg CCR3+ eosinophils to the lungs, which was accompanied by an increase in lung Epx (eosinophil peroxidase) mRNA levels. Mice deficient in the transcription factor dblGATA1, which exhibit a selective deficiency in eosinophils, demonstrated impaired A. fumigatus clearance and evidence of germinating organisms in the lung. Higher burden correlated with lower mRNA expression of Epx (eosinophil peroxidase) and Prg2 (major basic protein) as well as lower interleukin 1β (IL-1β), IL-6, IL-17A, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and CXCL1 levels. However, examination of lung inflammatory cell populations failed to demonstrate defects in monocyte/macrophage, dendritic cell, or neutrophil recruitment in dblGATA1-deficient mice, suggesting that the absence of eosinophils in dlbGATA1-deficient mice was the sole cause of impaired lung clearance. We show that eosinophils generated from bone marrow have potent killing activity against A. fumigtaus in vitro, which does not require cell contact and can be recapitulated by eosinophil whole-cell lysates. Collectively, our data support a role for eosinophils in the lung response after A. fumigatus exposure.
INTRODUCTION
Exposure to the opportunistic mold Aspergillus fumigatus may result in a wide range of infectious diseases, with the type and severity largely determined by the status of the host immune system. Among these aspergillosis infections, individuals may develop allergic bronchopulmonary aspergillosis, chronic pulmonary aspergillosis (chronic necrotizing [CNPA] or chronic cavitary pulmonary [CCPA] aspergillosis), and invasive aspergillosis (IA). Development of the last infection (IA) may be a result of multiple predisposing factors, yet immunosuppression leading to neutropenia remains the predominant risk factor (1, 2). Collectively, the most susceptible populations for the development of IA include those undergoing solid-organ or hematopoietic stem cell (HSC) transplants (3). The most recent data from the Transplant Associated Infections Surveillance Network (March 2001 to March 2006) reflect incidence rates for invasive fungal infections (IFIs) of ∼7% in both solid-organ (4) (19% due to IA) and HSC (5) (43% due to IA) transplants. Although IA is a known infectious complication of these conditions, there is a growing concern for the development of nosocomial IA in nonneutropenic patients in intensive care units (ICUs), particularly those on high-dose steroids (those with chronic obstructive pulmonary disease [COPD], for instance) and those with cirrhosis and solid cancers (6). Subsequent studies have identified “new” nonneutropenic conditions associated with the development of IA as COPD, chronic steroid use, and liver failure (7, 8, 9, 10).
Chronic pulmonary aspergillosis (CNPA and CCPA), which may be referred to as aspergilloma in some cases, is thought to be a result of an underlying or preexisting pulmonary disease, such as tuberculosis, pneumoconiosis, or COPD (11). Diagnosis of CNPA and CCPA relies heavily on histologic and radiographic examinations. The extra scrutiny regarding the diagnosis of chronic pulmonary aspergillosis has led to the discovery of an eosinophilic component in some cases (12). Fungal pneumonia and IA in chronic granulomatous disease (CGD) can also sometimes present as an eosinophilic pneumonia (13, 14).
Allergic bronchopulmonary aspergillosis (ABPA) is a hypersensitivity disorder caused by A. fumigatus (and other Aspergillus species) and is often marked by an eosinophilic immune response in humans (15). Moreover, in experimental repetitive-exposure models of A. fumigatus, we (16) and others (17, 18) have demonstrated that chronic exposure to A. fumigatus resulted in substantial recruitment of eosinophils to the airways. Assessment of the role allergic inflammatory responses play in fungal clearance has shown that mice deficient in the Th2 cytokine interleukin 13 (IL-13) or mice in which the Th2 and proeosinophil cytokine IL-5 was neutralized resulted in attenuated lung clearance of Aspergillus niger (19). Collectively, these results unexpectedly suggest that Th2 responses and eosinophils play a role in immune-mediated clearance of Aspergillus spp. from the lungs during asthma. As eosinophils have been identified in clinical cases of chronic pulmonary aspergillosis and in some cases of IA in CGD patients, the goal of the current study was to further evaluate the exact role of eosinophils during lung exposure to A. fumigatus. Employing mice deficient in the eosinophil lineage and in vitro eosinophil cultures, we found that this cell type is a potent lung effector cell against A. fumigatus.
MATERIALS AND METHODS
Mice.
Wild-type (WT) BALB/c and dblGATA1-deficient mice, 6 to 8 weeks of age, were obtained from The Jackson Laboratory (Bangor, ME) and bred at the University of Alabama at Birmingham (UAB). All animals were housed in a specific-pathogen-free, Association for Assessment and Accreditation of Laboratory Animal Care-certified facility and handled according to Public Health Service Office of Laboratory Animal Welfare policies after review by the UAB Institutional Animal Care and Use Committee.
Preparation of A. fumigatus, in vivo challenge, and lung fungal burden assessment and lung histology.
A. fumigatus isolate ATCC 13073 (ATCC, Manassas, VA) was maintained on potato dextrose agar for 5 to 7 days at 37°C. Conidia were harvested by washing the culture flask with 50 ml of sterile phosphate-buffered saline (PBS) supplemented with 0.1% Tween 20. The conidia were then passed through a sterile 40-μm nylon membrane to remove hyphal fragments and enumerated on a hemacytometer. For challenge, mice were lightly anesthetized with isoflurane and administered 7 × 107 A. fumigatus conidia in a volume of 50 μl intratracheally as previously described (20, 21). Briefly, mice were held in a vertical, upright position and the tongue was withdrawn from the mouth using forceps. A pipette was used to deliver the 50-μl inoculum to the caudal oropharynx, in which normal breathing results in fluid aspiration into the lungs (22). For lung fungal burden analysis, the left lungs were collected at 24 or 48 h postexposure and homogenized in 1 ml of PBS. Total RNA was extracted from 0.1 ml of unclarified lung homogenate using a MasterPure yeast RNA purification kit (Epicentre Biotechnologies, Madison, WI), which includes a DNase treatment step to eliminate genomic DNA as previously reported (23). Total RNA was also extracted from serial 1:10 dilutions of live A. fumigatus conidia (101 to 109) and DNase treated to form a standard curve. Lung A. fumigatus burden was analyzed with real-time PCR measurement of the A. fumigatus 18S rRNA (GenBank accession number AB008401 [24]) and quantified using a standard curve of A. fumigatus conidia as previously described (23). As a validation of the real-time PCR method, heat-killed A. fumigatus did not yield a signal by real-time PCR and was unable to grow on potato dextrose agar plates (23). In addition, no amplification controls (i.e., no reverse transcriptase included in the cDNA reaction) yielded a signal of <0.001% by real-time PCR, indicating that the DNase treatment step was efficient at eliminating contaminating A. fumigatus DNA (as DNA is not predicative of organism viability [25]). For lung histology, the left lungs were collected and fixed in 4% formalin. The fixed lungs were paraffin embedded and then processed and stained by the Comparative Pathology Laboratory at the University of Alabama at Birmingham. Imaging was performed using a Nikon Eclipse 90i microscope and Nikon NIS-Elements imaging analysis software.
Bronchoalveolar lavage cell surface marker flow cytometry.
Bronchoalveolar lavage cell isolations were performed as previously described (26, 27). Cells were washed and Fc receptors blocked with Mouse BD Fc Block (BD Biosciences, San Diego, CA) at 4°C for 20 min. Thereafter, cells were stained with a single-color LIVE/DEAD fixable dead cell stain (Invitrogen), followed by labeling with specific immune cell surface markers. The following staining parameters were employed: macrophages were identified as CD11blo/neg CD11c+, eosinophils as CD11b+ Siglec F+ Ly-6Glo Ly-6Cneg CCR3+, neutrophils as CD11b+ Ly-6G+, and dendritic cells as CD11b+ CD11c+ (all antibodies purchased from eBiosciences or BD Biosciences). Samples were acquired using a four-laser, 20-parameter analytic BD LSR II flow cytometer, and data were analyzed using the FlowJo software program (Tree Star, Ashland, OR).
Real-time PCR analysis for Epx, Prg2, Ear1, and Ear2 expression in lung tissue.
WT BALB/c and dblGATA1-deficient mice were challenged with A. fumigatus as described above; 24 h after exposure, lungs were collected and homogenized in TRIzol reagent (Invitrogen), and total RNA was isolated as per the manufacturer's instructions. Thereafter, RNA was transcribed to cDNA (iScript cDNA synthesis kit; Bio-Rad), and real-time PCR for Epx (Mm00514768_m1; Applied Biosystems), Prg2 (Mm00435905_m1; Applied Biosystems), Ear1 (Mm03059811_g10; Applied Biosystems), and Ear2 (Mm04207376_gH; Applied Biosystems) was performed (iQ Supermix; Bio-Rad). mRNA levels were normalized to Gapdh mRNA levels (primers and probe from Applied Biosystems) using the threshold cycle [2−ΔΔCT] method.
Whole-lung cytokine and chemokine analysis.
BALB/c WT and dblGATA1-deficient mice were challenged with A. fumigatus as described above; 24 or 48 h after exposure, the right lungs were isolated and homogenized in PBS supplemented with cOmplete Mini protease inhibitor tablets (Roche), clarified by centrifugation, and stored at −80°C. Supernatants from lung homogenates were analyzed for protein levels of 23 cytokines and chemokines using a Bio-Plex multiplex suspension cytokine array (Bio-Rad Laboratories), according to the manufacturer's instructions (20, 21). The data were analyzed using Bio-Plex Manager software (Bio-Rad Laboratories).
Derivation of eosinophils from bone marrow and assessment of A. fumigatus killing and cytokine production.
Bone marrow-derived eosinophils were generated using a previously described protocol (28). Briefly, bone marrow was isolated from naive BALB/c mice and cells plated at 1 × 106 cells/ml in RPMI 1640 containing 20% fetal bovine serum (FBS; Irvine Scientific, Santa Ana, CA), 2 mM glutamine, 25 mM HEPES, 1× minimal essential medium (MEM) nonessential amino acids, 1 mM sodium pyruvate (all from Life Technologies BRL, Rockville, MD), 50 μM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), and 100 ng/ml of stem cell factor (SCF) and 100 ng/ml of FLT3-L (both from Peprotech). After 4 days, cells were replated in the above medium supplemented with 10 ng/ml of IL-5. After 10 days, bone marrow cells were fully differentiated into eosinophils. As previously reported (28), samples of 1 × 105 cells were taken for RNA analysis each time medium was changed for real-time PCR analysis of Epx (Mm00514768_m1; Applied Biosystems) for eosinophil development and Mpo (Mm01298424_m1; Applied Biosystems) for neutrophil development. In addition, cells were cytospun onto glass slides, Giemsa stained, and analyzed for morphology and purity by a murine pathologist in the Comparative Pathology Laboratory at the University of Alabama at Birmingham. On the 10th day, bone marrow-derived eosinophils were enumerated and utilized in experiments. To assess eosinophil-mediated killing of A. fumigatus, 1 × 105 eosinophils in a volume of 50 μl were cultured in triplicate with 3 × 105 A. fumigatus conidia in a volume of 50 μl for 24 h at 37°C and 5% CO2. Controls included 3 × 105 A. fumigatus conidia cultured alone in triplicate in a volume of 100 μl for 24 h. Thereafter, total RNA was extracted from each well using a MasterPure yeast RNA purification kit (Epicentre Biotechnologies, Madison, WI) and real-time PCR was performed as described above. In other studies, the eosinophil-A. fumigatus cocultures were performed in Corning Transwell plates (24 wells; 0.4-μm pore size) to assess the requirement for cell contact. For this, 1 × 106 eosinophils in a volume of 100 μl were cultured in the top chamber in the presence of 3 × 106 eosinophils in a volume of 300 μl cultured in the bottom chamber. Controls included 100 μl of medium in the top chamber in the presence of 3 × 106 eosinophils in a volume of 300 μl cultured in the bottom chamber. Parallel studies determined the effects of eosinophil whole-cell lysates on A. fumigatus viability (as assessed by real-time PCR). For this, 1 × 106 bone marrow-derived eosinophils were lysed in 100 μl of PhosphoSafe extraction buffer (Novagen/EMD/Millipore) for 10 min at room temperature, followed by centrifugation at 12,000 × g for 10 min. Lysates (100 μl) were added in triplicate to 3 × 105 A. fumigatus conidia in a volume of 100 μl for 24 h at 37°C and 5% CO2. Controls included PhosphoSafe extraction buffer (100 μl) added in triplicate to 3 × 105 A. fumigatus conidia in a volume of 50 μl for 24 h at 37°C and 5% CO2. To assess eosinophil cytokine and chemokine production, eosinophils and A. fumigatus were cultured at the ratio above (except that 1 × 106 eosinophils and 3 × 106 conidia were employed). Twenty-four hours after culture, the contents of each well were collected and centrifuged, and clarified supernatants were assessed using the Bio-Plex multiplex suspension cytokine array (Bio-Rad Laboratories) as detailed above.
Statistics.
Data were analyzed using GraphPad Prism, version 5.0, statistical software (GraphPad Software, San Diego, CA). Comparisons between groups when data were normally distributed were made with the two-tailed unpaired Student t test. In real-time PCR analyses, the two-tailed paired Student t test was employed. Significance was accepted at a P value of <0.05.
RESULTS
Eosinophils are recruited to the lungs after A. fumigatus challenge.
Eosinophils are well documented to play a role in allergic disorders, including the allergic form of aspergillosis, allergic bronchopulmonary aspergillosis (15). In contrast, a previous study employing chemically immunosuppressed mice challenged with a low dose of A. fumigatus suggested that eosinophils likely played a minimal role in defense (29). Therefore, we sought to clarify their role during A. fumigatus exposure in immunocompetent mice. We first show that in normal BALB/c mice challenged with A. fumigatus, there is a significant increase in lung expression of the eosinophil marker Epx (eosinophil peroxidase) 24 h after exposure (Fig. 1A). We subsequently verified the presence of eosinophils by detecting CD11b+ Siglec F+ Ly-6GloLy-6Cneg CCR3+ cells in lung lavage fluid 24 and 48 h after A. fumigatus exposure (Fig. 1B). The absolute eosinophil numbers are roughly a log less than what we have previously reported for neutrophils in lung lavage fluid after A. fumigatus challenge in C57BL/6 mice (20, 21). Assessment of Epx mRNA levels and eosinophils by flow cytometry at 12 h did not reveal a significant presence in the lungs (data not shown). Thus, gene expression and flow cytometric analysis reveal that eosinophils are recruited to the lungs during an A. fumigatus infection.
FIG 1.
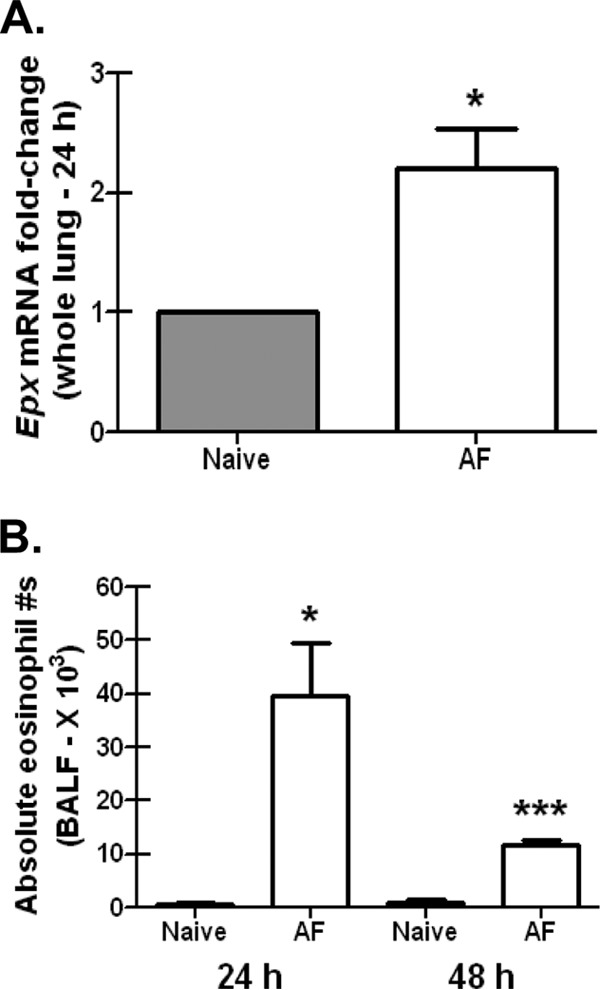
Eosinophils are recruited to lung after A. fumigatus challenge. (A) BALB/c wild-type mice were challenged intratracheally with 7 × 107 A. fumigatus conidia (AF), and 24 h postexposure, whole lungs were collected, total RNA was isolated and transcribed to cDNA, and quantitative real-time PCR was performed for Epx. Gene expression was normalized to Gapdh, and fold changes between naive mice (set at 1) and A. fumigatus-exposed BALB/c mice were determined using the 2−ΔΔCT method. Cumulative data from three independent studies (n = 3 to 5 mice per group per study) are illustrated. *, P value of <0.05 (paired two-tailed Student t test). (B) BALB/c wild-type mice were challenged intratracheally with 7 × 107 A. fumigatus conidia, and 24 or 48 h postexposure, lung cells were isolated via bronchoalveolar lavage, Fc blocked, stained with a LIVE/DEAD staining kit, and thereafter stained with fluorochrome-conjugated CD11c, CD11b, Gr-1, and Siglec F. Cumulative data from three independent studies (n = 2 or 3 mice per group per time point per study) are illustrated. Data are expressed as absolute numbers of live cells in lung lavage fluid. * and ***, P values of <0.05 and <0.0001, respectively (unpaired two-tailed Student t test).
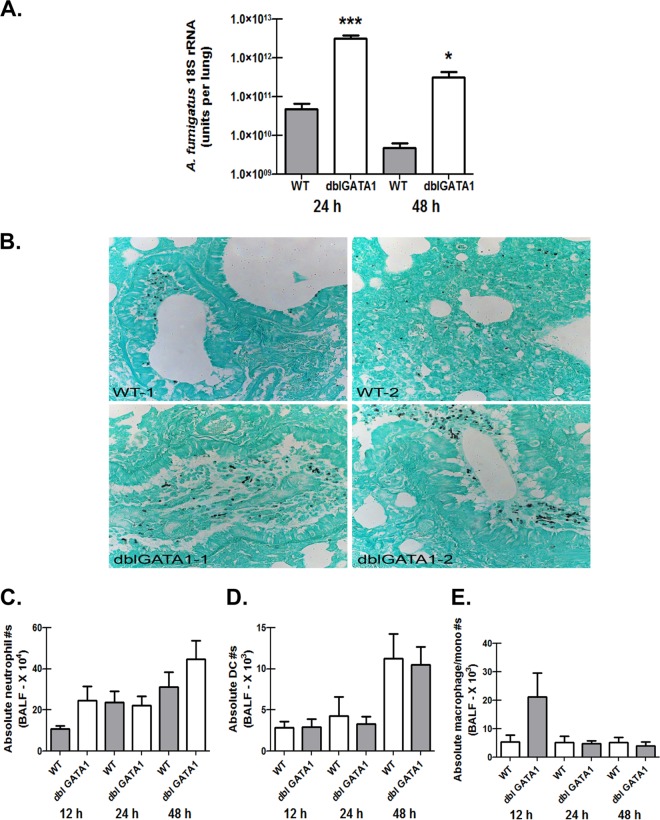
Enhanced susceptibility of eosinophil-deficient mice after challenge with A. fumigatus.
As eosinophils are recruited to the lungs during infection, we questioned whether they played a role in clearance an A. fumigatus challenge from the lung. To determine this, we challenged BALB/c WT and dblGATA1-deficient mice with A. fumigatus conidia and assessed fungal burden 24 h and 48 h postinfection. dblGATA1-deficient mice are deficient in a high-affinity GATA-binding site in the GATA-1 promoter and have a selective depletion of eosinophils but not mast cells or platelets (30). dblGATA1-deficient mice demonstrated a nearly 2-log-higher A. fumigatus burden 24 h postchallenge (Fig. 2A). Although there was some level of fungal clearance in dblGATA1-deficient mice between 24 and 48 h postchallenge, these mice nevertheless maintained a 2-log-higher burden at 48 h than did A. fumigatus-challenged BALB/c WT mice (Fig. 2A). Additional studies carried out for 7 days did not demonstrate increased mortality of dblGATA1-deficient mice (data not shown). However, Gomori-Grocott methenamine silver (GMS) staining of lung tissue from dblGATA1-deficient mice revealed the presence of germinating conidia and some hyphae, which was not observed in BALB/c WT mice (Fig. 2B). Neutrophils are widely acknowledged to be the most important effector cell for clearing A. fumigatus from the lungs (31, 32, 33). Assessing cell recruitment via flow cytometry of lung lavage cells 12, 24, and 48 h post-A. fumigatus challenge did not demonstrate a difference in neutrophil recruitment between BALB/c WT and dblGATA1-deficient mice (Fig. 2C). Moreover, we verified that thioglycolate-elicited neutrophils from dblGATA1-deficient mice stimulated with A. fumigatus conidia did not demonstrate impaired reactive oxygen species (ROS) production (data not shown). Dendritic cells (Fig. 2D) and monocyte/macrophage populations (Fig. 2E) were also not affected by A. fumigatus exposure in dblGATA1-deficient mice. Thus, mice deficient in eosinophils demonstrate impaired clearance of A. fumigatus from the lungs, which is not a result of defects in inflammatory cell recruitment or function.
FIG 2.
Enhanced susceptibility of eosinophil-deficient mice after challenge with A. fumigatus. (A) BALB/c WT and dblGATA1-deficient (dblGATA1) mice were challenged intratracheally with 7 × 107 A. fumigatus conidia, and 24 and 48 h after exposure, lung fungal burden was assessed by real-time PCR analysis of A. fumigatus 18S rRNA levels. Cumulative data from two independent studies (n = 4 or 5 mice per group per time point per study) are illustrated. Data are expressed as means + standard errors of the means. * and ***, P values of <0.05 and <0.0001, respectively (unpaired two-tailed Student t test). (B) Representative GMS-stained lung sections of two individual WT (top) and dblGATA1-deficient (bottom) mice 48 h post-A. fumigatus challenge. (C to E) WT and dblGATA1-deficient mice were challenged intratracheally with 7 × 107 A. fumigatus conidia, and lung cells were isolated via bronchoalveolar lavage, Fc blocked, stained with a LIVE/DEAD staining kit, and thereafter stained with fluorochrome-conjugated antibodies differentiating neutrophils (C), dendritic cells (D), and monocytes/macrophages (E). Cumulative data from three independent studies (n = 3 or 4 mice per group per study) are illustrated. Data are expressed as absolute numbers of live cells in lung lavage fluid.
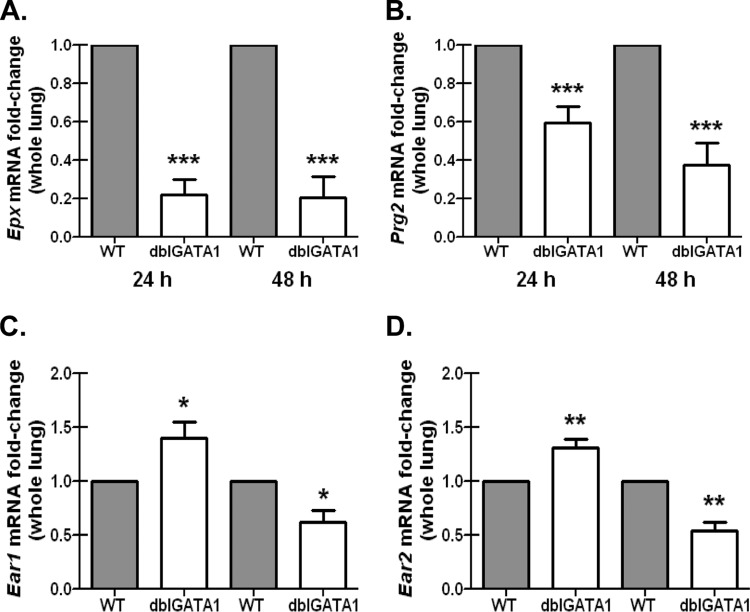
Eosinophil-deficient mice challenged with A. fumigatus demonstrate reductions in specific eosinophil antimicrobial factors.
Like neutrophils, eosinophils are equipped with potent granule proteins that possess antimicrobial activity, including basic protein (MBP), MBP2, eosinophil cationic protein (ECP), eosinophil peroxidase (EPX), and eosinophil-derived neurotoxin (EDN) (34). However, although highly expressed in eosinophils, some eosinophil-associated factors, such as various ribonucleases, may be expressed by additional cell types, such as macrophages and epithelial cells (35). Therefore, we sought to determine which of these eosinophil antimicrobial components were differentially expressed in the lungs of BALB/c WT and dblGATA1-deficient mice after A. fumigatus challenge. Results show that despite demonstrating significantly higher A. fumigatus lung burden at 24 and 48 h postchallenge (Fig. 2), mRNA levels of eosinophil peroxidase, Epx, were significantly lower in dblGATA1-deficient mice (Fig. 3A). Similar results were observed for major basic protein, Prg2 (Fig. 3B). In contrast, mRNA levels of eosinophil-associated RNase 1, Ear1, were significantly elevated in A. fumigatus-exposed dblGATA1-deficient mice at 24 h but significantly reduced at 48 h (Fig. 3C). mRNA levels of eosinophil-associated RNase 2, Ear2, followed the same trend (Fig. 3D). Thus, lower eosinophil peroxidase and major basic protein expression levels, but not eosinophil-associated RNase 1 or 2, correlate with impaired A. fumigatus lung clearance in dblGATA1-deficient mice and thus may contribute to antifungal host defense during infection.
FIG 3.
Eosinophil-deficient mice challenged with A. fumigatus demonstrate reductions in specific eosinophil antimicrobial factors. BALB/c WT and dblGATA1-deficient (dblGATA1) mice were challenged intratracheally with 7 × 107 A. fumigatus conidia, and 24 and 48 h after exposure, whole lungs were collected, total RNA was isolated and transcribed to cDNA, and quantitative real-time PCR was performed for Epx (A), Prg2 (B), Ear1 (C), and Ear2 (D). Gene expression was normalized to Gapdh, and fold changes between WT mice (set at 1) and dblGATA1-deficient mice were determined using the 2−ΔΔCT method. Cumulative data from two independent studies (n = 4 or 5 mice per group per time point per study) are illustrated. *, **, and ***, P values of <0.05, <0.01, and <0.0001, respectively (paired two-tailed Student t test).
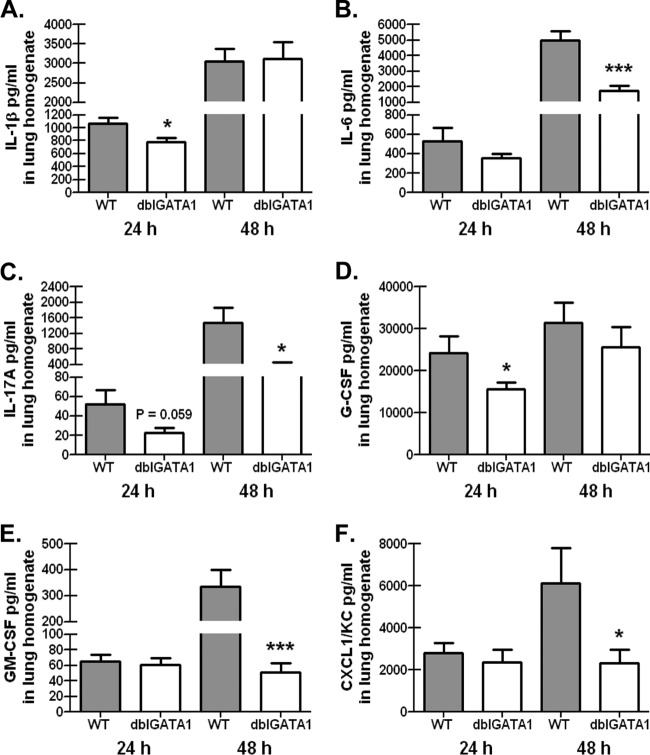
Eosinophil-deficient mice challenged with A. fumigatus demonstrate differences in lung proinflammatory cytokine and chemokine levels.
Despite observing no defects in neutrophil recruitment to the lungs of dblGATA1-deficient mice (Fig. 2), we nevertheless determined whether the absence of eosinophils during A. fumigatus exposure affected the magnitude of the lung inflammatory response. We analyzed the protein levels of 23 cytokines and chemokines in clarified whole-lung homogenates from BALB/c WT and dblGATA1-deficient mice 24 and 48 h postinfection. Results in Fig. 4 show that despite the significantly higher A. fumigatus lung burden at 24 h and 48 h postchallenge (Fig. 2), the proinflammatory cytokines IL-1β (Fig. 4A), IL-6 (Fig. 4B), and IL-17A (Fig. 4C) were significantly lower in dblGATA1-deficient mice at 24 h (IL-1β) and 48 h (IL-6 and IL-17A) postchallenge, respectively. Likewise, the growth factors granulocyte colony-stimulating factor (G-CSF) (Fig. 4D) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Fig. 4E) and the chemokine CXCL1/KC (Fig. 4F) were significantly lower in dblGATA1-deficient mice at 24 h (G-CSF) and 48 h (GM-CSF and CXCL1) postchallenge, respectively. Thus, attenuated lung cytokine and chemokine production in the absence of eosinophils correlates with impaired A. fumigatus lung clearance in dblGATA1-deficient mice.
FIG 4.
Eosinophil-deficient mice challenged with A. fumigatus demonstrate differences in lung proinflammatory cytokine and chemokine levels. BALB/c WT and dblGATA1-deficient (dblGATA1) mice were challenged intratracheally with 7 × 107 A. fumigatus conidia, and 24 and 48 h after exposure, levels of IL-1β (A), IL-6 (B), IL-17A (C), G-CSF (D), (E) GM-CSF (E), and CXCL1 (F) were quantified in lung homogenates by Bio-Plex. Cumulative data from two or three independent studies (n = 4 to 6 mice per group per time point per study) are illustrated. Data are expressed as mean concentrations + standard errors of the means. * and ***, P values of <0.05 and 0.001, respectively (unpaired two-tailed Student t test).
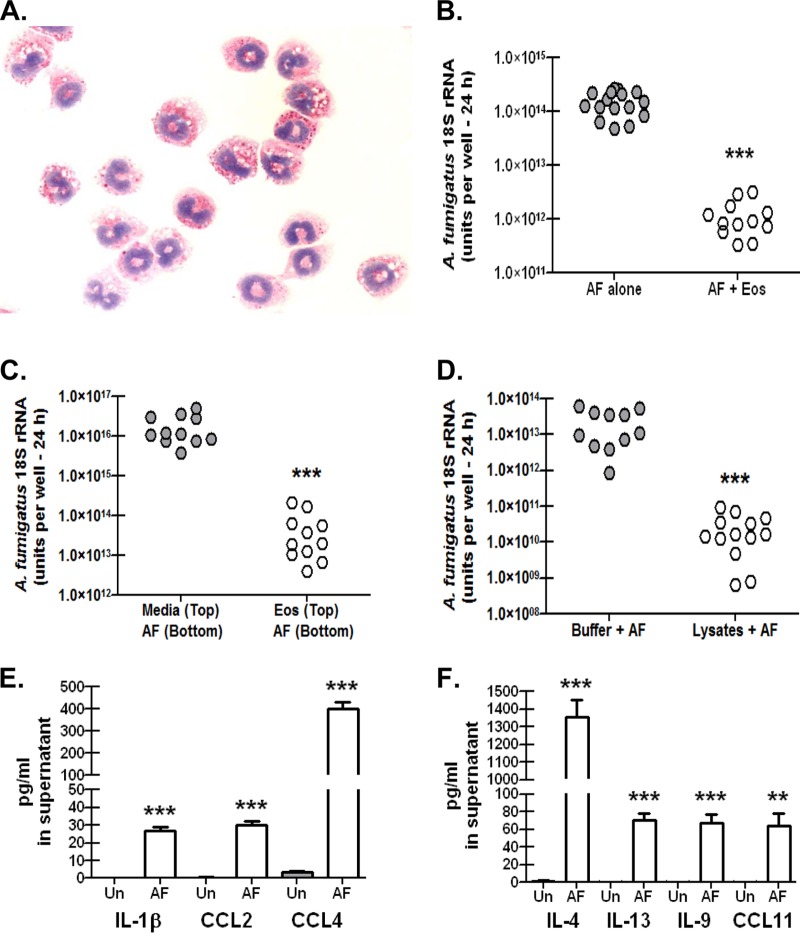
Eosinophils recognize and respond to A. fumigatus.
Data thus far have shown that mice deficient in the generation of eosinophils have a profound defect in clearing A. fumigatus from the lungs, and despite impairments in proinflammatory responses, there were no defects in neutrophil recruitment or function in the absence of eosinophils. These results suggest that eosinophils themselves are active inhibitors of A. fumigatus growth in the lungs. To address this, we generated eosinophils from the bone marrow of WT BALB/c mice (28) and determined whether they had the ability to affect A. fumigatus viability in vitro. As previously reported (28), we verified that culturing bone marrow cells in the presence of stem cell factor and FLT3-L for 4 days followed by the addition of IL-5 for an additional 6 days resulted in a significant enhancement of Epx mRNA expression, with a >300-fold increase by day 10 of the culture (data not shown). In contrast, throughout the 10-day culture period, we saw a parallel decrease in mRNA expression of Mpo (myeloperoxidase), a widely employed biomarker of neutrophils (data not shown). A representative image of a Giemsa-stained cytospin preparation of bone marrow-derived eosinophils at day 10 shows characteristic nuclear morphology and the presence of secretory granules (Fig. 5A). We next determined whether eosinophils limited the growth of A. fumigatus in vitro. The previously mentioned A. niger study employed ratios of eosinophils to conidia of 6:1 up to 100:1, which inhibited A. niger growth ∼25% and 100%, respectively (19). We assessed killing by employing a significantly lower ratio, as the low numbers of eosinophils detected in lung lavage fluid (Fig. 2) suggested a scenario whereby the organisms outnumbered the eosinophils. With a ratio of eosinophils to conidia of 1:3, results showed that culturing A. fumigatus in the presence of eosinophils decreased viability by more than 2 logs after 24 h of culture (Fig. 5B), a killing rate of >99%. This killing was not cell contact dependent, as separating eosinophils from A. fumigatus via a 0.4-μm-pore-containing Transwell membrane still resulted in significant killing of A. fumigatus (Fig. 5C). As this supports the hypothesis that secreted or soluble factors derived from eosinophils are toxic to A. fumigatus, we confirmed that eosinophil lysates containing granule proteins inhibited the growth of A. fumigatus (Fig. 5D). As eosinophils were capable of recognizing and killing A. fumigatus in vitro, we next determined whether this interaction also resulted in the elaboration of cytokines and chemokines. Employing a culture design identical to that in Fig. 5B, we observed significant production of the chemokine CCL4/MIP-1β (Fig. 5E) and the Th2 cytokine IL-4 (Fig. 5F). Eosinophils cultured in the presence of A. fumigatus also produced IL-1β and CCL2/MCP-1 (Fig. 5E), as well as the Th2 cytokines IL-13 and IL-9 and the well-recognized eosinophil chemoattractant CCL11/eotaxin (Fig. 5F). Thus, eosinophils respond to A. fumigatus with the production of proinflammatory cytokines and chemokines as well as Th2 cytokines, an event that also leads to destruction of the organism.
FIG 5.
Eosinophils recognize and respond to A. fumigatus. Bone marrow cells were isolated from naive BALB/c mice and cultured with 100 ng/ml of stem cell factor and 100 ng/ml of FLT3-L. After 4 days, cells were replated in medium supplemented with 10 ng/ml of IL-5 for an additional 6 days. (A) Representative Giemsa-stained cytospin of bone marrow-derived eosinophils at day 10 of culture showing characteristic nuclear structure and secretory granules. (B) Bone marrow-derived eosinophils (1 × 105 cells) were cultured in the presence of A. fumigatus conidia (3 × 105) in a total volume of 100 μl for 24 h (AF + Eos). Controls included A. fumigatus conidia (3 × 105) cultured alone in a volume of 100 μl for 24 h (AF alone). Cumulative data from four independent studies are illustrated. Each dot represents a single well. ***, P value of <0.0001 (unpaired two-tailed Student t test). (C) Bone marrow-derived eosinophils (1 × 106 cells; 100 μl) were cultured in the top chamber of a 0.4-μm-pore-containing Transwell membrane in the presence of A. fumigatus conidia (3 × 106; 300 μl) for 24 h [Eos (Top)/AF (Bottom)]. Controls included 100 μl of medium in the top chamber in the presence of A. fumigatus conidia (3 × 106; 300 μl) cultured alone in the bottom chamber for 24 h [Media (Top)/AF (Bottom)]. Cumulative data from three independent studies are illustrated. Each dot represents a single well. ***, P value of <0.0001 (unpaired two-tailed Student t test). (D) Bone marrow-derived eosinophils (1 × 106 cells) were lysed in 100 μl of PhosphoSafe extraction buffer (10 min at room temperature, followed by centrifugation at 12,000 × g for 10 min). Lysates (100 μl) were added in triplicate to 3 × 105 A. fumigatus conidia in a volume of 100 μl for 24 h. Controls included PhosphoSafe extraction buffer (100 μl) added in triplicate to 3 × 105 A. fumigatus conidia in a volume of 50 μl for 24 h. Cumulative data from three independent studies are illustrated. Each dot represents a single well. ***, P value of <0.0001 (unpaired two-tailed Student t test). (E and F) Bone marrow-derived eosinophils were cultured as for panel B, and supernatants were collected after 24 h and clarified by centrifugation. IL-1β, CCL2, and CCL4 (E) and IL-4, IL-13, IL-9, and CCL11 (F) were quantified by Bio-Plex. Controls included bone marrow-derived eosinophils cultured in medium alone (gray columns). Cumulative data from two independent studies are illustrated. ** and ***, P values of <0.01 and <0.0001, respectively (unpaired two-tailed Student t test). Un, unstimulated.
DISCUSSION
Eosinophils are recognized to play important roles in two main immune responses, antiparasitic responses and proallergic or asthmatic responses. However, the development of unique eosinophil-deficient mouse strains (30, 36) and additional mouse strains in which eosinophil recruitment or function is diminished (37, 38, 39) has provided novel insights into the role of eosinophils in T helper responses, interactions with immune cells such as macrophages and dendritic cells, and antimicrobial defense (40). With respect to the last, the identification of Toll-like receptor (TLR) expression and responsiveness by human eosinophils (41, 42) has renewed interest in their host defense capabilities, which is supported by findings such as a role for eosinophils in host defense against the Gram-negative bacterium Pseudomonas aeruginosa (43) and respiratory syncytial virus (RSV) (44, 45). Consequently, the goal of the current study was to determine whether the eosinophil antimicrobial activity extended to fungi, specifically A. fumigatus.
We have reported that eosinophils are the predominant cell type observed in the lungs of mice repetitively challenged with A. fumigatus in an experimental model of fungal asthma (16). However, we have reported that during acute A. fumigatus challenge, neutrophils represent the dominant lung cell type, although eosinophils were present as well (20). Curiously, eosinophils were significantly higher, and neutrophils significantly lower, in mice deficient in the beta-glucan receptor dectin-1, which are highly susceptible to A. fumigatus (20). This observation suggested the possibility that the ratio of neutrophils to eosinophils could determine the outcome of A. fumigatus infection, i.e., that eosinophils may actually be harmful for host defense. We first confirmed that eosinophils were recruited to the lungs of normal BALB/c mice after acute A. fumigatus challenge (as opposed to our previous study with normal 129/SvEv mice [20]). We assessed this via the expression of the gene for eosinophil peroxidase, Epx, which showed significant upregulation in the lungs after A. fumigatus challenge. Eosinophils are sometimes difficult to identify via flow cytometry, yet we corroborated the Epx analysis by employing a thorough staining procedure that identified eosinophils in lung lavage fluid via the absence of CD11c expression and moderate Gr-1 expression, which were subsequently identified by high expression of CD11b, Siglec F, and CCR3 (46). Moreover, this population was absent in dblGATA1-deficient mice (data not shown).
Having established that eosinophils are a component of the acute inflammatory response after A. fumigatus exposure, we next determined their relevance in A. fumigatus lung defense. As mentioned earlier, the development of mice with specific deficiencies in eosinophils has afforded the ability to better understand the roles these cells play in immune responses. We chose mice deficient in the dblGATA1 transcription factor, as they have been extensively employed for assessing eosinophil-mediated responses in models of asthma and infectious diseases (36, 47, 48, 49). Although bone marrow progenitors from dblGATA1-deficient mice may be forced to differentiate into eosinophils in vitro (50) (via various cytokine stimulations and differential promoter usage), this does not occur in dblGATA1-deficient mice in vivo during homeostatic (30) or disease or pathological (51) settings. Moreover, chronic A. fumigatus exposure, employed as a model of chronic experimental allergic airway inflammation, showed a >99% reduction in Siglec F+ CCR3+ cells in lung lavage fluid from dblGATA1-deficient mice (52). Employing the same dblGATA1-deficient strain, we observed a profound yet somewhat unexpected impairment in their ability to clear A. fumigatus from the lungs. In fact, the difference between WT and eosinophil-deficient mice at 24 and 48 h was more than 2 logs, much greater than the observed differences we have previously reported for Clec7a−/− (dectin-1-deficient) mice (20) and Il22−/− (IL-22-deficient) mice (21) (both approximately a 5- to 6-fold higher lung burden, although both of these strains are on the C57BL/6 background). Although dblGATA1-deficient mice had difficulty clearing A. fumigatus from the lungs, this did not result in increased mortality in these animals, which may be expected based on our observation of intact neutrophil recruitment in these mice. However, the observation of germinating A. fumigatus organisms in the lungs of dblGATA1-deficient mice implies that eosinophils target inhaled conidia and, along with inflammatory monocytes (53), potentially aid in limiting germination.
The susceptibility of dblGATA1-deficient mice to A. fumigatus lung infection appeared to be solely a result of eosinophil deficiency. However, we cannot exclude the possibility that other eosinophil-dependent or -independent mechanisms may have contributed to our observations. Indeed, eosinophils have been reported to modulate dendritic cell and T cell responses during allergen challenge (54) and have been shown to be required for the maintenance of plasma cells in the bone marrow (55). However, acknowledging that the time points employed in the current body of work are within 48 h after a single A. fumigatus exposure, in which innate immune responses are prominently active, it is unlikely that adaptive/T cell/B cell responses contributed significantly to the observed clearance defect in dblGATA1-deficient mice. We therefore concentrated our efforts on identifying specific eosinophilic inflammatory mediators. Whole-lung mRNA analysis revealed significantly reduced expression of Epx and Prg2 24 h and 48 h postinfection in dblGATA1-deficient mice. The eosinophil-associated ribonucleases Ear1 and Ear2 were unexpectedly observed to be significantly higher in dblGATA1-deficient mice 24 h post-A. fumigatus exposure, yet the opposite was true at 48 h. These data suggest that during a lung infection with A. fumigatus, Epx (EPO) and Prg2 (MBP) may play a role in host defense, whereas Ear1 and Ear2 are dispensable. Multiple mechanisms for killing by eosinophils have been proposed, including antibody-mediated release and/or complement-mediated release of eosinophil granule proteins, such as EPO, MBP, and ECP (56, 57, 58, 59). Eosinophils may also produce nitric oxide (NO) and Ear2 as antiviral factors, as demonstrated for RSV (44, 45). Lipopolysaccharide (LPS) stimulation of IL-5- or gamma interferon (IFN-γ)-primed eosinophils (60) or with platelet activating factor (PAF) in the presence of IL-5 or GM-CSF (61) promote the release of eosinophil extracellular traps (EETs) which contain mitochondrial DNA as well as granule proteins. Epx encodes EPO, a heme-containing cationic protein which comprises the bulk of protein found within eosinophilic granules (62) and has been shown to selectively kill bacteria, parasites, and viruses (63, 64). To the best of our knowledge, a potential requirement of EPO in fungal clearance has not been previously reported, and the substrate that drives fungal clearance by EPO remains to be identified. Although very little is known regarding EPO and eosinophil-mediated antifungal responses, a previous study reported that exposing macrophages to eosinophil-derived EPO enhanced their respiratory burst, leading to enhanced killing of the opportunistic yeast Candida albicans (65). This study suggests that a function of eosinophils and EPO may be to activate the antimicrobial activity of other innate cell types. Another study reported that stimulating human eosinophils with GM-CSF, IL-1, IL-3, and IL-5 augmented phagocytosis of C. albicans, yet this paradoxically did not result in increased killing of the organism (66). In contrast, while these studies suggest a possible role for eosinophils in anti-Candida defense, studies with the pathogenic yeast Cryptococcus neoformans are decidedly opposite. Mice deficient in scavenger receptor A demonstrate increased resistance to C. neoformans lung infection, which correlated with reduced Th2 responses and significantly lower eosinophil numbers in the lungs (67). In fact, C. neoformans infection in dblGATA-deficient mice is associated with lower lung fungal burden in the presence of enhanced Th1 and Th17 responses (68).
Our data suggest that the susceptibility of dblGATA1-deficient mice to A. fumigatus is directly due to the specific loss of eosinophils; thus, we felt it was imperative to demonstrate that eosinophils are capable of recognizing and responding to A. fumigatus conidia. Indeed, eosinophils derived from bone marrow cells with SCF, FLT3-L, and IL-5 were adept at inhibiting A. fumigatus viability in vitro, an interaction that also led to the production of proinflammatory mediators, chemokines, and Th2 cytokines. These observations indicate the use of pattern recognition receptors (PRRs) in eosinophil anti-Aspergillus responses. Studies with human eosinophils have identified expression of numerous TLRs, including TLR1, TLR4, TLR7, TLR9, and TLR10 (69), whereas murine eosinophils have been documented to express TLR2, TLR4, and TLR7 (45, 70). With respect to fungus-associated PRRs, the beta-glucan receptor dectin-1 has been observed in human eosinophils (71, 72). Identifying which PRR is utilized for A. fumigatus recognition is the subject of current investigations. Our data showing that cell contact was not required for eosinophil-mediated killing of A. fumigatus coupled with potent antifungal activity of eosinophil cell lysates suggest that innate recognition of A. fumigatus leads to the rapid degranulation of eosinophils and subsequent antifungal responses.
In summary, we show here that eosinophils are recruited to the lung after A. fumigatus exposure and play a necessary role in the elimination of this pathogenic mold from the lungs. Our data further suggest that the eosinophil antimicrobial factors eosinophil peroxidase and major basic protein putatively mediate lung clearance. Moreover, we show that eosinophils possess potent activity against A. fumigatus in vitro and serve as a source of numerous cytokines and chemokines. Future studies targeted at understanding which PRRs play a role in eosinophil-mediated recognition of A. fumigatus as well as specific mechanisms of antifungal activity will further define the importance of eosinophils in host defense against the spectrum of diseases caused by exposure to A. fumigatus.
ACKNOWLEDGMENTS
This work was supported was supported by Public Health Service grants HL096702 and HL117090.
Footnotes
Published ahead of print 30 December 2013
REFERENCES
- 1.Herbrecht R, Bories P, Moulin JC, Ledoux MP, Letscher-Bru V. 2012. Risk stratification for invasive aspergillosis in immunocompromised patients. Ann. N. Y. Acad. Sci. 1272:23–30. 10.1111/j.1749-6632.2012.06829.x [DOI] [PubMed] [Google Scholar]
- 2.De Le Rosa GR, Champlin RE, Kontoyiannis DP. 2002. Risk factors for the development of invasive fungal infections in allogeneic blood and marrow transplant recipients. Transpl. Infect. Dis. 4:3–9. 10.1034/j.1399-3062.2002.00010.x [DOI] [PubMed] [Google Scholar]
- 3.Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. 2012. The PATH (Prospective Antifungal Therapy) Alliance(R) registry and invasive fungal infections: update 2012. Diagn. Microbiol. Infect. Dis. 73:293–300. 10.1016/j.diagmicrobio.2012.06.012 [DOI] [PubMed] [Google Scholar]
- 4.Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. 2010. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin. Infect. Dis. 50:1101–1111. 10.1086/651262 [DOI] [PubMed] [Google Scholar]
- 5.Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW, Brown JM, Brumble LM, Freifeld AG, Hadley S, Herwaldt LA, Kauffman CA, Knapp K, Lyon GM, Morrison VA, Papanicolaou G, Patterson TF, Perl TM, Schuster MG, Walker R, Wannemuehler KA, Wingard JR, Chiller TM, Pappas PG. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin. Infect. Dis. 50:1091–1100. 10.1086/651263 [DOI] [PubMed] [Google Scholar]
- 6.Meersseman W, Lagrou K, Maertens J, Wilmer A, Hermans G, Vanderschueren S, Spriet I, Verbeken E, Van Wijngaerden E. 2008. Galactomannan in brochoalveolar lavage fluid—a tool for diagnosing aspergillosis in intensive care unit patients. Am. J. Respir. Crit. Care Med. 177:27–34. 10.1164/rccm.200704-606OC [DOI] [PubMed] [Google Scholar]
- 7.Trof RJ, Beishuizen A, YJbets-Ossenkopp Girbes AR, Groeneveld AB. 2007. Management of invasive pulmonary aspergillosis in non-neutropenic critically ill patients. Intensive Care Med. 33:1694–1703. 10.1007/s00134-007-0791-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandewoude KH, Blot SI, Depuydt P, Benoit D, Temmerman W, Colardyn F, Vogelaers D. 2006. Clinical relevance of Aspergillus isolation from respiratory tract samples in critically ill patients. Crit. Care 10:R31. 10.1186/cc4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnacho-Montero J, Rmaya-Villar Ortiz-Leyba C, Leon C, Fvarez-Lerma Nolla-Salas J, Iruretagoyena JR, Barcenilla F. 2005. Isolation of Aspergillus spp. from the respiratory tract in critically ill patients: risk factors, clinical presentation and outcome. Crit. Care 9:R191–R199. 10.1186/cc3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barberan J, Sanz F, Hernandez JL, Merlos S, Malmierca E, Garcia-Perez FJ, Sanchez-Haya E, Segarra M, Garcia de la Llana F, Granizo JJ, Gimenez MJ, Aguilar L. 2012. Clinical features of invasive pulmonary aspergillosis vs. colonization in COPD patients distributed by gold stage. J. Infect. 65:447–452. 10.1016/j.jinf.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 11.Schweer KE, Bangard C, Hekmat K, Cornely OA. 2 December 2013. Chronic pulmonary aspergillosis. Mycoses 10.1111/myc.12152 [DOI] [PubMed] [Google Scholar]
- 12.Yousem SA. 1997. The histological spectrum of chronic necrotizing forms of pulmonary aspergillosis. Hum. Pathol. 28:650–656. 10.1016/S0046-8177(97)90173-8 [DOI] [PubMed] [Google Scholar]
- 13.Trawick D, Kotch A, Matthay R, Homer RJ. 1997. Eosinophilic pneumonia as a presentation of occult chronic granulomatous disease. Eur. Respir. J. 10:2166–2170. 10.1183/09031936.97.10092166 [DOI] [PubMed] [Google Scholar]
- 14.Moskaluk CA, Pogrebniak HW, Pass HI, Gallin JI, Travis WD. 1994. Surgical pathology of the lung in chronic granulomatous disease. Am. J. Clin. Pathol. 102:684–691 [DOI] [PubMed] [Google Scholar]
- 15.Patterson K, Strek ME. 2010. Allergic bronchopulmonary aspergillosis. Proc. Am. Thorac Soc. 7:237–244. 10.1513/pats.200908-086AL [DOI] [PubMed] [Google Scholar]
- 16.Lilly LM, Gessner MA, Dunaway CW, Metz AE, Schwiebert L, Weaver CT, Brown GD, Steele C. 2012. The beta-glucan receptor Dectin-1 promotes lung immunopathology during fungal allergy via IL-22. J. Immunol. 189:3653–3660. 10.4049/jimmunol.1201797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fei M, Bhatia S, Oriss TB, Yarlagadda M, Khare A, Akira S, Saijo S, Iwakura Y, Fallert Junecko BA, Reinhart TA, Foreman O, Ray P, Kolls J, Ray A. 2011. TNF-alpha from inflammatory dendritic cells (DCs) regulates lung IL-17A/IL-5 levels and neutrophilia versus eosinophilia during persistent fungal infection. Proc. Natl. Acad. Sci. U. S. A. 108:5360–5365. 10.1073/pnas.1015476108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murdock BJ, Falkowski NR, Shreiner AB, Sadighi Akha AA, McDonald RA, White ES, Toews GB, Huffnagle GB. 2012. Interleukin-17 drives pulmonary eosinophilia following repeated exposure to Aspergillus fumigatus conidia. Infect. Immun. 80:1424–1436. 10.1128/IAI.05529-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter P, Susarla SC, Polikepahad S, Qian Y, Hampton J, Kiss A, Vaidya S, Sur S, Ongeri V, Yang T, Delclos GL, Abramson S, Kheradmand F, Corry DB. 2009. Link between allergic asthma and airway mucosal infection suggested by proteinase-secreting household fungi. Mucosal Immunol. 2:504–517. 10.1038/mi.2009.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner J, Metz AE, Horn D, Faro-Trindade I, Schoeb TR, Hewitt MM, Schwiebert LM, Brown GD, Steele C. 2009. Requisite role for the Dectin-1 beta-glucan receptor in pulmonary defense against Aspergillus fumigatus. J. Immunol. 182:4938–4946. 10.4049/jimmunol.0804250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gessner MA, Werner JL, Lilly LM, Nelson MP, Metz AE, Dunaway CW, Chan YR, Ouyang W, Brown GD, Weaver CT, Steele C. 2012. Dectin-1-dependent interleukin-22 contributes to early innate lung defense against Aspergillus fumigatus. Infect. Immun. 80:410–417. 10.1128/IAI.05939-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemzek JA, Ebong SJ, Kim J, Bolgos GL, Remick DG. 2002. Keratinocyte growth factor pretreatment is associated with decreased macrophage inflammatory protein-2alpha concentrations and reduced neutrophil recruitment in acid aspiration lung injury. Shock 18:501–506. 10.1097/00024382-200212000-00003 [DOI] [PubMed] [Google Scholar]
- 23.Mattila PE, Metz AE, Rapaka RR, Bauer LD, Steele C. 2008. Dectin-1 Fc targeting of Aspergillus fumigatus beta-glucans augments innate defense against invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 52:1171–1172. 10.1128/AAC.01274-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowman JC, Abruzzo GK, Anderson JW, Flattery AM, Gill CJ, Pikounis VB, Schmatz DM, Liberator PA, Douglas CM. 2001. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob. Agents Chemother. 45:3474–3481. 10.1128/AAC.45.12.3474-3481.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel M, Spiess B, Roder J, KGvon Durken M, Kentouche K, Laws HJ, Morz H, Hehlmann R, Buchheidt D. 2009. Detection of Aspergillus DNA by a nested PCR assay is able to improve the diagnosis of invasive aspergillosis in paediatric patients. J. Med. Microbiol. 58:1291–1297. 10.1099/jmm.0.007393-0 [DOI] [PubMed] [Google Scholar]
- 26.Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, Kolls JK, Brown GD. 2005. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 1:e42. 10.1371/journal.ppat.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson MP, Metz AE, Li S, Lowell CA, Steele C. 2009. The absence of Hck, Fgr, and Lyn tyrosine kinases augments lung innate immune responses to Pneumocystis murina. Infect. Immun. 77:1790–1797. 10.1128/IAI.01441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyer KD, Moser JM, Czapiga M, Siegel SJ, Percopo CM, Rosenberg HF. 2008. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J. Immunol. 181:4004–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cenci E, Mencacci A, Del SG, Bacci A, Montagnoli C, d'Ostiani CF, Mosci P, Bachmann M, Bistoni F, Kopf M, Romani L. 1999. Interleukin-4 causes susceptibility to invasive pulmonary aspergillosis through suppression of protective type I responses. J. Infect. Dis. 180:1957–1968. 10.1086/315142 [DOI] [PubMed] [Google Scholar]
- 30.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. 2002. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195:1387–1395. 10.1084/jem.20020656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnes PD, Marr KA. 2007. Risks, diagnosis and outcomes of invasive fungal infections in haematopoietic stem cell transplant recipients. Br. J. Haematol. 139:519–531. 10.1111/j.1365-2141.2007.06812.x [DOI] [PubMed] [Google Scholar]
- 32.Fukuda T, Boeckh M, Carter RA, Sandmaier BM, Maris MB, Maloney DG, Martin PJ, Storb RF, Marr KA. 2003. Risks and outcomes of invasive fungal infections in recipients of allogeneic hematopoietic stem cell transplants after nonmyeloablative conditioning. Blood 102:827–833. 10.1182/blood-2003-02-0456 [DOI] [PubMed] [Google Scholar]
- 33.Stephens-Romero SD, Mednick AJ, Feldmesser M. 2005. The pathogenesis of fatal outcome in murine pulmonary aspergillosis depends on the neutrophil depletion strategy. Infect. Immun. 73:114–125. 10.1128/IAI.73.1.114-125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kita H. 2013. Eosinophils: multifunctional and distinctive properties. Int. Arch. Allergy Immunol. 161(Suppl 2):3–9. 10.1159/000350662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cormier SA, Yuan S, Crosby JR, Protheroe CA, Dimina DM, Hines EM, Lee NA, Lee JJ. 2002. T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 27:678–687. 10.1165/rcmb.4882 [DOI] [PubMed] [Google Scholar]
- 36.Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O'Neill KR, Protheroe C, Pero R, Nguyen T, Cormier SA, Lenkiewicz E, Colbert D, Rinaldi L, Ackerman SJ, Irvin CG, Lee NA. 2004. Defining a link with asthma in mice congenitally deficient in eosinophils. Science 305:1773–1776. 10.1126/science.1099472 [DOI] [PubMed] [Google Scholar]
- 37.Kopf M, Brombacher F, Hodgkin PD, Ramsay AJ, Milbourne EA, Dai WJ, Ovington KS, Behm CA, Kohler G, Young IG, Matthaei KI. 1996. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity 4:15–24. 10.1016/S1074-7613(00)80294-0 [DOI] [PubMed] [Google Scholar]
- 38.Pope SM, Fulkerson PC, Blanchard C, Akei HS, Nikolaidis NM, Zimmermann N, Molkentin JD, Rothenberg ME. 2005. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J. Biol. Chem. 280:13952–13961. 10.1074/jbc.M406037200 [DOI] [PubMed] [Google Scholar]
- 39.Humbles AA, Lu B, Friend DS, Okinaga S, Lora J, Al-Garawi A, Martin TR, Gerard NP, Gerard C. 2002. The murine CCR3 receptor regulates both the role of eosinophils and mast cells in allergen-induced airway inflammation and hyperresponsiveness. Proc. Natl. Acad. Sci. U. S. A. 99:1479–1484. 10.1073/pnas.261462598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg HF, Dyer KD, Foster PS. 2013. Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13:9–22. 10.1038/nri3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driss V, Legrand F, Hermann E, Loiseau S, Guerardel Y, Kremer L, Adam E, Woerly G, Dombrowicz D, Capron M. 2009. TLR2-dependent eosinophil interactions with mycobacteria: role of alpha-defensins. Blood 113:3235–3244. 10.1182/blood-2008-07-166595 [DOI] [PubMed] [Google Scholar]
- 42.Wong CK, Cheung PF, Ip WK, Lam CW. 2007. Intracellular signaling mechanisms regulating Toll-like receptor-mediated activation of eosinophils. Am. J. Respir. Cell Mol. Biol. 37:85–96. 10.1165/rcmb.2006-0457OC [DOI] [PubMed] [Google Scholar]
- 43.Linch SN, Kelly AM, Danielson ET, Pero R, Lee JJ, Gold JA. 2009. Mouse eosinophils possess potent antibacterial properties in vivo. Infect. Immun. 77:4976–4982. 10.1128/IAI.00306-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domachowske JB, Dyer KD, Bonville CA, Rosenberg HF. 1998. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis. 177:1458–1464. 10.1086/515322 [DOI] [PubMed] [Google Scholar]
- 45.Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, Foster PS, Matthaei KI. 2007. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 110:1578–1586. 10.1182/blood-2007-01-071340 [DOI] [PubMed] [Google Scholar]
- 46.Shreiner AB, Murdock BJ, Sadighi Akha AA, Falkowski NR, Christensen PJ, White ES, Hogaboam CM, Huffnagle GB. 2012. Repeated exposure to Aspergillus fumigatus conidia results in CD4+ T cell-dependent and -independent pulmonary arterial remodeling in a mixed Th1/Th2/Th17 microenvironment that requires interleukin-4 (IL-4) and IL-10. Infect. Immun. 80:388–397. 10.1128/IAI.05530-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castilow EM, Legge KL, Varga SM. 2008. Cutting edge: Eosinophils do not contribute to respiratory syncytial virus vaccine-enhanced disease. J. Immunol. 181:6692–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jessup HK, Brewer AW, Omori M, Rickel EA, Budelsky AL, Yoon BR, Ziegler SF, Comeau MR. 2008. Intradermal administration of thymic stromal lymphopoietin induces a T cell- and eosinophil-dependent systemic Th2 inflammatory response. J. Immunol. 181:4311–4319 [DOI] [PubMed] [Google Scholar]
- 49.Gebreselassie NG, Moorhead AR, Fabre V, Gagliardo LF, Lee NA, Lee JJ, Appleton JA. 2012. Eosinophils preserve parasitic nematode larvae by regulating local immunity. J. Immunol. 188:417–425. 10.4049/jimmunol.1101980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyer KD, Czapiga M, Foster B, Foster PS, Kang EM, Lappas CM, Moser JM, Naumann N, Percopo CM, Siegel SJ, Swartz JM, Ting-De RS, Rosenberg HF. 2007. Eosinophils from lineage-ablated Delta dblGATA bone marrow progenitors: the dblGATA enhancer in the promoter of GATA-1 is not essential for differentiation ex vivo. J. Immunol. 179:1693–1699 [DOI] [PubMed] [Google Scholar]
- 51.Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, McKenna EE, Ghiran S, Gerard NP, Yu C, Orkin SH, Gerard C. 2004. A critical role for eosinophils in allergic airways remodeling. Science 305:1776–1779. 10.1126/science.1100283 [DOI] [PubMed] [Google Scholar]
- 52.Fulkerson PC, Fischetti CA, McBride ML, Hassman LM, Hogan SP, Rothenberg ME. 2006. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc. Natl. Acad. Sci. U. S. A. 103:16418–16423. 10.1073/pnas.0607863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, Pamer EG. 2009. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6:470–481. 10.1016/j.chom.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacobsen EA, Zellner KR, Colbert D, Lee NA, Lee JJ. 2011. Eosinophils regulate dendritic cells and Th2 pulmonary immune responses following allergen provocation. J. Immunol. 187:6059–6068. 10.4049/jimmunol.1102299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu VT, Frohlich A, Steinhauser G, Scheel T, Roch T, Fillatreau S, Lee JJ, Lohning M, Berek C. 2011. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat. Immunol. 12:151–159. 10.1038/ni.1981 [DOI] [PubMed] [Google Scholar]
- 56.David JR, Butterworth AE, Vadas MA. 1980. Mechanism of the interaction mediating killing of Schistosoma mansoni by human eosinophils. Am. J. Trop. Med. Hyg. 29:842–848 [DOI] [PubMed] [Google Scholar]
- 57.Haque A, Ouaissi A, Joseph M, Capron M, Capron A. 1981. IgE antibody in eosinophil- and macrophage-mediated in vitro killing of Dipetalonema viteae microfilariae. J. Immunol. 127:716–725 [PubMed] [Google Scholar]
- 58.Hamann KJ, Gleich GJ, Checkel JL, Loegering DA, McCall JW, Barker RL. 1990. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 144:3166–3173 [PubMed] [Google Scholar]
- 59.Lehrer RI, Szklarek D, Barton A, Ganz T, Hamann KJ, Gleich GJ. 1989. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 142:4428–4434 [PubMed] [Google Scholar]
- 60.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14:949–953. 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]
- 61.Ueki S, Melo RC, Ghiran I, Spencer LA, Dvorak AM, Weller PF. 2013. Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood 121:2074–2083. 10.1182/blood-2012-05-432088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JJ, Jacobsen EA, Ochkur SI, McGarry MP, Condjella RM, Doyle AD, Luo H, Zellner KR, Protheroe CA, Willetts L, Lesuer WE, Colbert DC, Helmers RA, Lacy P, Moqbel R, Lee NA. 2012. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red.” J. Allergy Clin. Immunol. 130:572–584. 10.1016/j.jaci.2012.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malik A, Batra JK. 2012. Antimicrobial activity of human eosinophil granule proteins: involvement in host defence against pathogens. Crit. Rev. Microbiol. 38:168–181. 10.3109/1040841X.2011.645519 [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Slungaard A. 2006. Role of eosinophil peroxidase in host defense and disease pathology. Arch. Biochem. Biophys. 445:256–260. 10.1016/j.abb.2005.10.008 [DOI] [PubMed] [Google Scholar]
- 65.Lefkowitz DL, Lincoln JA, Howard KR, Stuart R, Lefkowitz SS, Allen RC. 1997. Macrophage-mediated candidacidal activity is augmented by exposure to eosinophil peroxidase: a paradigm for eosinophil-macrophage interaction. Inflammation 21:159–172. 10.1023/A:1027366119901 [DOI] [PubMed] [Google Scholar]
- 66.Fabian I, Kletter Y, Mor S, Geller-Bernstein C, Ben-Yaakov M, Volovitz B, Golde DW. 1992. Activation of human eosinophil and neutrophil functions by haematopoietic growth factors: comparisons of IL-1, IL-3, IL-5 and GM-CSF. Br. J. Haematol. 80:137–143. 10.1111/j.1365-2141.1992.tb08890.x [DOI] [PubMed] [Google Scholar]
- 67.Qiu Y, Dayrit JK, Davis MJ, Carolan JF, Osterholzer JJ, Curtis JL, Olszewski MA. 2013. Scavenger receptor A modulates the immune response to pulmonary Cryptococcus neoformans infection. J. Immunol. 191:238–248. 10.4049/jimmunol.1203435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piehler D, Stenzel W, Grahnert A, Held J, Richter L, Kohler G, Richter T, Eschke M, Alber G, Muller U. 2011. Eosinophils contribute to IL-4 production and shape the T-helper cytokine profile and inflammatory response in pulmonary cryptococcosis. Am. J. Pathol. 179:733–744. 10.1016/j.ajpath.2011.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, Matsushima K, Ohta K, Yamamoto K, Hirai K. 2003. Expression and function of Toll-like receptors in eosinophils: activation by Toll-like receptor 7 ligand. J. Immunol. 171:3977–3982 [DOI] [PubMed] [Google Scholar]
- 70.D'Avila H, Almeida PE, Roque NR, Castro-Faria-Neto HC, Bozza PT. 2007. Toll-like receptor-2-mediated C-C chemokine receptor 3 and eotaxin-driven eosinophil influx induced by Mycobacterium bovis BCG pleurisy. Infect. Immun. 75:1507–1511. 10.1128/IAI.01326-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willment JA, Marshall ASJ, Reid DM, Williams DL, Wong SYC, Gordon S, Brown GD. 2005. The human β-glucan receptor is widely expressed and functionally equivalent to murine Dectin-1 on primary cells. Eur. J. Immunol. 35:1539–1547. 10.1002/eji.200425725 [DOI] [PubMed] [Google Scholar]
- 72.Ahrén IL, Eriksson E, Egesten A, Riesbeck K. 2003. Nontypeable Haemophilus influenzae activates human eosinophils through β-glucan receptors. Am. J. Respir. Cell Mol. Biol. 29:598–605. 10.1165/rcmb.2002-0138OC [DOI] [PubMed] [Google Scholar]