Abstract
Leptospira interrogans is a global zoonotic pathogen and is the causative agent of leptospirosis, an endemic disease of humans and animals worldwide. There is limited understanding of leptospiral pathogenesis; therefore, further elucidation of the mechanisms involved would aid in vaccine development and the prevention of infection. HtpG (high-temperature protein G) is the bacterial homolog to the highly conserved molecular chaperone Hsp90 and is important in the stress responses of many bacteria. The specific role of HtpG, especially in bacterial pathogenesis, remains largely unknown. Through the use of an L. interrogans htpG transposon insertion mutant, this study demonstrates that L. interrogans HtpG is essential for virulence in the hamster model of acute leptospirosis. Complementation of the htpG mutant completely restored virulence. Surprisingly, the htpG mutant did not appear to show sensitivity to heat or oxidative stress, phenotypes common in htpG mutants in other bacterial species. Furthermore, the mutant did not show increased sensitivity to serum complement, reduced survival within macrophages, or altered protein or lipopolysaccharide expression. The underlying cause for attenuation thus remains unknown, but HtpG is a novel leptospiral virulence factor and one of only a very small number identified to date.
INTRODUCTION
Leptospira interrogans is a global zoonotic pathogen that infects a wide variety of hosts, including humans, domestic animals, and rodents (1, 2). Leptospirosis is a systemic infection transmitted to susceptible hosts via contact with contaminated water or soil, or by direct contact with urine or other tissues of infected animals. The leptospires enter the host via skin abrasions or mucous membranes, disseminate rapidly via the blood, and invade the host tissues and organs (3, 4). Symptoms of leptospirosis include general malaise and fever and can lead to more-severe disease, including lung hemorrhages, multiorgan failure, and death (2–4). The understanding of leptospiral pathogenesis is limited, but headway has been made by the identification of a small number of factors required for disease (5–12); this advance was facilitated by the development of a transposon mutagenesis system for pathogenic Leptospira (13) and the generation of a library of mutants (14). However, the specific mechanisms of leptospiral pathogenesis still remain unknown.
Heat shock protein 90 (Hsp90) is a well-conserved molecular chaperone found in eukaryotes and bacteria; the prokaryotic Hsp90 homolog is more commonly known as HtpG (high-temperature protein G). Hsp90 is thought to act on its substrates by assisting in slight conformational changes that promote the activity of the substrate (15). Hsp90 is approximately 90 kDa and contains three domains: the N domain, the M domain, and the C domain. The N domain is located in the first ∼216 residues of Hsp90 and contains an ATP binding site; ATP hydrolysis facilitates conformational changes within Hsp90 to assist in substrate binding (16–18). The M domain is predicted to be the site of substrate binding and to provide the catalytic arginine residue for ATP hydrolysis (15, 19, 20). Hsp90 functions as a homodimer, and the C domain is responsible for dimerization (16, 21).
Bacterial HtpG is approximately 68 to 70 kDa and shares the three domains of Hsp90 but differs by the absence of two regions: a 50-residue linker between the N and M domains and a 35- to 40-residue extension at the C-terminal end of Hsp90 involved in binding to cochaperones (22). No bacterial HtpG cochaperones have been identified (23, 24).
Hsp90 is an essential protein in eukaryotes (25) and has many different roles in facilitating protein folding, along with a growing number of cochaperones (outlined in reference 18). On the other hand, in bacteria, the loss of HtpG is tolerated. The phenotypes resulting from htpG disruption can range from a slight growth defect at high temperatures and during cold shock recovery to increased sensitivity to oxidative stress (26–29). Two studies have revealed that HtpG from Francisella tularensis and Edwardsiella tarda is important in the pathogenesis of these bacteria (27, 30); however, the precise role of HtpG in bacterial pathogenesis is not well characterized.
A high-throughput method for screening Leptospira transposon mutants for attenuation in vivo has been developed; the application of this method revealed that a Leptospira interrogans serovar Manilae htpG mutant was attenuated in the hamster model of acute leptospirosis (31). In this study, we characterize the role of htpG in acute leptospirosis. The htpG mutant was complemented, resulting in the restoration of virulence.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study are listed in Table 1. Transposon mutagenesis was performed on L. interrogans serovar Manilae strain L495 previously (13, 14), and the location of TnSC189 insertion in the mutants was determined by direct sequencing of genomic DNA (32). All L. interrogans strains were grown in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Becton, Dickinson) at 30°C, unless otherwise stated.
TABLE 1.
Leptospiral strains used in this study
| Strain | Description | Comments/featuresa |
|---|---|---|
| Wild type | Parent strain | L. interrogans serovar Manilae strain L495 |
| M1233 | htpG mutant | TnSC189 inserted 46 bp from start codon of htpG |
| M1233+htpG | Complemented mutant | TnSC189 Spcr inserted in la0166 |
| M1004 | la1231 mutant | TnSC189 Kanr inserted in la1231 |
| M895 | LPS mutant | TnSC189 inserted in la1641b |
LA0166 encodes a protein of unknown function; la1231 is an htpG paralog.
See reference 9.
Complementation of the htpG mutant.
htpG was amplified with primers BAP6974 (5′-AAAAGGGCCCATGAGCGAAGAAATTAAAGGA-3′) (forward) and BAP6975 (5′-AAAAGTCGACTTACGTACCGCTCAACTCTT-3′) (reverse) (restriction enzyme sites are underlined), and the product was digested with restriction enzymes ApaI and SalI before insertion into TnSC189 containing a spectinomycin resistance cassette (TnSC189 Spcr) (9). The promoter region of lipL32 (la2637) was amplified with primers BAP7151 (5′-AAAAGGTACCACTTCGGAAGAACAAGAAAG-3′) (forward) and BAP7152 (5′-TTTTGGGCCCAGACTCTCCTTAGTTAGGAA-3′) (reverse), and the product was first digested with restriction enzymes KpnI and ApaI and then inserted into TnSC189 Spcr::htpG upstream of htpG. The htpG complementation construct was introduced by conjugation from Escherichia coli into the htpG mutant M1233, as described previously (33).
Evaluation of virulence in the hamster model of infection.
The virulence of wild-type L. interrogans serovar Manilae strain L495, htpG mutant M1233, and the M1233+htpG strain was tested in golden hamsters of either sex, aged 4 to 6 weeks, as described previously (14). The animals were monitored for 21 days, and moribund animals were euthanized in accordance with animal ethics requirements. The frequency (number) and severity (size) of the lung hemorrhages were assessed, and kidney tissue was collected postmortem for culture. PCR was used to confirm the genotype of the mutants reisolated from the hamsters. Animal experiments were approved by the Animal Ethics committees of Khon Kaen University and the Institut Pasteur. Histopathology examinations were performed on hamster kidney, liver, and lung tissue as described previously (10).
qPCR quantification of leptospires in hamster tissues.
Groups of four golden hamsters were infected intraperitoneally with 105 wild-type, M1233, or M1233+htpG leptospires. The animals were euthanized 5 days postinfection, and the kidneys, livers, and lungs were collected. The tissues were weighed and were homogenized in phosphate-buffered saline (PBS; pH 7.4) to a final concentration of 148 μg/μl. DNA was extracted from 300 μl of tissue suspension by using a Promega Maxwell 16 tissue DNA purification kit with a Promega Maxwell 16 instrument. The concentration of extracted DNA was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). Quantitative PCR (qPCR) was performed as described previously (34), by targeting lipL32 with primers 5′-AAG CAT TAC CGC TTG TGG TG-3′ (forward) and 5′-GAA CTC CCA TTT CAG CGA TT-3′ (reverse) (35) and targeting gapdh (the normalizing gene) with primers GAPDHF (5′-GGTTCACACCCATCACAAACAT-3′) (forward) and GAPDHR (5′-GGTGGAGCCAAGAGGGTCAT-3′) (reverse). A total of 60.8 ng of template DNA was used per qPCR, and PCR conditions (with ramp rates of 20°C s−1) were as follows: initial denaturation at 95°C for 600 s, followed by 45 cycles of amplification at 95°C for 10 s, 57°C for 8 s, and 72°C for 10 s, and fusion at 95°C for 360 s. Statistical analysis was performed using one-way analysis of variance (ANOVA) and subsequent t tests.
Motility, heat, and osmotic stress assays.
The motility of leptospiral strains was assessed by inoculating semisolid EMJH medium, containing 0.5% agar, with approximately 105 cells in quadruplicate. The plates were incubated at 30°C for 10 days, and the diameter of the zone of spread was measured. For heat stress, growth curves of the wild type and htpG mutant M1233 were conducted in triplicate in 15 ml EMJH medium. The medium was inoculated with 2 × 107 cells/ml and was incubated at 30°C, 37°C, or 39°C for 16 to 18 days. Measurements of optical density at 420 nm were recorded. Leptospires were tested for sensitivity to increased osmolarity. EMJH medium was supplemented with NaCl (240 mM, 180 mM, 120 mM, or 60 mM) or sucrose (480 mM, 360 mM, 240 mM, or 120 mM) to generate an osmolarity of 600, 450, 300, or 150 mosmol, respectively, and was aliquoted across 96-well trays. A 100-μl volume of leptospiral culture at 5 × 107 cells/ml was added to each well in biological triplicate, and the plates were incubated at 30°C for 4 days. The inhibitory osmolarity was the lowest concentration tested that resulted in no viable, intact cells (as determined by dark-field microscopy).
Growth under oxidative stress.
Hydrogen peroxide and cumene hydroperoxide were serially diluted in EMJH medium across 96-well trays from 8 mM to 7 μM and from 2.5 mM to 2 μM, respectively. A 100-μl volume of leptospiral culture at 5 × 107 cells/ml was added to each well in biological triplicate, and the plates were incubated at 30°C for 4 days. The MIC was determined by dark-field microscopy as the lowest concentration tested that resulted in no viable, intact cells.
Two-dimensional (2D) gel electrophoresis.
Wild-type and M1233 cells either were grown to 5 × 108 cells/ml in 5 ml EMJH medium at 30°C or were first grown at 30°C and then incubated at 39°C for 90 min to induce heat shock. Cells were harvested at 8,000 × g for 10 min. The cell pellets were resuspended in 280 μl rehydration buffer {7 M urea, 2 M thiourea, 4% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 13 mM dithiothreitol [DTT], 2% [vol/vol] Bio-Lyte 3/10 Ampholyte [Bio-Rad], and 0.001% [wt/vol] bromophenol blue} with vigorous mixing at room temperature for 2 h. Insoluble material was removed by centrifugation at 13,000 × g for 5 min. Isoelectric focusing was performed as described previously (36) using 7-cm Immobiline DryStrip pH 3–10 gels (immobilized pH gradient [IPG] strips) (Amersham Biosciences) that were rehydrated overnight with 125 μl of solubilized whole-cell lysate. The IPG strips were equilibrated in 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) sodium dodecyl sulfate (SDS), 0.25% (wt/vol) DTT, and 50 mM Tris-HCl (pH 6.8) for 15 min. The strips were placed in fresh equilibration buffer, with DTT replaced by 4.5% (wt/vol) iodoacetamide and 0.001% (wt/vol) bromophenol blue, for an additional 15 min. The second dimension was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). The gels were fixed with 50% ethanol and 3% phosphoric acid and were stained with colloidal Coomassie blue. Each sample was analyzed in duplicate. Proteins differentially expressed in the wild type and M1233 were excised and were identified by mass spectrometry as described previously (36) and by utilizing the MASCOT peptide mass fingerprint database (Matrix Science).
Analysis of LPS profiles by SDS-PAGE.
Lipopolysaccharide (LPS) samples were prepared as described previously (9). Briefly, mid-log-phase cultures were first centrifuged and then resuspended in 100 μl of sample buffer (50 mM Tris-HCl [pH 6.8], 14.4 mM β-mercaptoethanol, 2% SDS, and 0.1% bromophenol blue in 20% glycerol) at 107 leptospires/μl. The samples were boiled for 5 min and were cooled on ice; then 2 μl proteinase K (20 mg/ml; Roche) was added, and samples were incubated at 60°C for 2 h. The LPS samples were boiled for 5 min before analysis by SDS-PAGE. Silver staining was performed using the Pierce Silver Stain kit (Thermo Scientific) according to the manufacturer's instructions.
SDS-PAGE and immunoblotting.
To prepare whole-cell lysates, 109 cells were centrifuged (13,000 × g, 5 min), washed once in PBS, resuspended in 100 μl sample buffer, and then boiled for 5 min. The whole-cell lysates and LPS samples were analyzed by 12.5% SDS-PAGE and immunoblotting using standard methods (37). The immunoblots were blocked and were probed with a rabbit antiserum against either Loa22 (11) (dilution, 1:2,000), whole cells of serovar Manilae (dilution, 1:1,000) (9), or recombinant HtpG from Porphyromonas gingivalis (dilution, 1:1,000) (38) in Tris-buffered saline–Tween (TBS-T). The membranes were washed three times in TBS-T, probed with horseradish peroxidase-conjugated anti-rabbit IgG (diluted 1:5,000) (Millipore), developed using Amersham ECL Western blotting detection reagents (GE Healthcare), and exposed to a LAS3000 chemiluminescence imager (Fujifilm).
Complement resistance.
The resistance of the wild type and M1233 to complement was determined by incubating 4 × 106 cells with 90% pooled human serum (39). The cells were incubated at 37°C for 60 min before survival was determined by dark-field microscopy comparing the proportion of motile cells to that of nonmotile cells (see Results and Discussion).
Macrophage association assay.
The association of leptospires with murine bone marrow-derived macrophages was determined as described previously (40). Briefly, leptospires were incubated with murine bone marrow-derived macrophages at a multiplicity of infection of 50:1 for 2 and 48 h. The number of intracellular leptospires was quantified by qPCR using leptospiral 16S rRNA primers. Statistical analysis was performed using Student's t test on biological triplicates.
RESULTS AND DISCUSSION
Bioinformatic analysis of htpG.
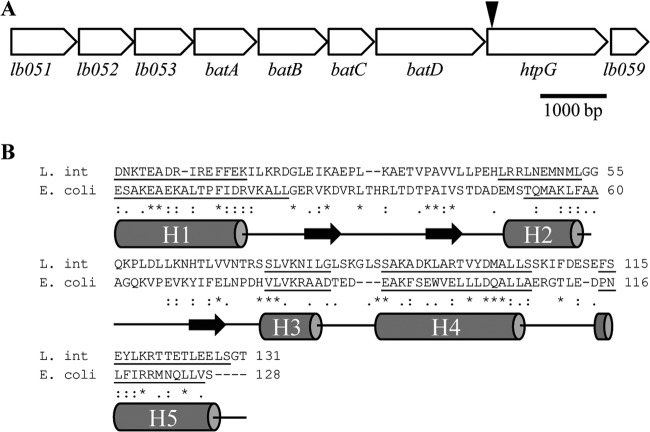
HtpG (LB058 in Leptospira interrogans serovar Lai) is a protein of 608 amino acids that is highly conserved (>92% identity) across L. interrogans and Leptospira borgpetersenii serovars (41–43). The saprophytic species Leptospira biflexa also contains an HtpG homolog with 65% identity to HtpG of L. interrogans serovar Manilae (LBF4008 in Leptospira biflexa serovar Patoc [44]). htpG is located in a putative operon (lb051 to lb059 in L. interrogans serovar Lai) consisting of nine open reading frames, either overlapping or with very small intergenic regions (Fig. 1). The first gene, lb051, encodes a putative MoxR-like ATPase. MoxR is associated with chaperone activities under stress conditions but has various roles in different bacteria (45). LB052 is a protein of unknown function containing a metal-binding von Willebrand factor A (VWA) domain. Proteins containing VWA domains are commonly found downstream of MoxR ATPases (45). LB053 is a protein of unknown function with no significant similarity to other proteins within the genus Leptospira or other bacteria; no conserved domains were identified. LB054 to LB057 share strong similarity with BatABCD from the batI operon in Bacteroides fragilis, involved in the aerotolerance of this obligate anaerobe (46). A recent study characterized the batI operon in L. biflexa but found no involvement in oxidative stress (47). Additionally, despite the operon structure surrounding htpG, this gene and lb059, located downstream of batD, are independently transcribed (47). The last gene in the operon, lb059 encodes a hypothetical protein with only limited similarity to a hypothetical protein in Leptonema illini, Lepil_4068 (28% identity; 55% similarity).
FIG 1.
Analysis of the htpG operon and protein homology. (A) Genomic organization of the putative htpG operon in L. interrogans. The gap between the coding regions of lb051 and lb052 is 6 bp. The gap between htpG and lb059 is 21 bp. The remaining genes are predicted to have overlapping open reading frames. The transposon insertion in htpG mutant M1233, located 46 bp from the start codon of htpG, is indicated by a black arrowhead. (B) C-terminal alignment of HtpG from L. interrogans (L. int) and HtpG from E. coli. The predicted secondary structure of L. interrogans HtpG shows close similarity to that of E. coli HtpG (16). Underlining indicates helical regions (H1 to H5), and black horizontal arrows represent strands, as predicted by PSIPRED (49).
BLASTp analysis of L. interrogans HtpG revealed that the protein shares significant similarity with other HtpG and Hsp90 proteins. There is a putative ATPase domain at the N terminus, where all of the 17 conserved residues identified for ATP binding are present (E34, N38, D41, L76, D78, G80, G82, M83, G121, L122, G124, F125, L136, T138, G155, T156, F158), including two G-X-G motifs (80GIG82 and 129GLG131) (17, 27). Analysis of the C terminus of L. interrogans HtpG by protein domain prediction servers DomPred and PSIPRED showed that it has a predicted secondary structure very similar to that of the E. coli HtpG C-terminal domain (16, 48, 49) (Fig. 1B).
htpG mutant M1233 is attenuated for acute infection.
L. interrogans serovar Manilae strain M1233 is an htpG mutant generated by transposon mutagenesis (13, 14), with the transposon insertion 46 bp from the start codon of the gene (Fig. 1). Complementation of leptospiral mutants is extremely difficult due to the lack of replicating plasmids for pathogenic leptospires; there are only three reports of complementation in the literature (8, 11, 12). Complementation has been achieved by using a transposon carrying a complementing gene (11), although transposition efficiency remains low. Four complemented M1233 strains were obtained in this study, each with TnSC189 Spcr::htpG, containing an intact htpG gene inserted into different genes. Preliminary analysis indicated that all four strains had restored virulence, so one complemented strain (named M1233+htpG), with the complementing construct inserted on the chromosome in the la0166 gene (encoding a protein of unknown function), was chosen for this study. The original insertion in htpG was retained in M1233+htpG.
Immunoblot analysis of the wild type, the htpG mutant M1233, and the complemented mutant M1233+htpG was performed with an HtpG antiserum (Fig. 2). A 68-kDa band corresponding to HtpG was present in the wild type and M1233+htpG but absent in M1233, indicating that complementation of M1233 restored the expression of HtpG in M1233+htpG (Fig. 2).
FIG 2.
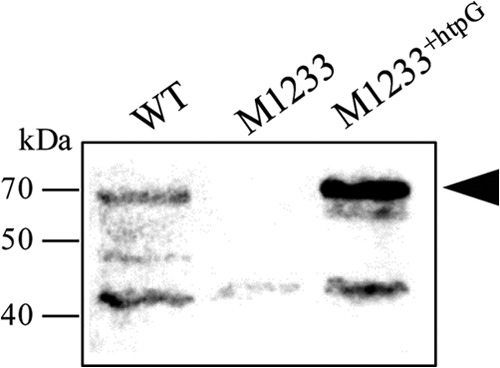
Anti-HtpG immunoblot analysis of wild-type (WT), htpG mutant M1233, and complemented mutant M1233+htpG whole-cell lysates. The black arrowhead indicates the HtpG-specific band observed at 68 kDa. The positions of molecular mass markers are shown on the left.
The virulence of M1233 was characterized in the hamster model of acute leptospirosis. M1233 was highly attenuated, with 100% survival of infected animals at a dose of 103 leptospires. All hamsters also survived infections at higher doses of 105 and 107 leptospires (Table 2); this corresponds to more than 106 times the 50% infective dose (ID50) of <10 for L. interrogans serovar Manilae (50). In contrast, all animals infected with 103 wild-type leptospires succumbed to infection by days 8 to 9. Likewise, all animals infected with M1233+htpG succumbed to infection, demonstrating full restoration of virulence (Table 2). Macroscopic lung hemorrhages were observed at similar frequencies and severities in animals infected with the wild type and the htpG mutant, and leptospires were cultured from the kidneys of all animals. The genotype of M1233 was confirmed by PCR upon reisolation from the hamsters.
TABLE 2.
Survival of hamsters infected with strains of L. interrogans serovar Manilae
| Strain | Description | Expt 1 |
Expt 2 |
||
|---|---|---|---|---|---|
| Dosea | Survivalb | Dosea | Survivalb | ||
| Wild type | Parent strain | 103 | 0/10 | 103 | 0/8 |
| M1233 | htpG mutant | 103 | 10/10* | 103 | 8/8# |
| 105 | 10/10* | ||||
| 107 | 10/10* | ||||
| M1233+htpG | Complemented strain | 103 | 0/8 | ||
| 105 | 0/4† | ||||
| M1004 | la1231 mutant | 103 | 0/4† | ||
| EMJH medium | Negative control | 10/10 | 2/2 | ||
Expressed as the number of leptospires.
Expressed as the number of hamsters surviving/total number of hamsters. The significance of differences from results for the wild-type control by Fisher's exact test is indicated as follows: *, P < 0.0001; #, P = 0.0002; †, P = 0.002.
Histopathological analysis of tissues from animals infected with M1233 revealed multifocal tubular and glomerular hemorrhage and necrosis of the kidney (Fig. 3). Diffuse mild dissociation and single-cell necrosis of hepatocytes with multifocal hemorrhages were observed in the liver tissue, and multifocal alveolar hemorrhages were observed in the lung tissue (Fig. 3). Macroscopic and histological lesions were indistinguishable from those in hamsters infected with wild-type leptospires. This finding suggests that serovar Manilae has a mechanism of lethality distinct from these pathologies, a possibility that warrants further investigation to define the mechanism of attenuation in M1233.
FIG 3.

Histopathological examination of tissues from hamsters infected with the htpG mutant M1233. Organs were fixed in 10% formalin and were then stained with hematoxylin and eosin. (A) Kidney tissue (magnification, ×20); (B) liver tissue (magnification, ×10); (C) lung tissue (magnification, ×10).
The numbers of leptospires present in hamster kidney, liver, and lung tissue at 5 days postinfection were quantified by qPCR (Fig. 4). The numbers of htpG mutant leptospires in kidney and liver tissue were significantly lower than the numbers of wild-type and M1233+htpG leptospires, whereas there was no difference in the number of leptospires present in lung tissue (Fig. 4). Of interest, the reduced bacterial burdens in the kidney and liver did not result in reduced tissue pathology. Despite the reduced number of M1233 leptospires in hamster kidney tissue, the leptospires were not completely cleared from hamster kidneys at 21 days postinfection; kidneys were culture positive at the conclusion of the infection experiment. A recent study showed that the leptospiral htpG mutant M1233 was unable to colonize the kidneys of BALB/c mice (31). Nothing is known about the mechanisms by which pathogenic Leptospira colonizes the kidneys of carrier host species or the molecular basis that determines whether an infection is an acute lethal infection (hamsters) or results in asymptomatic renal colonization (mice). The ability of M1233 to colonize hamster kidneys, but not mouse kidneys, is intriguing; defining the specific role of HtpG in leptospiral virulence may provide insight into the bases of lethal acute leptospirosis and renal colonization.
FIG 4.
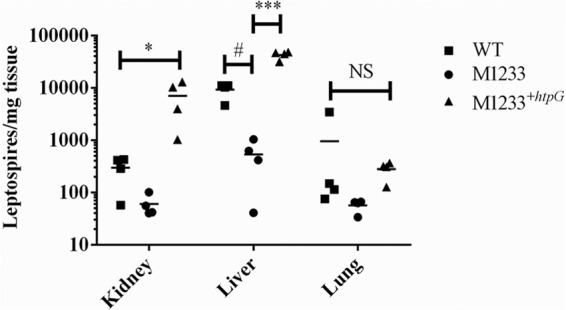
Leptospiral burdens in the kidneys, livers, and lungs of hamsters infected with 105 wild-type, M1233, or M1233+htpG bacteria. Bacterial numbers were determined by qPCR, targeting lipL32, 5 days postinfection. *, P < 0.05; #, P = 0.0015; ***, P < 0.0001; NS, not significant.
The precise role of HtpG in bacterial pathogenesis is unclear and appears to differ for different bacterial species. An htpG mutant in F. tularensis was attenuated in a mouse model of infection, with 1,000-fold fewer mutant than wild-type bacteria isolated in competition experiments. The mutant also showed defects in intracellular growth in macrophages (30). An E. tarda htpG mutant was attenuated for virulence in Japanese flounder, as demonstrated by diminished recovery of bacteria from blood and reduced fish mortality; the strain also had defects in growth at elevated temperatures (27). In contrast, analysis of a Porphyromonas gingivalis htpG mutant in cell adhesion and invasion assays found that its virulence was not impaired (38). Accordingly, we performed a number of phenotypic assays to investigate possible mechanisms responsible for the attenuation of the leptospiral htpG mutant.
Resistance to heat, oxidative, and osmotic stress.
HtpG is a well-conserved molecular chaperone, found in all bacteria, that assists in protein folding under stress conditions (15). The loss of HtpG can result in various effects on bacterial cells, including a slight growth defect at high temperatures and increased sensitivity to oxidative stress (27, 28, 51). To assess the role of HtpG during heat, oxidative, and osmotic stresses, the growth and survival of M1233 were measured.
The growth of M1233 was not impaired at the optimal laboratory growth temperature of 30°C; there was likewise no difference in growth between M1233 and the wild type at 37°C (Fig. 5). Neither strain grew at 39°C (data not shown). The cell morphology of M1233 remained normal at higher temperatures, as determined by dark-field microscopy. These results are in contrast to those of other studies on htpG mutants; Synechococcus elongatus PCC7942 and E. coli htpG mutants both showed growth rates lower than those of wild-type strains at 45°C and 44°C, respectively (28, 51). An E. tarda htpG mutant also exhibited a lowered growth rate at an elevated temperature of 37°C but reached a cell density equivalent to that of the wild type after 14 h (27). There are, however, htpG mutants that do not exhibit reduced growth at high temperatures; a Bacillus subtilis htpG mutant was able to grow unimpaired at 48°C (29).
FIG 5.
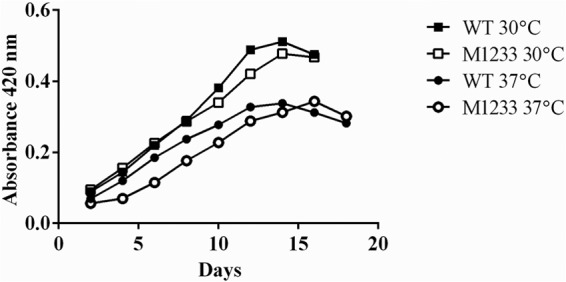
Growth of the wild type and htpG mutant M1233 at 30°C and 37°C in EMJH medium. Bacteria were grown in EMJH medium without aeration, and absorbance was measured at 420 nm.
In L. interrogans, the genomic location of htpG in a putative batI operon suggested that HtpG may play a role in protection against oxidative stress. The sensitivity of htpG mutant M1233 to hydrogen peroxide and cumene hydroperoxide was tested by MIC assays. The MICs determined for the wild type and M1233 were the same, at 250 μM hydrogen peroxide and 39 μM cumene hydroperoxide (Fig. 6A), indicating that the loss of HtpG does not impact the ability of M1233 to survive oxidative stress. In contrast, E. tarda and S. elongatus PCC7942 htpG mutants did exhibit increased sensitivity to oxidative stress and were less fit than their wild-type strains when grown in the presence of 1 mM hydrogen peroxide or 5 μM methyl viologen, respectively (27, 52).
FIG 6.
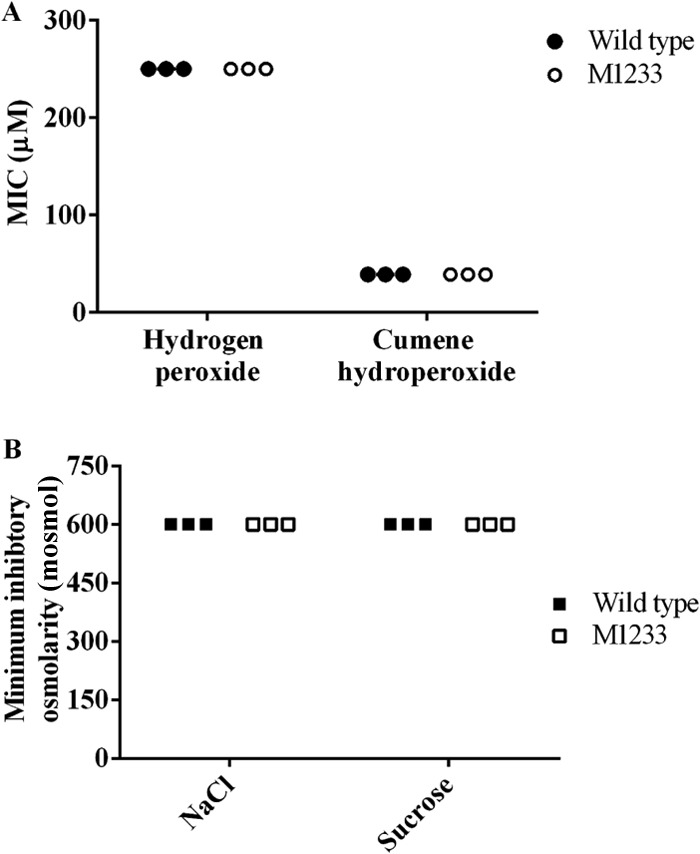
Resistance of L. interrogans serovar Manilae and M1233 to oxidative and osmotic stresses. Compounds were serially diluted in EMJH medium across microtiter trays. Cell viability was assessed by dark-field microscopy after 4 days. (A) Sensitivity to oxidative stress. Shown are MICs of hydrogen peroxide and cumene hydroperoxide for wild-type L. interrogans and htpG mutant M1233. (B) Sensitivity to osmotic stress. Shown are the minimum inhibitory osmolarities (mosmol) for wild-type L. interrogans and htpG mutant M1233 in EMJH medium supplemented with NaCl or sucrose.
Osmotic stress is a stress condition to which leptospires must adapt during infection, and studies have shown that expression of heat stress-related molecular chaperones can be induced by osmotic shock (53). EMJH medium has an approximate osmolarity of 70 mosmol, whereas physiological osmolarity is approximately 300 mosmol (54). Wild-type and M1233 leptospires were grown in EMJH medium or in EMJH medium with increased osmolarity (150 mosmol to 600 mosmol), supplemented with NaCl or sucrose. The growth of wild-type and M1233 cells was inhibited at 600 mosmol, indicating no difference in sensitivity to osmotic stress (Fig. 6B).
Based on the role of HtpG in tolerating stress in other bacteria, it was surprising that the htpG mutant M1233 did not exhibit increased sensitivity to heat, oxidative, or osmotic stress. However, as discussed above, htpG mutation results in a diverse range of phenotypes in different bacterial species. Additionally, L. interrogans has an HtpG paralog, LA1231 (serovar Lai), with 23% identity and 43% similarity to HtpG. LA1231 does not compensate for the loss of virulence observed in M1233; an la1231 mutant retained virulence in hamsters (Table 2). This paralog may have a primary or compensatory role in combating heat, oxidative, or osmotic stress in the absence of HtpG. A previous microarray study, in which the expression of htpG remained stable while its paralog la1231 was upregulated at a higher temperature (2.6-fold upregulation at 39°C over expression at 30°C) (55), supports this view.
Analysis of in vitro protein expression.
The loss of a molecular chaperone such as HtpG can result in protein aggregation (56) and could affect the expression levels of proteins associated with leptospiral virulence. To assess this possibility, the protein profiles of the wild type and M1233 were compared by 2-dimensional (2D) gel electrophoresis. The protein profiles under normal conditions (growth in EMJH medium at 30°C) and under heat shock (growth under normal conditions followed by a heat shock, 39°C for 90 min) were compared. Only two proteins were found to be downregulated in the htpG mutant under both normal and heat shock conditions; these proteins were identified as LipL48 and LA0505 by mass spectrometry (Mascot scores, 90 and 84, respectively). LipL48 (LA3240 in serovar Lai) is a Leptospira outer membrane lipoprotein. To determine if the reduction in the level of LipL48 protein expression resulted from htpG mutation, LipL48 immunoblot analysis was performed on a number of unrelated leptospiral strains. Variable levels of LipL48 expression were observed (data not shown; D. Haake, personal communication). Some strains, including L. interrogans serovar Manilae strain M874 and L. interrogans serovar Pomona strain P52, produced no detectable LipL48 yet retained virulence (57), indicating that reduced expression of LipL48 is not associated with the loss of htpG and does not attenuate virulence. The mechanism for variable expression of LipL48 is unknown.
LA0505 (serovar Lai) is a conserved hypothetical protein that is surface exposed (58). The reduced expression of LA0505, like that of LipL48, is unlikely to contribute to the attenuation of M1233, since a la0505 mutant, M1020, retained virulence (14). The analysis of in vitro protein profiles thus found no obvious differences in protein expression that could account for the complete loss of virulence observed for htpG mutant M1233.
Analysis of known leptospiral virulence factors: LPS, motility, and Loa22.
LPS (9), outer membrane protein Loa22 (11), and motility (6, 7) are three factors essential for leptospiral virulence. To determine if the attenuation of htpG mutant M1233 was due to the modification or loss of these, each was assessed in this mutant. The LPS profiles of proteinase K-treated whole-cell lysates of the wild type and M1233 were analyzed by SDS-PAGE and carbohydrate silver staining. M1233 retained a normal LPS profile by silver staining (Fig. 7A), with no obvious alterations in profile, in contrast to the lower-molecular-mass LPS exhibited by the attenuated control strain M895 (9). By immunoblot analysis, the reactivity of an anti-serovar Manilae antiserum to M1233 LPS was indistinguishable from that against wild-type LPS (Fig. 7A).
FIG 7.
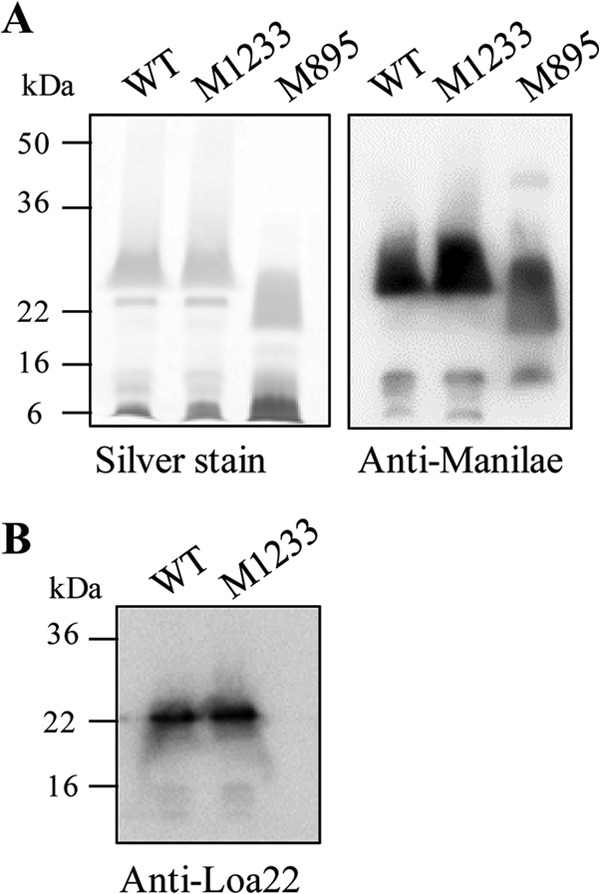
LPS profiles and Loa22 protein expression of wild-type L. interrogans and htpG mutant M1233. (A) LPS silver staining and anti-serovar Manilae immunoblotting of wild-type (WT), M1233, and LPS mutant M895 whole-cell lysates treated with proteinase K. (B) Anti-Loa22 immunoblot of wild-type and M1233 whole-cell lysates. The positions of molecular mass standards are shown on the left.
The motilities of wild-type serovar Manilae and M1233 were assessed by a plate assay using semisolid agar (0.5% agar). Motility was the same for the two strains; the wild type had a zone of spread of 15.6 ± 1.3 mm, and M1233 had a zone of spread of 15.2 ± 2.2 mm (P, >0.05 by Student's t test). The expression of Loa22 was measured by immunoblotting. A comparison of Loa22 expression in the wild type and htpG mutant M1233 revealed no difference (Fig. 7B). We therefore conclude that the attenuation of htpG mutant M1233 was not due to altered motility, LPS expression, or Loa22 expression.
Resistance to innate immune defenses.
Increased susceptibility to host innate immune factors could explain the attenuation of htpG mutant M1233. Two key defenses against pathogens are macrophages and serum complement. L. interrogans is resistant to killing by serum complement (39, 59), whereas the saprophytic species L. biflexa is sensitive. To assess whether the disruption of htpG in M1233 affected resistance to human complement, wild-type and M1233 cells were incubated with 90% pooled normal human serum at 37°C for 60 min; the viability of the cells was then determined by direct microscopic counts. Nonmotile cells are considered nonviable (60), with direct microscopic counts employed as a measure of viability in several studies (9, 59, 61, 62), including assessment of sensitivity to complement (59, 61). Additionally, studies with luminescent leptospires have shown motility to be an accurate measure of cell viability (39). Leptospires tend to aggregate in culture and have diffuse colony morphologies (60) that make viable plate counts problematic and highly inaccurate. There was no difference in the survival of wild-type and M1233 leptospires in the presence of human serum (Fig. 8); the saprophytic species L. biflexa was included as a positive control and was killed rapidly. Heat-inactivated serum did not kill wild-type, M1233, or L. biflexa cells; all strains exhibited >90% survival (Fig. 8). The disruption of htpG did not alter the serum resistance of M1233. LPS is an important mechanism used to evade complement activation (63), and we have shown that the LPS of M1233 does not appear to be altered. Pathogenic leptospires also possess a number of proteins that bind to complement factors, such as factor H and C4b-binding protein, and inhibit the classical and alternative pathways of complement; these include LenA/LfhA (64, 65), Lsa30 (66), LcpA (67), and Lig proteins (61, 68). Our 2D analysis of the protein profile of M1233 found changes in the expression of only two proteins (see above) following htpG disruption; the fact that M1233 retained complement resistance factors is consistent with the expression of these proteins. Of course, we cannot exclude the possibility that the expression profiles of these or other proteins may differ under in vivo conditions.
FIG 8.
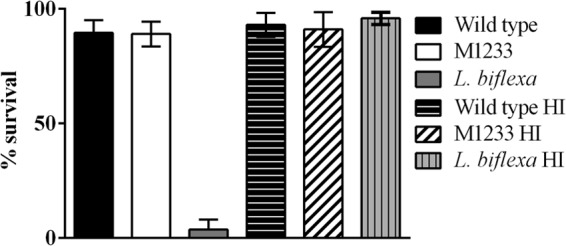
Resistance to serum complement. Bacteria were incubated in 90% pooled human serum or heat-inactivated 90% pooled human sera (HI). Survival was measured by direct counts of viable, motile cells. Error bars represent SD.
The function of HtpG in F. tularensis has been linked to intracellular survival in macrophages, where an htpG mutant of F. tularensis exhibited significantly reduced intracellular growth and a reduction in the rate of induced macrophage death (30). Pathogenic Leptospira has exhibited delayed clearance from murine macrophages, with the potential to replicate intracellularly (40). To determine if leptospiral HtpG has a role in leptospiral survival in macrophages, the association of wild-type and M1233 leptospires with murine macrophages was assessed (Fig. 9). After 2 h and 48 h, there was no difference between the number of intracellular wild-type and htpG mutant leptospires in the murine macrophages; M1233 did not exhibit reduced survival within murine macrophages. This result is supported by the virulence trials, which indicated that the htpG mutant was able to disseminate throughout the hamsters; leptospires were detected in the liver, lung, and kidneys (Fig. 4). Despite these signs of disease, the infection was not lethal, even at extremely high doses.
FIG 9.
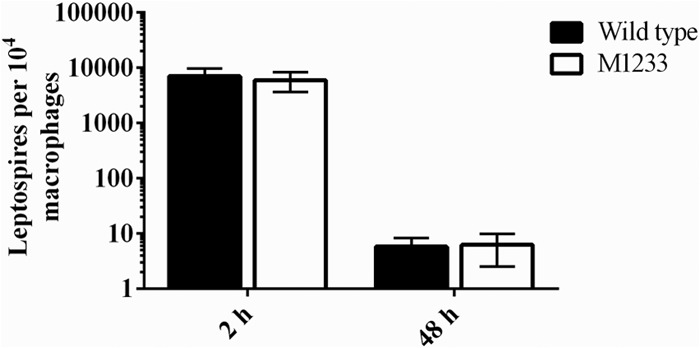
Leptospiral survival within macrophages. Bacteria were incubated with monolayers of murine primary bone marrow-derived macrophages, treated with gentamicin, and then incubated for 2 or 48 h. The presence of intracellular leptospires (expressed per 104 macrophages) was determined by qPCR after 2 h or 48 h of incubation. Error bars represent SD.
Role of HtpG in pathogenesis.
Recently, another molecular chaperone, ClpB, was shown to be required for leptospiral virulence; the leptospiral clpB mutant was attenuated in the gerbil model of acute leptospirosis (8). The clpB mutant showed impaired growth at 37°C compared to 30°C and increased sensitivity to oxidative stress when grown on plates in the presence of 10 mM and 20 mM butyl peroxide or 20 mM hydrogen peroxide (8). It is clear that impaired stress responses can lead to attenuation of virulence. This was observed in the leptospiral clpB molecular chaperone mutant (reduced virulence, impaired growth at elevated temperatures, increased sensitivity to oxidative stress) and in htpG mutants of F. tularensis (reduced growth in macrophages) and E. tarda (growth delay at 37°C, increased sensitivity to oxidative stress). The distinction between these three mutants and the leptospiral htpG mutant is that the leptospiral htpG mutant does not appear to be impaired under heat, oxidative, or osmotic stress, nor does it appear to have any in vitro proteins differentially expressed that account for attenuation. The leptospiral htpG mutant is not more susceptible to clearance by macrophages or more sensitive to human complement than the wild type. However, it is very clear that HtpG plays a critical role in leptospiral infection, making it one of only a very few confirmed virulence factors in Leptospira (see the introduction). Since the pathogenesis of Leptospira is poorly characterized, further analysis of this mutant may define novel and specific pathogenic mechanisms.
HtpG, as a molecular chaperone, is thought to act by assisting in slight conformational changes to its substrates (23); it may not affect the protein expression of its substrates directly but may affect the substrate activity. Only two HtpG client proteins have been identified: 50S ribosomal protein L2 from E. coli (69) and a 30-kDa linker polypeptide from the phycobilisome of the cyanobacterium Synechococcus elongatus (24). HtpG substrates may not be conserved in different bacteria due to variations occurring in the N domain and the M domain of HtpG, where substrates bind (24, 69), thus accounting for differing phenotypes when this gene is disrupted. Identification of the leptospiral HtpG substrates could help to elucidate the role of this protein in leptospiral pathogenesis. This study demonstrated that HtpG is essential for leptospiral virulence, and our results are consistent with a novel, as yet undetermined role for leptospiral HtpG in pathogenesis.
ACKNOWLEDGMENTS
This work was supported by grants from the Australian Research Council, the National Health and Medical Research Council, and the French Ministry of Research (ANR-08-MIE-018). G.P. was supported by a Bernardo Houssay grant.
We thank Kunkun Zhang and Stuart Cordwell for assistance with 2D gel electrophoresis and mass spectrometry and Suwimol Taweechaisupapong for assistance with histopathology interpretation. The HtpG antiserum was a generous gift from D. G. Sweier.
Footnotes
Published ahead of print 23 December 2013
REFERENCES
- 1.Adler B, de la Peña Moctezuma A. 2010. Leptospira and leptospirosis. Vet. Microbiol. 140:287–296. 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM, Peru-United States Leptospirosis Consortium 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771. 10.1016/S1473-3099(03)00830-2 [DOI] [PubMed] [Google Scholar]
- 3.Faine S, Adler B, Bolin C, Perolat P. 1999. Leptospira and leptospirosis, 2nd ed. MediSci, Melbourne, Australia [Google Scholar]
- 4.McBride AJA, Athanazio DA, Reis MG, Ko AI. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18:376–386. 10.1097/01.qco.0000178824.05715.2c [DOI] [PubMed] [Google Scholar]
- 5.Eshghi A, Lourdault K, Murray GL, Bartpho T, Sermswan RW, Picardeau M, Adler B, Snarr B, Zuerner RL, Cameron CE. 2012. Leptospira interrogans catalase is required for resistance to H2O2 and for virulence. Infect. Immun. 80:3892–3899. 10.1128/IAI.00466-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert A, Picardeau M, Haake DA, Sermswan RW, Srikram A, Adler B, Murray GL. 2012. FlaA proteins in Leptospira interrogans are essential for motility and virulence but are not required for formation of the flagellum sheath. Infect. Immun. 80:2019–2025. 10.1128/IAI.00131-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao S, Sun A, Ojcius DM, Wu S, Zhao J, Yan J. 2009. Inactivation of the fliY gene encoding a flagellar motor switch protein attenuates mobility and virulence of Leptospira interrogans strain Lai. BMC Microbiol. 9:253. 10.1186/1471-2180-9-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lourdault K, Cerqueira GM, Wunder EA, Picardeau M. 2011. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect. Immun. 79:3711–3717. 10.1128/IAI.05168-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray GL, Srikram A, Henry R, Hartskeerl RA, Sermswan RW, Adler B. 2010. Mutations affecting Leptospira interrogans lipopolysaccharide attenuate virulence. Mol. Microbiol. 78:701–709. 10.1111/j.1365-2958.2010.07360.x [DOI] [PubMed] [Google Scholar]
- 10.Murray GL, Srikram A, Henry R, Puapairoj A, Sermswan RW, Adler B. 2009. Leptospira interrogans requires heme oxygenase for disease pathogenesis. Microbes Infect. 11:311–314. 10.1016/j.micinf.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 11.Ristow P, Bourhy P, da Cruz McBride FW, Figueira CP, Huerre M, Ave P, Girons IS, Ko AI, Picardeau M. 2007. The OmpA-like protein Loa22 is essential for leptospiral virulence. PLoS Pathog. 3:e97. 10.1371/journal.ppat.0030097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Zhang C, Ojcius DM, Sun D, Zhao J, Lin X, Li L, Li L, Yan J. 2012. The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Mol. Microbiol. 83:1006–1023. 10.1111/j.1365-2958.2012.07985.x [DOI] [PubMed] [Google Scholar]
- 13.Bourhy P, Louvel H, Saint Girons I, Picardeau M. 2005. Random insertional mutagenesis of Leptospira interrogans, the agent of leptospirosis, using a mariner transposon. J. Bacteriol. 187:3255–3258. 10.1128/JB.187.9.3255-3258.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray GL, Morel V, Cerqueira GM, Croda J, Srikram A, Henry R, Ko AI, Dellagostin OA, Bulach DM, Sermswan RW, Adler B, Picardeau M. 2009. Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77:810–816. 10.1128/IAI.01293-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearl LH, Prodromou C. 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75:271–294. 10.1146/annurev.biochem.75.103004.142738 [DOI] [PubMed] [Google Scholar]
- 16.Harris SF, Shiau AK, Agard DA. 2004. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure 12:1087–1097. 10.1016/j.str.2004.03.020 [DOI] [PubMed] [Google Scholar]
- 17.Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65–75. 10.1016/S0092-8674(00)80314-1 [DOI] [PubMed] [Google Scholar]
- 18.Wandinger SK, Richter K, Buchner J. 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283:18473–18477. 10.1074/jbc.R800007200 [DOI] [PubMed] [Google Scholar]
- 19.Huai Q, Wang H, Liu Y, Kim H-Y, Toft D, Ke H. 2005. Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding. Structure 13:579–590. 10.1016/j.str.2004.12.018 [DOI] [PubMed] [Google Scholar]
- 20.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. 2003. Structural and functional analysis of the middle segment of Hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11:647–658. 10.1016/S1097-2765(03)00065-0 [DOI] [PubMed] [Google Scholar]
- 21.Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K. 1995. Mechanism of dimer formation of the 90-kDa heat-shock protein. Eur. J. Biochem. 233:1–8. 10.1111/j.1432-1033.1995.001_1.x [DOI] [PubMed] [Google Scholar]
- 22.Prodromou C, Panaretou B, Chohan S, Siligardi G, O'Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. 2000. The ATPase cycle of Hsp90 drives a molecular ‘clamp' via transient dimerization of the N-terminal domains. EMBO J. 19:4383–4392. 10.1093/emboj/19.16.4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchner J. 2010. Bacterial Hsp90—desperately seeking clients. Mol. Microbiol. 76:540–544. 10.1111/j.1365-2958.2010.07140.x [DOI] [PubMed] [Google Scholar]
- 24.Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H. 2010. HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol. Microbiol. 76:576–589. 10.1111/j.1365-2958.2010.07139.x [DOI] [PubMed] [Google Scholar]
- 25.Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603–648. 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- 26.Choi S, Jang K, Yun HJ, Kang DH. 2012. Identification of the Vibrio vulnificus htpG gene and its influence on cold shock recovery. J. Microbiol. 50:707–711. 10.1007/s12275-012-2294-z [DOI] [PubMed] [Google Scholar]
- 27.Dang W, Hu YH, Sun L. 2011. HtpG is involved in the pathogenesis of Edwardsiella tarda. Vet. Microbiol. 152:394–400. 10.1016/j.vetmic.2011.05.030 [DOI] [PubMed] [Google Scholar]
- 28.Tanaka N, Nakamoto H. 1999. HtpG is essential for the thermal stress management in cyanobacteria. FEBS Lett. 458:117–123. 10.1016/S0014-5793(99)01134-5 [DOI] [PubMed] [Google Scholar]
- 29.Versteeg S, Mogk A, Schumann W. 1999. The Bacillus subtilis htpG gene is not involved in thermal stress management. Mol. Gen. Genet. 261:582–588. 10.1007/s004380051004 [DOI] [PubMed] [Google Scholar]
- 30.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104:6037–6042. 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcsisin RA, Bartpho T, Bulach DM, Srikram A, Sermswan RW, Adler B, Murray GL. 2013. Use of a high-throughput screen to identify Leptospira mutants unable to colonize the carrier host or cause disease in the acute model of infection. J. Med. Microbiol. 62:1601–1608. 10.1099/jmm.0.058586-0 [DOI] [PubMed] [Google Scholar]
- 32.Murray GL, Ellis KM, Lo M, Adler B. 2008. Leptospira interrogans requires a functional heme oxygenase to scavenge iron from hemoglobin. Microbes Infect. 10:791–797. 10.1016/j.micinf.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 33.Picardeau M. 2008. Conjugative transfer between Escherichia coli and Leptospira spp. as a new genetic tool. Appl. Environ. Microbiol. 74:319–322. 10.1128/AEM.02172-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lourdault K, Aviat F, Picardeau M. 2009. Use of quantitative real-time PCR for studying the dissemination of Leptospira interrogans in the guinea pig infection model of leptospirosis. J. Med. Microbiol. 58:648–655. 10.1099/jmm.0.008169-0 [DOI] [PubMed] [Google Scholar]
- 35.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. 2009. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 64:247–255. 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 36.Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311–2318. 10.1128/IAI.70.5.2311-2318.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cullen PA, Haake DA, Bulach DM, Zuerner RL, Adler B. 2003. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 71:2414–2421. 10.1128/IAI.71.5.2414-2421.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweier DG, Combs A, Shelburne CE, Fenno JC, Lopatin DE. 2003. Construction and characterization of a Porphyromonas gingivalis htpG disruption mutant. FEMS Microbiol. Lett. 225:101–106. 10.1016/S0378-1097(03)00506-8 [DOI] [PubMed] [Google Scholar]
- 39.Murray GL, King AM, Srikram A, Sermswan RW, Adler B. 2010. Use of luminescent Leptospira interrogans for enumeration in biological assays. J. Clin. Microbiol. 48:2037–2042. 10.1128/JCM.02541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toma C, Okura N, Takayama C, Suzuki T. 2011. Characteristic features of intracellular pathogenic Leptospira in infected murine macrophages. Cell. Microbiol. 13:1783–1792. 10.1111/j.1462-5822.2011.01660.x [DOI] [PubMed] [Google Scholar]
- 41.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, Davis J, Johnson M, Kuczek E, Alt DP, Peterson-Burch B, Coppel RL, Rood JI, Davies JK, Adler B. 2006. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. U. S. A. 103:14560–14565. 10.1073/pnas.0603979103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nascimento ALTO, Ko AI, Martins EAL, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CFM, Leite LCC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El-Dorry H, Ferro ES, Ferro MIT, Furlan LR, Gamberini M, Giglioti EA, Góes-Neto A, Goldman GH, Goldman MHS, Harakava R, Jerônimo SMB, Junqueira-de-Azevedo ILM, Kimura ET, Kuramae EE, Lemos EGM, Lemos MVF, Marino CL, Nunes LR, De Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJG, Ferro JA, Camargo LEA, Kitajima JP, Setubal JC, Van Sluys MA. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164–2172. 10.1128/JB.186.7.2164-2172.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888–893. 10.1038/nature01597 [DOI] [PubMed] [Google Scholar]
- 44.Picardeau M, Bulach DM, Bouchier C, Zuerner RL, Zidane N, Wilson PJ, Creno S, Kuczek ES, Bommezzadri S, Davis JC, McGrath A, Johnson MJ, Boursaux-Eude C, Seemann T, Rouy Z, Coppel RL, Rood JI, Lajus A, Davies JK, Médigue C, Adler B. 2008. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One 3:e1607. 10.1371/journal.pone.0001607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong KS, Houry WA. 2012. Novel structural and functional insights into the MoxR family of AAA+ ATPases. J. Struct. Biol. 179:211–221. 10.1016/j.jsb.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 46.Tang YP, Dallas MM, Malamy MH. 1999. Characterization of the BatI (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol. Microbiol. 32:139–149. 10.1046/j.1365-2958.1999.01337.x [DOI] [PubMed] [Google Scholar]
- 47.Stewart PE, Carroll JA, Dorward DW, Stone HH, Sarkar A, Picardeau M, Rosa PA. 2012. Characterization of the Bat proteins in the oxidative stress response of Leptospira biflexa. BMC Microbiol. 12:290. 10.1186/1471-2180-12-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryson K, Cozzetto D, Jones DT. 2007. Computer-assisted protein domain boundary prediction using the DomPred server. Curr. Protein Pept. Sci. 8:181–188. 10.2174/138920307780363415 [DOI] [PubMed] [Google Scholar]
- 49.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 50.Murray GL, Srikram A, Hoke DE, Wunder EA, Jr, Henry R, Lo M, Zhang K, Sermswan RW, Ko AI, Adler B. 2009. Major surface protein LipL32 is not required for either acute or chronic infection with Leptospira interrogans. Infect. Immun. 77:952–958. 10.1128/IAI.01370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bardwell JC, Craig EA. 1988. Ancient heat shock gene is dispensable. J. Bacteriol. 170:2977–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hossain MM, Nakamoto H. 2003. Role for the cyanobacterial HtpG in protection from oxidative stress. Curr. Microbiol. 46:70–76. 10.1007/s00284-002-3831-5 [DOI] [PubMed] [Google Scholar]
- 53.Hecker M, Schumann W, Völker U. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417–428. 10.1046/j.1365-2958.1996.396932.x [DOI] [PubMed] [Google Scholar]
- 54.Matsunaga J, Sanchez Y, Xu X, Haake DA. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 73:70–78. 10.1128/IAI.73.1.70-78.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo M, Bulach DM, Powell DR, Haake DA, Matsunaga J, Paustian ML, Zuerner RL, Adler B. 2006. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 74:5848–5859. 10.1128/IAI.00755-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas JG, Baneyx F. 2000. ClpB and HtpG facilitate de novo protein folding in stressed Escherichia coli cells. Mol. Microbiol. 36:1360–1370. 10.1046/j.1365-2958.2000.01951.x [DOI] [PubMed] [Google Scholar]
- 57.King AM, Bartpho T, Sermswan RW, Bulach DM, Eshghi A, Picardeau M, Adler B, Murray GL. 2013. Leptospiral outer membrane protein LipL41 is not essential for acute leptospirosis, but requires a small chaperone, Lep, for stable expression. Infect. Immun. 81:2768–2776. 10.1128/IAI.00531-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cullen PA, Xu XY, Matsunaga J, Sanchez Y, Ko AI, Haake DA, Adler B. 2005. Surfaceome of Leptospira spp. Infect. Immun. 73:4853–4863. 10.1128/IAI.73.8.4853-4863.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meri T, Murgia R, Stefanel P, Meri S, Cinco M. 2005. Regulation of complement activation at the C3-level by serum resistant leptospires. Microb. Pathog. 39:139–147. 10.1016/j.micpath.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 60.Faine S. 1994. Leptospira and leptospirosis. CRC Press, Boca Raton, FL [Google Scholar]
- 61.Choy HA. 2012. Multiple activities of LigB potentiate virulence of Leptospira interrogans: inhibition of alternative and classical pathways of complement. PLoS One 7:e41566. 10.1371/journal.pone.0041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patarakul K, Lo M, Adler B. 2010. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 10:31. 10.1186/1471-2180-10-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395–1406. 10.1046/j.1365-2958.2003.03383.x [DOI] [PubMed] [Google Scholar]
- 64.Stevenson B, Choy HA, Pinne M, Rotondi ML, Miller MC, DeMoll E, Kraiczy P, Cooley AE, Creamer TP, Suchard MA, Brissette CA, Verma A, Haake DA. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One 2:e1188. 10.1371/journal.pone.0001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verma A, Hellwage J, Artiushin S, Zipfel PF, Kraiczy P, Timoney JF, Stevenson B. 2006. LfhA, a novel factor H-binding protein of Leptospira interrogans. Infect. Immun. 74:2659–2666. 10.1128/IAI.74.5.2659-2666.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Souza NM, Vieira ML, Alves IJ, de Morais ZM, Vasconcellos SA, Nascimento ALTO. 2012. Lsa30, a novel adhesin of Leptospira interrogans binds human plasminogen and the complement regulator C4bp. Microb. Pathog. 53:125–134. 10.1016/j.micpath.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 67.Barbosa AS, Monaris D, Silva LB, Morais ZM, Vasconcellos SA, Cianciarullo AM, Isaac L, Abreu PAE. 2010. Functional characterization of LcpA, a surface-exposed protein of Leptospira spp. that binds the human complement regulator C4BP. Infect. Immun. 78:3207–3216. 10.1128/IAI.00279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castiblanco-Valencia MM, Fraga TR, Silva LB, Monaris D, Abreu PA, Strobel S, Jozsi M, Isaac L, Barbosa AS. 2012. Leptospiral immunoglobulin-like proteins interact with human complement regulators factor H, FHL-1, FHR-1, and C4BP. J. Infect. Dis. 205:995–1004. 10.1093/infdis/jir875 [DOI] [PubMed] [Google Scholar]
- 69.Motojima-Miyazaki Y, Yoshida M, Motojima F. 2010. Ribosomal protein L2 associates with E. coli HtpG and activates its ATPase activity. Biochem. Biophys. Res. Commun. 400:241–245. 10.1016/j.bbrc.2010.08.047 [DOI] [PubMed] [Google Scholar]