Abstract
MicroRNAs (miRs) are increasingly associated with metabolic liver diseases. We have shown that ursodeoxycholic acid, a hydrophilic bile acid, counteracts the miR-34a/sirtuin 1 (SIRT1)/p53 pathway, activated in the liver of nonalcoholic steatohepatitis (NASH) patients. In contrast, hydrophobic bile acids, particularly deoxycholic acid (DCA), activate apoptosis and are increased in NASH. We evaluated whether DCA-induced apoptosis of rat hepatocytes occurs via miR-34a-dependent pathways and whether they connect with c-Jun N-terminal kinase (JNK) induction. DCA enhanced miR-34a/SIRT1/p53 proapoptotic signaling in a dose- and time-dependent manner. In turn, miR-34a inhibition and SIRT1 overexpression significantly rescued targeting of the miR-34a pathway and apoptosis by DCA. In addition, p53 overexpression activated the miR-34a/SIRT1/p53 pathway, further induced by DCA. DCA increased p53 expression as well as p53 transcriptional activation of PUMA and miR-34a itself, providing a functional mechanism for miR-34a activation. JNK1 and c-Jun were shown to be major targets of DCA, upstream of p53, in engaging the miR-34a pathway and apoptosis. Finally, activation of this JNK1/miR-34a proapoptotic circuit was also shown to occur in vivo in the rat liver. These results suggest that the JNK1/p53/miR-34a/SIRT1 pathway may represent an attractive pharmacological target for the development of new drugs to arrest metabolism- and apoptosis-related liver pathologies.
INTRODUCTION
Intrahepatic accumulation of hydrophobic bile acids contributes to liver injury, which is associated with development of nonalcoholic steatohepatitis, cholestatic diseases, cholangiocarcinoma, and liver failure (1). Bile acid-induced apoptosis involves activation of the proapoptotic stress-activated kinase, c-Jun N-terminal kinase (JNK), leading to increased plasma membrane expression of the Fas and TRAIL death receptors and subsequent ligand-independent activation (2, 3). Caspase-8 is then activated, ultimately leading to apoptosis (2). Conversely, deoxycholic acid (DCA) may engage JNK via activation of death receptors, namely, Fas and TGR5 (4, 5). DCA-induced apoptosis may also result from disruption of the mitochondrial transmembrane potential, through increased reactive oxygen species (ROS) production, leading to translocation of proapoptotic Bax protein to the mitochondria and release of cytochrome c (6). Finally, we have shown that DCA-induced apoptosis is also mediated by a cyclin D1/p53-dependent pathway (7). Still, the mechanisms by which DCA induces apoptosis in the liver remain poorly known, as does the network of targets signaling its actions.
MicroRNAs (miRNAs or miRs) are known to modulate the expression of numerous genes. In particular, upregulation of members of the miR-34 family induces apoptosis and cell cycle arrest. One of the main targets of miR-34a is sirtuin 1 (SIRT1), a NAD-dependent deacetylase that regulates apoptosis in response to oxidative and genotoxic stress (8). Furthermore, SIRT1 is capable of deacetylating all major p53 acetylation sites. SIRT1-mediated deacetylation antagonizes p53-dependent transcriptional activation, inhibiting p53-dependent apoptosis (9). Still, the interplay of p53 with miR-34a remains complex; not only does miR-34a regulate p53 activity through SIRT1, but miR-34a-induced apoptosis and cell cycle arrest are also, at least in part, dependent on the presence of p53 (10). In fact, activation of p53 has been shown to increase miR-34a transcription in a positive-feedback loop. Nevertheless, induction of miR-34a expression can also occur independently of p53 (11).
We have recently shown that ursodeoxycholic acid (UDCA), a strong inhibitor of DCA-induced apoptotic signaling pathways (7, 12, 13), downregulates the miR-34a/SIRT1/p53 proapoptotic circuit in primary rat hepatocytes. In turn, this pathway is also associated with nonalcoholic fatty liver disease severity (14). Furthermore, DCA levels are significantly increased in steatohepatitis patients (15), likely as the result of alterations in gut microbiota induced by obesity (16). Altogether, in this study we aimed to evaluate whether DCA activates miR-34a-dependent apoptotic pathways, with concomitant outcomes in viability and apoptosis of primary rat hepatocytes, both in vitro and in vivo and whether JNK acts as a novel regulator of this pathway. Our results support a link between liver cell apoptosis, miR-34a/SIRT1/p53, and JNK1/c-Jun signaling, where JNK1-mediated activation of p53 is key to induction of miR-34a by DCA.
MATERIALS AND METHODS
Animals and DCA treatment.
Male Wistar rats (Harlan Laboratories Models, S.L., Spain) weighing ∼150 g were maintained on a 12-h light-dark cycle and fed standard laboratory chow ad libitum. Water (n = 6) or DCA (n = 6) at a dose of 250 mg/kg of body weight/day, given twice a day, was administered by oral gavage for 5 consecutive days (17). Body weights were measured each day. On day 5, animals were weighed and liver perfusion was performed under pentobarbital anesthesia. The liver was quickly removed, rinsed in normal saline, flash-frozen in liquid nitrogen, and stored at −80°C. Plasma was collected and frozen at −20°C for gas chromatography bile acid analysis (18). All experiments involving animals were performed by an investigator accredited for directing animal experiments (FELASA level C), in conformity with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals, incorporated in the Institute for Laboratory Animal Research (ILAR) Guide for Care and Use of Laboratory Animals. Experiments received prior approval from the Portuguese National Authority for Animal Health (DGAV).
Cell culture and treatments.
Primary rat hepatocytes were isolated from male rats (100 to 150 g) by collagenase perfusion as previously described (19, 20). After isolation, hepatocytes were resuspended in complete Williams E medium (Sigma-Aldrich Co., St. Louis, MO) (14) and plated on Primaria tissue culture dishes (BD Biosciences, San Jose, CA) at 5 × 104 cells/cm2. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 for 6 h, to allow attachment. Plates were then washed with medium to remove dead cells and incubated in complete Williams E medium treated with 10 to 400 μM DCA (Sigma-Aldrich Co.) or no addition (control) for 24 h before processing for total protein, RNA extraction, and cell viability. Alternatively, primary rat hepatocytes were treated with 100 μM DCA or no addition (control) for 16, 28, 40, 52, and 64 h before processing for total protein and RNA extractions, cell viability and caspase activity assays, and Hoechst staining. When indicated, cells were coincubated with 50 μM pan-caspase-inhibitor Z-VAD-fmk (Sigma-Aldrich Co.) for 30 min before DCA incubation.
To assess the transfection efficiency of primary rat hepatocytes, cells were transfected with the pRNAT-H1.1/neo plasmid, expressing a green fluorescent protein (GFP) (GeneScript, Piscataway, NJ), or with a Dy547-labeled miRIDIAN microRNA mimic transfection control (Thermo Fisher Scientific, Inc., Waltham, MA) using Lipofectamine (Life Technologies Corp., Carlsbad, CA) (21). Transfection efficiencies for plasmid DNA were typically around 50% while, for miRNAs, values ranged between 80 and 90%.
For functional analyses, primary rat hepatocytes were transfected at the moment of plating with 100 pM specific miR-34a precursor (pre-miR-34a; Life Technologies Corp.), or with a pre-miR negative control using Lipofectamine (Life Technologies Corp.). Alternatively, miR-34a silencing was performed by transfecting primary rat hepatocytes with 100 pM miR-34a-specific inhibitor (anti-miRNA-34a; Life Technologies Corp.) or with an anti-miRNA negative control. After 6 h, cells were incubated with 100 μM DCA or no addition (control). Hepatocytes were harvested at 40 h posttransfection and processed for RNA and total protein extraction, cell viability and caspase activity assays, and Hoechst staining.
To assess miR-34a-dependent SIRT1 inhibition, a reporter plasmid driven by the pMIR-REPORT and harboring either a wild-type (Luc-SIRT1 Wt 3′ untranslated region [UTR]) or mutated (Luc-SIRT1 Mut 3′ UTR) miR-34a target sequence within the 3′ UTR of SIRT1 was used (plasmid 20379 or 20380, respectively; Addgene, Cambridge, MA) (8). Upon plating, primary rat hepatocytes were cotransfected with 500 ng of either construct, together with 100 pM pre-miR-34a or anti-miR-34a, or respective controls, using Lipofectamine.
To assess if DCA interacts with the SIRT1 promoter, a pGL3 reporter plasmid harboring the SIRT1 promoter luciferase reporter gene was used (22). At the time of plating, primary rat hepatocytes were cotransfected with 500 ng of the luciferase reporter construct or with a control reporter using Lipofectamine.
To analyze whether DCA interacts with the miR-34a promoter via p53, pGL4 vectors harboring the putative promoter regions of human miR-34a containing either wild-type (Luc miR-34a Wt p53) or mutant (Luc miR-34a Mut p53) p53 binding sites were used (23). In addition, p53 was overexpressed using a pCMV-Neo-Bam vector encoding wild-type human p53 (pCMV p53 Wt) or an empty vector (pCMV-Neo-Bam) (plasmids 16440 and 16434, respectively; Addgene) (24).
At the time of plating, primary rat hepatocytes were cotransfected with 500 ng of the luciferase reporter constructs and 2 μg of the p53 expression vector using Lipofectamine. Cells were incubated with 100 μM DCA or no addition (control) 6 h after transfections. Reporter assays were performed 48 h posttransfection using the dual-luciferase reporter assay system (Promega Corp., Madison, WI) according to the manufacturer's instructions. Cells were also cotransfected with pRL-SV40 (Promega Corp.). Renilla luciferase activity was used as a transfection normalization control for all experiments involving luciferase reporter constructs.
To confirm JNK interaction with the miR-34a/SIRT1/p53 proapoptotic pathway, primary rat hepatocytes were transfected with 2 specific short interference RNA (siRNA) nucleotides (si-JNK1 and si-JNK2) designed to knock down jnk gene expression of each isoform of JNK (JNK1 and JNK2) in rats, purchased from Dharmacon (Waltham, MA) (25). A control siRNA containing a scrambled sequence that does not lead to the specific degradation of any known cellular mRNA was used as a control. For c-Jun silencing experiments, a specific Silencer Select predesigned siRNA was purchased from Life Technologies Corp., along with a Silencer negative-control siRNA. siRNA transfection efficiencies were determined by using the Block-iT Alexa Fluor red fluorescent control (Life Technologies Corp.) as a positive control. Transfection efficiencies in primary rat hepatocytes were typically between 60 and 70%, in agreement with previous results (7). Furthermore, the degree of siRNA effectiveness was not significantly different between JNK1 and JNK2 siRNAs, including in cotransfection experiments with luciferase constructs. Primary rat hepatocytes were transfected with siRNA at the final concentration of 125 pM using Lipofectamine. Cells were incubated with 100 μM DCA or no addition (control) 6 h after transfections. Hepatocytes were harvested at 24 h posttransfection and processed for RNA and total protein extraction and cell viability and caspase activity assays. To confirm whether DCA interacts with JNK to regulate the miR-34a/SIRT1/p53 proapoptotic pathway, the p53 binding site miR-34a promoter Luc miR-34a Wt p53 or Luc miR-34a Mut p53 was used (23). Moreover, in parallel experiments, miR-34a target sequence within the 3′ UTR of SIRT1 was used (Luc-SIRT1 Wt 3′ UTR or Luc-SIRT1 Mut 3′ UTR) (plasmid 20379 or 20380, respectively; Addgene) (8). Upon plating, primary rat hepatocytes were cotransfected with 500 ng of either construct together with 125 pM JNK siRNAs purchased from Dharmacon (Waltham, MA) or respective controls, using Lipofectamine.
To confirm the effects of JNK silencing in the miR-34a/SIRT1/p53 pathway, primary rat hepatocytes were transfected with either the binding domain of JNK-interacting protein 1 (JIP-1) (pCMV-Flag-JBD [JIP-1]) (26) or dominant negative DN-c-Jun FlagΔ169 plasmid (27), using Lipofectamine. Cells were incubated with 100 μM DCA or no addition (control) 6 h after transfections. Hepatocytes were harvested at 24 h posttransfection and processed for RNA, cell viability, and caspase activity assays. p53 activation was assessed based on natural reporter genes. We used pBV-Luc vector harboring the PUMA (PUMA Frag1-Luc or PUMA-Luc) promoter containing p53-responsive elements (plasmid 16451; Addgene). The pBV-Luc construct was used as a control (plasmid 16539; Addgene). At the time of plating, primary rat hepatocytes were cotransfected with 500 ng of the luciferase reporter constructs, using Lipofectamine.
Quantitative RT-PCR.
RNA was extracted from cell samples using the TRIzol reagent according to the manufacturer's instructions (Life Technologies Corp.). Real-time reverse transcription-PCR (RT-PCR) was performed in an Applied Biosystems 7300 system (Life Technologies Corp.) to quantitate the expression of miR-34a, miR-195, miR-200a, miR-122, miR-143, miR-145, and sirt1. U87 snRNA was used as the normalization control for miRNAs. The relative amounts of miRNAs were determined by the threshold cycle (2−ΔΔCT) method, where ΔΔCT = (CTtarget − CTU87) sample − (CTtarget − CTU87) calibrator. To assess SIRT1 and β-actin mRNA levels, the following primer sequences were used: for the SIRT1 gene, 5′ AGG GAA CCT CTG CCT CAT CTA C 3′ (forward) and 5′ GGC ATA CTC GCC ACC TAA CCT 3′ (reverse), and for the β-actin gene, 5′ AGG CCC CTC TGA ACC CTA AG 3′ (forward) and 5′ GGA GCG CGT AAC CCT CAT AG 3′ (reverse). Three independent reactions for each primer set were assessed in a total volume of 25 μl containing 2× Power SYBR green PCR master mix and 0.5 μM (each) primer. The relative amounts of SIRT1 mRNA were determined by the 2−ΔΔCT method, where ΔΔCT = (CTSIRT1mRNA − CTβ-actin) sample − (CTSIRT1mRNA − CTβ-actin) calibrator.
Northern blot analysis.
Northern blot experiments were performed as described in reference 28. Briefly, 20 μg of total RNA was loaded onto a 15% SequaGel (National Diagnostics, Atlanta, GA), electrophoresed, and transferred to nylon membranes at 15 V for 60 min using a Trans-Blot SD semidry transfer system (Bio-Rad Laboratories, Hercules, CA). Membranes were cross-linked to RNA at 60°C for 2 h using freshly prepared cross-linking reagent (28). Locked nucleic acid (LNA)-modified oligonucleotides for each miRNA were purchased from Exiqon (Exiqon A/S, Vedbaek, Denmark) and labeled with digoxigenin (DIG), using an end tailing kit (Roche Applied Science, Indianapolis, IN). Sequences of the oligonucleotide probes were ACAACCAGCTAAGACACTGCCA (miR-34a), GAGCTACAGTGCTTCATCTCA (miR-143), AGGGATTCCTGGGAAAACTGGAC (miR-145), GCCAATATTTCTGTGCTGCTA (miR-195), and ACATCGTTACCAGACAGTGTTA (miR-200a). After hybridization (ULTRAhyb; Life Technologies Corp.), membranes were washed with different-stringency buffer solutions and incubated in blocking buffer (DIG Wash and Block buffer set; Roche Applied Science) for 3 h at room temperature before incubation with a DIG antibody at room temperature for 30 min. Membranes were then washed in DIG washing buffer and incubated in substrate solution (CSPD; Roche Applied Science) before photoemissions were detected in a ChemiDoc-MP imaging system (Bio-Rad Laboratories). Blots were normalized to total RNA input.
Immunoblotting.
Seventy-five micrograms of total protein extracts was separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following electrophoretic transfer onto nitrocellulose membranes and blocking with 5% milk solution, blots were incubated overnight at 4°C with primary rabbit polyclonal antibodies against Ac-p53, Ac-H3 histone, and H3 histone (Cell Signaling Technology, Danvers, MA), primary goat polyclonal antibody against JNK2 and p21 (Santa Cruz Biotechnology, Santa Cruz, CA), or primary mouse monoclonal antibodies reactive to SIRT1 (Abcam PLC, Cambridge, United Kingdom); JNK, pJNK, Bax, c-Jun, and JNK1 (Santa Cruz Biotechnology); and p53 (Cell Signaling Technology) and finally with secondary antibodies conjugated with horseradish peroxidase (Bio-Rad Laboratories) for 3 h at room temperature. Membranes were processed for protein detection using Super Signal substrate (Pierce, Rockford, IL). β-Actin was used as a loading control. Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories) according to the manufacturer's specifications.
Immunocytochemistry and immunohistochemistry.
Immunocytochemistry was performed to detect SIRT1 localization in primary rat hepatocytes with overexpression of miR-34a incubated with 100 μM DCA or no addition (control). Cells were washed twice, fixed with paraformaldehyde (4%, wt/vol) in phosphate-buffered saline (PBS), and then blocked for 1 h at room temperature in PBS, containing 0.1% Triton X-100, 1% fetal bovine serum (FBS), and 10% normal donkey serum (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Cells were incubated with primary mouse monoclonal antibody reactive to SIRT1 (Abcam PLC) at a dilution of 0.5 μg/ml overnight at 4°C. After being washed twice, secondary DyLight 568-conjugated anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc.) was diluted 1:200 and added to cells for 2 h at room temperature. Immunohistochemistry was performed to detect active caspase-3 in liver sections from rats administered DCA or water (controls). A 4-μm liver section was incubated with a primary rabbit polyclonal antibody reactive to active caspase-3 (EMD Millipore, Billerica, MA) at a dilution of 0.5 μg/ml overnight at 4°C. After two washes, secondary DyLight 594-conjugated anti-rabbit antibody (Jackson ImmunoResearch Laboratories, Inc.) was diluted 1:200 and added to the sections overnight at 4°C. For both staining procedures, hepatocyte nuclei were stained with Hoechst 33258 (Sigma-Aldrich Co.) at 50 μg/ml in PBS for 6 min at room temperature. Samples were mounted using Fluoromount-G (Beckman Coulter, Inc., Brea, CA). Detection of SIRT1 and active caspase-3 puncta in cells was visualized using an Axioskop fluorescence microscope (Carl Zeiss GmbH, Jena, Germany) with a magnification of ×630.
Cell viability, cytotoxicity, and caspase activity measurements.
The ApoTox-Glo triplex assay (Promega Corp.) was used according to the manufacturer's protocol. Alternatively, cells were incubated with Caspase-Glo-8 or -9 reagent. Lactate dehydrogenase (LDH) activity and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt (MTS) metabolism were also assessed as measures of cell death and viability, respectively. Briefly, to assess LDH release, supernatants resulting from a soft centrifugation of the cell culture medium at 250 × g were combined in microplates with lactate (substrate), tetrazolium salt (coloring solution), and NAD (cofactor), previously mixed in equal proportions, according to the manufacturer's instructions (Sigma-Aldrich Corp.). Multiwell plates were protected from light and incubated for 10 min at room temperature. Finally, absorbance was measured at 490 nm, with 690 nm as reference, using a Bio-Rad model 680 microplate reader (Bio-Rad Laboratories). Cell viability was evaluated with the CellTiter96 aqueous nonradioactive cell proliferation assay (Promega), according to the manufacturer's instructions. Absorbance was measured at 490 nm, with 620 nm as reference, using a Bio-Rad model 680 microplate reader (Bio-Rad Laboratories). Hoechst labeling of attached cells was used to detect apoptotic nuclei by morphological analysis, as previously described (14). Finally, fluorescent transferase-mediated dUTP-digoxigenin nickend labeling (TUNEL) (EMD Millipore) was performed in rat liver sections according to the manufacturer's protocol.
Mitochondrial transmembrane potential and ROS.
Primary rat hepatocytes were transfected with anti-miR-34a and incubated with or without DCA before loading with 50 nM 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3); Molecular Probes, Life Technologies Corp.] or 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) for 30 min at 37°C for analysis of mitochondrial permeabilization or ROS production, respectively. Cells were then treated with TrypLE (Life Technologies Corp.), harvested, and resuspended in Ca2+-free and Mg2+-free PBS with 2% FBS (Life Technologies Corp.). The emission of green fluorescence was analyzed in intact cells by cytometric analysis on the Guava EasyCyte 5HT flow cytometer (EMD Millipore). Data were statistically evaluated using the GuavaSoft software (EMD Millipore).
p53 activity.
For assaying p53 activity, the TransAM p53 transcription factor assay kit (Active Motif, Carlsbad, CA) and the p53/MDM2 ImmunoSet assay (Enzo Life Sciences, Farmingdale, NY) were used according to the manufacturer's protocols.
Densitometry and statistical analysis.
The relative intensities of protein bands were analyzed using the Quantity One Version 4.6 densitometric analysis program (Bio-Rad Laboratories). Statistical analysis was performed using GraphPad InStat version 3.00 (GraphPad Software, San Diego, CA) for the analysis of variance and Bonferroni's multiple comparison tests. Values of P of <0.05 were considered significant.
RESULTS
DCA induces the miR-34a/SIRT1/p53 proapoptotic pathway in primary rat hepatocytes.
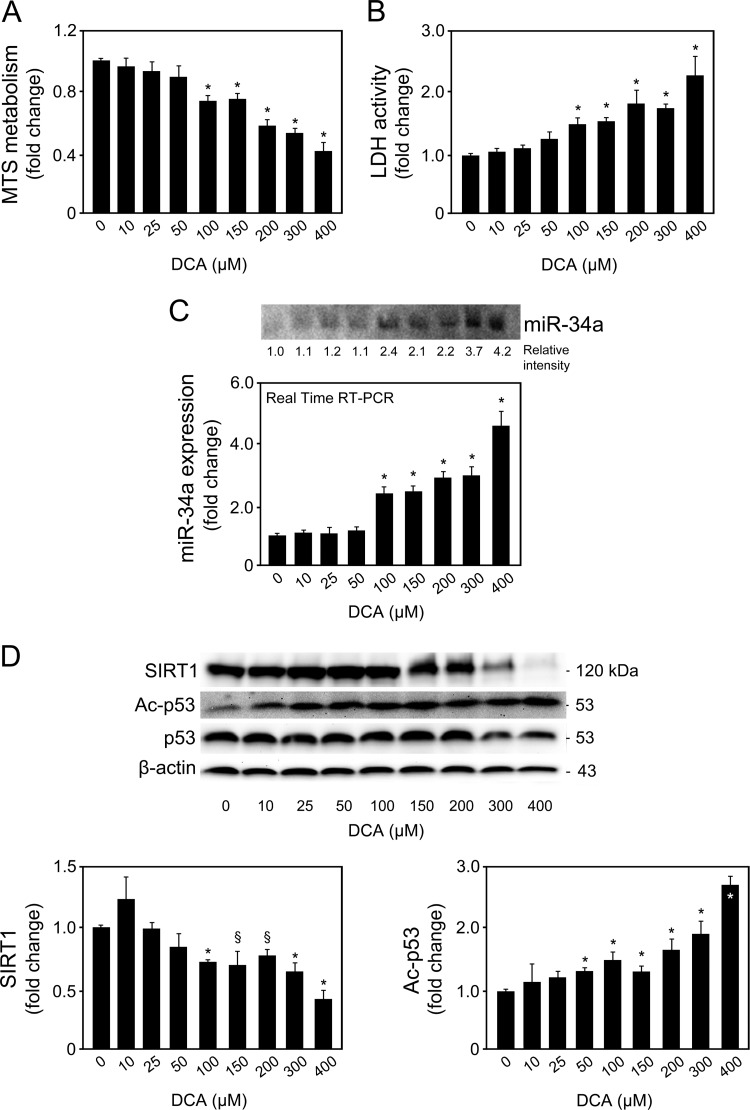
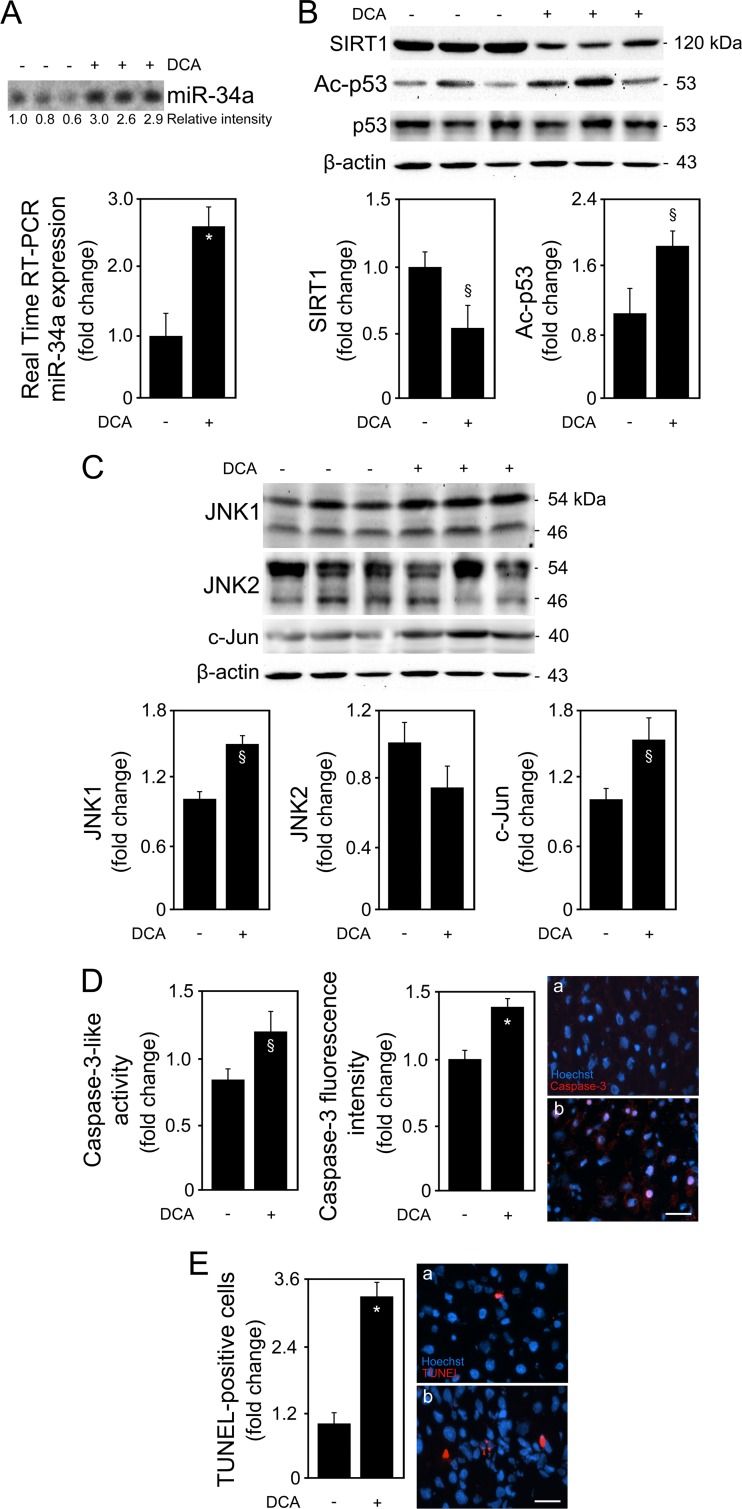
We first analyzed whether DCA could induce apoptosis through the miR-34a signaling pathway in primary rat hepatocytes. As a proof of principle, we started by evaluating the dose-dependent effects of DCA on the miR-34a/SIRT1/p53 proapoptotic pathway. Primary rat hepatocytes were incubated with 10 to 400 μM DCA for 24 h. Our results indicated that hepatocytes incubated with DCA showed a progressive and significant decrease in cell viability from ∼30% to 60% when incubated with 100 μM and 400 μM DCA, respectively (P < 0.01) (Fig. 1A). Conversely, DCA-induced cell death, as measured by LDH release, increased from ∼45% to more than 2-fold (P < 0.01) (Fig. 1B). We next evaluated whether the miR-34a/SIRT1/p53 pathway was specifically activated by DCA in cultured primary rat hepatocytes. miR-34a induces apoptosis by repressing SIRT1, which then leads to p53 acetylation and activation, with induction of proapoptotic genes (8, 29). In fact, induction of miR-34a expression by DCA was also shown to be dose dependent (Fig. 1C). Concentrations as low as 50 μM DCA resulted in slightly induced miR-34a expression. DCA further, and significantly, induced miR-34a expression levels by almost 2.5-fold at 100 μM and up to ∼5-fold at 400 μM (P < 0.01), compared to control.
FIG 1.
DCA induces apoptosis and the miR-34a/SIRT1/p53 pathway in primary rat hepatocytes in a dose-dependent manner. Cells were isolated as described in Materials and Methods and treated with 10 to 400 μM DCA or no addition (control) for 24 h. (A) MTS metabolism. (B) LDH activity. (C) Northern blot (top) and real-time RT-PCR (bottom) analysis of miR-34a. A representative blot is shown. Blots were normalized for total RNA input. (D) Immunoblotting of SIRT1 and acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to endogenous β-actin or total p53, respectively. Results are expressed as mean (±standard error of the mean) fold change of at least 4 independent experiments. §, P < 0.05, and *, P < 0.01, from control.
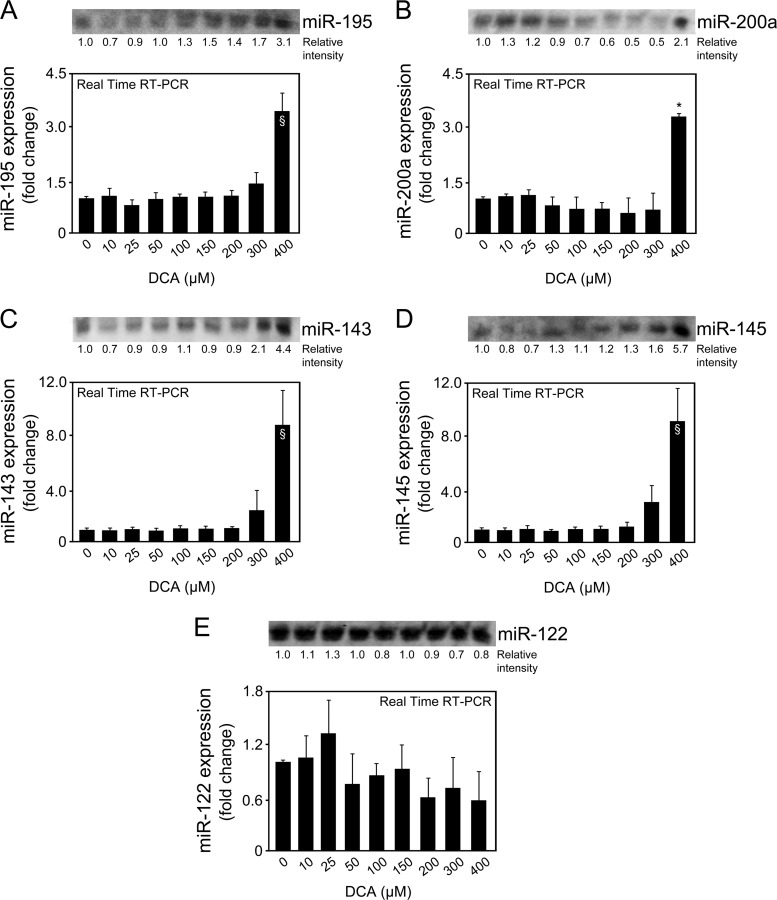
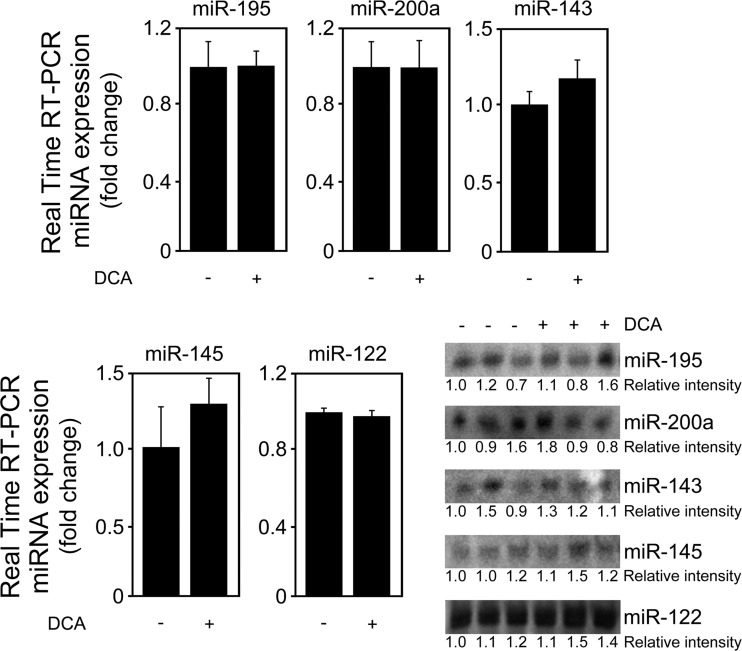
Importantly, DCA also significantly decreased SIRT1 expression while increasing acetylated p53 levels in a dose-dependent manner for doses higher than 50 μM (at least P < 0.05) (Fig. 1D). Of note, several other miRNAs have been shown to target SIRT1. For instance, miR-195 promotes palmitate-induced apoptosis in cardiomyocytes by downregulating SIRT1 (30). Also, miR-200a has been shown to regulate SIRT1 expression in mammary epithelial cells, regulating epithelial-to-mesenchymal transition-like transformation (31). Therefore, to confirm the specific regulation of SIRT1 by DCA via miR-34a, we evaluated miR-195 and miR-200a expression levels by real-time RT-PCR and Northern blot analyses (Fig. 2A and B). In addition, miR-143, -145, and -122 expression analyses also demonstrated a more selective preference of DCA for miR-34a (Fig. 2C to E). miR-122 is the most abundant liver-specific miRNA and was already shown to play a key role in hepatic function and liver disease pathogenesis (32–34). In fact, very recent miR-122 knockout studies have further expanded our view on the critical metabolic, anti-inflammatory, and antitumorigenic functions of miR-122 in the liver (35, 36). In addition, miR-143 and -145 are upregulated in the liver of genetic and dietary mouse models of obesity (37), and we have previously shown that UDCA modulates miR-143a expression during liver regeneration (20). All miRNA expression levels were relatively unchanged in the presence of 10 to 300 μM DCA. Only in the presence of 400 μM DCA, and with the exception of miR-122, did miRNA expression increase, as observed from real-time RT-PCR results and, to a lesser extent, from Northern blot analysis. These results may reflect an unspecific effect of this high, nonphysiological DCA concentration. Further, they suggest that the higher increase seen in the real-time PCR results may not be truly specific and, rather, result from a PCR artifact, common when analyzing small miRNAs. At the end, it cannot be completely excluded that SIRT1 suppression by DCA results from modulation of additional miRNAs for concentrations higher than 300 to 400 μM.
FIG 2.
DCA does not modulate miR-195, -200a, -143, -145, and -122 expression in primary rat hepatocytes. Cells were isolated as described in Materials and Methods and treated with 10 to 400 μM DCA or no addition (control) for 24 h. Northern blot analysis (top) and real-time RT-PCR analysis (bottom) of miR-195 (A), miR-200a (B), miR-143 (C), miR-145 (D), and miR-122 (E). Representative blots are shown. Blots were normalized for total RNA input. Results are expressed as mean (±standard error of the mean) fold change of at least 3 independent experiments. §, P < 0.05, and *, P < 0.01, from control.
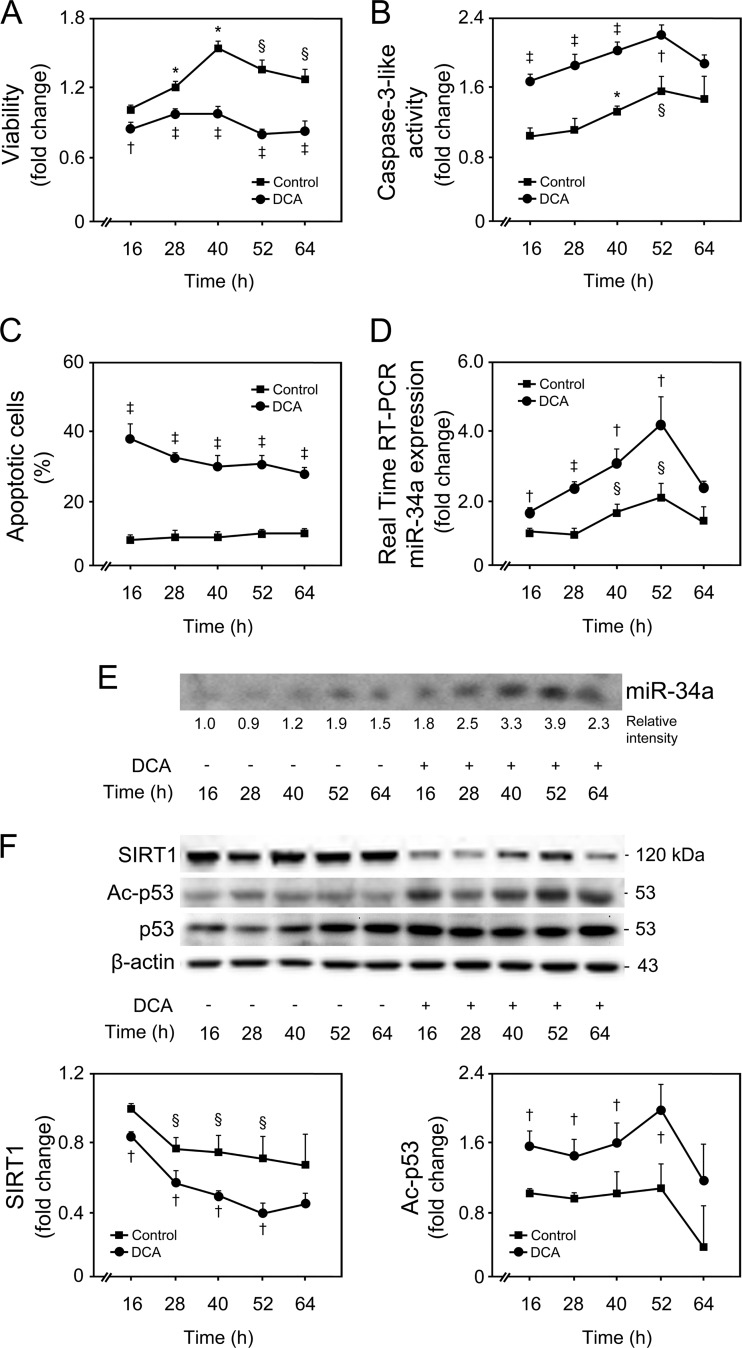
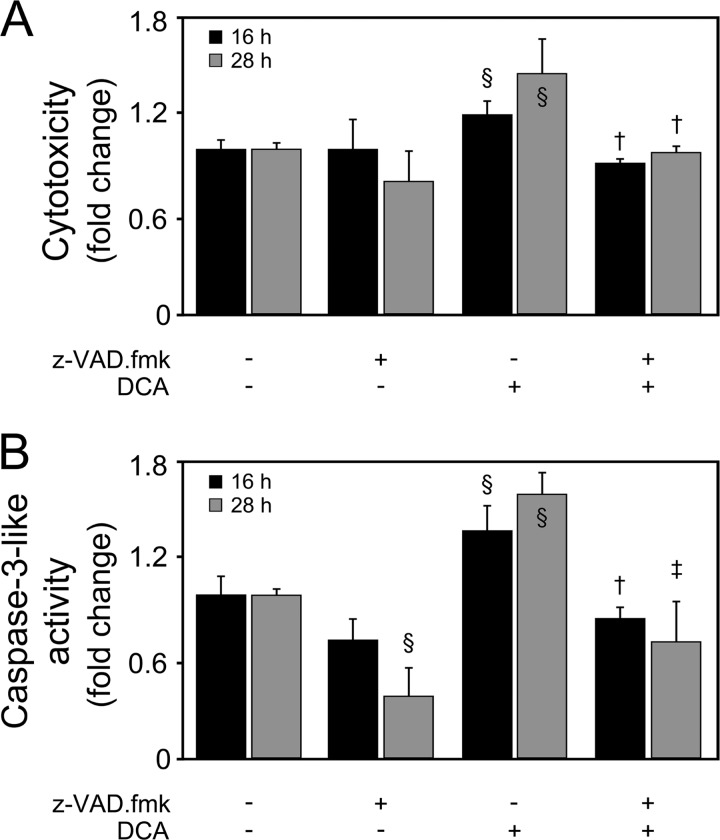
All together, these findings suggest that the miR-34a/SIRT1/p53 pathway is more selectively activated by DCA in primary rat hepatocytes, in a dose-dependent manner. Taking these results into consideration, we mimicked a strong and significant increase in the levels of DCA in vitro, using 100 μM DCA for subsequent studies, a value above physiological concentrations (38). Cells were incubated with 100 μM DCA for 16 to 64 h (Fig. 3). Primary rat hepatocytes displayed a progressive decrease in cell viability from 20%, after 16 h of incubation, to a maximum of ∼60%, after 40 to 52 h of incubation (P < 0.01) (Fig. 3A). This effect was concomitant with a >60% increase in caspase-3-like activity by DCA up until 52 h of incubation (at least P < 0.05), as measured by the ApoTox-Glo triplex assay (Fig. 3B) and Western blot analysis (data not shown). Coincubation of cells with DCA and pan-caspase inhibitor Z-VAD-fmk completely abrogated DCA-induced cytotoxicity (Fig. 4), evidencing that apoptosis is a major cell death pathway induced by DCA. In fact, DCA also increased caspase-8- and -9-like activities, up until 52 h of incubation (P < 0.05) (data not shown), implying the engagement of both the death receptor and the mitochondrial pathways of apoptosis (39). End-stage apoptosis, evaluated using Hoechst staining, confirmed DCA-induced nuclear fragmentation in ∼30% of cultured hepatocytes throughout the time course (P < 0.01) (Fig. 3C). For exposure periods longer than 64 h, DCA-induced cell death shifted more to a necrotic nature, likely reflecting an increase of its toxicity, independently of activation of programmed apoptosis.
FIG 3.
DCA induces apoptosis and the miR-34a/SIRT1/p53 pathway in primary rat hepatocytes in a time-dependent manner. Cells were isolated as described in Materials and Methods and treated with 100 μM DCA or no addition (control) for 16, 28, 40, 52, and 64 h. (A and B) Viability (A) and caspase-3-like activity (B) were measured using the ApoTox-Glo triplex assay. (C) Apoptotic cells were detected by Hoechst staining, and results are expressed as percentages of apoptotic cells. (D and E) Real-time RT-PCR (D) and Northern blot (E) analysis of miR-34a. A representative blot is shown. Blots were normalized for total RNA input. (F) Immunoblotting of SIRT1 and acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to endogenous β-actin or total p53, respectively. Results are expressed as mean (±standard error of the mean) fold change of at least 4 independent experiments. §, P < 0.05, and *, P < 0.01, from 16-h control; †, P < 0.05, and ‡, P < 0.01, from respective time point control.
FIG 4.
DCA induces caspase-dependent cell death in primary rat hepatocytes. Cells were isolated as described in Materials and Methods and treated with 100 μM DCA or no addition (control) in the presence or absence of 50 μM Z-VAD-fmk for 16 and 28 h. Cytotoxicity (A) and caspase-3-like activity (B) measured using the ApoTox-Glo triplex assay. Results are expressed as mean (±standard error of the mean) fold change of 3 independent experiments. §, P < 0.05, from control; † P < 0.05, and ‡, P < 0.05, from DCA alone.
miR-34a basal expression levels slightly increased in primary rat hepatocytes with time in culture (P < 0.05), whereas DCA significantly induced miR-34a expression up until 52 h of incubation (P < 0.05) (Fig. 3D and E). Both SIRT1 expression and p53 acetylation, normalized with total p53 levels, reflected miR-34a expression changes in control hepatocytes (Fig. 3F). Importantly, DCA significantly decreased SIRT1 expression (P < 0.05), while increasing acetylated p53 levels (P < 0.05) up until 52 h of incubation. Histone H3 acetylation levels were also evaluated, as an additional target of SIRT1 modulation. In fact, DCA also increased acetylated histone H3 levels but only up until 40 h of incubation, suggesting that p53 acetylation is a major effect of DCA-induced inhibition of SIRT1 expression (data not shown).
Activation of miR-34a is an important event during DCA-induced apoptosis in primary rat hepatocytes.
Hydrophilic bile acids, namely, ursodeoxycholic and tauroursodeoxycholic acids, significantly inhibit miR-34a/SIRT1/p53-dependent apoptosis (14). Therefore, our results suggest that activation of the miR-34a/SIRT1/p53 proapoptotic pathway is a specific effect of DCA, and not of other bile acids, resulting in apoptosis of primary rat hepatocytes, at least in the early stages.
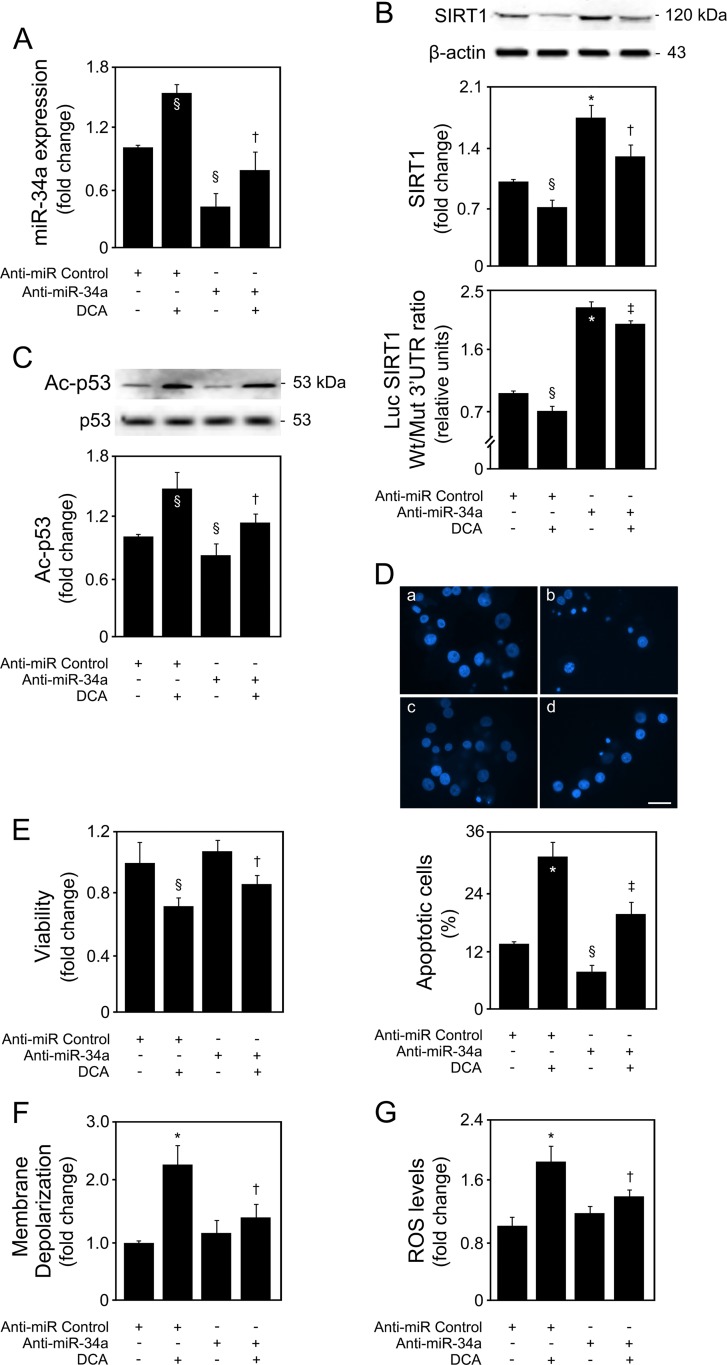
To clarify this and to determine to what extent miR-34a is essential to DCA-mediated cell death, primary rat hepatocytes were transfected with an miR-34a inhibitor in the presence or absence of DCA. As expected, miR-34a was markedly decreased in anti-miR-34a-transfected cells, compared with anti-miR Control (P < 0.05) (Fig. 5A). DCA significantly counteracted miR-34a downregulation (P < 0.05). In addition, miR-34a inhibition increased SIRT1 protein levels by >80% (P < 0.01), with DCA preventing this increase (P < 0.05) (Fig. 5B, top). These results were validated in cells cotransfected with a luciferase reporter construct containing the wild-type miR-34a binding site within the SIRT1 3′ UTR (Luc-SIRT1 Wt 3′ UTR) or a mutated sequence (Luc-SIRT1 Mut 3′ UTR), together with anti-miR-34a (Fig. 5B, bottom). p53 acetylation was decreased by almost 30% following miR-34a inhibition (P < 0.05), an effect completely reversed by DCA (P < 0.05) (Fig. 5C). Anti-miR-34a also mediated downregulation of histone H3 and PGC-1α acetylation, two additional SIRT1 targets, although to a smaller extent than p53 acetylation. Nevertheless, these effects were also counteracted by DCA (data not shown).
FIG 5.
miR-34a inhibition impairs the ability of DCA to inhibit SIRT1 and induce acetyl-p53, as well as apoptotic features. Primary rat hepatocytes were transfected with an miR-34a inhibitor (anti-miR-34a) or control (anti-miR control) and treated with 100 μM DCA or no addition (control) for 40 h as described in Materials and Methods. (A) Real-time RT-PCR analysis of miR-34a inhibition. (B) Immunoblotting of SIRT1 in cells transfected with the miR-34a inhibitor (top) and ratio between Wt and Mut miR-34a luciferase activity (bottom). Representative immunoblots are shown. Blots were normalized to endogenous β-actin. Cells were cotransfected with a reporter vector consisting of a luciferase cDNA fused to the 3′ UTR of SIRT1, containing either a wild-type (Wt) or mutant (Mut) miR-34a binding site. The cytomegalovirus-renilla luciferase vector served as an internal standard control. (C) Immunoblotting of acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to total p53. (D) Apoptotic cells were detected by Hoechst staining, and results are expressed as percentages of apoptotic cells (bottom). Representative images of control (a), DCA (b), miR-34a inhibition (c), and miR-34a inhibition with DCA treatment (d) are shown (top). Bar, 10 μm. (E) Cell viability was determined by the ApoTox-Glo triplex assay. (F) Mitochondrial permeabilization was determined by the loss in the retention of the 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3)] dye by flow cytometry. (G) ROS levels were determined as the oxidation of the H2DCFDA dye by flow cytometry. Results are expressed as mean (±standard error of the mean) fold change from 4 to 6 different experiments. §, P < 0.05, and *, P < 0.01, from anti-miR control; †, P < 0.05, and ‡, P < 0.01, from anti-miR control with DCA.
The effects of miR-34a inhibition in modulating cellular viability and apoptosis by DCA were also evaluated. miR-34a inhibition alone slightly decreased apoptosis (Fig. 5D) and increased cellular viability (Fig. 5E). More importantly, it significantly impaired the ability of DCA to induce primary rat hepatocyte apoptosis (P < 0.01) and the decrease of cellular viability (P < 0.01). Because DCA-induced apoptosis is known to result, at least in part, from disruption of mitochondrial transmembrane potential and increased ROS production (6), we also evaluated the effect of miR-34a inhibition on membrane depolarization (Fig. 5F) and oxidative stress (Fig. 5G). Interestingly, both DCA-induced membrane depolarization (P < 0.01) and ROS production (P < 0.01) were significantly inhibited in the presence of anti-miR-34a (P < 0.05 for both). Cell cycle analysis indicated that DCA slightly increased the number of cells in G1, with anti-miR-34a alone having little effect on the cell cycle. In the presence of anti-miR-34a, DCA-induced G1 was reduced, and a small increase in the number of cells in S phase was observed (data not shown).
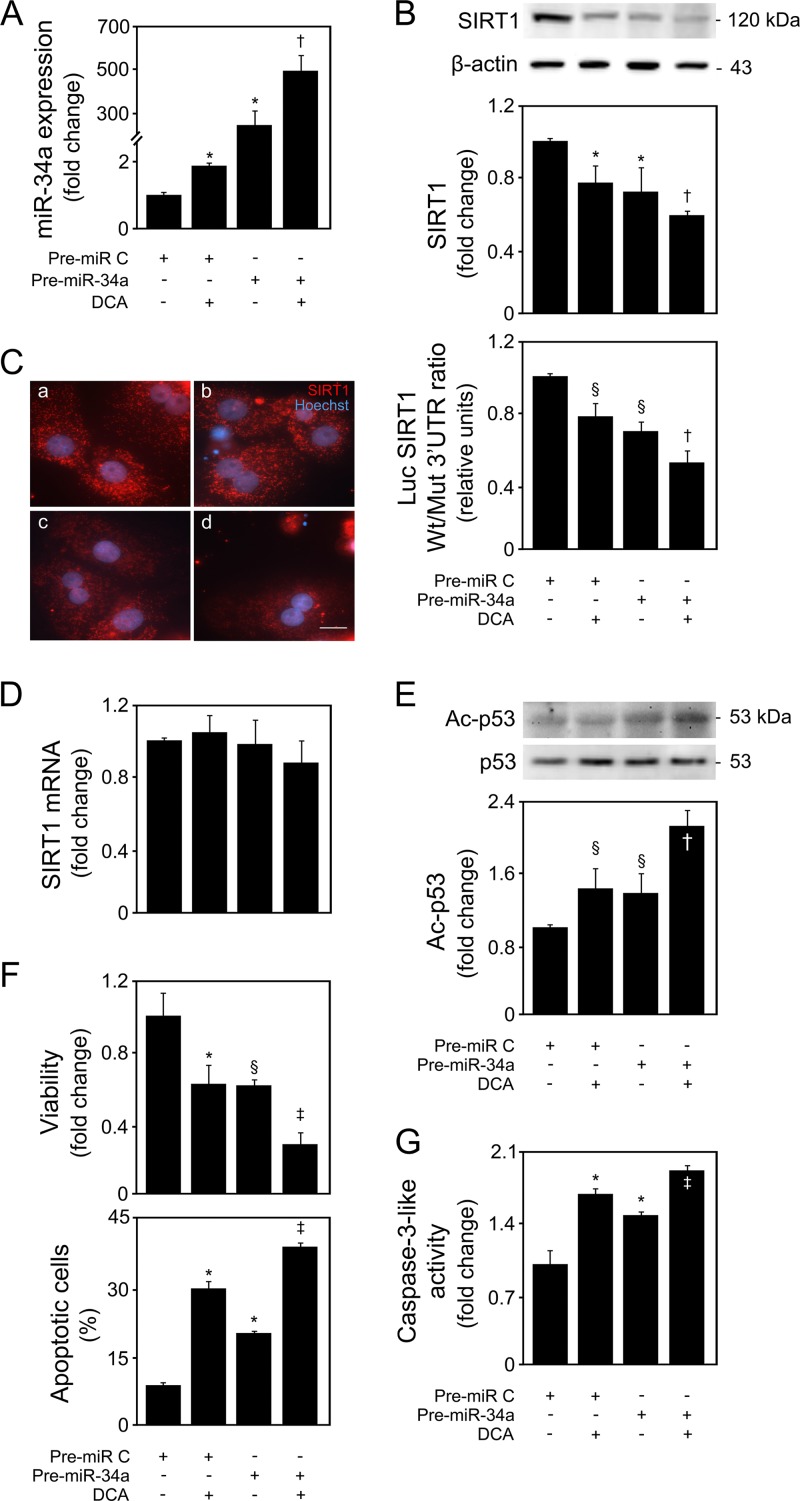
To clearly establish that DCA specifically modulates the miR-34a-dependent proapoptotic pathway, cells were alternatively transfected with an miR-34a precursor, in the presence or absence of DCA. DCA potentiated the effects of miR-34a overexpression (P < 0.05) (Fig. 6A), including those on SIRT1 inhibition (P < 0.05) (Fig. 6B). Curiously, the magnitudes of the two effects were quite different, probably indicating that the system becomes so saturated with miR-34a levels that its downstream effects are not further modulated in the same proportion as its increased expression. SIRT1 expression and localization were also assessed by immunocytochemistry in parallel with Hoechst staining. SIRT1 expression was strong in control cells, being localized in large punctae in both the cytoplasm and the nucleus (Fig. 6C). Pre-miR-34a- and DCA-treated cells displayed a decreased number and intensity of SIRT1 punctae, with lower accumulation of nuclear SIRT1. Curiously, modulation of SIRT1 by DCA appears to be mostly posttranscriptional, as DCA had little effect in cells transfected with a reporter plasmid harboring the SIRT1 promoter (data not shown). In fact, neither miR-34a overexpression nor DCA was able to significantly modulate sirt1 mRNA levels (Fig. 6D), further indicating that DCA inhibits SIRT1 protein expression via miR-34a. As for p53 acetylation, its levels were increased following miR-34a overexpression (P < 0.05) and further by DCA (P < 0.05) (Fig. 6E).
FIG 6.
DCA exacerbates miR-34a-dependent signaling and apoptosis in primary rat hepatocytes. Cells were transfected with an miR-34a precursor (pre-miR-34a) or control (pre-miR control) and treated with 100 μM DCA or no addition (control) for 40 h as described in Materials and Methods. (A) Real-time RT-PCR analysis of miR-34a overexpression. (B) Immunoblotting of SIRT1 in cells transfected with the miR-34a precursor (top) and ratio between Wt and Mut SIRT1 3′ UTR luciferase activity (bottom). Representative immunoblots are shown. Blots were normalized to endogenous β-actin. Primary rat hepatocytes were cotransfected with a reporter vector consisting of a luciferase cDNA fused to the 3′ UTR of SIRT1, containing either a wild-type (Wt) or mutant (Mut) miR-34a binding site. The cytomegalovirus-renilla luciferase vector served as an internal standard control. (C) SIRT1 localization determined by immunocytochemistry. SIRT1 staining (red) and Hoechst staining (blue) are shown as control (a), DCA (b), miR-34a overexpression (c), and miR-34a overexpression with DCA treatment (d). Bar, 10 μm. Magnification, ×630. (D) SIRT1 mRNA levels were measured by real-time RT-PCR. (E) Immunoblotting of acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to total p53. (F) Cell viability (top), determined by the ApoTox-Glo triplex assay, and apoptosis (bottom), determined by Hoechst staining. (G) Caspase-3-like activity determined by the ApoTox-Glo triplex assay. Results are expressed as mean (±standard error of the mean) percentage from 6 different experiments. §, P < 0.05, and *, P < 0.01, from pre-miR control; †, P < 0.05, and ‡, P < 0.01, from pre-miR-34a.
Finally, miR-34a overexpression significantly decreased cell viability (P < 0.05) (Fig. 6F, top) while increasing apoptosis (P < 0.01) (Fig. 6F, bottom) and caspase-3-like activity (P < 0.01) (Fig. 6G). miR-34a-overexpressing cells treated with DCA were less viable (P < 0.01) and had higher levels of caspase-3 activity (P < 0.01) and apoptosis (P < 0.01), compared with miR-34a-overexpressing cells alone, further reinforcing the notion that induction of apoptosis by DCA is largely dependent on miR-34a.
Targeting of SIRT1 by DCA via miR-34a plays a key role in its ability to activate p53 and apoptosis.
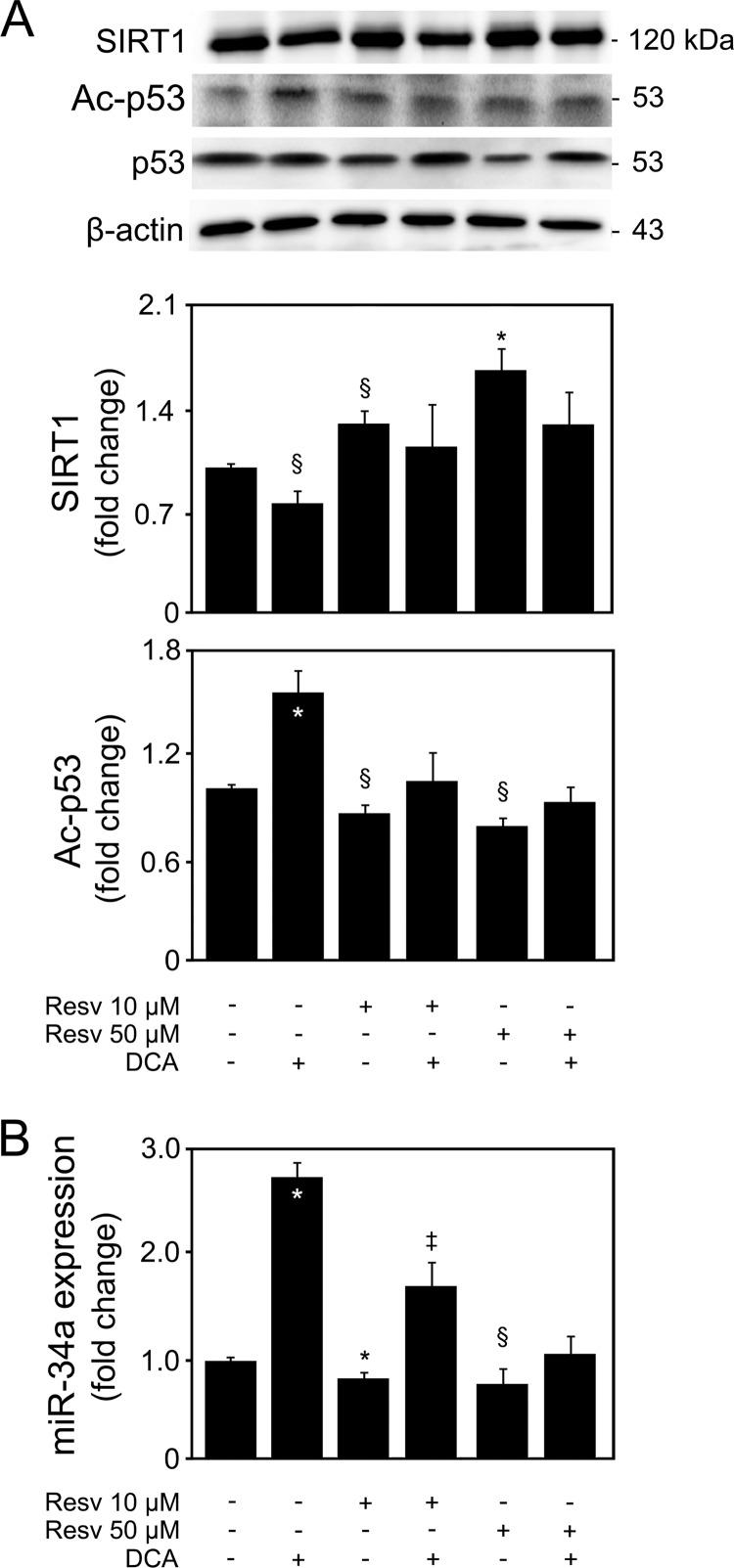
In order to determine to what extent miR-34a-dependent SIRT1 downregulation by DCA is critical for its apoptotic effects, primary rat hepatocytes were incubated with resveratrol in the presence or absence of DCA in order to overexpress SIRT1. Cells incubated with 10 and 50 μM resveratrol displayed an ∼30% (P < 0.05) and 65% (P < 0.01) increase in SIRT1 protein levels, respectively (Fig. 7A). Interestingly, DCA was no longer capable of significantly reducing SIRT1 in the presence of resveratrol (P < 0.05). Moreover, DCA-induced cell death and caspase-3 activity were also inhibited (data not shown). A word of caution is necessary when analyzing these results, as resveratrol has multiple downstream targets, including PGC-1α, peroxisome proliferator-activated receptor γ (PPARγ), NF-κB, tumor necrosis factor alpha (TNF-α), SREBP1, CIDEA, FOXO, ROS, and endothelial nitric oxide synthase (eNOS) (40). Furthermore, one cannot fully exclude the possibility that a high resveratrol concentration has off-target effects, as there is still a lot of controversy on the mechanism and specificity of direct SIRT1 activation by resveratrol. Nevertheless, SIRT1 overexpression by resveratrol also impacted the ability of DCA to acetylate p53 (Fig. 7A). In fact, DCA-induced p53 acetylation was abrogated in the presence of resveratrol (at least P < 0.05), indicating that targeting of SIRT1 by DCA mediates p53 activation. It was recently shown that in vivo silencing or genetic deletion of miR-34a reduces cell death and fibrosis following acute myocardial infarction (41). However, it may not always impair p53-induced cell cycle arrest or apoptosis (42). Still, we have also shown that ursodeoxycholic acid inhibits miR-34a-dependent apoptosis by reducing p53 transactivation (14) and that DCA-induced apoptosis is associated with p53-dependent mechanisms (7). In addition, SIRT1 also regulates p53-dependent apoptosis through deacetylating and stabilizing p53. Furthermore, p53 induces expression of miR-34a, which suppresses SIRT1, resulting in a positive-feedback loop (8, 29). Therefore, we next investigated whether SIRT1 overexpression was also affecting miR-34a activation and whether this impacted the ability of DCA to induce miR-34a. SIRT1 overexpression decreased miR-34a expression levels (P < 0.01) (Fig. 7B). More importantly, DCA-induced miR-34a was significantly reduced in the presence of resveratrol (P < 0.01), suggesting that DCA-induced p53 activation may in fact be an important regulatory step in engaging the miR-34a/SIRT1 pathway of apoptosis in primary rat hepatocytes. Again, this appears to be a specific effect, as miR-195 and miR-200a expression was not significantly affected by SIRT1 overexpression in either the presence or the absence of DCA (data not shown).
FIG 7.
Overexpression of SIRT1 impairs DCA induction of the miR-34a/SIRT1/p53 pathway in primary rat hepatocytes. Cells were isolated as described in Materials and Methods and treated with 10 μM or 50 μM resveratrol or no addition (control) for 24 h. When indicated, cells were coincubated with 100 μM DCA. (A) Immunoblotting of SIRT1 and acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to endogenous β-actin or total p53, respectively. (B) Real-time RT-PCR analysis of miR-34a. Results are expressed as mean (±standard error of the mean) fold change of at least 4 independent experiments. Resv, resveratrol. §, P < 0.05, and *, P < 0.01, from control; †, P < 0.05, and ‡, P < 0.01, from DCA alone.
DCA engages the miR-34a/SIRT1-dependent proapoptotic pathway via p53.
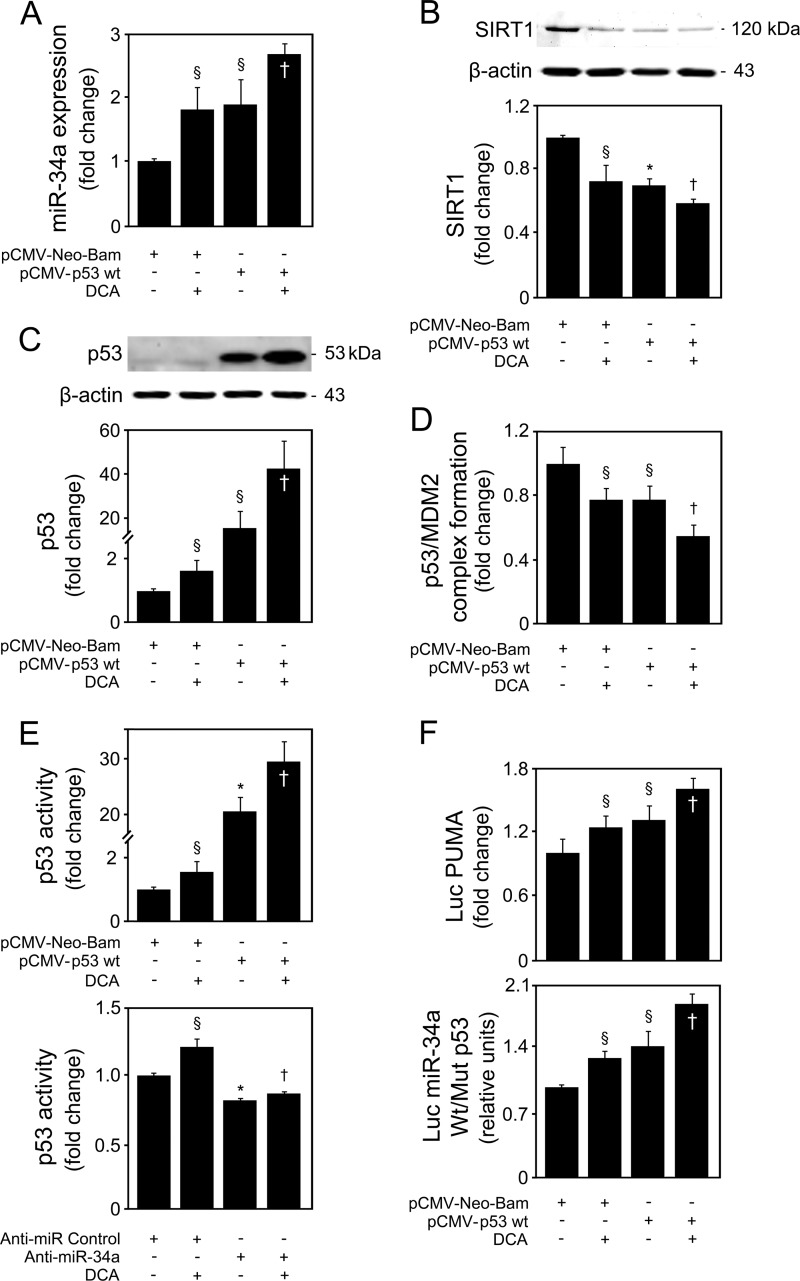
Since p53 arose as a likely target of DCA in activating the miR-34a/SIRT1 pathway, we next investigated whether p53-induced miR-34a expression was enhanced by DCA. Primary rat hepatocytes were transfected with a construct overexpressing p53 in the presence or absence of DCA. Both DCA and p53 overexpression led to a 2-fold increase in miR-34a expression (P < 0.05) (Fig. 8A). Incubation of p53-overexpressing cells with DCA further increased miR-34a expression by almost 3-fold (P < 0.05), compared with empty-vector-transfected cells. In agreement, inhibition of SIRT1 by p53 overexpression was potentiated in the presence of DCA (P < 0.05) (Fig. 8B). To elucidate whether DCA activation of the miR-34a/SIRT1/p53 proapoptotic pathway was dependent on p53, we first analyzed total p53 levels in cells incubated with DCA with or without p53 overexpression. In control-transfected cells, DCA increased total p53 levels by >50% (P < 0.05) (Fig. 8C) and, more strikingly, further increased p53 levels in p53-overexpressing cells (P < 0.05). We also investigated whether DCA was simultaneously activating p53 at the posttranscriptional level by analyzing p53/MDM2 complex formation. DCA and p53 overexpression alone inhibited complex formation by almost 30% (P < 0.05) (Fig. 8D). In addition, both synergistically decreased p53/MDM2 association by ∼50% (P < 0.05), compared with control-transfected cells. To assess whether the reduced p53/MDM2 complex formation was related to increased p53 transactivity, we used a p53 transcription factor assay kit. DCA was shown to induce p53 activity, either alone (P < 0.05) or after p53 overexpression (P < 0.05) (Fig. 8E, top). In addition, and attesting to the targeting of the p53/miR-34a/SIRT1 positive-feedback loop by DCA, our results also showed that DCA-induced p53 activity was completely abrogated in cells transfected with anti-miR-34a (P < 0.01), compared with anti-miR control-treated cells (Fig. 8E, bottom).
FIG 8.
DCA induces p53-dependent activation of the miR-34a apoptotic pathway in primary rat hepatocytes. Cells were transfected using a pCMV-Neo-Bam vector harboring the wild-type p53 human sequence (pCMV-p53 Wt) or an empty vector (pCMV-Neo-Bam) and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. (A) Real-time RT-PCR analysis of miR-34a expression. (B) Immunoblotting of SIRT1. (C) Immunoblotting of p53. Representative immunoblots are shown. Protein blots were normalized to endogenous β-actin. (D) p53/MDM2 binding as determined by the ImmunoSet p53/MDM2 complex-specific immunometric enzyme immunoassay and expressed as fold change relative to the control. (E) Levels of nuclear p53 able to bind to its DNA consensus recognition sequence, as determined by the TransAM p53 enzyme-linked immunosorbent assay with p53 overexpression (top) and miR-34a inhibition (bottom). (F) p53-dependent PUMA and miR-34a promoter activation. Primary rat hepatocytes were cotransfected with a luciferase construct with the PUMA promoter containing consensus p53 binding sites upstream of the transcription start site (Luc PUMA) (top). Alternatively, cells were cotransfected with a pGL4 reporter vector consisting of a luciferase cDNA fused to the miR-34a promoter containing either wild-type (Wt) or mutant (Mut) p53 binding sequence (bottom). Cells were also cotransfected with a cytomegalovirus-renilla luciferase vector as an internal standard. Results are expressed as mean (±standard error of the mean) fold change from 5 different experiments. §, P < 0.05, and *, P < 0.01, from pCMV-Neo-Bam alone; †, P < 0.05, from pCMV-Neo-Bam with DCA.
As an additional measure of p53 activation, we next analyzed the ability of DCA to modulate transcriptional activation of p53 target PUMA (Fig. 8F, top). p53 overexpression or DCA treatment alone increased the promoter activity of PUMA (P < 0.05) compared with control. Maximum activation was seen in cells overexpressing p53 and incubated with DCA (P < 0.05). Finally, to clearly establish that activation of the miR-34a/SIRT1/p53 pathway by DCA is largely dependent on its ability to increase p53 expression and activity, cells were cotransfected with luciferase reporter constructs under the transcriptional control of human miR-34a promoter elements containing either wild-type or mutant p53 binding sites. p53 overexpression increased wild-type miR-34a promoter activity by almost 40% (P < 0.05) (Fig. 8F, bottom). Significantly, DCA treatment increased wild-type miR-34a promoter activity either alone (P < 0.05) or in p53-overexpressing cells (P < 0.05). In addition, DCA also increased apoptosis induced by p53 overexpression (data not shown). Altogether, these results indicate that DCA activates p53 by inducing both its expression and its transcriptional activity, as well as reducing p53/MDM2 complex formation, resulting in a strong, functional activation of the miR-34a/SIRT1/p53 proapoptotic pathway.
p53/miR-34a/SIRT1-dependent apoptosis by DCA is activated by JNK1.
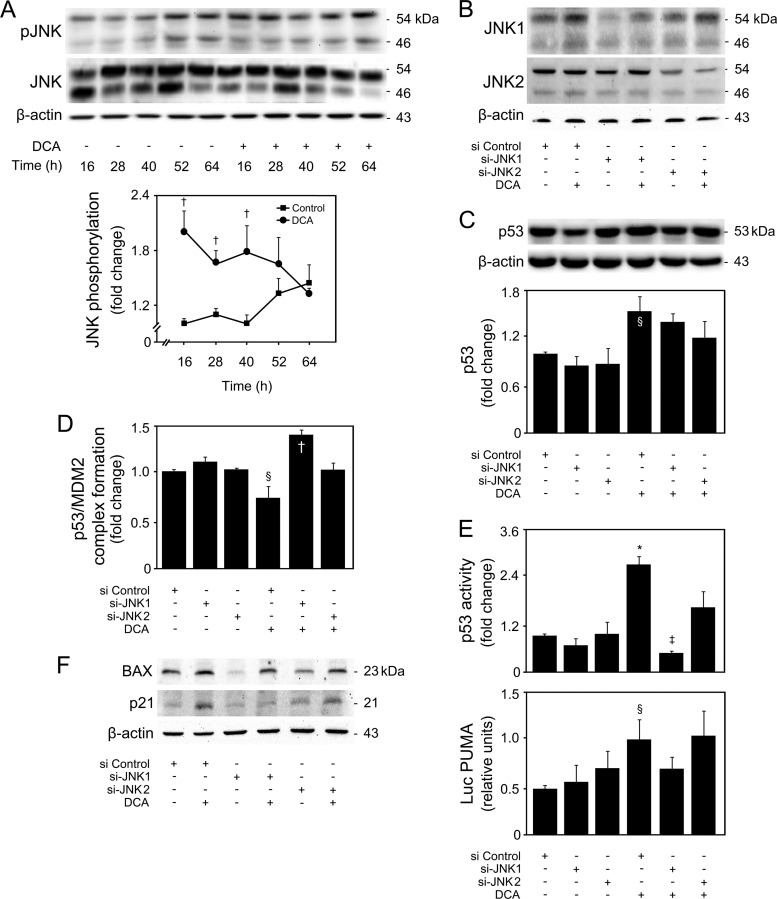
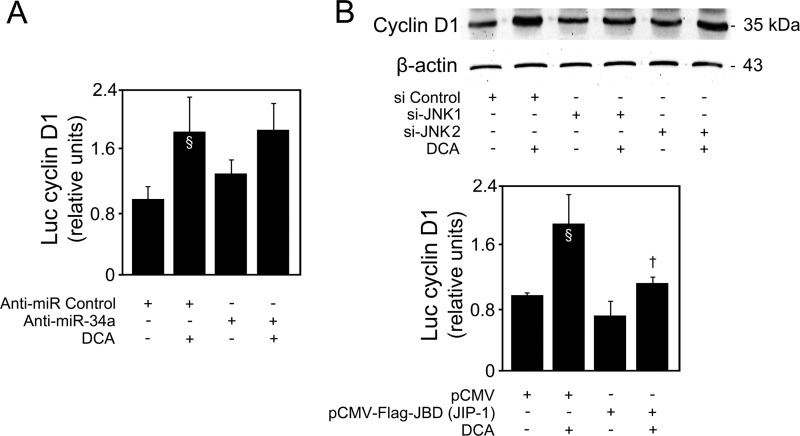
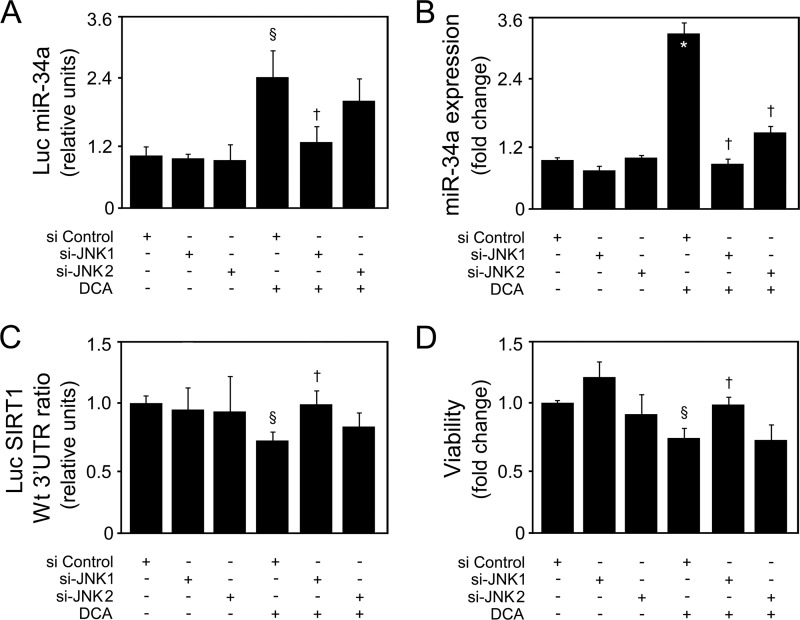
Bile acid-induced apoptosis was already shown to involve JNK1/2 activation (3, 4). In fact, JNK overactivation is one of the most common effector mechanisms of liver injury, including for DCA (43). In our model, DCA significantly induced JNK1/2 phosphorylation up until 40 h of incubation (P < 0.05) (Fig. 9A). Curiously, JNK can also, directly or indirectly, modulate p53 expression and positively influence apoptosis. In fact, JNK signaling may stabilize p53 and enhance its ability to elicit cellular apoptosis by inducing p53 phosphorylation and leading to attenuation of the p53/MDM2 interaction (44, 45). Finally, we have recently shown that JNK phosphorylation levels increase with nonalcoholic fatty liver disease severity, in parallel with miR-34a expression (14, 46). Therefore, we next analyzed whether DCA-induced JNK was the mechanistic link responsible for p53/miR-34a activation. Primary rat hepatocytes were transfected with small interfering RNAs (siRNAs) targeting either JNK1 or JNK2, as these JNK isoforms appear to have very different and opposite functions in hepatocyte death (47). Upon silencing, JNK1 levels decreased by ∼70% (P < 0.01), while JNK2 levels were inhibited by ∼65% (P < 0.01) (Fig. 9B). Importantly, siJNK1 and siJNK2 had little effect on JNK2 and JNK1 expression, respectively, attesting to their specificity. Further, DCA induced JNK1 expression (P < 0.05) but had little effect on JNK2 expression levels. JNK1 phosphorylation was also increased by DCA (data not shown). DCA-induced p53 expression (Fig. 9C) but, more significantly, p53/MDM2 dissociation (Fig. 9D) and activation (Fig. 9E and F) were completely or almost abolished in the absence of JNK1 (P < 0.05) but much less affected when JNK2 was silenced, suggesting that JNK1 plays a more significant role than does JNK2 in DCA-mediated activation of p53. In more detail, a greater effect of si-JNK1 was seen in p53 target genes PUMA (Fig. 9E, bottom) and BAX and p21 (Fig. 9F). Interestingly, we have previously reported that DCA-induced apoptosis of primary rat hepatocytes is associated with cyclin D1-dependent Bax translocation, through p53-dependent mechanisms (7). Curiously, cyclin D1 is an miR-34a target (10), and in fact, modulation of cyclin D1 by DCA appears to be miR-34a independent (Fig. 10A). Still, cyclin D1 is also a downstream target of the JNK pathway in the liver (48). In particular, our results show that JNK1 modulates activation of cyclin D1 by DCA (Fig. 10B, top and bottom), reinforcing the critical role of this kinase for the global apoptotic mechanisms induced by DCA in liver cells. Therefore, we next investigated to what extent modulation of the miR-34a/SIRT1 proapoptotic pathway by DCA was dependent on JNK. Our results showed that DCA-induced miR-34a expression, as seen in cells transfected with an miR-34a promoter Luc construct (Fig. 11A) and by real-time RT-PCR (Fig. 11B), was also inhibited by JNK1 silencing (P < 0.05). In addition, the loss of SIRT1 (Fig. 11C) and cell viability (Fig. 11D) induced by DCA was almost completely restored when JNK1, but not JNK2, was silenced (P < 0.05). Altogether, these results suggest that activation of JNK1 by DCA is responsible for engaging p53-dependent apoptotic pathways, particularly the miR-34a signaling pathway.
FIG 9.
JNK1 is responsible for DCA-induced p53 activation in primary rat hepatocytes. Cells were treated with 100 μM DCA or no addition (control) for 16, 28, 40, 52, and 64 h. Alternatively, cells were transfected with specific short interference RNA (siRNA) nucleotides designed to knock down jnk1 (si-JNK1) or jnk2 (si-JNK2) gene expression and a control siRNA containing a scrambled sequence and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. (A to C) Immunoblotting of JNK phosphorylation (pJNK) (A), JNK1 and JNK2 (B), and p53 expression (C). Representative immunoblots are shown. Blots were normalized to total JNK (pJNK) or β-actin (JNK1, JNK2, and p53). (D) p53/MDM2 binding as determined by the ImmunoSet p53/MDM2 complex-specific immunometric enzyme immunoassay and expressed as fold change relative to control. (E) Levels of nuclear p53 able to bind to its DNA consensus recognition sequence, as determined by the TransAM p53 enzyme-linked immunosorbent assay (top) and p53-dependent PUMA promoter activation (bottom). Primary rat hepatocytes were cotransfected with a luciferase construct with the PUMA promoter containing consensus p53 binding sites upstream of the transcription start site (Luc PUMA). Results were normalized for the cytomegalovirus-renilla luciferase activity. (F) Immunoblotting of BAX and p21. Representative immunoblots are shown. Blots were normalized to β-actin. Results are expressed as mean (±standard error of the mean) fold change from 3 to 7 different experiments. §, P < 0.05, and *, P < 0.01, from control or si Control; †, P < 0.05, and ‡, P < 0.01, from respective time point control or from si Control with DCA.
FIG 10.
DCA regulates cyclin D1 expression via JNK1. (A) Primary rat hepatocytes were transfected with an miR-34a inhibitor (anti-miR-34a) or control (anti-miR control) and treated with 100 μM DCA or no addition (control) for 40 h as described in Materials and Methods. Cells were cotransfected with a reporter vector consisting of a luciferase cDNA fused to the cyclin D1 promoter containing the p53 binding site. The cytomegalovirus-renilla luciferase vector served as an internal standard control. (B) Cells were transfected with specific short interference RNA (siRNA) nucleotides designed to knock down jnk1 (si-JNK1) or jnk2 (si-JNK2) gene expression and a control siRNA containing a scrambled sequence and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. Immunoblotting of cyclin D1 (top). A representative immunoblot is shown. Blots were normalized to endogenous β-actin. In parallel, cells were cotransfected with the binding domain of the JNK-interacting protein 1 (JIP-1) plasmid [pCMV-Flag-JBD (JIP-1)] and the luciferase cDNA fused to the cyclin D1 promoter construct and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods (bottom). Results were normalized for the cytomegalovirus-renilla luciferase activity. Results are expressed as mean (±standard error of the mean) fold change from at least 3 different experiments. §, P < 0.05, from control; †, P < 0.05, from DCA alone.
FIG 11.
DCA-induced p53/miR-34a signaling and apoptosis of primary rat hepatocytes are JNK1 dependent. Cells were transfected with specific short interference RNA (siRNA) nucleotides designed to knock down jnk1 or jnk2 gene expression and a control siRNA containing a scrambled sequence and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. (A) Cells were cotransfected with a pGL4 reporter vector consisting of a luciferase cDNA fused to an miR-34a promoter containing either wild-type (Wt) or mutant (Mut) p53 binding sequence. Cells were also cotransfected with a cytomegalovirus-renilla luciferase vector as an internal standard. (B) Real-time RT-PCR analysis of miR-34a expression. (C) Cells were cotransfected with a reporter vector consisting of a luciferase cDNA fused to the 3′ UTR of SIRT1, containing either a wild-type (Wt) or mutant (Mut) miR-34a binding site. Ratios of Wt to Mut miR-34a luciferase activities are displayed. Cells were also cotransfected with a cytomegalovirus-renilla luciferase vector as an internal standard. (D) Viability was measured using the ApoTox-Glo triplex assay. Results are expressed as mean (±standard error of the mean) fold change from 7 different experiments. §, P < 0.05, and *, P < 0.01, from si Control; †, P < 0.05, from si Control with DCA.
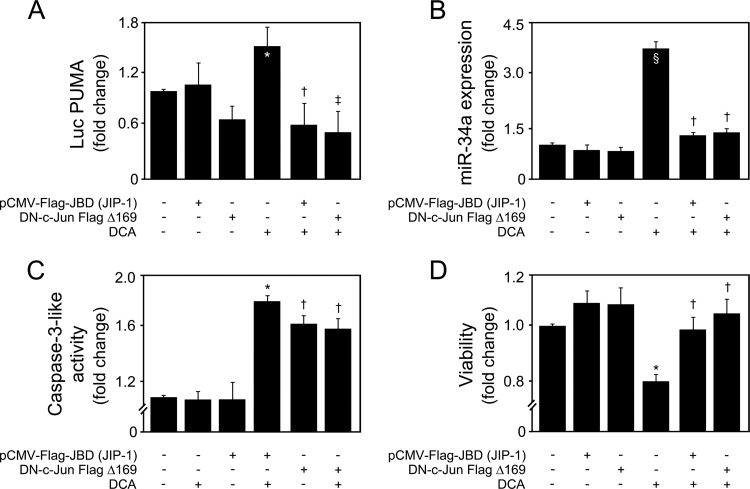
To better clarify the mechanisms by which DCA-induced JNK activation was promoting p53/miR-34a-dependent apoptosis, we additionally used two dominant interfering forms of the JNK signaling pathway, pCMV-Flag-JBD (JIP-1) (26) and DN-c-Jun FlagΔ169 (27). The binding domain fragment of scaffolding protein JIP-1 has been shown to bind JNK1/2, leading to its cytoplasmic retention and inhibition of JNK-regulated gene expression. As for DN-c-Jun FlagΔ169, it lacks the c-Jun transactivation domain. When overexpressed, DN-c-Jun FlagΔ169 competes with endogenous c-Jun for binding to DNA regulatory elements, thus inhibiting c-Jun-dependent transcription. As expected, cells transfected with constructs encoding either pCMV-Flag-JBD (JIP-1) or DN-c-Jun FlagΔ169, in the presence or absence of DCA, had no effects on JNK levels (data not shown). However, pCMV-Flag-JBD (JIP-1) was able to inhibit DCA-induced p53 activation and miR-34a expression (P < 0.05) (Fig. 12A and B, respectively), as well as caspase-3 activation and cell death (P < 0.05) (Fig. 12C and D, respectively). Interestingly, DN-c-Jun FlagΔ169 was as effective as pCMV-Flag-JBD (JIP-1) in inhibiting DCA-induced p53/miR-34a-dependent signaling and apoptosis. Therefore, these results further suggest that c-Jun also plays a key role in mediating DCA-induced p53/miR-34a/SIRT1, leading to increased cellular caspase activation and apoptosis.
FIG 12.
JNK and c-Jun act as important triggers of the miR-34a/SIRT1/p53 proapoptotic pathway by DCA. Primary rat hepatocytes were transfected with either the binding domain of JNK-interacting protein 1 (JIP-1) [pCMV-Flag-JBD (JIP-1)] or dominant negative DN-c-Jun FlagΔ169 plasmid and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. (A) Cells were cotransfected with a luciferase construct with the PUMA promoter containing consensus p53 binding sites upstream of the transcription start site (Luc PUMA). Results were normalized for the cytomegalovirus-renilla luciferase activity. (B) Real-time RT-PCR analysis of miR-34a expression. (C and D) Caspase-3-like activity (C) and viability (D) measured using the ApoTox-Glo triplex assay. Results are expressed as mean (±standard error of the mean) fold change from 5 different experiments. §, P < 0.05, and *, P < 0.01, from control; †, P < 0.05, and ‡, P < 0.01, from DCA alone.
c-Jun is a critical target of JNK in mediating activation of the p53/miR-34a/SIRT1 pathway and apoptosis by DCA.
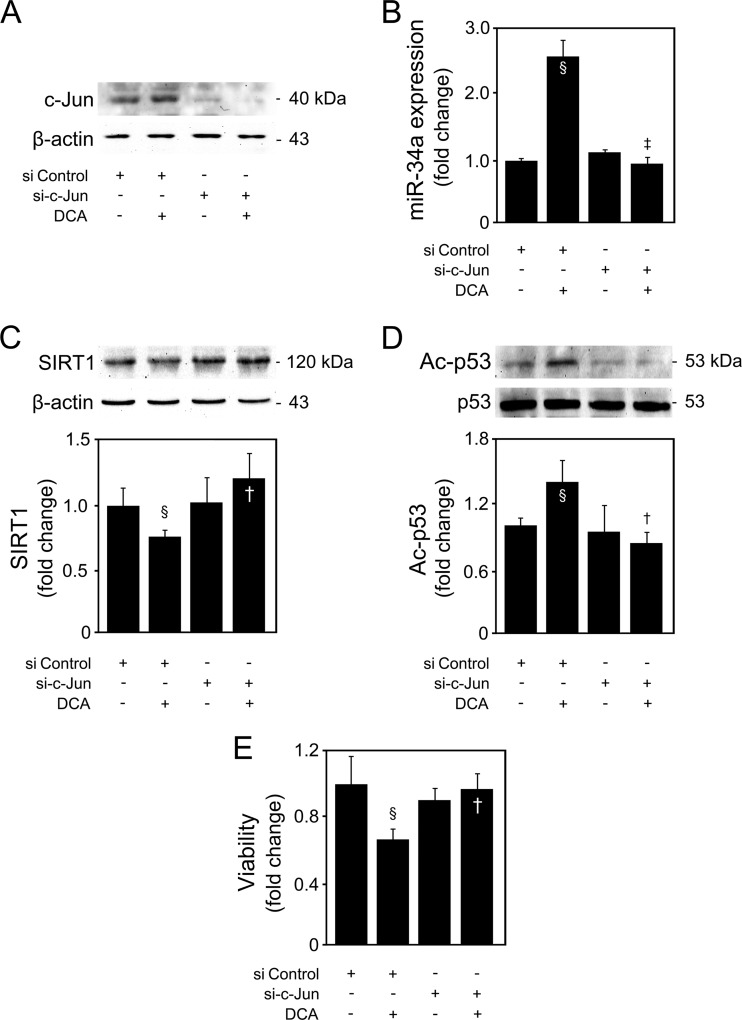
c-Jun represents one of the main targets of JNK. Interestingly, the apparent opposite or, rather, differential effects of JNK1 and JNK2 on cell death and proliferation have been proposed to be the consequence of opposite actions on c-Jun (49). While JNK2 appears to bind to c-Jun and target it for degradation, JNK1 appears to be more effective in activating and stabilizing c-Jun (50). In addition, p53 can be activated by JNK via c-Jun (51). Primary rat hepatocytes were transfected with a c-Jun siRNA, in the presence or absence of DCA. Silencing suppressed c-Jun protein expression levels by 75% (P < 0.05), compared with control cells transfected with a negative-control siRNA (Fig. 13A). Furthermore, DCA-induced c-Jun was completely abolished in cells transfected with c-Jun siRNA. Importantly, DCA-induced miR-34a (Fig. 13B) and p53 acetylation (Fig. 13C), as well as SIRT1 inhibition (Fig. 13D), were completely abolished in the absence of c-Jun (at least P < 0.05). Finally, DCA-reduced cell viability was also rescued after c-Jun inhibition (P < 0.05) (Fig. 13E). These results corroborate our previous findings using the DN-c-Jun FlagΔ169 dominant negative, further establishing c-Jun as a critical target for JNK1 during DCA-induced activation of the p53/miR-34a/SIRT1 pathway and apoptosis in primary rat hepatocytes.
FIG 13.
c-Jun is a key target of JNK1 during DCA-induced p53/miR-34a proapoptotic signaling in primary rat hepatocytes. Cells were transfected with a specific siRNA designed to knock down c-jun gene expression (si-c-Jun) or a negative-control siRNA and treated with 100 μM DCA or no addition (control) for 24 h as described in Materials and Methods. (A) Immunoblotting of c-Jun. A representative immunoblot is shown. Blots were normalized to β-actin. (B) Real-time RT-PCR analysis of miR-34a expression. (C and D) Immunoblotting of SIRT1 (C) and acetyl-p53 (Ac-p53) (D). Representative immunoblots are shown. Blots were normalized to endogenous β-actin or total p53, respectively. (E) Viability was measured using the ApoTox-Glo triplex assay. Results are expressed as mean (±standard error of the mean) fold change from 4 different experiments. §, P < 0.05, from si Control; †, P < 0.05, and ‡, P < 0.01, from si Control with DCA.
The JNK1/p53/miR-34a/SIRT1 pathway is activated by DCA in the rat liver in vivo, correlating with induction of apoptosis.
To clearly establish the physiological relevance of the JNK1/p53/miR-34a/SIRT1 pathway in mediating apoptosis by DCA, rats were administered this bile acid. Total plasma bile acid levels increased >2-fold in DCA-treated animals (P < 0.01), while DCA increased ∼15-fold (P < 0.01). This correlates with previous data showing that DCA plasma levels in rats, under physiological conditions, are low (∼5 μM) (38) but can strikingly increase severalfold in the context of different liver diseases (15, 52). Our results show that liver miR-34a expression was increased by >2.5-fold in the group administered DCA (P < 0.01) (Fig. 14A). In accordance with our in vitro results, DCA-treated animals do not present statistically significant changes in other miRNAs, such as miR-122, miR-143, miR-145, miR-195, and miR-200a (Fig. 15). Consistent with downstream modulation of the pathway, SIRT1 protein expression levels decreased by almost 50% (P < 0.05) while, inversely, acetylated p53 levels increased ∼80% (P < 0.05) (Fig. 14B). Of note, SIRT1 mRNA expression levels did not differ between DCA and control groups (data not shown). We next evaluated whether changes in the JNK pathway recapitulated those observed in vitro. In fact, JNK1 expression levels were found increased by 50% in the DCA group, compared with controls (P < 0.05) (Fig. 14C). No significant changes were observed between the two animal groups at the JNK2 level, although protein expression was slightly decreased in the DCA group. More importantly, c-Jun expression levels were induced by ∼50% in the DCA group (P < 0.05), further reinforcing the role of this transcription factor in downstream JNK1 activation. Finally, apoptosis was evaluated by caspase-3 activation and the number of TUNEL-positive cells. Active caspase-3 was increased by 40% in the liver of DCA-treated animals, as determined in total protein extracts (Fig. 14D, left) and by immunohistochemistry (Fig. 14D, right) (P < 0.05 for both). As expected, in agreement with our previous findings (53), the number of TUNEL-positive cells was significantly increased in the liver of DCA-treated animals (P < 0.01) (Fig. 14E). These results strongly suggest that the JNK1/p53/miR-34a/SIRT1 pathway constitutes a mechanistic circuit activated by DCA in vivo, contributing to induction of apoptosis.
FIG 14.
DCA activates the JNK1/c-Jun/p53/miR-34a/SIRT1 proapoptotic pathway in vivo. Wistar male rats were treated with 250 mg/kg/day of DCA by oral gavage for 5 days before sacrifice. In parallel, rats in the control group were administered water as described in Materials and Methods. (A) Northern blot (top) and real-time RT-PCR analysis of miR-34a. A representative blot is shown. Blots were normalized for total RNA input. (B) Immunoblotting of SIRT1 and acetyl-p53 (Ac-p53). Representative immunoblots are shown. Blots were normalized to endogenous β-actin or total p53, respectively. (C) Immunoblotting of JNK1, JNK2, and c-Jun. Representative immunoblots are shown. Blots were normalized to endogenous β-actin. (D) Caspase-3-like activity (left) measured using the ApoTox-Glo triplex assay and active caspase-3 localization (right), determined in liver sections by immunohistochemistry. Active caspase-3 staining (red) and Hoechst staining (blue) are shown in liver sections from representative control (a) and DCA-treated (b) animals. Bar, 10 μm. Magnification, ×630. (E) Fluorescent TUNEL assay in liver slices. TUNEL-positive cells (red) and Hoechst staining (blue) are shown in liver slices from representative control (a) and DCA-treated (b) animals. Bar, 10 μm. Results are expressed as mean (±standard error of the mean) fold change from 6 control rats and 6 DCA-treated rats. §, P < 0.05, and *, P < 0.01, from control.
FIG 15.
DCA does not modulate miR-195, -200a, -143, -145, and -122 expression in vivo. Wistar male rats were treated with 250 mg/kg/day of DCA by oral gavage for 5 days before sacrifice. In parallel, rats in the control group were administered water as described in Materials and Methods. Real-time RT-PCR and Northern blot analysis of miR-195, miR-200a, miR-143, miR-145, and miR-122 are shown. Results are expressed as mean (±standard error of the mean) fold change from 6 control rats and 6 DCA-treated rats.
DISCUSSION
Bile acids are essential to facilitate the digestion and absorption of fat. Despite the physiological properties, excessive accumulation of bile acids is associated with cytotoxic effects. In fact, previous studies have argued that conjugated bile acids, including DCA, may induce apoptosis in hepatocytes upon activation of death receptors (2). In addition, DCA induces apoptosis by impairing mitochondrial function and leading to cytochrome c release into the cytosol (12). Both JNK and p53 may also represent important mediators of DCA-induced cytotoxicity (7, 43). However, the exact network of pathways involved in DCA-induced hepatocyte apoptosis remains poorly known. In particular, no studies have yet explored miRNA expression changes and function during cell death by cytotoxic bile acids. In this study, we investigated whether DCA modulates the miR-34a/SIRT1/p53 proapoptotic pathway in primary rat hepatocytes and in the rat liver in vivo and whether JNK1 may link this pathway to already-described mechanistic actions of DCA. Our results show that by activating p53, DCA induces miR-34a transcription and inhibition of SIRT1, which translates into increased caspase activation and apoptosis of primary rat hepatocytes. Importantly, JNK1 and c-Jun act as crucial targets of DCA in mediating p53/miR-34a activation and downstream apoptosis.
miR-34a-dependent apoptosis has already been shown to occur through both p53-dependent and -independent mechanisms (54). In addition, miR-34a inhibits translation of SIRT1, a NAD-dependent deacetylase, with antiapoptotic properties (9). Our results show that DCA induces miR-34a expression, abrogates SIRT1 expression, and increases p53 acetylation in primary rat hepatocytes, providing a new mechanistic link for its proapoptotic properties. Of note, because several other miRNAs are not affected by low doses of DCA, both in vitro and in vivo, it appears that global miRNA biogenesis and processing are not greatly affected by this bile acid. Still, the exact mechanisms responsible for the differential modulation of miRNAs by DCA remain incomplete.
Interestingly, the effect of DCA on the miR-34a-dependent pathway appears to be both dose and time dependent, as DCA-induced miR-34a and downstream targets are significant only for concentrations higher than 50 μM DCA and start to attenuate after 52 h of incubation. This is consistent with the intrinsic nature of miRNA-mediated regulation and with the fact that bile acids, including DCA, sustain apoptosis in primary hepatocytes within a relatively short time (55). In fact, after 64 h of incubation, DCA-induced cell death was predominantly necrotic in nature. Finally, activation of miR-34a during apoptosis of primary hepatocytes by DCA appears to be an early event, as inhibition of miR-34a also impairs the ability of DCA to induce membrane depolarization and ROS production. As we have recently shown, miR-34a overexpression leads to cell death and apoptosis in primary rat hepatocytes (14). In this study, we unequivocally characterized not only miR-34a but also its target SIRT1 as a determinant player during DCA-induced apoptosis in primary rat hepatocytes. In fact, the functional role and significance of the p53/miR-34a axis have been expanding in these last years, not only in apoptosis (56) but in cancer as well (57–59).
In addition, we have also previously shown that p53-induced apoptosis involves impairment of MDM2-dependent shuttling of p53 to the cytoplasm in hepatocytes (60). This results in higher levels of nuclear p53 capable of transactivating target genes, which could include miR-34a. Moreover, p53 acetylation, which usually occurs in response to DNA damage and genotoxic stress and which we show to also occur in hepatocytes in response to DCA, is indispensable for p53 transcriptional activity. Upon p53 acetylation, the p53-MDM2 interaction is disrupted and p53 is able to transcribe genes involved in apoptosis (61). Our results show that DCA increases p53 expression, transactivity, and DNA binding activities, in parallel with p53 acetylation, in a positive-feedback loop contributing to augmented p53 activation. More importantly, it appears that by doing so, DCA is engaging miR-34a-dependent apoptotic signaling. In fact, this may explain why DCA is more capable of inducing miR-34a expression when miR-34a itself is overexpressed, compared with DCA alone. As for the mechanisms by which DCA induces p53 expression and transactivity, our results show that they are largely dependent on JNK. In fact, JNK has been previously shown to be able to activate p53 (44, 51), and our results demonstrate that treatment of primary rat hepatocytes with DCA significantly induces JNK phosphorylation. Interestingly, in the absence of DCA, JNK phosphorylation, activation of the miR-34a/SIRT1/p53 pathway, and cell death all increased in primary hepatocytes cultured from 52 to 64 h. This is consistent with the notion that hepatocytes start to dedifferentiate upon isolation and culture. After several days in culture, oxidative stress-induced apoptosis becomes a major event (62). Under these conditions, extramitochondrial glutathione depletion may lead to a sustained activation of JNK, which is then capable of activating the p53/miR-34a proapoptotic pathway.
JNK has three isoforms: JNK1, -2, and -3. While JNK1 and JNK2 are extensively expressed in mammalian tissues, including hepatocytes, expression of JNK3 is restricted to the brain and testis (63). Each isoform has overlapping or distinct roles in liver pathophysiology. In particular, studies using primary mouse hepatocytes have suggested that DCA-induced JNK1 signaling is mostly cytotoxic, while DCA-induced JNK2 plays more of a protective role against apoptosis (43). In fact, our results showed that JNK1 silencing abrogated DCA-induced p53/miR-34a expression and activation, thereby diminishing apoptosis. In contrast, JNK2 silencing did not significantly repress the DCA-induced p53/SIRT1/miR-34a proapoptotic pathway, despite a tendency to inhibit miR-34a expression in cells incubated with DCA, or DCA inhibition of the p53/MDM2 complex formation. Curiously, under specific settings, JNK2 may activate apoptotic players in the liver (64). Therefore, it is possible that JNK2 plays a redundant role in activating p53/miR-34a, but only when in the presence of an apoptotic stimulus; while its silencing alone results in a very slight inhibition of p53/miR-34a/SIRT1-associated apoptosis, it no longer does so in cells incubated with DCA. From a mechanistic point of view, the differential activation of JNK1 versus JNK2 by DCA remains unknown. Some possibilities exist; in JNK2 but not JNK1, a rigid β-strand conformation exists in its activation loop, rendering this protein much less activated by MEKK1 and MKK4 signals (65). Therefore, DCA could be more selectively activating JNK1 through MEKK1 and MKK4. Of note, JNK1 may also be activated through stimulation of specific upstream kinases, perturbation of the protein ubiquitination cascade, and inactivation of the dual-specific JNK1 phosphatases (66), where DCA could be playing a role. Studies looking in more detail at the greater selectivity of DCA for JNK1 are warranted.
Nevertheless, our findings suggest that DCA-induced JNK1 activation is a critical apoptotic mechanism of DCA, upstream of p53 and miR-34a activation and SIRT1 inhibition. In addition, JNK1, but not JNK2, phosphorylates c-Jun, a critical member of the activator protein 1 transcription factor complex, which can then induce expression of several death mediators (67). In fact, c-Jun appeared to be the next immediate target transducing JNK1 effects, as cells transfected with the DN-c-Jun FlagΔ169 plasmid showed an almost complete abrogation of DCA-induced p53/miR-34a apoptotic signaling. c-Jun itself plays a critical role during apoptosis (68), and studies in genetically modified mice have indicated that the effects of JNK activation can be recapitulated by c-Jun activation (66). Our results show that c-Jun represents a critical target of JNK1 during DCA activation of the p53/miR-34a/SIRT1 pathway and apoptosis. Still, it remains possible that JNK1 regulates miR-34a directly, as a recent study has shown that the miR-34a promoter contains an activator protein 1 site, which appears to be required for maximal transactivation of miR-34a (11, 69). Nevertheless, and all together, DCA-induced JNK1 phosphorylation appears to induce both p53 expression and activation, converging in a strong and functional engagement of the miR-34a-dependent apoptotic pathway in primary rat hepatocytes.
Importantly, our findings also support a role for the JNK1/c-Jun-mediated activation of the p53/miR-34a/SIRT1 circuit during apoptosis by DCA in vivo, attesting to the physiological relevance of this pathway. Based on these findings, it may be hypothesized that activation of miR-34a-dependent cytotoxicity by DCA may play an important role in liver disease. We have previously shown that patients with steatohepatitis display higher levels of DCA than do control patients (15). These may arise from a secretory dysfunction associated with hepatic injury in steatohepatitis patients. Whatever the cause, increased bile acids may aggravate injury, thus creating a vicious cycle. Remarkably, in cellular models, free fatty acids activate the JNK1/c-Jun pathway, inducing PUMA transcriptional upregulation with subsequent Bax activation as integral steps promoting saturated free fatty acid-mediated apoptosis (70). In addition, we have also shown that apoptosis is a prominent feature in patients with alcoholic steatohepatitis and nonalcoholic steatohepatitis (71). Thus, hepatocytes in steatohepatitis and many other liver diseases exhibit receptor-dependent and -independent forms of cell death, both of which are associated with mitochondrial dysfunction, in the same fashion as DCA-induced apoptosis in hepatocytes. Furthermore, we have recently demonstrated that JNK could be a mechanistic link between insulin resistance and apoptosis during nonalcoholic fatty liver disease progression (46) and that the miR-34a/SIRT1/p53 proapoptotic pathway is increased in more severe stages of nonalcoholic fatty liver disease (14). In addition, SIRT1 protein is degraded in response to JNK1 activation, thus contributing to hepatic steatosis in obese mice (72). In the light of this finding, it is possible that DCA-induced SIRT1 degradation is also occurring through JNK1 directly, in parallel with miR-34a-mediated inhibition. Finally, a deficiency of JNK1 but not JNK2 has been shown to improve insulin sensitivity and decrease adiposity in different animal models of obesity (73). Therefore, the functional relevance of the JNK1/p53/miR-34a/SIRT1 proapoptotic pathway in steatohepatitis in vivo and other liver diseases should be further exploited. Strategies aimed at antagonizing JNK1/p53- or miR-34a-dependent pathways may prove useful in ameliorating pathologies involving bile acid-associated cytotoxicity.
In conclusion, the specific targeting of miR-34a by DCA in primary rat hepatocytes results in decreased SIRT1 expression and increased p53 acetylation and apoptosis. The mechanism by which DCA induces miR-34a expression appears to occur, at least in part, through JNK1/c-Jun activation that increases p53 expression and transactivity. By adding more pieces to the puzzle, these findings underscore new targets for the development of novel drugs to ameliorate liver diseases, particularly those involving bile acid-, apoptosis-, or inflammation-associated cytotoxicity.
ACKNOWLEDGMENTS
This work was supported by grants PTDC/SAU-OSM/102099/2008, PTDC/SAU-ORG/111930/2009, and Pest-OE/SAU/UI4013/2011 and fellowships SFRH/BD/60521/2009 (D.M.S.F.), SFRH/BD/91119/2012 (M.B.A.), and SFRH/BD/88212/2012 (P.M.R.) from FCT, Lisbon, Portugal.
We thank R. W. A. Wayne for SIRT1-luciferase reporters, C. J. Lowenstein for SIRT1-3′-UTR-luciferase reporters, G. J. Hannon for miR-34a-luciferase reporters, and B. Vogelstein for p53 expression and PUMA luciferase vectors. We thank João Rocha for his expertise in animal oral gavage. We also thank all members of the laboratory for insightful discussions.
Footnotes
Published ahead of print 13 January 2014
REFERENCES
- 1.Schmucker DL, Ohta M, Kanai S, Sato Y, Kitani K. 1990. Hepatic injury induced by bile salts: correlation between biochemical and morphological events. Hepatology 12:1216–1221. 10.1002/hep.1840120523 [DOI] [PubMed] [Google Scholar]
- 2.Higuchi H, Gores GJ. 2003. Bile acid regulation of hepatic physiology: IV. Bile acids and death receptors. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G734–G738. 10.1152/ajpgi.00491.2002 [DOI] [PubMed] [Google Scholar]
- 3.Higuchi H, Grambihler A, Canbay A, Bronk SF, Gores GJ. 2004. Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J. Biol. Chem. 279:51–60. 10.1074/jbc.M309476200 [DOI] [PubMed] [Google Scholar]
- 4.Gupta S, Natarajan R, Payne SG, Studer EJ, Spiegel S, Dent P, Hylemon PB. 2004. Deoxycholic acid activates the c-Jun N-terminal kinase pathway via FAS receptor activation in primary hepatocytes. Role of acidic sphingomyelinase-mediated ceramide generation in FAS receptor activation. J. Biol. Chem. 279:5821–5828. 10.1074/jbc.M310979200 [DOI] [PubMed] [Google Scholar]
- 5.Yang JI, Yoon JH, Myung SJ, Gwak GY, Kim W, Chung GE, Lee SH, Lee SM, Kim CY, Lee HS. 2007. Bile acid-induced TGR5-dependent c-Jun-N terminal kinase activation leads to enhanced caspase 8 activation in hepatocytes. Biochem. Biophys. Res. Commun. 361:156–161. 10.1016/j.bbrc.2007.07.001 [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues CM, Sola S, Sharpe JC, Moura JJ, Steer CJ. 2003. Tauroursodeoxycholic acid prevents Bax-induced membrane perturbation and cytochrome C release in isolated mitochondria. Biochemistry 42:3070–3080. 10.1021/bi026979d [DOI] [PubMed] [Google Scholar]
- 7.Castro RE, Amaral JD, Sola S, Kren BT, Steer CJ, Rodrigues CM. 2007. Differential regulation of cyclin D1 and cell death by bile acids in primary rat hepatocytes. Am. J. Physiol. Gastrointest. Liver Physiol. 293:G327–G334. 10.1152/ajpgi.00093.2007 [DOI] [PubMed] [Google Scholar]
- 8.Yamakuchi M, Ferlito M, Lowenstein CJ. 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 105:13421–13426. 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks CL, Gu W. 2009. How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 9:123–128. 10.1038/nrc2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermeking H. 2010. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17:193–199. 10.1038/cdd.2009.56 [DOI] [PubMed] [Google Scholar]
- 11.Ichimura A, Ruike Y, Terasawa K, Shimizu K, Tsujimoto G. 2010. MicroRNA-34a inhibits cell proliferation by repressing mitogen-activated protein kinase kinase 1 during megakaryocytic differentiation of K562 cells. Mol. Pharmacol. 77:1016–1024. 10.1124/mol.109.063321 [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues CM, Fan G, Wong PY, Kren BT, Steer CJ. 1998. Ursodeoxycholic acid may inhibit deoxycholic acid-induced apoptosis by modulating mitochondrial transmembrane potential and reactive oxygen species production. Mol. Med. 4:165–178 [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues CM, Ma X, Linehan-Stiers C, Fan G, Kren BT, Steer CJ. 1999. Ursodeoxycholic acid prevents cytochrome c release in apoptosis by inhibiting mitochondrial membrane depolarization and channel formation. Cell Death Differ. 6:842–854. 10.1038/sj.cdd.4400560 [DOI] [PubMed] [Google Scholar]
- 14.Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM. 2013. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 58:119–125. 10.1016/j.jhep.2012.08.008 [DOI] [PubMed] [Google Scholar]
- 15.Aranha MM, Cortez-Pinto H, Costa A, da Silva IB, Camilo ME, de Moura MC, Rodrigues CM. 2008. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur. J. Gastroenterol. Hepatol. 20:519–525. 10.1097/MEG.0b013e3282f4710a [DOI] [PubMed] [Google Scholar]
- 16.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499:97–101. 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues CM, Kren BT, Steer CJ, Setchell KD. 1995. The site-specific delivery of ursodeoxycholic acid to the rat colon by sulfate conjugation. Gastroenterology 109:1835–1844. 10.1016/0016-5085(95)90750-5 [DOI] [PubMed] [Google Scholar]
- 18.Kren BT, Rodrigues CM, Setchell KD, Steer CJ. 2001. Modulation of steady-state messenger RNA levels in the regenerating rat liver with bile acid feeding. Liver Transpl. 7:321–334. 10.1053/jlts.2001.23062 [DOI] [PubMed] [Google Scholar]
- 19.Mariash CN, Seelig S, Schwartz HL, Oppenheimer JH. 1986. Rapid synergistic interaction between thyroid hormone and carbohydrate on mRNAS14 induction. J. Biol. Chem. 261:9583–9586 [PubMed] [Google Scholar]
- 20.Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CM. 2010. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am. J. Physiol. Gastrointest. Liver Physiol. 299:G887–G897. 10.1152/ajpgi.00216.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park JS, Surendran S, Kamendulis LM, Morral N. 2011. Comparative nucleic acid transfection efficacy in primary hepatocytes for gene silencing and functional studies. BMC Res. Notes 4:8. 10.1186/1756-0500-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong S, Salazar G, Patrushev N, Alexander RW. 2011. FoxO1 mediates an autofeedback loop regulating SIRT1 expression. J. Biol. Chem. 286:5289–5299. 10.1074/jbc.M110.163667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447:1130–1134. 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912–915. 10.1126/science.2144057 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Luo W, Reiser G. 2007. Proteinase-activated receptor-1 and -2 induce the release of chemokine GRO/CINC-1 from rat astrocytes via differential activation of JNK isoforms, evoking multiple protective pathways in brain. Biochem. J. 401:65–78. 10.1042/BJ20060732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. 1997. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science 277:693–696. 10.1126/science.277.5326.693 [DOI] [PubMed] [Google Scholar]
- 27.Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. 1995. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron 14:927–939. 10.1016/0896-6273(95)90331-3 [DOI] [PubMed] [Google Scholar]
- 28.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. 2010. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 38:e98. 10.1093/nar/gkp1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamakuchi M, Lowenstein CJ. 2009. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle 8:712–715. 10.4161/cc.8.5.7753 [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. 2011. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc. Res. 92:75–84. 10.1093/cvr/cvr145 [DOI] [PubMed] [Google Scholar]
- 31.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. 2011. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J. Biol. Chem. 286:25992–26002. 10.1074/jbc.M111.229401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. 2012. MiR-122 in hepatic function and liver diseases. Protein Cell 3:364–371. 10.1007/s13238-012-2036-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. 2013. MicroRNAs in metabolic disease. Arterioscler. Thromb. Vasc. Biol. 33:178–185. 10.1161/ATVBAHA.112.300144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuhara T, Matsuura Y. 2013. Role of miR-122 and lipid metabolism in HCV infection. J. Gastroenterol. 48:169–176. 10.1007/s00535-012-0661-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. 2012. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122:2871–2883. 10.1172/JCI63539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. 2012. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122:2884–2897. 10.1172/JCI63455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T, Braun T, Seibler J, Bruning JC. 2011. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 13:434–446. 10.1038/ncb2211 [DOI] [PubMed] [Google Scholar]
- 38.Kren BT, Rodrigues CM, Setchell KD, Steer CJ. 1995. Posttranscriptional regulation of mRNA levels in rat liver associated with deoxycholic acid feeding. Am. J. Physiol. 269:G961–G973 [DOI] [PubMed] [Google Scholar]
- 39.Ignacio Barrasa J, Olmo N, Perez-Ramos P, Santiago-Gomez A, Lecona E, Turnay J, Antonia Lizarbe M. 2011. Deoxycholic and chenodeoxycholic bile acids induce apoptosis via oxidative stress in human colon adenocarcinoma cells. Apoptosis 16:1054–1067. 10.1007/s10495-011-0633-x [DOI] [PubMed] [Google Scholar]
- 40.Poulsen MM, Jorgensen JO, Jessen N, Richelsen B, Pedersen SB. 2013. Resveratrol in metabolic health: an overview of the current evidence and perspectives. Ann. N. Y. Acad. Sci. 1290:74–82. 10.1111/nyas.12141 [DOI] [PubMed] [Google Scholar]
- 41.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. 2013. MicroRNA-34a regulates cardiac ageing and function. Nature 495:107–110. 10.1038/nature11919 [DOI] [PubMed] [Google Scholar]
- 42.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. 2012. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 8:e1002797. 10.1371/journal.pgen.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao L, Han SI, Fang Y, Park JS, Gupta S, Gilfor D, Amorino G, Valerie K, Sealy L, Engelhardt JF, Grant S, Hylemon PB, Dent P. 2003. Bile acid regulation of C/EBPbeta, CREB, and c-Jun function, via the extracellular signal-regulated kinase and c-Jun NH2-terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol. Cell. Biol. 23:3052–3066. 10.1128/MCB.23.9.3052-3066.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuchs SY, Adler V, Pincus MR, Ronai Z. 1998. MEKK1/JNK signaling stabilizes and activates p53. Proc. Natl. Acad. Sci. U. S. A. 95:10541–10546. 10.1073/pnas.95.18.10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ljungman M. 2000. Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia 2:208–225. 10.1038/sj.neo.7900073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira DM, Castro RE, Machado MV, Evangelista T, Silvestre A, Costa A, Coutinho J, Carepa F, Cortez-Pinto H, Rodrigues CM. 2011. Apoptosis and insulin resistance in liver and peripheral tissues of morbidly obese patients is associated with different stages of non-alcoholic fatty liver disease. Diabetologia 54:1788–1798. 10.1007/s00125-011-2130-8 [DOI] [PubMed] [Google Scholar]
- 47.Amir M, Liu K, Zhao E, Czaja MJ. 2012. Distinct functions of JNK and c-Jun in oxidant-induced hepatocyte death. J. Cell. Biochem. 113:3254–3265. 10.1002/jcb.24203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwabe RF, Bradham CA, Uehara T, Hatano E, Bennett BL, Schoonhoven R, Brenner DA. 2003. c-Jun-N-terminal kinase drives cyclin D1 expression and proliferation during liver regeneration. Hepatology 37:824–832. 10.1053/jhep.2003.50135 [DOI] [PubMed] [Google Scholar]
- 49.Sabapathy K, Wagner EF. 2004. JNK2: a negative regulator of cellular proliferation. Cell Cycle 3:1520–1523. 10.4161/cc.3.12.1315 [DOI] [PubMed] [Google Scholar]
- 50.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15:2760–2770 [PMC free article] [PubMed] [Google Scholar]
- 51.Saha MN, Jiang H, Yang Y, Zhu X, Wang X, Schimmer AD, Qiu L, Chang H. 2012. Targeting p53 via JNK pathway: a novel role of RITA for apoptotic signaling in multiple myeloma. PLoS One 7:e30215. 10.1371/journal.pone.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589:47–65. 10.1016/j.mrrev.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 53.Rodrigues CM, Fan G, Ma X, Kren BT, Steer CJ. 1998. A novel role for ursodeoxycholic acid in inhibiting apoptosis by modulating mitochondrial membrane perturbation. J. Clin. Invest. 101:2790–2799. 10.1172/JCI1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26:745–752. 10.1016/j.molcel.2007.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiao L, Studer E, Leach K, McKinstry R, Gupta S, Decker R, Kukreja R, Valerie K, Nagarkatti P, El Deiry W, Molkentin J, Schmidt-Ullrich R, Fisher PB, Grant S, Hylemon PB, Dent P. 2001. Deoxycholic acid (DCA) causes ligand-independent activation of epidermal growth factor receptor (EGFR) and FAS receptor in primary hepatocytes: inhibition of EGFR/mitogen-activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol. Biol. Cell 12:2629–2645. 10.1091/mbc.12.9.2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hau A, Ceppi P, Peter ME. 2012. CD95 is part of a let-7/p53/miR-34 regulatory network. PLoS One 7:e49636. 10.1371/journal.pone.0049636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim NH, Cha YH, Kang SE, Lee Y, Lee I, Cha SY, Ryu JK, Na JM, Park C, Yoon HG, Park GJ, Yook JI, Kim HS. 2013. p53 regulates nuclear GSK-3 levels through miR-34-mediated Axin2 suppression in colorectal cancer cells. Cell Cycle 12:1578–1587. 10.4161/cc.24739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. 2011. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 195:417–433. 10.1083/jcb.201103097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siemens H, Jackstadt R, Kaller M, Hermeking H. 2013. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget 4:1399–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sola S, Ma X, Castro RE, Kren BT, Steer CJ, Rodrigues CM. 2003. Ursodeoxycholic acid modulates E2F-1 and p53 expression through a caspase-independent mechanism in transforming growth factor beta1-induced apoptosis of rat hepatocytes. J. Biol. Chem. 278:48831–48838. 10.1074/jbc.M300468200 [DOI] [PubMed] [Google Scholar]
- 61.Brooks CL, Gu W. 2011. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2:456–462. 10.1007/s13238-011-1063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elaut G, Henkens T, Papeleu P, Snykers S, Vinken M, Vanhaecke T, Rogiers V. 2006. Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures. Curr. Drug Metab. 7:629–660. 10.2174/138920006778017759 [DOI] [PubMed] [Google Scholar]
- 63.Yan F, Wang XM, Liu ZC, Pan C, Yuan SB, Ma QM. 2010. JNK1, JNK2, and JNK3 are involved in P-glycoprotein-mediated multidrug resistance of hepatocellular carcinoma cells. Hepatobiliary Pancreat. Dis. Int. 9:287–295 [PubMed] [Google Scholar]
- 64.Wang Y, Singh R, Lefkowitch JH, Rigoli RM, Czaja MJ. 2006. Tumor necrosis factor-induced toxic liver injury results from JNK2-dependent activation of caspase-8 and the mitochondrial death pathway. J. Biol. Chem. 281:15258–15267. 10.1074/jbc.M512953200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takatori A, Geh E, Chen L, Zhang L, Meller J, Xia Y. 2008. Differential transmission of MEKK1 morphogenetic signals by JNK1 and JNK2. Development 135:23–32. 10.1242/dev.007120 [DOI] [PubMed] [Google Scholar]
- 66.Sabapathy K. 2012. Role of the JNK pathway in human diseases. Prog. Mol. Biol. Transl. Sci. 106:145–169. 10.1016/B978-0-12-396456-4.00013-4 [DOI] [PubMed] [Google Scholar]
- 67.Czaja MJ. 2003. The future of GI and liver research: editorial perspectives. III. JNK/AP-1 regulation of hepatocyte death. Am. J. Physiol. Gastrointest. Liver Physiol. 284:G875–G879. 10.1152/ajpgi.00549.2002 [DOI] [PubMed] [Google Scholar]
- 68.Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13:607–619. 10.1101/gad.13.5.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen F, Hu SJ. 2012. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J. Biochem. Mol. Toxicol. 26:79–86. 10.1002/jbt.20412 [DOI] [PubMed] [Google Scholar]
- 70.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Werneburg NW, Akazawa Y, Kahraman A, Garrison SP, Zambetti GP, Charlton MR, Gores GJ. 2009. JNK1-dependent PUMA expression contributes to hepatocyte lipoapoptosis. J. Biol. Chem. 284:26591–26602. 10.1074/jbc.M109.022491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ribeiro PS, Cortez-Pinto H, Sola S, Castro RE, Ramalho RM, Baptista A, Moura MC, Camilo ME, Rodrigues CM. 2004. Hepatocyte apoptosis, expression of death receptors, and activation of NF-kappaB in the liver of nonalcoholic and alcoholic steatohepatitis patients. Am. J. Gastroenterol. 99:1708–1717. 10.1111/j.1572-0241.2004.40009.x [DOI] [PubMed] [Google Scholar]
- 72.Gao Z, Zhang J, Kheterpal I, Kennedy N, Davis RJ, Ye J. 2011. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 286:22227–22234. 10.1074/jbc.M111.228874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. 2002. A central role for JNK in obesity and insulin resistance. Nature 420:333–336. 10.1038/nature01137 [DOI] [PubMed] [Google Scholar]