Abstract
IκB kinase ε (IKK-ε) has an essential role as a regulator of innate immunity, functioning downstream of pattern recognition receptors to modulate NF-κB and interferon (IFN) signaling. In the present study, we investigated IKK-ε activation following T cell receptor (TCR)/CD28 stimulation of primary CD4+ T cells and its role in the stimulation of a type I IFN response. IKK-ε was activated following TCR/CD28 stimulation of primary CD4+ T cells; however, in T cells treated with poly(I·C), TCR/CD28 costimulation blocked induction of IFN-β transcription. We demonstrated that IKK-ε phosphorylated the transcription factor IFN regulatory factor 1 (IRF-1) at amino acid (aa) 215/219/221 in primary CD4+ T cells and blocked its transcriptional activity. At the mechanistic level, IRF-1 phosphorylation impaired the physical interaction between IRF-1 and the NF-κB RelA subunit and interfered with PCAF-mediated acetylation of NF-κB RelA. These results demonstrate that TCR/CD28 stimulation of primary T cells stimulates IKK-ε activation, which in turn contributes to suppression of IFN-β production.
INTRODUCTION
The interferon (IFN) regulatory factor (IRF) family of transcription factors includes key regulators of type I IFN expression that are also involved in several immunological and growth-related cellular functions (1, 2). IRF-1, the first member of the IRF family to be identified, was considered initially to be a positive regulator of the IFN-β gene but was subsequently shown to not be essential for IFN-β gene induction, based on studies in IRF-1−/− cells (1, 2). IRF-1 stimulates other cellular target genes as well as the viral HIV-1 promoter long terminal repeat (LTR) (5–10) and exerts predominantly antiproliferative and antitumor activities (2, 11) as well as immune-modulatory functions (12–14).
The IFN-β promoter-enhancer is a 60-bp DNA element composed of four positive regulatory domains (PRDI to PRDIV) that bind IRF-3/7 (PRDI and PRDIII), NF-κB (PRDII), and ATF-2/c-Jun (PRDIV) transcription factors; additionally, the high-mobility-group protein HMG I(Y) binds to the minor DNA groove; these transcription factors all together form a transcriptionally active structure known as the enhanceosome (15). The acetylation of a nucleosome that blocks the formation of the preinitiation complex by the PCAF histone acetyltransferase (HAT) is also required for full gene activation (16, 17); PCAF is subsequently removed from the promoter and replaced by another HAT, CBP (17). The RelA subunit of NF-κB also undergoes reversible acetylation by HATs at multiple sites, which differentially affects its transcriptional activity (4, 18–20), where acetylation of Lys310 determines full transcriptional activity of RelA (19).
IRF-3 and IRF-7 are crucial players in the induction of the IFN-β gene following virus infection (21, 22), and the transcriptional activity is controlled through COOH-terminal phosphorylation at different serine/threonine clusters (33, 37), mediated by the IκB-related kinases TANK-binding kinase 1 (TBK-1) and IκB kinase ε (IKK-ε) (24, 25), following virus infection or Toll-like receptor triggering. These kinases were originally characterized as activators of NF-κB but have also been implicated in malignant transformation and inflammation (26). Toll-like receptor-independent mechanisms also activate TBK-1 and IKK-ε, and a bioinformatics analysis recently identified many potential substrates of these kinases (27). The mechanisms of activation of IRF-1 have not been well characterized, and to date, IRF-1 activation through phosphorylation has been poorly studied, despite evidence that IRF-1 activity may indeed involve phosphorylation (40–42).
The IKK-ε kinase is predominantly expressed and active in peripheral blood leukocytes, pancreas, thymus, and spleen (29) and contributes to IRF as well as NF-κB activation (3); furthermore, IKK-ε has been implicated in the stimulation of specific NF-κB-controlled genes following T cell receptor (TCR) cross-linking in T cells (30). In the present study, we report that IKK-ε is activated following TCR/CD28 stimulation of primary CD4+ T cells; TCR/CD28 costimulation actively suppressed IFN-β induction after poly(I·C) treatment. IRF-3 was not activated in CD4+ T cells following TCR/CD28 costimulation, whereas IRF-1 was COOH-terminally phosphorylated by IKK-ε. At the mechanistic level, IKK-ε-mediated phosphorylation of IRF-1 interfered with PCAF-mediated acetylation of NF-κB RelA, resulting in the suppression of genes regulated by IRF-1/RelA synergism.
MATERIALS AND METHODS
Cell cultures and treatments.
HEK 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Bio-Whittaker, Cambrex Bio Science, Verviers, Belgium) supplemented with 10% fetal calf serum (FCS) and antibiotics (growth medium). Human peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated by Ficoll-Hypaque gradient centrifugation, and the CD4+ T cell population was purified by negative selection using magnetic beads (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany). Recovered cells were >96% CD4+, as determined by fluorescence-activated cell sorter (FACS) analysis. Cells were cultured in RPMI 1640 medium (Bio-Whittaker, Cambrex Bio Science) containing 20% FCS and antibiotics and activated with anti-CD3 and anti-CD28 monoclonal antibodies (MAbs) (R&D Systems, Minneapolis, MN) and poly(I·C) (Amersham Biosciences Corp., Piscataway, NJ), as indicated. HEK 293 cells were treated with dimethyl sulfoxide (DMSO) or 50 μM Mc 1783 or anacardic acid (28) dissolved in DMSO.
Plasmids.
CMVBL, CMVBL IRF-1, and pFLAG pCDNA3.1 Zeo IKK-ε were described previously (24, 31). pMSCV neo CMV IKK-ε 1-361 was obtained from pFLAG pCDNA3.1 Zeo IKK-ε, after subcloning using the XhoI restriction site in the IKK-ε sequence to generate the deletion mutant, followed by blunt ligation with the pMSCV neo vector. IFN-β–Luc was a gift of Giridhar R. Akkaraju (Department of Biology, Texas Christian University, Fort Worth, TX), interleukin-8 (IL-8)–Luc was a gift of Michael Kracht (Rudolf Buchheim Institute of Pharmacology, Institute of Biochemistry, Justus Liebig University, Giessen, Germany), and pRLβactin was a gift of T. Matsuyama (Nagasaki University School of Medicine, Nagasaki, Japan). CMVBLRelA was described previously (10). The RelA 310A DNA sequence was obtained by de novo gene synthesis (GenScript USA Inc., Piscataway, NJ) and subsequently inserted into CMVBL through directional BamHI/HindIII subcloning.
CMVBL IRF-1 151-325 was obtained by PCR amplification from CMVBL IRF-1 and then inserted back again into CMVBL by directional EcoRI/XbaI subcloning. CMVBL IRF-1 2A, CMVBL IRF-1 3A, and CMVBL IRF-1 3D were obtained from CMVBL IRF-1 by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. The glutathione S-transferase (GST)–IRF-1 NH2 terminus (amino acids [aa] 109 to 163) and the GST–IRF-1 COOH terminus (aa 181 to 240) were obtained by PCR amplification. Inserts were then digested with EcoRI-XhoI and cloned in frame into pGEX-4T1. pGEX-4T1 IRF-1 181-240 2A and pGEX-4T1 IRF-1 181-240 3A were obtained by PCR amplification from CMVBL IRF-1 2A and CMVBL IRF-1 3A. CMVBL IRF-1 151-325 was obtained by PCR amplification from CMVBL IRF-1 by using the following primers: IRF-1 delta150 For (5′-CCGGAATTCGTCGCCACCATGGGACTCAGCAGCTCCACTCT-3′) and IRF-1 delta150Rev (5′-GCTCTAGACTACGGTGCACAGGGAATGGCCTGG-3′). CMVBL IRF-1 2A, CMVBL IRF-1 3A, and CMVBL IRF-1 3D were obtained from CMVBL IRF-1 by using the following oligonucleotides: 2A for (5′-CCCATGCCCTCCATCTCTGAAGCTGCAGCAGATGAGGATGAGGAAGGG-3′) and 2A rev (5′-CCCTTCCTCATCCTCATCTGCTGCAGCTTCAGAGATGGAGGGCATGGG-3′) (CMVBL IRF-1 2A); 1A for (5′-GTACAACTTCCAGGTGGCACCCATGCCCGCCATC-3′), 1A rev (5′-GATGGCGGGCATGGGTGCCACCTGGAAGTTGTAC-3′), 2A′ for (5′-CCAGGTGGCACCCATGCCCGCCATCGCAGAAGCTACAACAG-3′), and 2A′ rev (5′-CTGTTGTAGCTTCTGCGATGGCGGGCATGGGTGCCACCTGG-3′) (CMVBL IRF-1 3A, in two steps from CMVBL IRF-1); and 1D for (5′-GTACAACTTCCAGGTGGATCCCATGCCCGCCATC-3′), 1D rev (5′-GATGGCGGGCATGGGATCCACCTGGAAGTTGTAC-3′), 2D for (5′-CCAGGTGGCACCCATGCCCGATATCGATGAAGCTACAACAG-3′), and 2D rev (5′-CTGTTGTAGCTTCATCGATATCGGGCATGGGTGCCACCTGG-3′) (CMVBL IRF-1 3D, in two steps from CMVBL IRF-1). The GST–IRF-1 NH2 terminus (residues 109 to 163) and GST–IRF-1 COOH terminus (residues 181 to 240) were obtained by PCR amplification by using the following primers: IRF-1(109-163) For (5′-CCGGAATTCTACCGGATGCTTCCACCT-3′), IRF-1(109-163) Rev (5′-TTAGCTGCTGTGGTCATCAGG-3′), IRF-1(181-240) For (5′-CCCAGCACTGTCGCCATGTGC-3′), and IRF-1(181-240) Rev (5′-TTAGAGCTTCATGATGTCCTG-3′). pGEX-4T1 IRF-1 181-240 2A and pGEX-4T1 IRF-1 181-240 3A were obtained by PCR amplification from CMVBL IRF-1 2A and CMVBL IRF-1 3A by using the following primers: IRF-1(181-240) For and IRF-1(181-240) Rev.
Primary CD4+ T cell transfection.
CD4+ T cells (5 × 106) were transfected using nucleofection (nucleofected) with 4 μg of the pMSCV CMV neo empty vector or with pMSCV CMV neo IKK-ε 1-361 by using the Human T Cell Nucleofector kit according to the manufacturer's instructions (Amaxa, Cologne, Germany). After 4 h, cells were stimulated with anti-CD3/CD28 for 30 min, and whole-cell extracts (WCEs) were obtained and used for in vitro kinase assays and to perform immunoblotting using anti-IKK-ε and antiactin antibodies (Abs) as a control for IKK-ε 1-361 expression.
Transient-transfection and reporter gene assays.
Transfection experiments were performed by using JetPei reagent (Polyplus Transfection SA, Illkirch, France) according to the manufacturer's instructions. Amounts of transfected DNA were normalized by using empty vectors. Reagents from Promega Corp. (Madison, WI) were used to assay extracts for dual-luciferase activity in a Lumat LB9501 luminometer (E&G Berthold, Bad Wildbad, Germany).
Coimmunoprecipitation and immunoblot analysis.
WCEs (300 μg) from HEK 293 cells, prepared as described previously (32), were incubated overnight with 1 μg of polyclonal anti-IRF-1 antibody. Cell extracts were then incubated with Ultralink immobilized protein A/G-Sepharose (Pierce Biotechnology, Rockford, IL) for 2 h at room temperature (25°C). After extensive washing, immunoprecipitates were eluted by boiling the beads for 5 min in 2× sodium dodecyl sulfate (SDS) sample buffer. Eluted proteins were separated by SDS–10% polyacrylamide gel electrophoresis (PAGE) and subjected to immunoblotting analysis. Immunoblotting analysis on CD4+ T cells was performed by using extracts derived from each donor, and blots from a representative donor from each experiment are shown. Aliquots of whole-cell extracts, prepared as previously described (32) were separated by SDS–10% PAGE and transferred onto a nitrocellulose membrane (Amersham Pharmacia Biotech, United Kingdom). Nitrocellulose blots were probed with polyclonal antibodies against IRF-1, IRF-3, IKK-ε, and actin (Santa Cruz Biotechnology Inc., Santa Cruz, CA); acetyl-NF-κB p65 (RelA) Lys310 and IRF-3 Ser396 (Cell Signaling Technology, Beverly, MA); and IKK-ε Ser172 (Merck Millipore, Darmstadt, Germany) and with monoclonal Abs against IRF-1, IRF-7, and IRF-5 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) and then with anti-rabbit, -goat, or -mouse horseradish peroxidase-coupled secondary antibody (1:2,000 dilution) (Calbiochem, EMD Biosciences Inc., Merck KGaA, Darmstadt, Germany). Immunoprecipitated proteins were probed with a polyclonal Ab against RelA (sc-109) and a monoclonal Ab against IRF-1 (sc-74530), from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Input extracts were blotted with polyclonal antibodies against IRF-1 (sc-497), RelA (sc-109), PCAF (sc-8999), and actin (sc-1616). Detection was performed by using ECL Western blotting reagents (Amersham Pharmacia Biotech).
SeV infection.
A total of 6 × 106 CD4+ T cells were infected with 2,000 hemagglutinating units (HAU) of Sendai virus (SeV) for 18 h in complete growth medium containing 20% fetal bovine serum.
DNA affinity binding assay.
For DNA affinity binding assays, a biotinylated oligonucleotide corresponding to the PRDIII/I region of the IFN-β promoter (sequence, 5′-GAAAACTGAAAGGGAGAAGTGAAAGT-3′) was synthesized (Eurofins MWG/Operon, Ebersberg, Germany). DNA affinity binding assays were performed by annealing the sense and the antisense oligonucleotides in 1× sodium chloride-Tris-EDTA (STE) buffer, containing 10 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 2 mM EDTA. Thirty picomoles of biotinylated DNA was mixed with 200 μg of WCE in 200 ml of binding buffer containing 20 mM Tris-HCl (pH 7.5), 75 mM KCl, 1 mM dithiothreitol (DTT), and 5 mg/ml BSA in the presence of 10% glycerol and 6 μg of poly(dI·dC) and incubated for 25 min at room temperature. The complexes were pulled down with magnetic beads (Streptavidin MagneSphere; Promega) for 30 min at 4°C and for 10 min at room temperature by mixing with rotation. The collected beads were washed, and bound material was eluted by boiling in sample buffer. Eluted material was separated onto 10% SDS-PAGE gels, followed by immunoblotting with anti-PCAF (sc-8999), anti-IRF-3 (sc-9082), anti-IRF-1 (sc-74530), and antiactin (sc-1616), from Santa Cruz Biotechnology.
Quantitative real-time reverse transcription-PCR.
Total cellular RNA was isolated from 2 × 106 cells by using the RNeasy total RNA extraction kit (Qiagen). RNA was treated with RNase-free DNase (Qiagen), and first-strand cDNA was synthesized with Superscript II (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Real-time PCR was performed by using an ABI 7000 sequence detection system (PE Applied Biosystems, Warrington, United Kingdom) and SYBR green PCR master mix (Applied Biosystems). The optimization of the real-time PCR was performed according to the manufacturer's instructions but scaled down to 25 μl per reaction mixture. The relative quantification of mRNA expression was analyzed by using ΔΔCT methods (means ± standard deviations). Primers used for quantitative real-time reverse transcription-PCR (qRT-PCR) were IFN-β forward primer 5′-GCAGCAGTTCCAGAAGGAG-3′ and reverse primer 5′-GCCAGGAGGTTCTCAACAAT-3′. The transcript levels were normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-GGGTGTGAACCATGAGAAG-3′; reverse, 5′-GCTAAGCAGTTGGTGGTGC-3′).
In vitro kinase assay.
A total of 0.5 μg of WCE was added to kinase buffer (10 mM HEPES [pH 7.4], 5 mM MgCl2, 50 μM orthovanadate, 10 mM beta-glycerophosphate, 1 mM DTT, 50 mM NaCl, 10 mM p-nitrophenylphosphate [PNPP]). An in vitro kinase assay was performed by adding the GST-purified GST–IRF-1 109-163, 181-240, 181-240 2A, 181-240 3A, or GST–IRF-3 380-427 or 380-427 7A substrate (1 μg), 10 μM cold ATP, and 10 μCi of [γ-32P]ATP in kinase buffer. The kinase reaction was performed at 30°C for 30 min and stopped by the addition of SDS sample buffer. Samples were analyzed by 10% SDS-PAGE followed by Coomassie staining. The dried gels were exposed to film at −70°C for 5 h. Preparation of baculovirus-expressed, IKK-ε-purified kinase was performed as previously described (33).
Statistical analysis.
Significant differences between experimental points measured by qRT-PCR and luciferase assays were assessed by using the Student-Neuman-Keuls posttest following significant (P < 0.05, P < 0.01, and P < 0.001) repeated-measures analysis of variance (ANOVA).
RESULTS
Anti-CD3/CD28 treatment of primary CD4+ T cells induces IKK-ε activation and correlates with IRF-1 posttranslational modification.
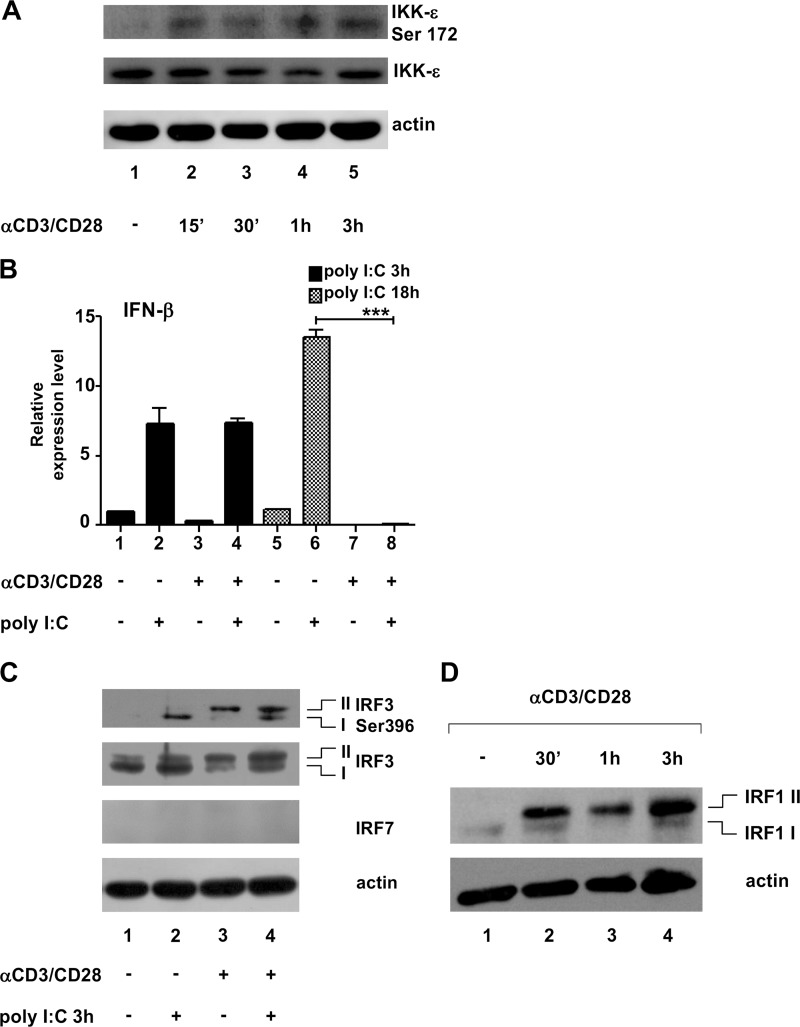
We set out to determine whether T cell receptor (TCR) stimulation activated IKK-ε in primary T lymphocytes. To investigate the role of IKK-ε upon T cell receptor stimulation (30), primary CD4+ T cells were treated with anti-CD3/CD28, and IKK-ε activation was monitored by using antibodies that specifically detect IKK-ε and its activated Ser172 form (34). We chose to stimulate cells with anti-CD3 and anti-CD28 to strengthen the activation signal mediated by the TCR (anti-CD3), a stimulation that mimics the physiological encounter between a CD4+ T cell and an antigen-presenting cell. As shown in Fig. 1A, CD4+ T cells expressed IKK-ε constitutively, and the activated form of the kinase appeared beginning at 15 min after treatment (Fig. 1, lanes 2 to 5). This IKK-ε activation, however, did not result in IFN-β synthesis (Fig. 1B, lane 3), whereas IFN-β mRNA was induced in poly(I·C)-stimulated cells (Fig. 1B, lane 2) and in Sendai virus (SeV)-infected cells (data not shown). Interestingly, TCR/CD28 activation was sufficient to block activation of IFN-β by poly(I·C)-stimulated (Fig. 1B, lanes 6 and 8) and SeV-infected (data not shown) cells. Using the phosphospecific Ser396 antibody to detect IRF-3 activation, different patterns of Ser396 phosphorylation were observed in T cells treated with poly(I·C) and/or anti-CD3/CD28: poly(I·C) treatment stimulated phosphorylation of form I IRF-3, while anti-CD3/CD28 induced phosphorylation of the slower-migrating isoform (form II) (Fig. 1C, lanes 2 and 3, respectively). Both IRF-3 isoforms were instead phosphorylated in T cells treated with poly(I·C) and stimulated with anti-CD3/CD28 (Fig. 1C, lane 4).
FIG 1.
Anti-CD3/CD28 treatment of primary CD4+ T cells results in IKK-ε activation, which correlates with IRF-1 posttranslational modification. (A) A total of 2 × 106 CD4+ T cells were left untreated or treated for 15 min, 30 min, 1 h, and 3 h with 2 μg/ml of anti-CD3/CD28, and 50 μg of WCEs was subjected to immunoblotting using specific Abs recognizing activated IKK-ε (Ser172), IKK-ε, and actin, as indicated. (B) CD4+ T cells (2 × 106) were prestimulated with 2 μg/ml of anti-CD3/CD28 for 3 h and then treated with 100 μg/ml of poly(I·C) for the indicated times. Total RNA was extracted and retrotranscribed, and IFN-β RNA expression was analyzed by qRT-PCR. Expression levels were normalized to GAPDH and unstimulated samples (means ± standard deviations from three separate experiments are shown [∗∗∗, P < 0.001; P = 0.00024]). (C) CD4+ T cells (4 × 106) were stimulated as described above for panel B, and 100 μg of WCEs was subjected to immunoblotting using specific Abs recognizing IRF-3 Ser396, IRF-3, IRF-7, and actin. IRF-3 isoforms I and II are indicated. (D) CD4+ T cells (10 × 106) were stimulated for 30 min, 1 h, and 3 h with 2 μg/ml of anti-CD3/CD28, and 50 μg of WCEs was subjected to immunoblotting using specific Abs recognizing IRF-1 (monoclonal) and actin. IRF-1 isoforms I and II are indicated.
IRF-7 was not expressed in nonstimulated or poly(I·C)-treated cells (Fig. 1C). Further evaluation revealed a constitutive level of IRF-1 in untreated cells (Fig. 1D, lane 1), and following anti-CD3/CD28 treatment of T cells, slower-migrating form II of IRF-1 was detected at 30 min and increased until 3 h (Fig. 1D, lanes 2 to 5); compared to form I in untreated cells, the percentage of form II versus total IRF-1 (form I plus form II) increased >20-fold (Fig. 1D). These results indicate that stimulation of CD4+ T cells by anti-CD3/CD28 resulted in IKK-ε activation and correlated with a modification of IRF-1 mobility. Because IRF-3 phosphorylation may involve both isoform-specific positive and negative regulation of IFN gene expression (35), we focused on the novel observation of IRF-1 phosphorylation by IKK-ε.
A COOH-terminal phosphorylation cluster present in IRF-1 is the target of IKK-ε phosphorylation.
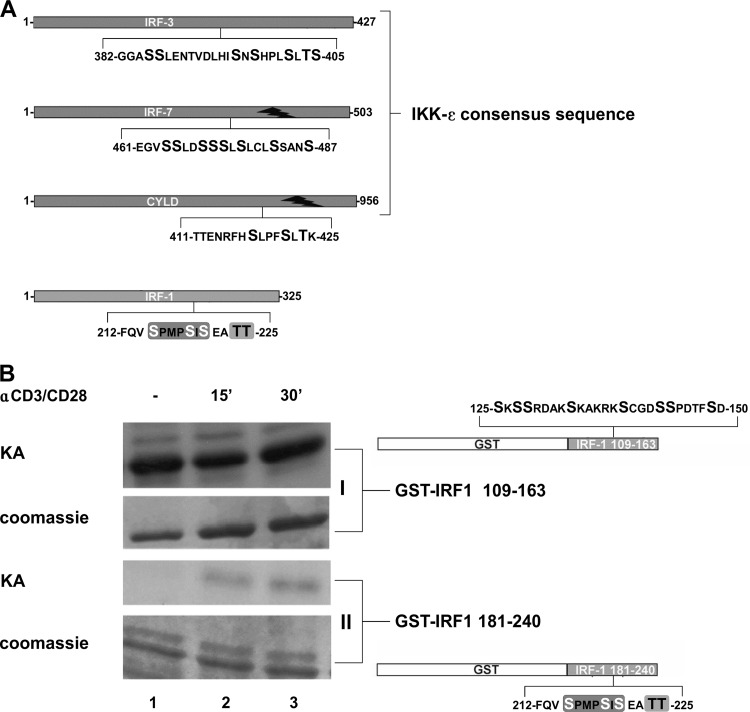
Next, to determine if IRF-1 posttranslational modification was due to IKK-ε kinase-mediated phosphorylation, we initially used the NetPhos 2.0 server (36) to scan the IRF-1 amino acid sequence, and we identified two putative phosphorylation clusters in the protein, spanning amino acids (aa) 125 to 150 (an NH2-terminal cluster) and aa 212 to 225 (a COOH-terminal cluster) (Fig. 2A), that resembled the IKK-ε consensus sequence NH2-ENRFHS418LPFSLTK-COOH, identified in the tumor suppressor CYLD (27), and the sequence NH2-SXSXXXS-COOH, present in IRF-3 and IRF-7 (37). In particular, the COOH-terminal cluster of IRF-1 bears a motif, 215SXXXSXS221, with the same orientation as the 418SXXXSXT424 motif in human CYLD (Fig. 2A) and is closely related to the SXSXXXS motif present in IRF-3 and IRF-7. Two threonine residues, at positions 224 and 225, of IRF-1 that may function as a potential regulatory site in the cluster were also identified (Fig. 2A).
FIG 2.
Activation of primary CD4+ T cells by anti-CD3/CD28 stimulates IRF-1 phosphorylation. (A) Comparison of IKK-ε consensus sequences of IRF-3, IRF-7, and CYLD and the putative IKK-ε consensus sequence present at the COOH terminus of IRF-1. (B) CD4+ T cells (2 × 106) were stimulated with 2 μg/ml of anti-CD3/CD28 for 15 min and 30 min or left untreated. WCEs (0.5 μg) were utilized to perform in vitro kinase assays (KA) using recombinant GST–IRF-1 fusion proteins containing fragments of the NH2-terminal (GST–IRF-1 109-163) (form I) and of the COOH-terminal (GST–IRF-1 181-240) (form II) phosphorylation clusters as substrates. Coomassie staining is also shown.
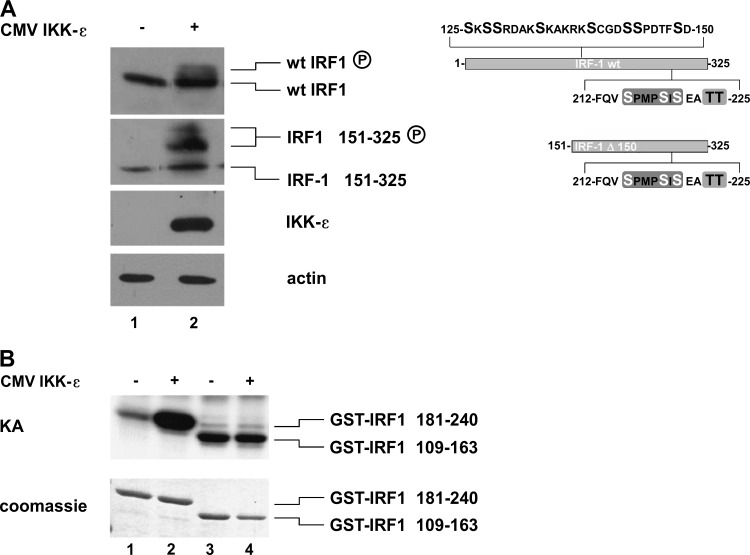
To assess if the putative COOH-terminal IRF-1 motif was a target of phosphorylation after anti-CD3/CD28 treatment, an in vitro kinase assay was performed in primary CD4+ T cells, comparing the NH2-terminal (GST–IRF-1 aa 109 to 163) and COOH-terminal (GST–IRF-1 aa 181 to 240) fragments as substrates. Inducible phosphorylation of the GST-IRF-1 fragment at aa 181 to 240 was detected, whereas no inducible phosphorylation was detected with the NH2-terminal fragment (Fig. 2B). Additionally, wild-type (wt) IRF-1 or the COOH-terminal portion of IRF-1 (IRF-1 aa 181 to 240), together with IKK-ε, was coexpressed in HEK 293 cells; IKK-ε expression resulted in a modest shift in migration of wt IRF-1 and a significant shift in migration of IRF-1 aa 151 to 325 (Fig. 3A). IKK-ε-mediated phosphorylation of IRF-1 was further confirmed by in vitro kinase assays using GST–IRF-1 aa 109 to 163, GST–IRF-1 aa 181 to 240, and WCEs derived from IKK-ε-expressing cells. A clear increase in phosphorylation of the COOH-terminal GST–IRF-1 fragment at aa 181 to 240 but not the NH2-terminal GST–IRF-1 fragment at aa 109 to 163 was detected (Fig. 3B). Taken together, these data provide evidence that IRF-1 is a target of IKK-ε kinase and becomes COOH-terminally phosphorylated when coexpressed with IKK-ε.
FIG 3.
IKK-ε induces IRF-1 COOH-terminal phosphorylation. (A) HEK 293 cells were transiently cotransfected with 1 μg of vectors expressing wt IRF-1 or IRF-1 151-325 and IKK-ε. WCEs (50 μg) were used to perform immunoblotting using anti-IRF-1, anti-IKK-ε, and antiactin. A schematic representation of expressed proteins, wt IRF-1 (aa 1 to 325) and IRF-1 Δ150 (aa 151 to 325), is also shown. (B) A WCE (0.5 μg) in vitro kinase assay (KA) was performed by using purified GST–IRF-1 181-240 and GST–IRF-1 109-163 recombinant fusion proteins as substrates upon HEK 293 cell transfection with 1 μg of the IKK-ε expression vector. Coomassie staining of the recombinant proteins is also shown. CMV, cytomegalovirus.
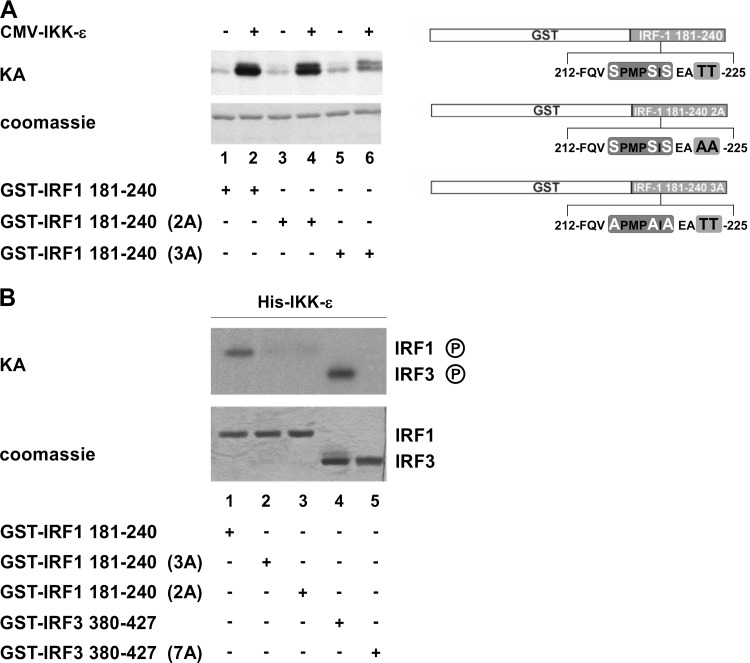
To validate the targeting of the phosphoacceptor site present in the COOH-terminal cluster of IRF-1 by IKK-ε, an in vitro kinase assay was performed by using the GST–wt IRF-1 fragment at aa 181 to 240 and point-mutated substrates termed IRF-1 181-240 2A and IRF-1 181-240 3A, containing Thr224/225Ala and Ser215/219/221Ala substitutions, respectively (Fig. 4A, right), together with extracts from IKK-ε-expressing HEK 293 cells or recombinant purified IKK-ε kinase. A 10-fold increase in IRF-1 COOH-terminal phosphorylation was obtained with IKK-ε-expressing cellular extracts (Fig. 4A, lanes 1 and 2); no significant change in phosphorylation was observed when the Thr224/225Ala mutations (IRF-1 181-240 2A) were introduced (Fig. 4A, lanes 3 and 4), whereas a dramatic decrease in phosphorylation was observed with the Ser 215/219/221Ala mutations (IRF-1 181-240 3A) (Fig. 4A, lanes 5 and 6). These results were further supported by in vitro kinase assays using the IRF-1 substrates and purified recombinant IKK-ε kinase; phosphorylation of wt IRF-1 but not mutated IRF-1 fragments was observed (Fig. 4B, lanes 1 and 2); however, in contrast to the results with IKK-ε cell extracts, IRF-1 181-240 2A was not phosphorylated by purified IKK-ε kinase (Fig. 4B, lane 3). As a positive control, the GST–wt IRF-3 380-427 substrate, but not the GST–IRF-3 7A substrate, was also phosphorylated by purified IKK-ε (Fig. 4B, lanes 4 and 5).
FIG 4.
IRF-1 COOH-terminal phosphoacceptor sites are targets of IKK-ε. (A) WCEs (0.5 μg) from HEK 293 cells overexpressing IKK-ε were used in in vitro kinase assays (KA) using wt IRF-1 and mutant substrates (GST–IRF-1 181-240 2A and 3A). A schematic representation of IRF-1 substrates is shown on the right side. (B) An in vitro kinase assay was performed by using a His-tagged recombinant IKK-ε kinase using wt IRF-1 and IRF-3 and mutated substrates (GST–IRF-1 181-240 2A and 3A and GST–IRF-3 380-427 7A).
The IRF-1 phosphorylation cluster is the target of IKK-ε in primary CD4+ T cells.
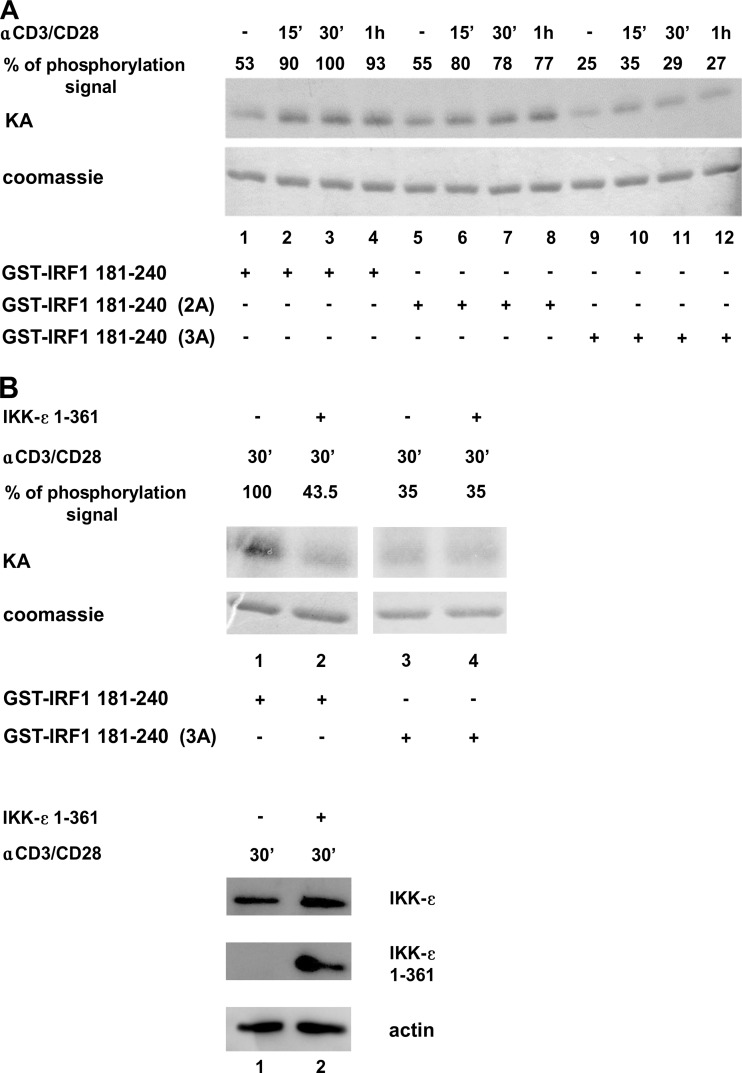
To determine if the COOH-terminal residues of IRF-1 were phosphorylated in vivo upon TCR/CD28-mediated stimulation of primary T cells, a kinase assay using WCEs together with wt IRF-1 and mutated substrates was performed (Fig. 5A). A 40 to 50% increase in IRF-1 COOH-terminal phosphorylation was observed with the wt IRF-1 substrate at all time points (Fig. 5A, lanes 1 to 4), compared to unstimulated cells. With the 2A mutant substrate, no significant difference in phosphorylation was detected (Fig. 5A, lanes 6 to 8), whereas with the 3A mutant, a 70 to 80% decrease in phosphorylation signal was detected (Fig. 5A, lanes 10 to 12), arguing that the Ser215/219/221 cluster is important for IKK-ε-mediated regulation of activity. To demonstrate that IRF-1 phosphorylation was indeed dependent on IKK-ε activation, a kinase assay was performed by using extracts from primary CD4+ T cells nucleofected with a control vector or with a vector expressing a dominant negative form of IKK-ε, IKK-ε 1-361, and stimulated for 30 min with anti-CD3/CD28 (Fig. 5B). A >50% decrease in COOH-terminal phosphorylation was obtained in the presence of IKK-ε 1-361. Specific expression of the IKK-ε dominant negative form upon nucleofection is shown in Fig. 5B (bottom).
FIG 5.
Anti-CD3/CD28-mediated phosphorylation of IRF-1 involves Ser215, Ser219, and Ser221 and depends on IKK-ε. (A) CD4+ T cells (2 × 106) were stimulated with 2 μg/ml of anti-CD3/CD28 for 15 min, 30 min, and 1 h or left untreated. WCEs (0.5 μg) were utilized to perform in vitro kinase assays (KA) using recombinant GST–IRF-1 fusion proteins containing fragments of the COOH-terminal wt and mutated IRF-1 phosphorylation clusters (GST–IRF-1 181-240 wt, 2A, and 3A) as the substrates. The amount of phosphorylation signal was calculated by densitometry and is expressed as a percentage, setting the strongest signal to 100%. (B, top) In vitro kinase assay performed with WCEs (0.5 μg) derived from CD4+ T cells (2.5 × 106) stimulated with anti-CD3/CD28 (2 μg/ml) for 30 min and 4 h after nucleofection with 4 μg of an empty vector or with an expression vector for a dominant negative form of IKK-ε (IKK-ε 1-361). The amount of the phosphorylation signal was calculated by densitometry and is expressed as a percentage, setting the strongest signal to 100%. (B, bottom) WCEs (30 μg) were subjected to immunoblotting using specific Abs recognizing IKK-ε and actin.
IRF-1 but not a phosphomimetic of IRF-1 (3D) interacts with RelA at Lys310 in a PCAF-dependent manner.
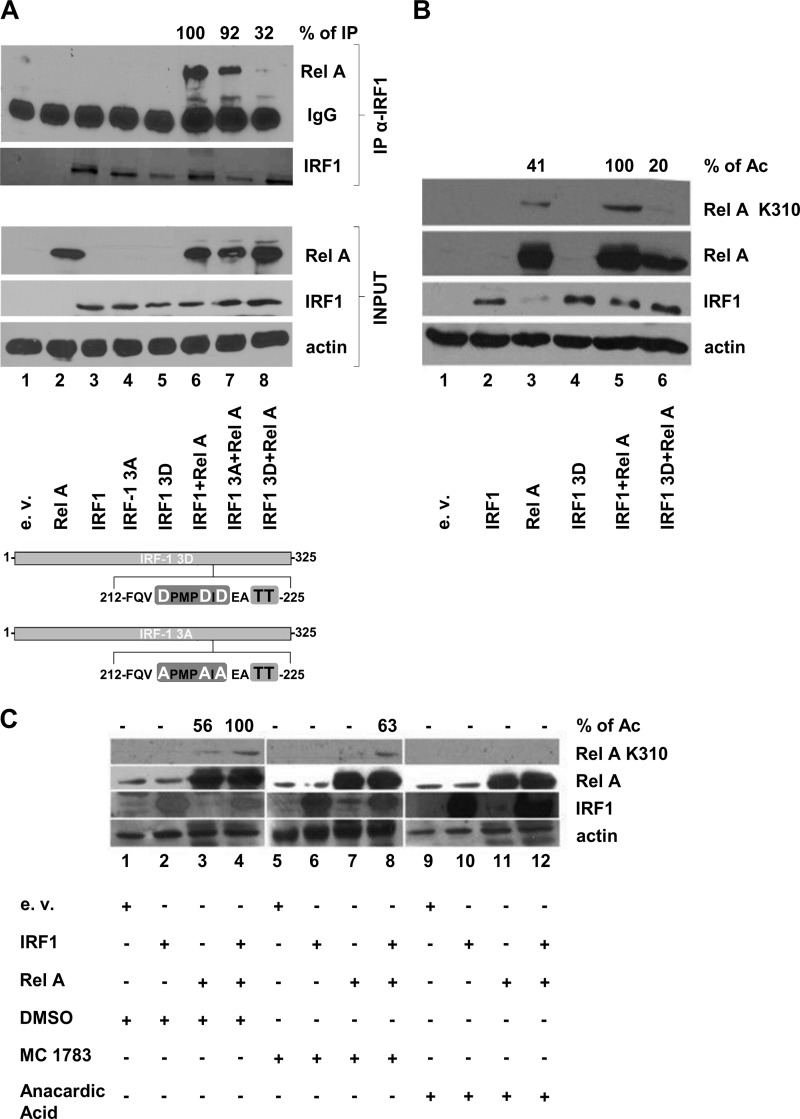
IRF-1 has been shown to associate with the NF-κB subunit RelA in tumor necrosis factor alpha (TNF-α)-stimulated T cells, resulting in enhanced transcriptional activation of the HIV-1 LTR enhancer (10). To determine if IRF-1 phosphorylation impaired the IRF-1 interaction with RelA and hampered IRF-1/RelA synergism, we coimmunoprecipitated IRF-1 from HEK 293 extracts expressing RelA and either wt IRF-1 or mutated forms of the protein (IRF-1 3D and 3A, where Ser215, Ser219, and Ser221 were modified to the phosphomimetic residue aspartic acid [D] and the inert residue alanine [A], respectively), followed by immunoblotting with a specific RelA antibody (Fig. 6A). RelA was detected by immunoprecipitation when coexpressed with wt IRF-1 or with IRF-1 3A, but not with IRF-1 3D, with a 70% decrease (Fig. 6A, lanes 6 to 8). No RelA was coimmunoprecipitated in the absence of IRF-1 (Fig. 6A, lane 2). This result indicates that IRF-1, once phosphorylated, is no longer able to interact with RelA.
FIG 6.
IRF-1 determines acetylation of RelA at lysine 310 in a PCAF-dependent manner. (A) HEK 293 cells were transiently transfected with 5 μg of the control empty vector (e.v.) or IRF-1 wt or mutated vectors (3A and 3D), alone or in combination with the RelA expression vector. After 24 h, WCEs were prepared, and immunoprecipitation (IP) was performed by using a polyclonal anti-IRF-1 antibody. Both inputs and immunoprecipitated proteins were revealed by immunoblotting using anti-RelA and monoclonal anti-IRF-1. An antiactin blot was used as an input loading control. The percentage of immunoprecipitated RelA, measured by densitometry, is shown and was obtained by setting the largest amount of immunoprecipitated protein of the RelA band as 100% (lane 6), after normalization to RelA content, the amount of IRF-1 immunoprecipitated, and actin levels. A schematic representation of expressed IRF-1 3D and IRF-1 3A proteins is shown at the bottom. (B) HEK 293 cells were transiently transfected, as described above for panel A. After 24 h, WCEs were prepared, and immunoblotting using anti-acetylated (Ac) RelA Lys310, anti-IRF-1, and anti-RelA polyclonal Abs was performed. An actin blot was used as the input loading control. The percentage of RelA Lys310 acetylation is shown at the top and was obtained by densitometry as the percentage of RelA K310 signal intensity over the total RelA signal intensity (RelA plus RelA K310) normalized to actin. (C) HEK 293 cells were treated for 1 h with DMSO or 50 μM of the HAT inhibitors Mc 1783 and anacardic acid in DMSO and removed prior to transfection. Pretreated HEK 293 cells were transiently transfected with 5 μg of the control vector or IRF-1 expression vector alone or together with the RelA expression vector. After 16 h, cells were treated again with DMSO or 50 μM Mc 1783 and anacardic acid and harvested 8 h later, and WCEs were obtained. Immunoblotting using anti-acetylated RelA Lys310, anti-IRF-1, and anti-RelA was performed, while an antiactin blot was used as the input loading control. The percentage of RelA Lys310 acetylation is shown at the top and was obtained by densitometry as described above for panel B.
Since IRF-1 physically interacts with PCAF and CBP/p300 HATs (23) and facilitates CBP/p300-mediated coactivation of IFN-β (38) as well as of other promoters (39), we next sought to determine if the failure of IRF-1 3D to interact with RelA also affected RelA acetylation. Using a RelA K310-specific antibody, IRF-1 but not IRF-1 3D coexpression resulted in a substantial increase in RelA-specific acetylation at Lys310 (Fig. 6B, lanes 5 and 6). Moreover, when specific inhibitors of CBP/p300 or both PCAF and CBP/p300 HATs (Mc 1783 and anacardic acid, respectively) were used (28), RelA acetylation was completely abolished by the PCAF-CBP/p300 inhibitor anacardic acid (Fig. 6C, lane 12) but not by the specific CBP/p300 inhibitor Mc 1783 (Fig. 6C, lane 8), indicating that PCAF but not CBP/p300 was involved in IRF-1-induced RelA acetylation.
RelA acetylation at Lys310 is essential for IRF-1/RelA synergism.
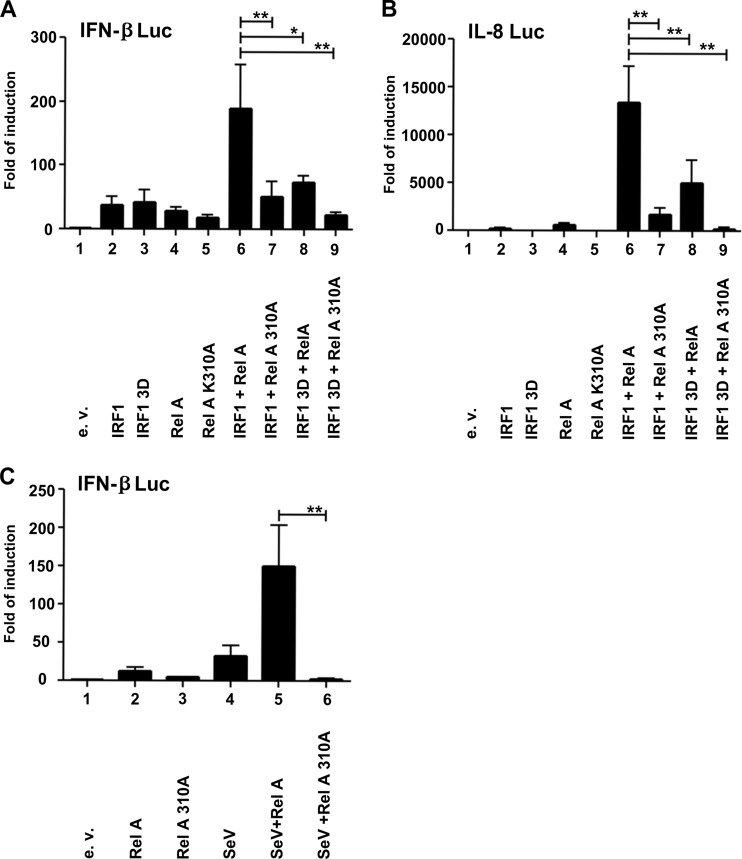
To determine the impact of RelA K310 acetylation on the expression of IRF-1-bound promoters, a RelA K310A expression construct was generated and used in luciferase assays with IFN-β and IL-8 promoter reporter constructs in various combinations with RelA, wt IRF-1, and IRF-1 3D. A synergistic effect was observed for both promoters when both wt IRF-1 and RelA proteins were expressed (Fig. 7A and B, lane 6), whereas synergism was abrogated when RelA K310A or IRF-1 3D was expressed in combination with the reciprocal wt protein (Fig. 7A and B, lanes 7 and 8). Furthermore, complete inhibition of promoter activity was obtained in the presence of both mutated activators (Fig. 7A and B, lane 9). A similar requirement for synergism was observed when the IFN-β promoter was stimulated by Sendai virus infection (Fig. 7C): activation was increased >150-fold when RelA was coexpressed, whereas expression of RelA K310A decreased promoter activity to baseline levels.
FIG 7.
RelA acetylation at Lys310 is essential for IRF-1/RelA synergism and for virus-induced IFN-β promoter-driven expression. (A) HEK 293 cells (1 × 105) were cotransfected with the IFN-β reporter luciferase construct (250 ng) together with a β-actin Renilla internal control vector (50 ng) and 250 ng of vectors expressing wt IRF-1, IRF-1 3D, or the wt or mutated version of the RelA protein (RelA 310A). (B) HEK 293 cells (1 × 105) were cotransfected with 250 ng of the IL-8 reporter luciferase construct and 250 ng of vectors expressing wt IRF-1, IRF-1 3D, RelA, and RelA 310A. (C) HEK 293 cells (1 × 105) were cotransfected with the IFN-β reporter luciferase construct (250 ng) together with the β-actin Renilla internal control vector (50 ng) and 250 ng of vectors expressing wt RelA or RelA 310A. After 10 h, cells were left uninfected or infected with 2.5 HAU of SeV for 18 h. A dual-luciferase assay was performed 24 h after transfection. Results are expressed as fold induction with respect to cells transfected with the empty vector (e.v.), upon normalization to the Renilla gene product activity. Means ± standard errors from three separate experiments are shown (∗, P < 0.05; ∗∗, P < 0.01).
Phosphorylated IRF-1 impairs RelA acetylation in anti-CD3/CD28-treated primary CD4+ T cells.
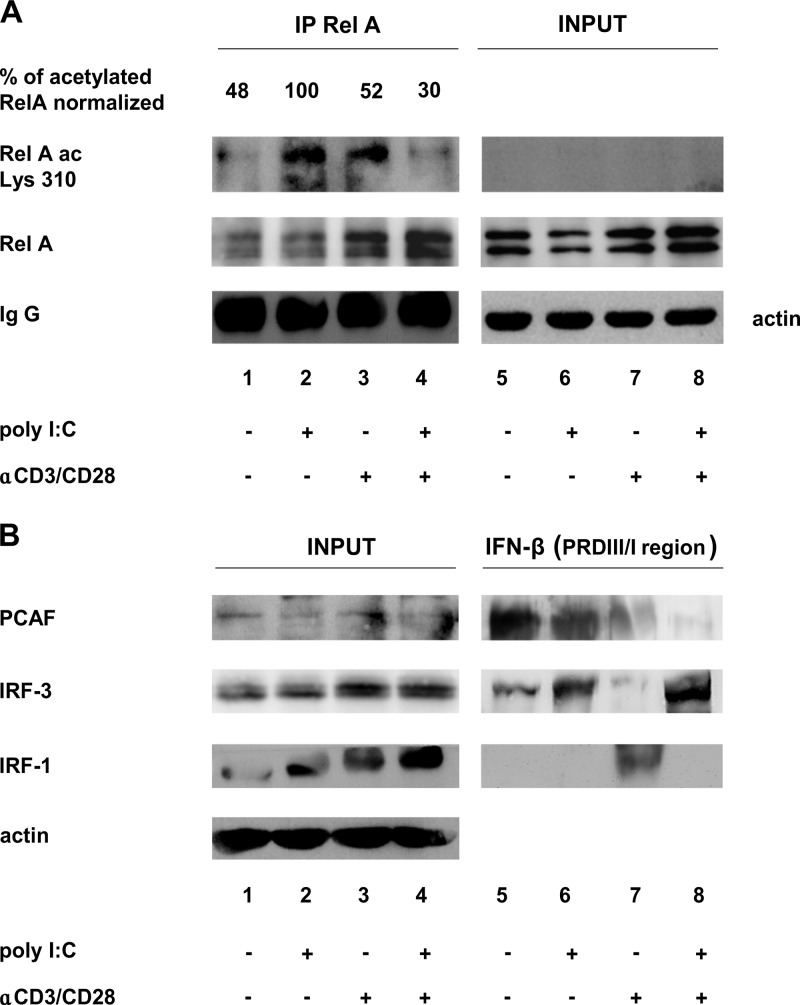
To confirm these data in the context of primary CD4+ T cells, we assessed RelA acetylation in T cells pretreated with anti-CD3/CD28 and then treated with poly(I·C), as shown in Fig. 1B. Cell extracts were immunoprecipitated with anti-RelA and immunoblotted with a RelA K310-specific antibody. As shown in Fig. 8A, RelA-specific acetylation was observed in poly(I·C)-treated cells, whereas acetylation was inhibited in anti-CD3/CD28-pretreated T cells. These results were mirrored by pulldown assays performed on the same cell extracts incubated with an oligonucleotide corresponding to the PRDIII/I region of the IFN-β promoter (Fig. 8B). The isolated complexes were then detected by immunoblotting against IRF-1, IRF-3, and PCAF. As shown in Fig. 8B, in poly(I·C)-treated cells, when IRF-1 is only partially phosphorylated (Fig. 8B, lane 2), IRF-3 binds DNA together with PCAF (Fig. 8B, lane 6), while in anti-CD3/CD28-stimulated and poly(I·C)-treated cells, when IRF-1 is fully phosphorylated (Fig. 8B, lane 4), PCAF does not bind despite IRF-3 binding (Fig. 8B, lane 8). Overall, these results indicate that in TCR-stimulated CD4+ T cells, phosphorylated IRF-1 inhibits PCAF-mediated RelA acetylation, thus contributing to the suppression of IFN-β transcription.
FIG 8.
Anti-CD3/CD28 pretreatment of CD4+ T cells determines inhibition of RelA acetylation at lysine 310 and decreased recruitment of PCAF to the IRF binding site on the IFN-β promoter sequence. (A) Immunoprecipitation assay. CD4+ T cells (20 × 106) were prestimulated with 2 μg/ml of anti-CD3/CD28 for 3 h and then treated with 100 μg/ml of poly(I·C) for 18 h. WCEs (1 mg) were prepared, and immunoprecipitation (IP) was performed by using a polyclonal anti-RelA antibody. Both inputs and immunoprecipitated proteins were revealed by immunoblotting using anti-RelA and anti-acetylated RelA Lys310. An antiactin blot was used as the input loading control. The percentage of acetylated RelA, measured by densitometry, is shown and was obtained by setting the largest amount of acetylated protein of the RelA band (lane 2) as 100%, after normalization with immunoprecipitated RelA. (B) DNA affinity purification assay. WCEs (200 μg), prepared as described above for panel A, were incubated with a biotinylated oligonucleotide corresponding to the IRF consensus binding site on the IFN-β promoter. The isolated complexes were detected by immunoblotting using antibodies against PCAF, IRF-3, and IRF-1.
DISCUSSION
IKK-ε was originally identified as being crucial for NF-κB-driven gene activation upon T cell receptor stimulation with anti-CD3 (30) but, together with the other noncanonical IκB kinase homolog TANK-binding kinase 1 (TBK1), was shown to stimulate IFN-β production through the phosphorylation and subsequent activation of IRF-3 and IRF-7 transcription factors (24, 25). Here we demonstrate that IKK-ε is also activated within minutes by TCR/CD28-mediated activation of T cells (Fig. 1), although TCR/CD28 stimulation did not induce IFN-β production. We present, for the first time, evidence that IKK-ε phosphorylates transcription factor IRF-1 at aa 215, 219, and 221, consistent with previous studies suggesting that IRF-1 regulation may indeed involve posttranslational modification (40–42). We identified a 5′-SXXXSXS-3′ motif (aa 215 to 221) in the C-terminal half of IRF-1, a sequence that resembles the IKK-ε target sequence in the tumor suppressor CYLD (SXXXSXT), with the exception of a different phosphoacceptor amino acid at position 424 (27). Mutation of Ser215, Ser219, and Ser221 affected the ability of a purified IKK-ε kinase to phosphorylate IRF-1 in an in vitro kinase assay and IRF-1 phosphorylation in kinase assays performed with extracts from stimulated CD4+ T cells. In close proximity to the SXXXSXS motif, two threonine residues in the IRF-1 sequence (aa 224 and aa 225) were identified, which behaved as regulatory sites, since their mutation profoundly affected the ability of purified IKK-ε kinase to phosphorylate IRF-1 in in vitro kinase assays. Instead, a modest effect on phosphorylation was observed upon mutation of Thr224 and Thr225 using extracts from cells overexpressing IKK-ε and in stimulated CD4+ T cells, suggesting that additional regulatory proteins bound to IKK-ε may compensate for the lack of phosphorylation at Thr224 and Thr225.
This result is reminiscent of IKK-ε phosphorylation of Ser36 and Ser32 of IκB-α: in vitro, Ser32 and Ser36 were phosphorylated, whereas in vivo, only Ser36 was phosphorylated (29). The observation that IKK-ε phosphorylates IRF-1 was further confirmed by the use of a dominant negative IKK-ε construct that reduced anti-CD3/CD28-mediated IRF-1 phosphorylation in primary CD4+ T cells.
The posttranslational modification of IRF-1, mediated by IKK-ε, completely blocked its transcriptional activity. Using a phosphomimetic mutant version of IRF-1 (IRF-1 3D), we demonstrated that in contrast to wild-type IRF-1, IRF-1 3D failed to physically interact with the NF-κB RelA subunit (Fig. 6A). Association of IRF-1 and NF-κB leading to the synergistic activation of several cellular genes has been extensively reported (5–8). In this respect, we demonstrated that in TNF-α-stimulated T cells, IRF-1 physically associates with RelA, resulting in a greater transcriptional activation of the HIV-1 LTR enhancer (10). RelA undergoes reversible acetylation at multiple sites after induction with several stimuli, a modification that differentially affects its transcriptional activity (4, 18–20). Specifically, acetylation of Lys310 determines full transcriptional activity of RelA (19). Interestingly, in the presence of wt IRF-1, RelA was also acetylated on Lys310, whereas with the phosphomimetic IRF-1 3D, RelA acetylation was blocked (Fig. 6B). Using inhibitors specific for CBP/p300 (Mc 1783) or for both CBP/p300 and PCAF (anacardic acid), we observed that acetylation at Lys310 was mediated by the histone acetyltransferase (HAT) activity of PCAF but not of CBP/p300 (Fig. 6C). This result is consistent with previous reports demonstrating that IRF-1 interacts with different HATs, including CBP, p300, and PCAF (23), and that PCAF participates in the assembly of the IFN-β enhanceosome prior to the recruitment of CBP (17). Thus, IRF-1 may bridge PCAF and RelA to permit acetylation of RelA. We further established a role for RelA acetylation at Lys310 in RelA/IRF-1 synergism through the use of a RelA K310A mutant that cannot be acetylated at Lys310. The synergistic activation of IFN-β and IL-8 promoters was inhibited by the expression of either IRF-1 3D or RelA 310A mutants and was completely abolished when the two mutants were coexpressed (Fig. 7A and B). RelA acetylation at Lys310 determines the recruitment of the cellular elongation factor PTEF-b by the binding of the bromodomain 4 (Brd4) protein to obtain full transcriptional processivity by RNA polymerase II (Pol II) (43, 44). We speculate that IRF-1, through its ability to promote RelA acetylation at Lys310, may facilitate both PTEF-b recruitment and transcriptional elongation of the IFN-β and IL-8 genes. In the physiological setting of primary CD4+ T cells, we also demonstrated that when IRF-1 is phosphorylated, RelA is not acetylated (Fig. 8A); in pulldown assays, despite the binding of IRF-3, PCAF is not recruited on the IRF-consensus sequence present on the IFN-β promoter (Fig. 8B). Thus, we conclude that in TCR-stimulated T cells, IRF-1 is phosphorylated and, upon poly(I·C) treatment, inhibits PCAF-mediated RelA acetylation.
With regard to other IRFs involved in IFN activation, IRF-3 phosphorylation was observed in activated T lymphocytes, but the phosphorylation pattern was consistent with Ser396 phosphorylation of different IRF-3 isoforms, including the phosphorylation of a splicing isoform that functions as an inhibitor of IFN-β expression (35). In our CD4+ T cell experiments, this IRF-3 isoform may be inactive rather than inhibitory (Fig. 1B and C, lane 4). This observation requires further analysis in the context of positive and negative regulation by IRF-3. IRF-7 appeared to be dispensable for IFN-β expression in CD4+ T cells, since no IRF-7 protein was present after poly(I·C) treatment, despite IFN-β induction (Fig. 1C). Similarly, the accumulation of another IRF factor, IRF-5 (45, 46), was not detected in unstimulated or activated/infected cells (data not shown). In conclusion, we provide evidence that TCR and CD28 engagement in primary T lymphocytes leads to the activation of IKK-ε. Activated IKK-ε in turn directly phosphorylates IRF-1; phosphorylated IRF-1 then blocks PCAF-mediated acetylation of NF-κB RelA and inhibits expression of genes synergistically regulated by IRF-1/RelA, such as IFN-β and IL-8. Experiments investigating in mechanistic detail the synergy between IRF1 and RelA in the activation or repression of immunoregulatory and antiviral gene programs in primary T lymphocytes are ongoing.
ACKNOWLEDGMENTS
We thank Paolo Silveri and Roberto Gilardi for technical assistance and artwork.
This work was supported by grants from the Italian AIDS Project and the Italian Ministry of Health (A.B. and M.S.) and the Canadian Institutes of Health Research (J.H.).
Footnotes
Published ahead of print 6 January 2014
REFERENCES
- 1.Honda K, Takaoka A, Taniguchi T. 2006. Type I inteferon [sic] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 2.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655. 10.1146/annurev.immunol.19.1.623 [DOI] [PubMed] [Google Scholar]
- 3.Nomura F, Kawai T, Nakanishi K, Akira S. 2000. NF-kappaB activation through IKK-i-dependent I-TRAF/TANK phosphorylation. Genes Cells 5:191–202. 10.1046/j.1365-2443.2000.00315.x [DOI] [PubMed] [Google Scholar]
- 4.Rothgiesser KM, Fey M, Hottiger MO. 2010. Acetylation of p65 at lysine 314 is important for late NF-kappaB-dependent gene expression. BMC Genomics 11:22. 10.1186/1471-2164-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azimi N, Shiramizu KM, Tagaya Y, Mariner J, Waldmann TA. 2000. Viral activation of interleukin-15 (IL-15): characterization of a virus-inducible element in the IL-15 promoter region. J. Virol. 74:7338–7348. 10.1128/JVI.74.16.7338-7348.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drew PD, Franzoso G, Becker KG, Bours V, Carlson LM, Siebenlist U, Ozato K. 1995. NF kappa B and interferon regulatory factor 1 physically interact and synergistically induce major histocompatibility class I gene expression. J. Interferon Cytokine Res. 15:1037–1045 [DOI] [PubMed] [Google Scholar]
- 7.Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. 1995. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol. Cell. Biol. 15:2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saura M, Zaragoza C, Bao C, McMillan A, Lowenstein CJ. 1999. Interaction of interferon regulatory factor-1 and nuclear factor kappaB during activation of inducible nitric oxide synthase transcription. J. Mol. Biol. 289:459–471. 10.1006/jmbi.1999.2752 [DOI] [PubMed] [Google Scholar]
- 9.Sgarbanti M, Borsetti A, Moscufo N, Bellocchi MC, Ridolfi B, Nappi F, Marsili G, Marziali G, Coccia EM, Ensoli B, Battistini A. 2002. Modulation of human immunodeficiency virus 1 replication by interferon regulatory factors. J. Exp. Med. 195:1359–1370. 10.1084/jem.20010753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, Ensoli B, Battistini A. 2008. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J. Virol. 82:3632–3641. 10.1128/JVI.00599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. 1996. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature 382:816–818. 10.1038/382816a0 [DOI] [PubMed] [Google Scholar]
- 12.Tamura T, Yanai H, Savitsky D, Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26:535–584. 10.1146/annurev.immunol.26.021607.090400 [DOI] [PubMed] [Google Scholar]
- 13.Lohoff M, Ferrick D, Mittrucker HW, Duncan GS, Bischof S, Rollinghoff M, Mak TW. 1997. Interferon regulatory factor-1 is required for a T helper 1 immune response in vivo. Immunity 6:681–689. 10.1016/S1074-7613(00)80444-6 [DOI] [PubMed] [Google Scholar]
- 14.Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, Mitsuyama M, Shin EH, Kojima S, Taniguchi T, Asano Y. 1997. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6:673–679. 10.1016/S1074-7613(00)80443-4 [DOI] [PubMed] [Google Scholar]
- 15.Merika M, Thanos D. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205–208. 10.1016/S0959-437X(00)00180-5 [DOI] [PubMed] [Google Scholar]
- 16.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667–678. 10.1016/S0092-8674(00)00169-0 [DOI] [PubMed] [Google Scholar]
- 17.Ford E, Thanos D. 2010. The transcriptional code of human IFN-beta gene expression. Biochim. Biophys. Acta 1799:328–336. 10.1016/j.bbagrm.2010.01.010 [DOI] [PubMed] [Google Scholar]
- 18.Chen LF, Greene WC. 2003. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J. Mol. Med. 81:549–557. 10.1007/s00109-003-0469-0 [DOI] [PubMed] [Google Scholar]
- 19.Chen LF, Mu Y, Greene WC. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539–6548. 10.1093/emboj/cdf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758–2766. 10.1074/jbc.M209572200 [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548. 10.1016/S1074-7613(00)00053-4 [DOI] [PubMed] [Google Scholar]
- 22.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- 23.Masumi A, Wang IM, Lefebvre B, Yang XJ, Nakatani Y, Ozato K. 1999. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol. Cell. Biol. 19:1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151. 10.1126/science.1081315 [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- 26.Shen RR, Hahn WC. 2011. Emerging roles for the non-canonical IKKs in cancer. Oncogene 30:631–641. 10.1038/onc.2010.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC, Cantley LC. 2009. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell 34:461–472. 10.1016/j.molcel.2009.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mai A, Rotili D, Tarantino D, Nebbioso A, Castellano S, Sbardella G, Tini M, Altucci L. 2009. Identification of 4-hydroxyquinolines inhibitors of p300/CBP histone acetyltransferases. Bioorg. Med. Chem. Lett. 19:1132–1135. 10.1016/j.bmcl.2008.12.097 [DOI] [PubMed] [Google Scholar]
- 29.Shimada T, Kawai T, Takeda K, Matsumoto M, Inoue J, Tatsumi Y, Kanamaru A, Akira S. 1999. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int. Immunol. 11:1357–1362. 10.1093/intimm/11.8.1357 [DOI] [PubMed] [Google Scholar]
- 30.Peters RT, Liao SM, Maniatis T. 2000. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol. Cell 5:513–522. 10.1016/S1097-2765(00)80445-1 [DOI] [PubMed] [Google Scholar]
- 31.Lin R, Mustafa A, Nguyen H, Gewert D, Hiscott J. 1994. Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J. Biol. Chem. 269:17542–17549 [PubMed] [Google Scholar]
- 32.Sgarbanti M, Arguello M, tenOever BR, Battistini A, Lin R, Hiscott J. 2004. A requirement for NF-kappaB induction in the production of replication-competent HHV-8 virions. Oncogene 23:5770–5780. 10.1038/sj.onc.1207707 [DOI] [PubMed] [Google Scholar]
- 33.tenOever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, Hemmi H, Yamamoto M, Akira S, Yeh WC, Lin R, Hiscott J. 2004. Activation of TBK1 and IKKepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 78:10636–10649. 10.1128/JVI.78.19.10636-10649.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark K, Plater L, Peggie M, Cohen P. 2009. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284:14136–14146. 10.1074/jbc.M109.000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Ma L, Chen X. 2011. Interferon regulatory factor 3-CL, an isoform of IRF3, antagonizes activity of IRF3. Cell. Mol. Immunol. 8:67–74. 10.1038/cmi.2010.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreegipuu A, Blom N, Brunak S. 1999. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 27:237–239. 10.1093/nar/27.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz S, Sun Q, Nakhaei P, Romieu-Mourez R, Goubau D, Julkunen I, Lin R, Hiscott J. 2006. Induction of IRF-3 and IRF-7 phosphorylation following activation of the RIG-I pathway. Cell. Mol. Biol. (Noisy-le-Grand) 52:17–28. 10.1170/T694 [DOI] [PubMed] [Google Scholar]
- 38.Merika M, Williams AJ, Chen G, Collins T, Thanos D. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277–287. 10.1016/S1097-2765(00)80028-3 [DOI] [PubMed] [Google Scholar]
- 39.Dornan D, Eckert M, Wallace M, Shimizu H, Ramsay E, Hupp TR, Ball KL. 2004. Interferon regulatory factor 1 binding to p300 stimulates DNA-dependent acetylation of p53. Mol. Cell. Biol. 24:10083–10098. 10.1128/MCB.24.22.10083-10098.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giroux M, Schmidt M, Descoteaux A. 2003. IFN-gamma-induced MHC class II expression: transactivation of class II transactivator promoter IV by IFN regulatory factor-1 is regulated by protein kinase C-alpha. J. Immunol. 171:4187–4194 http://www.jimmunol.org/content/171/8/4187.long [DOI] [PubMed] [Google Scholar]
- 41.Lin R, Hiscott J. 1999. A role for casein kinase II phosphorylation in the regulation of IRF-1 transcriptional activity. Mol. Cell. Biochem. 191:169–180. 10.1023/A:1006850009017 [DOI] [PubMed] [Google Scholar]
- 42.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T, Honda K. 2006. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc. Natl. Acad. Sci. U. S. A. 103:15136–15141. 10.1073/pnas.0607181103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. 2011. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 85:11752–11769. 10.1128/JVI.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. 2009. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29:1375–1387. 10.1128/MCB.01365-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. 2004. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J. Biol. Chem. 279:45194–45207. 10.1074/jbc.M400726200 [DOI] [PubMed] [Google Scholar]
- 46.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434:243–249. 10.1038/nature03308 [DOI] [PubMed] [Google Scholar]