Abstract
Often considered to be a “dead” kinase, erbB3 is implicated in escape from erbB-targeted cancer therapies. Here, heregulin stimulation is shown to markedly upregulate kinase activity in erbB3 immunoprecipitates. Intact, activated erbB3 phosphorylates tyrosine sites in an exogenous peptide substrate, and this activity is abolished by mutagenesis of lysine 723 in the catalytic domain. Enhanced erbB3 kinase activity is linked to heterointeractions with catalytically active erbB2, since it is largely blocked in cells pretreated with lapatinib or pertuzumab. erbB2 activation of erbB3 is not dependent on equal surface levels of these receptors, since it occurs even in erbB3-transfected CHO cells with disproportionally small amounts of erbB2. We tested a model in which transient erbB3/erbB2 heterointeractions set the stage for erbB3 homodimers to be signaling competent. erbB3 homo- and heterodimerization events were captured in real time on live cells using single-particle tracking of quantum dot probes bound to ligand or hemagglutinin tags on recombinant receptors.
INTRODUCTION
ErbB3 is a member of the epidermal growth factor receptor (EGFR)/erbB family of receptor tyrosine kinases that has been implicated in escape from targeted therapies (1, 2). In the classic view of erbB3 signaling, ligand binding to this “kinase-dead” receptor leads to dimerization with erbB2 or another kinase-competent family member (3, 4). The dimerizing partner is then solely responsible for phosphorylating erbB3 and any binding partners recruited to the erbB3 tail (5). When expressed alone, the erbB3 extracellular domain or full-length receptor is refractory to activation by ligand (6–9). These data, along with experiments using chimeric forms of erbB3 (10), led to the assumption that erbB3 does not homodimerize and that erbB2 is obligatory for erbB3 signaling. Interpretation of these findings must now be revisited in the context of the evolving understanding of both extracellular and intracellular erbB dimerization interfaces (11), as well as new evidence that the erbB3 cytoplasmic tail is capable of measurable kinase activity (12, 13). We reexplored erbB3's catalytic activity, using an in vitro peptide substrate and immunoisolated wild-type or mutant forms of the receptor. One goal of this work was to determine whether native, full-length erbB3 can engage in productive homointeractions (dimers or possibly higher-order oligomers). By conducting our work in live cells, these results strengthen the case that erbB3 signals in part through its modest catalytic activity, as proposed recently by the molecular dynamic simulations of Telesco et al. (14).
Several recent advances now make it possible to directly observe dimerization dynamics for receptors in live cell membranes. When conjugated to ligands or antibodies, quantum dots (QDs) serve as improved probes for single-particle tracking (SPT) (15–17). Our group recently used two-color QD-based probes to track the diffusion-limited interactions of EGFR/erbB1, including observations of homodimerization in real time (18). New analytical methods were developed that distinguish between membrane domain coconfinement and true dimerization events. Mathematical algorithms also permit the estimation of dimer off rates from these data sets. We now apply these novel techniques to the study of erbB3 homo- and heterointeractions, using QD probes conjugated to either erbB3 ligand (heregulin [HRG]) or Fabs that recognize hemagglutinin (HA) tags on recombinant erbB receptors. Note that the term “dimerization” refers herein to two labeled receptors interacting; our methods cannot detect nor rule out the presence of other unlabeled receptors within the complex. The sub-second dynamics captured by SPT complements our prior evidence for nanometer-scale erbB3 signaling patches, based upon immunoelectron microscopy techniques (19).
The results of these studies show that, while ligand-induced homointeractions readily occur on the surface of live cells, erbB3 activation remains dependent on its transient interactions with erbB2 (6, 9). We suggest a modified model for erbB3 signaling, where the first step toward kinase activation is heterodimerization and transphosphorylation. As in prior models, phosphorylated tyrosines on the erbB3 cytoplasmic tail would then serve as docking sites for the p85 subunit of phosphatidylinositol 3-kinase (PI3-kinase) and other adaptor molecules (20, 21). In the revised model, however, activated erbB3 homodimers should also be competent for both transphosphorylation (12) and phosphorylation of cytoplasmic partners. This work has strong implications for computational models of erbB signal transduction, which differ widely in their consideration of erbB3 dimer states and their associated signaling outcomes (14, 22–28).
MATERIALS AND METHODS
Cell lines and reagents.
SKBR3 cells and CHO cells were cultured in McCoy's media and Dulbecco modified Eagle medium, respectively, supplemented with 10% fetal bovine serum (HyClone), penicillin-streptomycin, and 2 mM l-glutamine. HRG-β1 was from US Biological (Swampscott, MA), staurosporine from Calbiochem (La Jolla, CA) and lapatinib from Eton Biosciences, Inc. (San Diego, CA). Recombinant human NRG1-β1/HRG1-β1 extracellular domain (R&D Systems, Minneapolis, MN) was singly biotinylated using NHS-ester conjugation chemistry (Biotin-XX, sulfosuccinimidyl ester; Invitrogen). HRG-biotin and anti-HA Fab-biotin (Roche) were conjugated to Qdot 655 or Qdot 585 streptavidin conjugate (Invitrogen) in phosphate-buffered saline plus 1% bovine serum albumin (BSA) to generate stock solutions of 30 nM 1:1 monovalent QD-conjugates. Anti-erbB1 SC-03 antibody was from Santa Cruz (Santa Cruz, CA); this reagent cross-reacts with erbB2. erbB2 antibodies RB9040-P and MS-325 were from Labvision (Fremont, CA). Mouse 2C4 anti-erbB2 antibody and the expression plasmids, pcDNA6-erbB3wt-mCitrine and pcDNA3-ErbB2wt-mYFP, were gifts from D. Arndt-Jovin (Max Planck Institute for Biophysical Chemistry). The humanized antibody (pertuzumab; Genentech) was purchased from the UNM pharmacy. erbB3 antibodies, SC-285 and SC-415, and p85 antibodies SC-1637 and SC-423 were from Santa Cruz; the p85 06-497 antibody was from UBI (Lake Placid, NY). Antibodies for phospho-Y1248-erbB2 and phospho-Y1289-erbB3 were from Cell Signaling; horseradish peroxidase (HRP)-conjugated PY20 antibodies were from BD Transduction.
erbB2 and erbB3 site-directed mutagenesis, modification, and transfection.
HA-tagged versions of erbB2 and erbB3 were produced using the pcDNA3.1 cytomegalovirus-based expression plasmid (15). The HA tag was introduced between the erbB3 signal peptide sequence and the sequence of the mature erbB3 or erbB2 protein by fusion PCR. Kinase domain mutations were produced using the QuikChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA) with PAGE-purified mutagenesis primers and their reverse complement from Integrated DNA Technologies (Coralville, IA). Plasmids were transfected into CHO cells using Lipofectamine 2000 (Invitrogen) or Amaxa Nucleofector reagents (Lonza). Blasticidin-resistant transfectants expressing erbB3-mCit were sorted by Moflo, followed by single cell growth in 96-well plates and selection of clones with membrane fluorescence. CHO cells expressing HA-tagged receptors were labeled with anti HA Fab-fluorescein isothiocyanate (FITC; Roche) and sorted by Moflo. SKBR3 cells were transiently transfected with HA-erbB3 by AMAXA nucleofection and imaged 2 days posttransfection. For heterodimer tracking, CHO cells stably transfected with HA-erbB2 were transiently transfected with untagged erbB3. Cells were imaged 2 days posttransfection. Membrane localization of the HA-tagged receptors was confirmed by labeling live cells with anti-HA Fab–FITC and imaging on a Nikon TE2000 fluorescence microscope.
Immunoblotting and immunoprecipitation.
Cells were serum starved for 4 h, stimulated with ligands with or without inhibitors as reported in the legends, and solubilized in cold NP-40 lysis buffer (19). Protein concentrations in cleared lysates were measured by BCA assay (Pierce, Rockford, IL). Where stated, amounts of erbB3-mCit protein samples were normalized by quantifying total mCitrine fluorescence using a Qubit fluorometer (Life Technologies) prior to immunoprecipitation. Supernatants were prewashed with protein A-beads (Amersham GE Healthcare, Chicago, IL), followed by incubation at 4°C with fresh beads and primary antibodies. Incubation with rabbit IgG (Cell Signaling) served as a negative control. Immune complexes were denatured and separated by SDS-PAGE, transferred to nitrocellulose membranes, probed with primary and HRP-conjugated secondary antibodies, and detected by the enhanced chemiluminescence method.
In vitro tyrosine kinase assay.
Kinase activity in immune complexes was measured using multiwell strips that were coated with 1 μg of EAY peptide/ml and blocked with BSA (K-LISA). Aliquots of precipitate/slurry were transferred in triplicate to EAY precoated wells, followed by incubation in kinase reaction buffer with 200 μM ATP for 30 min. at 30°C. Phosphorylated substrate was detected using a PY20-HRP antibody and a soluble TMB solution. The optical density at 450 nm was measured to assess the color change of the TMB after the addition of 0.5 N sulfuric acid. For the erbB2 titration assay, total protein from serum-starved SKBR3 lysate was quantified by a BCA assay. Dilutions of the lysate (30 μg to 1 mg of total protein) were immunoprecipitated with an anti-erbB2 antibody, RB9040-P, and the kinase activity was measured for each protein amount. These values were compared to the kinase activity of erbB3 immunocomplexes with or without HRG precipitated from 1 mg of total protein. Replicate samples for each protein amount were assayed for erbB2 or erbB3 kinase activity or subjected to SDS-PAGE and immunoblotted for semiquantitative evaluation. Significant differences in kinase activity were evaluated by using the Student t test on values from at least three trials.
Single particle tracking (SPT), image registration, and processing.
Cells plated for 24 h in eight-well Lab-Tek chambers (Nunc, Rochester, NY) were serum starved for 1 h and then imaged in Tyrode's buffer supplemented with 0.1% BSA–20 mM glucose. Where indicated, 500 nM lapatinib was added 30 min before and during imaging. Cells were labeled with QD anti-HA (100 pM) for 15 min at 37°C and/or QD-HRG (20 pM) for 5 min and washed three times with Tyrode's buffer. Labeled receptors on the apical surface of individual cells were then imaged. Images were acquired at 20 frames/s using an Olympus IX71 inverted microscope with a 60× 1.2-numerical-aperture water objective lens. An objective heater (Bioptechs, Butler, PA) maintained samples at 34 to 35°C. A mercury lamp with a 436/10-nm BP excitation filter provided wide-field excitation. Emission was collected by an electron multiplying charge-coupled device camera (Andor iXon 887) using a QuadView or DuoView image splitter (Optical Insights) to simultaneously image the QD585 (585/20 BP) and QD655 (655/40 BP) probes. Image processing was performed using Matlab (The MathWorks, Inc., Natick, MA) functions in conjunction with the image processing software DIPImage (Delft University of Technology). Single molecule localization, trajectory elongation, and two-channel image registration were performed as previously described (18, 29) with the addition of an affine transform to generate channel overlays. Pairwise trajectories were analyzed to determine the degree of correlated motion (16, 18). Diffusion was calculated based on the mean square displacement of all QD 655 tracks per condition per experiment. Average diffusion was calculated using data from at least three experiments (±the standard error of the mean).
Analysis of dimer states.
Dimer events were identified using a three-state hidden Markov model (HMM), adapted from earlier work (18). The observed parameter is the separation between candidate pairs. Distribution of the displacements between the QDs is modeled by a zero mean Gaussian distribution in each (x, y) dimension using σdimer and σdomain, respectively. The resulting distribution of displacements is a derivation of the actual displacements and the errors in measurement, which are also assumed to contribute as unbiased Gaussian distributions. The value σdimer is estimated by combining information from crystal structure measurements, homology modeling, and the size of QDs. For the “separated” state in the model, the probability density is calculated as a function of the observed distance in the previous frame and a characteristic diffusion constant. Rate constants are determined by maximizing the likelihood over all interactions of two QDs for a specific condition. Standard errors for parameters are calculated as (Hi,i−1)−0.5, where H is the Hessian matrix of the negative log likelihood and i denotes one of the estimated rate constants. The Viterbi algorithm (30) is used to identify the most likely state within individual QD interactions. For diffusion by state calculations, tracks from all candidate pairs from a given condition were partitioned based on interaction state (as determined by HMM analysis and the Viterbi algorithm) and all QD-655 track segments in a given state were analyzed.
RESULTS
Isolated erbB3 receptor complexes contain HRG-induced tyrosine kinase activity.
The isolated erbB3 intracellular domain was recently shown to be capable of weak autophosphorylation (12). To observe the activity of endogenous full-length erbB3, we measured kinase activity in erbB3 immune complexes using a nonradioactive, microplate-based peptide substrate assay (19). Our initial model system was the SKBR3 breast cancer cell line that does not express erbB4. Thus, any HRG-dependent changes can be attributed to stimulation of erbB3. Expression of the other erbB family members in SKBR3 cells was previously determined: <200,000 erbB1, 2 million erbB2, and 70,000 erbB3 per cell (16). As shown in Fig. 1A, isolated erbB3 immune complexes were able to phosphorylate an EAY peptide substrate in a ligand-dependent manner. This kinase assay provides the first concrete evidence that HRG treatment markedly upregulates erbB3 tyrosine kinase activity. Stimulation of erbB3 catalytic activity reached a maximum after 2 min exposure to 12 nM HRG and was strongly correlated with an increase in erbB3 tyrosine phosphorylation (PY blot, Fig. 1A). erbB2 immune complexes are shown alongside erbB3 immune complexes as a control for this assay (Fig. 1B). Note that erbB2 kinase activity is reduced upon HRG stimulation consistent with decreased erbB2 phosphorylation after HRG treatment in this cell line (19).
FIG 1.
Robust tyrosine kinase activity in erbB3 immune complexes correlates with erbB2-dependent HRG-induced erbB3 phosphorylation. (A) SKBR3 cells were stimulated with 12 nM HRG-β for the indicated treatment interval, lysed, and immunoprecipitated with an erbB3-specific antibody. Immune complexes were evaluated for kinase activity by in vitro K-LISA using an EAY peptide substrate (top panel). Relative kinase activity is reported as changes in the 450-nm absorbance of the chromogen substrate. Bars are the mean of three replicates ± the standard deviations (SD). (B) SKBR3 cells were stimulated with HRG-β as in panel A for 2 min, lysed, and immunoprecipitated with a rabbit IgG control or for erbB2 or erbB3 as indicated. (C) Other erbB family members are not present in erbB3 immunoprecipitates (IPs). SKBR3 total lysates or erbB3 IPs were immunoblotted and probed with an anti-EGFR antibody that recognizes both EGFR and erbB2, and erbB2-specific or erbB3-specific antibodies. (D) Dilutions of SKBR3 total lysate were immunoprecipitated with anti-erbB2 antibody and tested for kinase activity or immunoblotted and probed for erbB2 protein. K-LISA results are shown as the average activity of two to three trials ± the SD. erbB2 protein was detectable in all erbB2 immune complexes (representative blot below the graph). No erbB2 band was visible in the erbB3 IPs. (E) Kinase activity of erbB3 IPs with or without 1 μM staurosporine that was added directly to the kinase reaction. Rabbit IgG was used as a negative control. (F and G) Mutation of the erbB3 kinase domain modulates erbB3 phosphorylation and kinase activity. (F) Immunoblot shows the phosphorylation status detected by an anti-PY20 antibody of erbB2 or erbB3 IPs from CHO cells expressing erbB2 wt, erbB2 K753M, erbB3 wt, or erbB3 K723M with or without 2 min of stimulation with 12 nM HRG. (G) erbB kinase activity was measured in erbB IPs identical to those shown in panel F. The kinase activity was significantly higher in erbB3 wt plus HRG than without HRG. erbB3-K723M IPs showed no HRG-dependent increase in kinase activity. Values are normalized to erbB2 kinase activity and represent the mean value of three independent experiments ± the SD. P values are based on statistical analysis by using the Student t test. N.S., not significant.
Kinase activity in erbB3 immune complexes is not attributed to associated erbB1, erbB2, or Src kinases.
Because of the long-standing presumption that erbB3 kinase activity is negligible, it was important to rule out that the kinase activity in erbB3 immune complexes is not due to coprecipitation with its dimerizing partners in SKBR3 cells. erbB3 immunoprecipitates were subjected to SDS-PAGE and blotted for potential partners, erbB1 or erbB2 (Fig. 1C). As shown, erbB1 and erbB2 were not detectable in the erbB3 immunoprecipitates.
We considered the remote possibility that the kinase activity is attributable to trace amounts of coprecipitating erbB2 not detectable by Western blotting. We addressed this two ways. First, erbB2 immunoprecipitates were prepared from between 1 mg and 30 μg of total protein to immobilize various concentrations of erbB2 on the beads. Replicate samples under each condition were assayed for constitutive erbB2 kinase activity or subjected to SDS-PAGE and immunoblotted for semiquantitative evaluation. As shown in Fig. 1D, erbB2 protein levels sufficient to match the kinase activity of the erbB3 sample can clearly be detected by immunoblotting. Thus, while minute amounts of coprecipitating erbB2 cannot be completely ruled out, the titration experiment strongly supports the conclusion that erbB2 alone is not responsible for the kinase activity of erbB3 immune complexes.
As a second approach to rule out potential contribution by coprecipitating kinases, we applied a panel of ATP-binding competitive inhibitors directly to the in vitro kinase reaction. No inhibition was seen with erbB1-2 inhibitors AG1478, AG879 and PD153035 or with the Src inhibitor PP2 (data not shown). Inhibition of HRG-dependent erbB3 kinase activity was observed only with staurosporine (Fig. 1E); this inhibitor is known to have very broad specificity (31). Although these results are not definitive, we speculate that the inability to completely block erbB3 activity with staurosporine could be due to the unusual catalytic site proposed for erbB3 (12).
A TranSignal phosphotyrosine profiling array (Panomics, Redwood City, CA) was screened for potential erbB3 binding kinases. This array, spotted with SH2-domains derived from 13 cytoplasmic tyrosine kinases (Abl, Csk, BTK, Zap-70, and all Src-family members), was incubated with SKBR3 lysates then probed for erbB2 or erbB3. None of the panel of cytoplasmic tyrosine kinases bound to erbB3. erbB3-specific binding was only seen with the p85 regulatory subunit of PI3 kinase, which has no tyrosine kinase activity and is a known binding partner of erbB3 (data not shown).
The K723M mutation in the erbB3 kinase domain abrogates kinase activity.
Our next goal was to directly alter erbB3 kinase activity using a mutagenesis approach. The erbB3 kinase domain lacks a catalytic aspartic acid conserved in the other three erbB family members (D813 in erbB1). In place of the aspartic acid, erbB3 has an asparagine (N815). Shi et al. suggested the possibility that erbB3 compensates for loss of the catalytic base by using an alternate reaction pathway for phosphoryl transfer (12). Based upon this information, and prior knowledge of specific kinase domain mutations that inhibit erbB activity (32–35), we focused on a conserved lysine at position K723 in erbB3 as a site for mutagenesis. This lysine (K721 in erbB1) is required for ATP-binding in erbB1 (32), erbB2, and erbB3 (12) and, when substituted by a methionine in the recombinant erbB3 cytoplasmic tail, abolished in vitro erbB3 activity (12). We constructed expression vectors for both full-length wild-type erbB3 (erbB3wt) and the K723M mutant (erbB3K723M). Both constructs were fused in frame to mCitrine to facilitate flow sorting and to confirm comparable plasma membrane localization of the recombinant proteins when stably expressed in Chinese hamster ovary (CHO) cells. CHO cells lack endogenous erbB3 but express low levels of endogenous erbB2. Here, the cell lines expressing these erbB3 proteins are referred to as erbB3wt-mCit and erbB3K723M-mCit.
Results in Fig. 1F and G compare the relative phosphorylation and activity of wild-type and mutant erbB3. As shown in Fig. 1F, phosphorylation of erbB3wt-mCit was robust in response to 12 nM HRG treatment, indicating that the low levels of endogenous erbB2 are sufficient to activate erbB3. Phosphorylation of erbB3K723M-mCit in response to HRG treatment was dramatically lower despite the observation that the total amount of mutant protein was significantly higher than wild type. Lysates from erbB3wt-mCit and erbB3K723M-mCit CHO cells were prepared with or without HRG stimulation, normalized based upon mCitrine fluorescence, and the kinase activity of erbB3 immune complexes was compared. The results show that HRG fails to increase kinase activity in erbB3K723M-mCit immune complexes above background levels (Fig. 1G). Together, the data suggest that erbB3 is an autophosphorylation substrate, as well as a substrate of erbB2.
As an additional control, we evaluated the kinase activity of both the wild type and a K753M mutant of erbB2 expressed in CHO cells as fluorescent fusion proteins. Figure 1F shows that wild-type human erbB2-YFP was phosphorylated in live CHO cells. As shown in Fig. 1G, erbB2wt-YFP also had measurable in vitro kinase activity in anti-human erbB2 immune complexes (Fig. 1G). By comparison, activity was abolished in the human erbB2K753M-YFP mutant. These results validate the approach and demonstrate that mutation of the homologous crucial lysine in the ATP-binding domain of either erbB2 or erbB3 abolishes kinase activity. Furthermore, these data provide definitive evidence that kinase activity in erbB3 immune complexes, prepared from either SKBR3 or CHO cells expressing wild-type receptor, is a reflection of erbB3's own catalytic potential.
erbB2 mediates activation of erbB3.
To explore the link between erbB2-mediated phosphorylation of erbB3 (36) and erbB3 kinase activity, we pretreated cells with erbB2 inhibitors prior to stimulation with HRG and immunoprecipitation of erbB3. We chose two classes of erbB2 inhibitors: (i) lapatinib, a dual erbB1-erbB2 cell-permeable tyrosine kinase inhibitor, and (ii) the monoclonal 2C4 antibody that blocks the erbB2 dimerization arm (37). The humanized form of 2C4, pertuzumab, is used for treatment of erbB2-positive tumors (38).
Results for experiments in SKBR3 cells are shown in Fig. 2A and B. 2C4 had little effect on erbB2 phosphorylation, possibly due to the high basal phosphorylation of erbB2 in these cells. However, pretreatment with 2C4 reduced the HRG-dependent increase in erbB3 phosphorylation. Treatment with lapatinib abolished erbB2 phosphorylation and significantly blocked the increase in erbB3 tyrosine phosphorylation seen in response to HRG. Note that lapatinib increased the level of total erbB2 as reported previously (39). Lapatinib also appears to stabilize total erbB3 levels. The top panels in these figures show blotting results, while the graphs below report erbB3 tyrosine phosphorylation as a ratio to total erbB3. Importantly, pretreatment with the erbB1-2 inhibitor lapatinib also blocked upregulation of erbB3 kinase activity in response to HRG (Fig. 2C). erbB2 kinase activity was only partially reduced after pretreatment with lapatinib, likely due to the reversible nature of lapatinib inhibition. Pretreatment with 2C4 antibodies or lapatinib also blocked erbB3 activation in CHO cells expressing erbB3wt-mCit (Fig. 2D and E).
FIG 2.
erbB3 activation is erbB2 dependent. (A) The anti-erbB2 antibody, 2C4, reduces HRG-dependent erbB3 phosphorylation in SKBR3 cells. SKBR3 lysates from cells that were untreated (−), treated for 1 h with 100 nM 2C4 (2C4), stimulated 2 min with 3.2 nM HRG (+HRG), or treated for 1 h with 100 nM 2C4 and stimulated 2 min with 3.2 nM HRG (2C4+HRG) were separated by SDS-PAGE and immunoblotted with antibodies against phospho or total erbB3 and erbB2. The graphs in panels A and B report the ratio of p-erbB3 to total erbB3 based upon quantitative analysis of band intensity. (B) Lapatinib inhibition of erbB3 phosphorylation in SKBR3 cells. SKBR3 lysates from cells treated with or without 500 nM lapatinib for 4 h and then with or without 12 nM HRG for 2 min were immunoblotted as in panel A. (C) Pretreatment of SKBR3 cells with lapatinib reduces erbB2 and erbB3 kinase activity. SKBR3 cells were treated as in panel B, followed by immunoprecipitation with anti-erbB2 or erbB3 antibodies. Note that erbB2 kinase activity was only reduced by ∼50%, likely due to the reversible nature of lapatinib inhibition (54). (D) 2C4 pretreatment, as in panel A, reduces erbB3 phosphorylation after 3.2 nM HRG stimulation in CHO erbB3wt-mCit cells. (E) Lapatinib treatment inhibits erbB3 phosphorylation in CHO erbB3wt-mCit cells. Cells were serum starved, pretreated with 1 μM lapatinib for 30 min, and then stimulated with 12 nM HRG for 2 min. Western blot analyses were performed, and bands were quantified as in panel A.
Several important conclusions can be drawn from these experiments. First, since lapatinib does not directly affect erbB3 activity (12), erbB3's phosphorylation and activation status is linked to its encounters with catalytically active erbB2. Second, these encounters require the erbB2 dimerization arm that is blocked by pretreatment of cells with 2C4. Third, the ability of erbB2 to activate erbB3 is not tightly coupled to their levels of expression, since erbB3 can be activated in both SKBR3 cells that overexpress erbB2 and in CHO cells that have much lower levels of endogenous erbB2 compared to the recombinant erbB3. Thus, it is possible that serial engagements with a small number of erbB2 are able to activate a larger number of erbB3.
It is worth noting that, unlike lapatinib, 2C4 treatment had negligible effects on erbB2 autophosphorylation. We speculate that erbB2 homodimers may be stabilized by the bivalent antibody and that the dimer may be active for transphosphorylation despite the bulky antibody bridging the two dimerization arms.
QD probes permit tracking of erbB3 and erbB2 receptors.
Our next goal was to apply SPT methods to track the interactions of erbB3 and erbB2 on the surfaces of SKBR3 and CHO cells. Figure 3A depicts the structures of two hypothetical receptor dimer pairs bound to our QD probes: an erbB3 homodimer (top) and an erbB3/erbB2 heterodimer (bottom). For tracking of ligand-bound erbB3, HRG-β was biotinylated and conjugated to commercial streptavidin-QDs. We confirmed that conjugation of ligand to QDs does not affect its ability to activate erbB3 by treating SKBR3 cells with 12 nM unconjugated or QD-conjugated HRG (a concentration of ligand that yields robust erbB3 phosphorylation). The extent of erbB3 phosphorylation was comparable to unconjugated HRG (Fig. 3B, upper panel).
FIG 3.
Characterization of erbB3 diffusion by single particle tracking (SPT). (A) Configuration of QD probes bound to hypothetical dimer structures: an erbB3 homodimer tracked with two QD-HRG probes (top) and an erbB3/erbB2 heterodimer tracked with a QD655-HRG probe and a QD585 anti-HA Fab probe (bottom). Structural information from erbB1 dimers, erbB2, Fab fragments, and reported QD diameters (55) was used to estimate the distance between QDs in a dimer pair. Homology modeling indicates that erbB3 homodimer distances would be comparable to those of erbB1 (43). (B) For the top panel, SKBR3 cells were stimulated for 2 min with 12 nM unlabeled HRG, biotinylated HRG (bHRG), or biotinylated HRG conjugated to QDs (QD-HRG). erbB3 phosphorylation was assessed by immunoblotting with the phospho-specific PY1289 erbB3 antibody and normalizing to the total erbB3. The lower panel shows that binding of the QD-conjugated anti-HA Fab to HA-erbB3 does not interfere with erbB3 phosphorylation. To ensure that steric hindrance of the QDs do not disrupt erbB3 activation, serum-starved CHO-HA-erbB3 cells were incubated for 30 min with or without QD anti-HA Fab and then stimulated with 12 nM HRG for 2 min. (C) Diffusion coefficients of ligand-bound erbB3 tracked on the surface of SKBR3 cells with QD-HRG with or without 30 min pretreatment with 500 nM lapatinib. In SKBR3 cells transiently transfected with HA-erbB3, unliganded erbB3 was tracked with QD anti-HA and liganded erbB3 was tracked with QD anti-HA plus 100 nM unlabeled HRG or QD-HRG. (D) Diffusion coefficients of unliganded or liganded HA-erbB3 (wild type or kinase dead [K723M]) tracked on the surface of CHO cells. The diffusion coefficients shown in panels C and D were calculated by fitting the ensemble mean square displacement (MSD) of each experiment, and the average of at least three experiments was plotted (±the standard error of the mean). P values are based on statistical analysis using the Student t test.
To track erbB2 or unliganded erbB3, we expressed HA-tagged versions of these two receptors in both SKBR3 and CHO cells. The design of these constructs is described in Materials and Methods. A biotinylated anti-HA Fab antibody fragment was conjugated to quantum dots (Fig. 3A). Binding of the anti-HA Fab-QD probe did not effect phosphorylation of erbB3 after stimulation with 12 nM unlabeled HRG (Fig. 3B, lower panel). For all SPT experiments, cells were grown on chamber slides and mounted on an inverted Olympus microscope equipped with an objective heater. Picomolar concentrations of QD probes were added directly to the imaging buffer using pairs of QDs with distinct emission spectra (QD585 and QD655); the low labeling ensures the ability to resolve individual particles during analysis. All data reported were collected after focusing on the apical (upper) surface of adherent cells.
erb3 diffusion reflects its activation state.
Figure 3C shows the diffusion characteristics of QD-bound erbB3. Average diffusion coefficients were calculated by fitting the ensemble mean square displacement of all trajectories tracked within a single experiment. Values are the average of at least three experiments ± the standard error. Endogenous ligand-bound erbB3 on the surface of SKBR3 cells tracked using the HRG-QD probe had an average diffusion coefficient of 0.013 μm2/s (white bar). Importantly, preincubation of SKBR3 cells with the erbB2 inhibitor, lapatinib, increased the diffusion coefficient of liganded erbB3 to 0.021 μm2/s (hatched bar). This increased diffusion may be due to effects of blocking erbB signaling capabilities, remodeling of the local membrane environment or its associated cytoskeleton, or on the failure of lapatinib-bound erbB2 (which is locked in an “inactive” kinase domain configuration) to form a stable interaction with erbB3.
By expressing HA-tagged erbB3 in SKBR3 cells, the diffusion characteristics of unliganded and liganded receptor could be compared in the presence of high levels of endogenous erbB2. Unliganded erbB3 tracked with the QD anti-HA Fab probe was the fastest with a diffusion coefficient of 0.038 μm2/s (black bar). This is in agreement with diffusion of erbB3 (0.036 μm2/s) measured by FRAP in untreated A431 cells expressing erbB3-mCitrine (40). Ligand-bound receptor tracked with the QD-HRG probe (tracking both endogenous and exogenous erbB3) was significantly slower with an average diffusion coefficient of 0.010 μm2/s, which is comparable to that of ligand-bound endogenous receptor alone (compare white bars). This slower diffusion of ligand-bound receptors may be associated with receptor activation as seen with EGF-bound erbB1 (18). Tracking with the QD anti-HA Fab probe in the presence of excess dark HRG (100 nM) yielded intermediate diffusion values (gray bar), which may reflect a mixed population of liganded and unliganded receptors.
Since SKBR3 cells express endogenous erbB3 and overexpress erbB2, we compared the diffusion characteristics of wild-type versus K723M erbB3 in transfected CHO cells, where erbB3 expression is restricted to recombinant tagged forms (HA-erbB3wt or HA-erbB3K723M). The results are summarized in Fig. 3D. Of note, HA-erbB3wt showed a diffusion coefficient of 0.019 μm2/s in the absence of ligand (black bar). The slightly slower diffusion of unliganded erbB3 in CHO cells, compared to SKBR3 cells, may reflect cell-specific topographical features. In CHO cells, ligand-bound HA-erbB3wt tracked with the QD-HRG probe was significantly slower than unliganded receptor (P = 0.014) with an average diffusion coefficient of 0.010 μm2/s (white bar), similar to measurements in SKBR3 cells.
Finally, HA-erbB3 bearing the K723M mutation showed markedly faster diffusion than HA-erbB3wt even in the absence of ligand (Fig. 3D). This faster diffusion combined with the reduced phosphorylation of erbB3K723M when expressed in CHO cells (Fig. 1F) suggests that this mutation may alter the structure of the kinase domain in a way that affects N-lobe mediated receptor interactions (41). The fast diffusion of resting HA-erbB3K723M with a diffusion coefficient of 0.031 μm2/s was only slightly diminished by the addition of ligand (0.025 μm2/s) (white bar, Fig. 3D), a finding consistent with the very small change in phosphorylation (Fig. 1F, far right lane). Diffusion of ligand-bound HA-erbB3K723M was >2-fold faster than HA-erbB3wt (P = 0.05).
QD probe tracking captures erbB3-erb2 heterointeractions and erbB3-erbB3 homointeractions.
Previous work showed that single particle methods permit the visualization of erbB dimerization in real time (18, 42). In the present study, erbB3 homo- and heterointeractions were observed in both SKBR3 cells that overexpress erbB2 and in CHO cells that express low levels of endogenous erbB2. Figure 4A and B show representative distance traces with inset images of an erbB3/erbB2 heterodimerizing event (Fig. 4A) and an erbB3 homodimerizing event (Fig. 4B) captured on the surface of live cells. The individual QD-bound receptors are distinguished based on the distinct emission spectra of the two probes (insets in Fig. 4D to I). erbB3-erbB2 pairs were tracked in CHO cells expressing erbB3 and HA-tagged erbB2. Heterointeractions were not tracked in SKBR3 cells since the high expression of endogenous erbB2 overwhelmingly favors “dark” (unlabeled) interactions. At the start of imaging in Fig. 4A, one erbB3 (bound to QD-HRG) is separated by over a micron from an HA-erbB2 (bound to QD anti-HA Fab) on the surface of a CHO cell. These receptors diffuse, approach, and form a dimer at ∼41 s into the image acquisition; they dissociate after 5 s.
FIG 4.
Single particle tracking (SPT) detects short-lived erbB2-erbB3 heterodimers and longer-lived erbB3 homodimers. (A and B) Receptor dimerization behavior based on tracking of a pair of QD-tagged receptors over time. Shown are representative plots of separation distances for an erbB2-erbB3 heterodimer on CHO cells expressing exogenous HA-erbB2 and untagged erbB3 (A) and endogenous erbB3 receptors interacting on SKBR3 cells (B). Horizontal lines indicate state calls (right axis, dimer, coconfined, or separated receptors) as determined by a Viterbi algorithm. Insets show images of pairs at specific time frames. Scale bar, 1 μm. (C) Diagram illustrating interaction states of erbB3 tracked with two colors of QD-HRG and with different levels of unlabeled erbB2 (gray spheres). (D to I) Histograms plotting the distribution of dimer lifetimes for receptor pairs as indicated (diagrams). koff values are predicted from the HMM analysis.
erbB3 homointeractions were observed in both CHO cells expressing HA-erbB3 and SKBR3 cells tracking endogenous erbB3 or HA-erbB3. Figure 4B shows single particle analysis applied to an erbB3 homodimer on the surface of an SKBR3 cell using two different colors of HRG-conjugated QDs. A notable feature seen in the distance trace in Fig. 4B is that before the two receptors form a homodimer, and between dimerization events, they are transiently trapped together in a restricted membrane compartment (denoted with a magenta bar). This phenomenon is referred to as “coconfinement” (18, 43) and has been variously attributed to trapping by cytoskeletal corrals (16, 44) or short-term residency in lipid rafts (45) or protein islands (46). Coconfinement and dimerization events can be quantitatively evaluated by our three-state hidden Markov model (HMM) analysis that classifies the kinetics of pairs of receptors interacting in three states: distant (“separated”), coconfined, or interacting (“dimer”). Further, by applying a Viterbi algorithm, it is possible to assign these states to the changing relationships of receptor pairs during imaging (18). In the HMM analysis, the probability is optimized if the separation distance approaches the predicted dimer distance based on structural features of the dimer and the QD probe pair (47) (Fig. 3A). As previously observed for erbB1 (18), HMM analysis revealed that coconfinement in domains promotes repeated homodimerization events between individual pairs of erbB3 receptors. In this figure, the two tracked receptors dimerize, dissociate and redimerize four times over the 50 s of image acquisition. Movies of the interactions plotted in Fig. 4A and B are provided in the supplemental material.
Because two-color SPT requires underlabeling of receptors and can only resolve interactions between two receptors labeled with spectrally distinct QDs, it is important to acknowledge that unlabeled erbB2 may be influencing the interactions of labeled erbB3 receptors. Figure 4C depicts the interactions of a pair of QD-labeled erbB3 receptors within the three interaction states: “separated,” “coconfined,” or interacting as a “bound pair” in the presence of high levels (left) or low levels (right) of unlabeled erbB2. The advantage of tracking erbB3 interactions in both SKBR3 cells and CHO cells is that interactions with unliganded erbB2 are more likely to occur in SKBR3 cells that overexpress endogenous erbB2 compared to CHO cells. Similar erbB3 interactions seen in both cell lines could indicate that these interactions are not entirely dependent on erbB2.
Figure 4D to I illustrate dimer lifetimes obtained from tracking tens to hundreds of individual interaction events per probe set. The plots in these graphs show the distribution of hetero- and homodimer lifetimes, measured in seconds. The results are shown for erbB3 interactions when tracked on both cell types (SKBR3 and CHO) and using both QD probes for erbB3 (QD anti-HA Fab or QD-HRG). Since dissociation is a stochastic process, interactions persist from <1 s to >50 s. Importantly, increases in the number of long-lived events reflect a higher dimer stability and provide insight into receptor signaling capacity. For example, although erbB3 receptors interact in the absence of ligand, these interactions are remarkably short-lived. This is illustrated in Fig. 4D, where >75% of minimal dimers dissociate within 2.5 s. By comparison, almost 50% of liganded-erbB3 homointeractions persist longer than 2.5 s, with a significant number of events ranging from 5 to 50 s in duration (Fig. 4E, H, and I). On the whole, erbB3-erbB2 interactions are less stable than liganded erbB3 pairs, with the majority of events lasting <2.5 s (Fig. 4F).
An important output of HMM analysis is estimation of dimer stability (18). The off rates for heregulin-bound erbB3 minimal homodimers measured using the QD-HRG probe on two different cell lines are similar, with values ranging from 0.13 ± 0.04 s−1 (CHO-HA-erbB3) to 0.18 ± 0.02 s−1 (SKBR3). By comparison, erbB3 pairs that interact in the absence of ligand have fast dissociation rates (between 0.44 ± 0.08 s−1 [CHO-HA-erbB3] and 1.08 ± 0.1 s−1 [SKBR3-HA-erbB3]).
The off rates for erbB3-erbB2 minimal heterodimers are estimated to be 0.41 ± 0.06 s−1. Heterodimer dissociation was measured only in CHO cells, which were transfected with HA-erbB2 and have low endogenous erbB2 levels. This strategy was not applicable to the SKBR3 system because the abundant (unlabeled) endogenous erbB2 dominates the behavior of the system, and there is a low probability of capturing rare heterodimer events with the small fraction of tagged receptors.
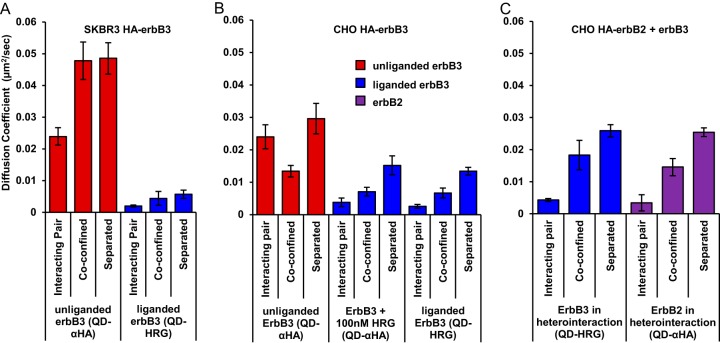
Slowed diffusion of interacting receptor pairs can be an indication of productive receptor interactions that lead to downstream signaling (18). Analyzing the mean square displacement of QD-labeled erbB3 and erbB2 receptors based on their interaction state shows that receptors, when they are part of an interacting pair, diffuse slower than receptors that are either coconfined or separated (Fig. 5). In both cell types, unliganded erbB3 interacting pairs have a similar diffusion coefficient of 0.024 ± 0.004 μm2/s. Liganded erbB3 diffusion is slower (0.002 to 0.004 μm2/s) when observed to pair as homo- or heterodimers. Although erbB2-erbB3 heterointeractions appear to be transient, reduced diffusion of interacting pairs suggests that these interactions may still be productive. In SKBR3 cells, QD-HRG-labeled erbB3 is uniformly slow, presumably due to interactions with abundant dark erbB2. The slowdown is less pronounced in CHO cells, where dark erbB2 is scarce. We assume that the slower diffusion of unliganded erbB3 in CHO cells even when separated (Fig. 5B, red bars) reflects unknown topological differences between CHO and SKBR3 membranes.
FIG 5.
Analysis of diffusion by state shows slowed diffusion when receptors form complexes (minimal dimers). Diffusion coefficients based on fitted mean square displacement are plotted for unliganded (QD-HA) and liganded (QD-HRG) erbB3 in SKBR3 HA-erbB3 cells (A), unliganded and liganded HA-erbB3 in CHO HA-erbB3 cells (B), and liganded erbB3 (QD-HRG) and HA-erbB2 (QD anti-HA) in CHO HA-erbB2 plus erbB3 cells (C). Error bars show the 95% confidence interval returned from the weighted least-squares fit.
Population analysis of all erbB3 trajectories confirms erbB3 homointeractions are significant events.
The importance of erbB3 homodimerization has been debated in the literature (10, 48, 49). To rigorously address this, Fig. 6 shows the results of a statistical evaluation of erbB3 homointeractions based upon the entire data set of trajectories on SKBR3 cells using combinations of two-color QD probes. Pairs of two-color, QD-labeled erbB3 receptors with a separation distance of <1 μm were analyzed for two criteria that reflect dimerization: uncorrelated jump distance and jump magnitude (18). Uncorrelated jump distance reflects the fact that trajectories for each candidate pair of receptors are uncorrelated unless the two receptors are tightly bound, in which case their motion becomes correlated. At long separation distances between pairs of receptors, this quantitative analysis will always report their uncorrelated motion, since they are too far apart to interact. At short distances, binding events may occur and are indicated as a sharp decrease in uncorrelated motion. Note that receptors, which are coconfined (in a signaling patch, for instance), may be physically close together but will not show correlated motion (16). Jump magnitude, the average displacement of receptors between imaging frames, can report changes in mobility observed as receptors form complexes. As shown in Fig. 6, both of these values are informative when plotted as a function of receptor proximity.
FIG 6.
Liganded erbB3 receptor pairs show correlated motion and reduced diffusion at small separation distances. Uncorrelated jump distance and jump magnitude of QD pairs on SKBR3 cells are plotted as a function of separation distance. Correlated motion is indicated by a reduction in uncorrelated jump distance (blue) at small separation distances. A similar reduction in jump magnitude (red) indicates reduced diffusion of interacting receptors. Error bars represent standard error of the mean. (A) Ligand-bound endogenous erbB3 pairs in SKBR3 cells show correlated motion. (B) Preincubation with 100 nM lapatinib reduces correlated motion of ligand bound erbB3 receptors, as shown by larger displacement at short separation distances. (C) In SKBR3 cells expressing HA-erbB3, unliganded HA-erbB3 receptors show no correlated motion. (D) The addition of 12 nM unlabeled HRG results in correlated motion at small separation distances. (E) Pairs of ligand-bound receptors (HA-erbB3 or endogenous erbB3) tracked with QD-HRG probes in SKBR3 HA-erbB3 cells show correlated motion similar to endogenous erbB3 in SKBR3 cells (see panel A).
Liganded erbB3 shows a reduction in uncorrelated jump distance and a concurrent reduction in jump magnitude at short separation distances consistent with stable homodimer formation (Fig. 6A). This correlated motion is less pronounced in the presence of lapatinib (Fig. 6B), suggesting the possibility that homointeractions are stabilized if one or both receptors have been phosphorylated by erbB2 in an earlier interaction. Unliganded HA-erbB3 shows limited correlated motion at short separation distances with no decrease in jump magnitude (Fig. 6C). These data are consistent with a fast off rate (0.44 s−1 [Fig. 4D]) for homointeractions observed between unliganded erbB3; these interactions are so transient that they may rarely result in transphosphorylation and recruitment of docking partners that contribute to receptor slowdown (18).
After the addition of 12 nM unlabeled HRG, sharp decreases in uncorrelated jump distance and jump magnitude are observed for HA-tagged erbB3, suggesting dimer formation of ligand-bound receptors (Fig. 6D). When events were tracked using HRG-conjugated QDs (Fig. 6E), the cumulative data also validate the formation of stable erbB3 minimal homodimers that occur while ligand is bound (off rate of 0.16 s−1 for SKBR3-HA-erbB3 cells). These events show both correlated motion at short distances and the slowed mobility expected for a larger protein complex comprised of erbB3 receptors and associated docking partners. Similar results were observed in CHO cells expressing HA-erbB3.
DISCUSSION
erbB3 was formerly thought to be an obligate heterodimer due to the difficulty of measuring homodimerization (8–10) and tyrosine kinase activity (3, 4, 36). It is important to note that early studies of erbB3 kinase activity relied on measurements using only the cytoplasmic tail or measured activity of erbB3 without ligand or in the absence of activation by other erbB family members (3, 4, 9). Indeed, our results agree with prior work concluding that erbB3 basal kinase activity is weak (Fig. 1, unstimulated cells). However, we now show that upregulation of kinase activity in the intact erbB3 receptor is markedly dependent on diffusion-limited interactions with a heterodimeric partner, such as the constitutively active erbB2. Ligand binding promotes this step. Low levels of erbB2, as found in CHO cells, were sufficient to phosphorylate wild-type erbB3 upon HRG stimulation (Fig. 1F). However, the kinase-dead erbB3 mutant, K723M, showed little phosphorylation in these cells, suggesting that erbB3 autophosphorylation may contribute to total erbB3 phosphorylation. Alternatively, the K723M mutant could alter the structure of the kinase domain, thereby limiting heterointeractions and reducing erbB3 phosphorylation by erbB2. Although the proposed catalytic mechanism for erbB3 is thought to be unique (12), mutation of a conserved lysine in the ATP-binding domain, K723M, can ablate HRG-dependent kinase activity (Fig. 1G). Taken together with the observations of Lemmon and coworkers that in vitro activity of the erbB3 cytoplasmic tail is highly dependent on achieving an appropriate local density on the surface of beads (12), it is not surprising that erbB3's role as a kinase has been undervalued.
In agreement with previous work, erbB3 phosphorylation was shown to be erbB2-dependent in our system (6, 9, 35, 50). We further suggest a role for erbB2 in the transactivation of erbB3, since erbB3 kinase activity was reduced after pretreatment with lapatinib (Fig. 2). Thus, transient binding of erbB3 to erbB2 may lead to activation of erbB3. By analogy with the integrin receptors that undergo “inside-out” upregulation (51), the transactivation step could potentially involve marked conformational changes in either the extracellular or intracellular domains that enable subsequent erbB3 homodimerization. Since erbB2-erbB3 interactions are accompanied by phosphorylation of erbB3, the simplest explanation may be that erbB2 directly phosphorylates erbB3 on a residue in the kinase domain to upregulate its catalytic activity. Interestingly, a Y849E mutation was introduced into the erbB3 activation loop by Shi et al. (12); rather than acting as a phospho-mimetic to activate kinase activity, this mutation reduced erbB3 autophosphorylation. If tyrosine phosphorylation of erbB3 is the activating stimulus for erbB3 kinase activity, it is likely to occur at another tyrosine site.
In the current asymmetric dimer model for activation of erbB kinase domains, one kinase domain serves as an “activator” for the other (52). The isolated kinase domain of erbB3 is able to bind as an activator but not an acceptor even when coincubated with the erbB2 kinase domain (41, 53). Interestingly, chimeras composed of erbB3 extracellular and erbB1 intracellular domains showed differential HRG-dependent activity depending on which transmembrane domain was used in the construct, stressing the importance of interdomain interactions (10, 50). If active erbB3 forms asymmetric dimers, then the transactivation of erbB3 by erbB2 is likely influenced by interactions outside the kinase domain. We note that lapatinib treatment increased erbB3 diffusion (from 0.013 to 0.021 μm2/s), which may be linked to the fact that this inhibitor stabilizes the “inactive” conformation of its target kinase domain (54).
We took advantage of our capabilities in single-particle tracking (16–18, 55) to show that erbB3 receptors slow down upon ligand binding, becoming nearly immobile in both SKBR3 and CHO cell lines. This is similar to changes in diffusion seen with ligand binding to erbB1 (18). erbB3 readily engages in both homointeractions and heterointeractions with erbB2. When measured in transfected CHO cells, erbB3 minimal homodimers are ∼3-fold-more stable than erbB3-erbB2 minimal heterodimers (off rates of 0.13 versus 0.41 s−1). This is consistent with our previous immunoelectron microscopy colocalization analyses, showing reorganization of erbB3 into large “signaling patches” and only partial overlap with erbB2 (19).
This work supports a reinterpretation of the traditional erbB3 signaling model. After ligand binding leads to stabilization of the extended form of the erbB3 extracellular domain, erbB3 more readily engages in both hetero- and homointeractions. Based upon inhibition by 2C4, which blocks the erbB2 dimerization arm, erbB2-erbB3 pairs are likely to form “conventional” dimers using both dimerization arms. However, our data still allow for the possibility of tetramers formed from two heterodimers (56). Although bound to erbB2, erbB3 is transactivated. Due to a relatively fast off rate (0.41 s−1), minimal heterodimers dissociate readily. Activated erbB3 monomers are available to rebind erbB2 or to form homodimers, where at least one of the erbB3 monomers has measurable kinase activity. This model is compatible with the lateral signaling hypothesis proposed by others (57, 58). One important aspect of this “transient activation” model is the reduced reliance on equivalent (or greater) levels of expression of erbB3 dimerizing partners, since multiple erbB3 could conceivably be activated by a single erbB2. This is supported by the robust erbB3 kinase activity seen in CHO cells that have ∼16 times less erbB2 than SKBR3 cells. Assuming that phosphorylation is the key event in activation, recruitment of a tyrosine phosphatase would be the essential step to limit the duration of erbB3 activation.
It is interesting that our single-particle tracking studies were able to capture a significant number of homodimer events between unliganded erbB3, albeit with a very short lifetime (off rate of 0.44 to 1.08 s−1). Our experimental protocol does not permit us to evaluate the structural basis for these interactions between resting receptors. Others have suggested that erbB3 could form “back-to-back” ligand-independent oligomers (59, 60), as suggested by the orientation of crystallized erbB1 (47). Based on homology modeling (not shown), more traditional dimerization using the erbB3 dimerization arms are also possible. Continued discovery of the unique features of erbB3 has important implications for both normal development and disease, particularly in carcinogenesis where erbB3 is a potential therapeutic target.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH-CA119232 (B.S.W.), NSF MCB-0845062 (D.S.L.), and P50GM085273 (B.S.W.). Use of the UNM CC microscopy and flow cytometry facilities, and NIH support for these cores, is gratefully acknowledged.
We thank Donna Arndt-Jovin for the erbB2-mYFP and erbB3-mCit constructs and 2C4 reagent, Michael Batenjany for assistance with the K-LISA assay, Alexandre Chigaev for consultation regarding flow measurements, and Jason Byars and Samantha Schwartz for assistance with SPT analysis.
Footnotes
Published ahead of print 30 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01605-13.
REFERENCES
- 1.Sithanandam G, Anderson LM. 2008. The ErbB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 15:413–448. 10.1038/cgt.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baselga J, Swain SM. 2009. Novel anticancer targets: revisiting ErbB2 and discovering ErbB3. Nat. Rev. Cancer 9:463–475. 10.1038/nrc2656 [DOI] [PubMed] [Google Scholar]
- 3.Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., III 1994. Insect cell-expressed p180 erbB3 possesses an impaired tyrosine kinase activity. Proc. Natl. Acad. Sci. U. S. A. 91:8132–8136. 10.1073/pnas.91.17.8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sierke SL, Cheng K, Kim HH, Koland JG. 1997. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem. J. 322(Pt 3):757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden Y, Sliwkowski MX. 2001. Untangling the ErbB signaling network. Nat. Rev. Mol. Cell. Biol. 2:127–137. 10.1038/35052073 [DOI] [PubMed] [Google Scholar]
- 6.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. 1996. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 15:2452–2467 [PMC free article] [PubMed] [Google Scholar]
- 7.Sliwkowski MX, Schaefer G, Akita RW, Lofgren JA, Fitzpatrick VD, Nuijens A, Fendly BM, Cerione RA, Vandlen RL, Carraway KL., III 1994. Coexpression of erbB2 and erbB3 proteins reconstitutes a high-affinity receptor for heregulin. J. Biol. Chem. 269:14661–14665 [PubMed] [Google Scholar]
- 8.Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. 2000. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 19:4632–4643. 10.1093/emboj/19.17.4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riese DJ, II, van Raaij TM, Plowman GD, Andrews GC, Stern DF. 1995. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol. Cell. Biol. 15:5770–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger MB, Mendrola JM, Lemmon MA. 2004. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 569:332–336. 10.1016/j.febslet.2004.06.014 [DOI] [PubMed] [Google Scholar]
- 11.Bublil EM, Pines G, Patel G, Fruhwirth G, Ng T, Yarden Y. 2010. Kinase-mediated quasi-dimers of EGFR. FASEB J. 24:4744–4755. 10.1096/fj.10-166199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. 2010. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U. S. A. 107:7692–7697. 10.1073/pnas.1002753107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, Janakiraman V, Scholz R-P, Bowman K, Lorenzo M, Li H, Wu J, Yuan W, Peters BA, Kan Z, Stinson J, Mak M, Modrusan Z, Eigenbrot C, Firestein R, Stern HM, Rajalingam K, Schaefer G, Merchant MA, Sliwkowski MX, de Sauvage FJ, Seshagiri S. 2013. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 23:603–617. 10.1016/j.ccr.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 14.Telesco SE, Shih AJ, Jia F, Radhakrishnan R. 2011. A multiscale modeling approach to investigate molecular mechanisms of pseudokinase activation and drug resistance in the HER3/ErbB3 receptor tyrosine kinase signaling network. Mol. Biosyst. 7:2066–2080. 10.1039/c0mb00345j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. 2004. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat. Biotechnol. 22:198–203. 10.1038/nbt929 [DOI] [PubMed] [Google Scholar]
- 16.Andrews NL, Lidke KA, Pfeiffer JR, Burns AR, Wilson BS, Oliver JM, Lidke DS. 2008. Actin restricts Fc[epsiv]RI diffusion and facilitates antigen-induced receptor immobilization. Nat. Cell Biol. 10:955–963. 10.1038/ncb1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews NL, Pfeiffer JR, Martinez AM, Haaland DM, Davis RW, Kawakami T, Oliver JM, Wilson BS, Lidke DS. 2009. Small, mobile FcεRI receptor aggregates are signaling competent. Immunity 31:469–479. 10.1016/j.immuni.2009.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low-Nam ST, Lidke KA, Cutler PJ, Roovers RC, van Bergen en Henegouwen PM, Wilson BS, Lidke DS. 2011. ErbB1 dimerization is promoted by domain co-confinement and stabilized by ligand binding. Nat. Struct. Mol. Biol. 18:1244–1249. 10.1038/nsmb.2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, Raymond-Stintz MA, Ying W, Zhang J, Lidke DS, Steinberg SL, Williams L, Oliver JM, Wilson BS. 2007. Mapping ErbB receptors on breast cancer cell membranes during signal transduction. J. Cell Sci. 120:2763–2773. 10.1242/jcs.007658 [DOI] [PubMed] [Google Scholar]
- 20.Schulze WX, Deng L, Mann M. 2005. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 1:1–13. 10.1038/msb4100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes NE, Lane HA. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5:341–354. 10.1038/nrc1609 [DOI] [PubMed] [Google Scholar]
- 22.Shankaran H, Wiley HS, Resat H. 2006. Modeling the effects of HER/ErbB1-3 coexpression on receptor dimerization and biological response. Biophys. J. 90:3993–4009. 10.1529/biophysj.105.080580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh MY, Yang S, Raymond-Stinz MA, Steinberg S, Vlachos DG, Shu W, Wilson B, Edwards JS. 2008. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET Syst. Biol. 2:256–272. 10.1049/iet-syb:20070073 [DOI] [PubMed] [Google Scholar]
- 24.Schoeberl B, Pace EA, Fitzgerald JB, Harms BD, Xu L, Nie L, Linggi B, Kalra A, Paragas V, Bukhalid R, Grantcharova V, Kohli N, West KA, Leszczyniecka M, Feldhaus MJ, Kudla AJ, Nielsen UB. 2009. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2:ra31. 10.1126/scisignal2000352 [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Opresko L, Shankaran H, Chrisler WB, Wiley HS, Resat H. 2009. HER/ErbB receptor interactions and signaling patterns in human mammary epithelial cells. BMC Cell Biol. 10:78. 10.1186/1471-2121-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faratian D, Goltsov A, Lebedeva G, Sorokin A, Moodie S, Mullen P, Kay C, Um IH, Langdon S, Goryanin I, Harrison DJ. 2009. Systems biology reveals new strategies for personalizing cancer medicine and confirms the role of PTEN in resistance to trastuzumab. Cancer Res. 69:6713–6720. 10.1158/0008-5472.CAN-09-0777 [DOI] [PubMed] [Google Scholar]
- 27.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, Lauffenburger DA, Sorger PK. 2009. Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol. Syst. Biol. 5:239. 10.1038/msb.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzara MJ, Lauffenburger DA. 2009. Quantitative modeling perspectives on the ErbB system of cell regulatory processes. Exp. Cell Res. 315:717–725. 10.1016/j.yexcr.2008.10.033 [DOI] [PubMed] [Google Scholar]
- 29.Smith CS, Joseph N, Rieger B, Lidke KA. 2010. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7:373–375. 10.1038/nmeth.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forney GD. 1973. The Viterbi algorithm. Proc. IEEE 61:268–278. 10.1109/PROC.1973.9030 [DOI] [Google Scholar]
- 31.Fabian MA, WHBiggs, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. 2005. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23:329–336. 10.1038/nbt1068 [DOI] [PubMed] [Google Scholar]
- 32.Honegger AM, Dull TJ, Felder S, Van Obberghen E, Bellot F, Szapary D, Schmidt A, Ullrich A, Schlessinger J. 1987. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell 51:199–209. 10.1016/0092-8674(87)90147-4 [DOI] [PubMed] [Google Scholar]
- 33.Qian X, LeVea CM, Freeman JK, Dougall WC, Greene MI. 1994. Heterodimerization of epidermal growth factor receptor and wild-type or kinase-deficient Neu: a mechanism of inter-receptor kinase activation and transphosphorylation. Proc. Natl. Acad. Sci. U. S. A. 91:1500–1504. 10.1073/pnas.91.4.1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WS, Lazar CS, Poenie M, Tsien RY, Gill GN, Rosenfeld MG. 1987. Requirement for intrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature 328:820–823. 10.1038/328820a0 [DOI] [PubMed] [Google Scholar]
- 35.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. 1995. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 14:4267–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Citri A, Skaria KB, Yarden Y. 2003. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp. Cell Res. 284:54–65. 10.1016/S0014-4827(02)00101-5 [DOI] [PubMed] [Google Scholar]
- 37.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. 2004. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 5:317–328. 10.1016/S1535-6108(04)00083-2 [DOI] [PubMed] [Google Scholar]
- 38.Adams CW, Allison DE, Flagella K, Presta L, Clarke J, Dybdal N, McKeever K, Sliwkowski MX. 2006. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol. Immunother. 55:717–727. 10.1007/s00262-005-0058-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, Smith DJ, Landolfi S, Ramon y Cajal S, Arribas J, Baselga J. 2009. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene 28:803–814. 10.1038/onc.2008.432 [DOI] [PubMed] [Google Scholar]
- 40.Hagen GM, Caarls W, Lidke KA, De Vries AH, Fritsch C, Barisas BG, Arndt-Jovin DJ, Jovin TM. 2009. Fluorescence recovery after photobleaching and photoconversion in multiple arbitrary regions of interest using a programmable array microscope. Microsc. Res. Technol. 72:431–440. 10.1002/jemt.20686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. 2009. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U. S. A. 106:21608–21613. 10.1073/pnas.0912101106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. 2005. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 170:619–626. 10.1083/jcb.200503140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82:274–284. 10.1016/S0006-3495(02)75393-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- 45.Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. 2012. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc. Natl. Acad. Sci. U. S. A. 109:137–142. 10.1073/pnas.1111316109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. 2006. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 103:18992–18997. 10.1073/pnas.0609009103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warren CM, Landgraf R. 2006. Signaling through ErbB receptors: multiple layers of diversity and control. Cell. Signal. 18:923–933. 10.1016/j.cellsig.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 48.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin BJ, Yarden Y. 1996. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 16:5276–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao R-H, Maruyama IN. 2008. All EGF(ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J. Cell Sci. 121:3207–3217. 10.1242/jcs.033399 [DOI] [PubMed] [Google Scholar]
- 50.Alimandi M, Wang LM, Bottaro D, Lee CC, Kuo A, Frankel M, Fedi P, Tang C, Lippman M, Pierce JH. 1997. Epidermal growth factor and betacellulin mediate signal transduction through coexpressed ErbB2 and ErbB3 receptors. EMBO J. 16:5608–5617. 10.1093/emboj/16.18.5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margadant C, Monsuur HN, Norman JC, Sonnenberg A. 2011. Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 23:607–614. 10.1016/j.ceb.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137–1149. 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 53.Collier TS, Diraviyam K, Monsey J, Shen W, Sept D, Bose R. 2013. Carboxyl group footprinting mass spectrometry and molecular dynamics identify key interactions in the HER2-HER3 receptor tyrosine kinase interface. J. Biol. Chem. 288:25254–25264. 10.1074/jbc.M113.474882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu C, Mi LZ, Grey MJ, Zhu J, Graef E, Yokoyama S, Springer TA. 2010. Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor. Mol. Cell. Biol. 30:5432–5443. 10.1128/MCB.00742-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lidke K, Rieger B, Jovin T, Heintzmann R. 2005. Super-resolution by localization of quantum dots using blinking statistics. Opt. Express 13:7052–7062. 10.1364/OPEX.13.007052 [DOI] [PubMed] [Google Scholar]
- 56.Zhang Q, Park E, Kani K, Landgraf R. 2012. Functional isolation of activated and unilaterally phosphorylated heterodimers of ErbB2 and ErbB3 as scaffolds in ligand-dependent signaling. Proc. Natl. Acad. Sci. U. S. A. 109:13237–13242. 10.1073/pnas.1200105109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graus-Porta D, Beerli RR, Daly JM, Hynes NE. 1997. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 16:1647–1655. 10.1093/emboj/16.7.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kancha RK, von Bubnoff N, Peschel C, Duyster J. 2009. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin. Cancer Res. 15:460–467. 10.1158/1078-0432.CCR-08-1757 [DOI] [PubMed] [Google Scholar]
- 59.Kani K, Warren CM, Kaddis CS, Loo JA, Landgraf R. 2005. Oligomers of ERBB3 have two distinct interfaces that differ in their sensitivity to disruption by heregulin. J. Biol. Chem. 280:8238–8247 [DOI] [PubMed] [Google Scholar]
- 60.Landgraf R, Eisenberg D. 2000. Heregulin reverses the oligomerization of HER3. Biochemistry 39:8503–8511. 10.1021/bi000953+ [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.