Abstract
To adapt to stress, cells must undergo major changes in their gene expression profiles. We have previously described a largely uncharacterized stress response pathway in Caenorhabditis elegans that acts through an evolutionarily conserved motif, termed ESRE, for ethanol and stress response element. We characterize here the requirements for ESRE gene expression and show that the ESRE network is regulated by a conserved SWI/SNF family nucleosome remodeling complex termed PBAF. Depletion of PBAF subunits SWSN-7/BAF200 and PBRM-1/BAF180 results in decreased expression of ESRE genes and increased sensitivity to thermal stress. When overexpressed, SWSN-7/BAF200 and PBRM-1/BAF180 led to increased ESRE transcription, enhanced thermotolerance, and induction of a nuclear ESRE-binding activity. Our data support a model in which PBAF is recruited by an ESRE-binding protein to genomic ESRE sites. We also show that the closely related SWI/SNF complex, BAF, which regulates stress induction through DAF-16/FOXO, does not contribute to ESRE gene expression or bind directly to ESRE sites. To our knowledge, this is the first report demonstrating direct and specific regulation of a stress response network by the PBAF nucleosome-remodeling complex in vivo in metazoa. In addition, we show that PBAF cooperates with the histone demethylase, JMJC-1/NO66, to promote expression of ESRE genes following stress.
INTRODUCTION
All living organisms require a stable internal environment to develop, survive, and reproduce. This internal homeostasis is constantly challenged by a variety of potentially harmful stressors emanating from the external environment. To reestablish internal homeostasis, cells must rapidly alter their gene expression profiles through the activation of stress response pathways or networks (1–10). These pathways typically lead to the synchronized short-term expression of a large number of stress-responsive target genes. The activation of stress-associated genes is triggered by the binding of transcriptional regulators to one or more regulatory elements located within the proximal promoter region of target genes. For example, DAF-16/FOXO and heat shock factor 1 (HSF-1)/HSF1, transcription factors that are part of the conserved insulin/insulin-like growth factor 1 signaling (IIS) pathway in Caenorhabditis elegans, regulate the expression of stress-responsive genes by binding to DNA elements within the promoters of their downstream targets (GTAAAC/TA and TTCTA/CGAA, respectively) (11).
In some cases, stress-responsive genes may be embedded within a repressed genomic environment, which may require chromatin reorganization prior to binding or activation by transcriptional regulators (12–14). In the Drosophila genome, for instance, active heat shock element (HSE) sites reside in chromatin areas marked by histone acetylation (H3K9, H3K18, H3K27, H4K5, H4K8, and H4K16) and methylation (H3K4me3 and H3K79me2), covalent modifications associated with transcriptional activation (12). Conversely, HSE motifs buried within an inactive unmarked chromatin environment are not bound by HSF. Moreover, although HSF binding can be detected at more than a hundred different loci, only a subset of these loci may be transcriptionally active, indicating the presence of additional layers of regulation. These latter findings can be explained by the general observation that genes may often require specific combinations of chromatin regulators and transcription factors for strong expression to occur (15, 16).
One broad class of chromatin-level transcriptional regulators is the nucleosome-remodeling complexes, which can occupy the same genomic loci as modifiers of histones (15, 16). Nucleosome-remodeling complexes use energy derived from ATP to remove histones, replace them with other histone variants, or move nucleosomes along the DNA strand to facilitate or inhibit the access of transcription factors and the basal transcriptional machinery (17). There are four major classes of evolutionarily conserved chromatin-remodeling complexes: SWI/SNF, ISWI, CHD, and INO80. Whereas these complexes have well-known roles in development and disease states, their importance in the adaptation to stress is less well understood (18–21). Although the depletion of certain chromatin-remodeling subunits from SWI/SNF, ISWI, and CHD families results in hypersensitivity to stress in yeast, evidence for their role in stress adaptation in multicellular organisms is very limited (22–24).
C. elegans serves as an important model for studying stress adaptation, with the majority of stress response pathways being highly conserved (25, 26). The most thoroughly studied stress-response network in C. elegans, the IIS pathway, regulates the expression of target genes by activating the transcription factor DAF-16/FOXO in response to a wide spectrum of stress conditions (27, 28). In addition, the IIS pathway leads to HSF-1/HSF1 activation under a more restricted subset of conditions (11, 29). The IIS pathway is activated by heat (11), heavy metals (30), UV radiation (31), hypertonic (32) and oxidative stress (32), and bacterial infection (33). Functional overlap of the IIS pathway with independent stress regulators, such as SKN-1/NRF-2 and PHA-4/FOXA, helps to ensure a robust transcriptional response and survival of the organism (9, 10).
We previously identified two factors that regulate a largely uncharacterized stress response pathway in C. elegans that functions independently of other pathways, including IIS (34). The pathway acts through an evolutionarily conserved motif, termed ESRE, for ethanol and stress response element (34–36). The ESRE pathway regulates the expression of hundreds of genes under a variety of stress conditions, including heat, ethanol, hypertonic, and oxidative stress. The ESRE response network includes SLR-2/ZTF-24, a Zn finger protein, and its downstream transcriptional target, JMJC-1/NO66, a conserved jumonji-C domain-containing histone demethylase (34). We report here the specific requirement for a subclass of the SWI/SNF nucleosome-remodeling family, PBAF, in the ESRE-mediated stress response in C. elegans. Loss of PBAF function resulted in decreased survival under stress conditions, whereas strains that overexpressed PBAF subunits had a markedly enhanced survival compared to wild type. In addition, we observed the presence of a sequence-specific stress-inducible ESRE-binding protein that may recruit PBAF to ESRE sites within promoters.
MATERIALS AND METHODS
Strains.
All strains were cultured on nematode growth medium (NGM) supplemented with Escherichia coli OP50 as a food source according to standard protocols (37) and were maintained at 20°C. The strains used in the present study included N2, WY651 (fdEx87 {3×ESRE::GFP; pRF4 [rol-6(gf)]}), WY664 [jmjc-1(tm3525); fdEx87], WY703 [fdIs2 (3×ESRE::GFP; pRF4)V], WY689 [fdEx99 (3×ESREM1a::GFP; pRF4)], WY693 [fdEx103 (3×ESREM1b::GFP; pRF4)], WY692 [fdEx102 (3×ESREM2::GFP; pRF4)], WY690 [fdEx100 (3×ESREM3::GFP; pRF4)], WY686 [fdEx97 (3×ESREM4::GFP; pRF4)], WY786 [fdEX156 (3×ESRE::mCherry; pRF4)], WY787 [fdEX157 (3×ESRE::mCherry; pRF4)], WY788 [fdEX158 (3×ESRE::mCherry; pRF4)], WY846 [fdIs4 (3×ESRE::mCherry; pRF4)], WY838 [fdEx178 (Prol-6::GFP; pRF4)], WY839 [fdEx179 (Prol-6::GFP; pRF4)], WY840 [fdEx180 (Prol-6::GFP; pRF4)], WY852 [fdEx184 (Prol-6::GFP; pRF4)], WY699 [unc-119(ed3); fdEx109 (3×ESRE::GFP; unc-119+)], WY866 [unc-119(ed3); fdEx190 (3×ESRE::GFP; unc-119+)], WY867 [unc-119(ed3); fdEx191 (3×ESRE::GFP; unc-119+)], WY868 [unc-119(ed3); fdEx192 (3×ESRE::GFP; unc-119+)], WY877 {fdEx193 [pDP#MMUGF12 (UNC-119::GFP)]}, WY878 [fdEx194 (pDP#MMUGF12)], WY879 [fdEx195 (pDP#MMUGF12)], WY880 [fdEx196 (pDP#MMUGF12)], CL2070 {dvIs70 [pCL25 (Phsp-16.2::GFP); pRF4]}, WY753 {fdEx142 [pJY323 (Phsp-16.1::GFP); pRF4]}, WY756 {fdEx139 [pJY312 (Phsp-16.1(dd)::GFP); pRF4]}, VC2538 [swsn-7(gk1041)/mIn1II], WY882 [swsn-7(gk1041)/mIn1II; dvIs70; pRF4)], WY891 [swsn-7(gk1041)/mIn1II; fdEx142], HS184 [swsn-4(os13)IV], WY903 [swsn-4(os13)IV; fdIs2V], WY899 [swsn-4(os13)IV; dvIs70], WY883 [swsn-2.1(n1654); zdIs13(tph-1::GFP)], WY901 [swsn-2.1(n1654); dvIs70], WY946 [swsn-2.1(n1654); fdIs2V], HS1257 [unc-76(e911)V; osEx219 (PBRM-1::GFP + unc-76+)], HS845 [osEx138 (SWSN-8::GFP + pRF4)], WY1025 [osEx219; fdEx139], WY945 [fdEx224 (SWSN-7::GFP; pRF4)], WY988 [swsn-7(gk1041); fdEx224], WY890 [fdIs2V; dfEx199 (swsn-7/pcr; sur-5::RFP)], WY909 {fdIs2V; fdEx204 [pDF155 (swsn-7 genomic locus in pGEM-T Easy); sur-5::RFP]}, WY915 [fdIs2V; fdEx210 (pbrm-1/pcr; sur-5::RFP)], WY895 [fdIs2V; fdEx200 (pDF155; pbrm-1/pcr); sur-5::RFP], RW11148 [unc-119(ed3)III; stIs11148 (Ppbrm-1::H1-mCherry + unc-119+)], and RW10190 [unc-119(ed3)III; stIs10190 (Pswsn-7::HIS-24::mCherry + unc-119+)].
DNA constructs.
The 3×ESRE::GFP construct was generated as described previously (34). Primers used for generating 3×ESRE mutational variants were as follows (mutated residues are underlined): M1a, 5′-aaaagcttTGTGCGTCTCTTGTGCGTCTCTTGTGCGTCTCTatgcctgcaggtcgactct-3′; M1b, 5′-aaaagcttTCTCCGTCTCTTCTCCGTCTCTTCTCCGTCTCTatgcctgcaggtcgactct-3′; M2, 5′-aaaagcttTGTCCGTCTCTTGTCCGTCTCTTGTCCGTCTCTatgcctgcaggtcgactct-3′; M3, 5′-aaaagcttTGTCCGTCACTTGTCCGTCACTTGTCCGTCACTatgcctgcaggtcgactct-3′; and M4, 5′-aaaagcttAGTCCGTCACAGTCCGTCACAGTCCGTCACTatgcctgcaggtcgactct-3′. The reverse primer used for all of the variant constructs was pPD_R (5′-CGCTCAGTTGGAATTCTACG-3′).
To create an mCherry version of the 3×ESRE reporter, green fluorescent protein (GFP) was switched with mCherry using KpnI and EcoRI sites. The mCherry gene was amplified from pJA304 (Addgene) using the primers 5′-AAAAAGGTACCATGGTCTCAAAGGGTGAAGAAGATAAC-3′ and 5′-AAAAAGAATTCTGAGACTTTTTTCTTGGCGG-3′. These were digested with KpnI and EcoRI and cloned into the corresponding sites in 3×ESRE::GFP reporter. The Prol-6::GFP construct was generated by amplifying ∼1.7 kb of the rol-6 promoter sequence from N2 genomic DNA using the primers 5′-CGCCCCTCTAGATTATCATCTTCGGTTTTG-3′ and 5′-AACCCCGGTACCCTGGAAATTTTCAGTTAGATCTAAAG-3′. After digestion with XbaI and KpnI, the PCR amplicon was cloned into pPD95.77 (Addgene). The SWSN-7::GFP translational fusion reporter was generated by amplifying an ∼6.2-kb PCR fragment containing the complete swsn-7 promoter, coding, and 3′ untranslated region (3′UTR) regions from genomic DNA. The PCR fragment was cloned into the T/A cloning vector pGEM-T Easy (pDF155), and an in-frame XbaI site was introduced immediately downstream of the swsn-7 start codon by site-directed mutagenesis (QuikChange II XL site-directed mutagenesis kit; Stratagene) to generate plasmid pDF156. A PCR fragment containing the GFP gene (from pPD95.77) was amplified using the primers 5′-AAACAGTCTAGAGGCGCTATGAGTAAAGGAGAAGAACTTTTCACTGG-3′ and 5′-AAAGTTTCTAGAGCTGCTGCTTTTGTATAGTTCATCCATGCCATGTG-3′. The amplicon was digested with XbaI and cloned into the generated XbaI site of pDF156. Positive clones containing the insert were sequence verified. To generate animals that overexpress SWSN-7 and PBRM-1, we used plasmid pDF155 and an ∼9.4-kb PCR fragment containing the pbrm-1 promoter, coding, and 3′UTR region, respectively. pbrm-1 was amplified from WRM0635dB10 using 5′-TCTACACGCACACATGCACAC-3′ and 5′-GAATCGCTGACGAGAAGCAGG-3′ primers. The plasmid and PCR fragments were injected along with the sur-5::RFP injection marker.
Recombinant protein.
A cDNA fragment encoding full-length SLR-2 was cloned into pET30a plasmid (Novagen) and His-tagged SLR-2 protein was expressed using the E. coli BL21(DE3) pLysS strain. Cells were grown in Luria-Bertani (LB) medium at 37°C until log phase (optical density at 600 nm = 0.6) and induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h. His-tagged SLR-2 expression was purified by using a Probond purification system (Invitrogen).
RNAi.
RNA interference (RNAi) studies were carried out as described previously (38). Control RNAi assays were carried out using a bacterial strain carrying the RNAi vector pDF129.36, which produces an ∼200-bp dsRNA that is not homologous to any C. elegans genes (39). For studies using the 3×ESRE::GFP reporter, L4 hermaphrodites were placed on RNAi plates and incubated for ∼24 h at 20°C, followed by heat shock at 30°C for 12 h. In cases where standard RNAi plates resulted in substantial embryonic lethality (swsn-7 and swsn-8), dilutions of the targeting RNAi were carried out using a strain containing the vector control RNAi plasmid (pPD129.36). For viability assays and for studies using Phsp-16.1::GFP, Phsp-16.1(dd)::GFP, and Phsp-16.2::GFP reporters, strains were maintained for two generations at 20°C on RNAi plates, followed by heat shock at 30°C for 22 h. RNAi clones from the Source Bioscience library were sequence verified.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were derived from stressed mixed-stage worm populations exposed to 30°C heat shock for 12 h and prepared using an NE-PER nuclear and cytoplasmic extraction reagent kit (Pierce). Radioactive probes were generated by 5′ end labeling of the “top” strand (5′-AAAAGTATCTGCGTCTCTTCAGTACG-3′) using γ-32P, followed by annealing to a 10-fold excess of the unlabeled complementary strand (40). Nuclear extract was preincubated for 10 min on ice in HEPES-based binding buffer [10 mM HEPES-NaOH, pH 7.6; 1 mM dithiothreitol; 1 mM EDTA, pH 8; 5% glycerol; 2.5 mM MgCl2; heparin, 25 fg/μl; 0.05% bovine serum albumin; poly(dI-dC), 6.25 ng/μl] and then incubated on ice with a 600-fold excess of unlabeled specific (WT) or mutant (M1a, M1b, M2, M3, and M4) competitors for 20 min before labeled probe was added. The final incubation mixture was incubated at room temperature for 1 h before loading (in the absence of loading buffer) onto a 5 or 6% Tris-borate-EDTA gel. Gels were visualized using PharosFX molecular imager system (Bio-Rad). EMSA reactions with the recombinant SLR-2 protein were carried out using the LightShift chemiluminescent EMSA kit (Pierce). For quantification purposes, densities of individual bands were measured using ImageJ (National Institutes of Health).
Quantitative real-time PCR (qPCR).
N2 animals were grown on plates with a 1:4 swsn-7 RNAi dilution or on vector-only plates at 20°C for two generations. Embryos were isolated using bleaching methods, cultured overnight in M9 medium, and placed on fresh RNAi plates. Once populations reached the L3 stage, worms were exposed to 30°C for 12 h before being collected for RNA isolation. Total RNA from each sample was isolated using TRIzol and purified on RNeasy minicolumns (Qiagen). cDNA was prepared from 1 μg of RNA using a SuperScript II first-strand synthesis system (Invitrogen), and qPCR was performed using SYBR green PCR mastermix (Bio-Rad). Quantitative PCR data represent the mean of three independent biological replicates, each performed in triplicate. CT values were normalized using primers specific to the housekeeping gene act-1.
ChIP assays.
Mixed-stage populations of worms containing GFP-tagged SWSN-7, SWSN-8, and PBRM-1 were exposed to 30°C heat shock for 12 h before cross-linking with 2% formaldehyde for 20 min at room temperature. SWSN-8 and PBRM-1 strains were also tested in the absence of stress (20°C). Samples were quenched using 1 M Tris (pH 7.5), pelleted, snap-frozen in liquid nitrogen, and stored at −80°C. After sonication, 2.5 mg of extract was immunoprecipitated using 15 μg of anti-GFP (ab290; Abcam) or anti-IgG (sc-2004; Santa Cruz Biotechnology) antibodies. After reversal of cross-linking, DNA was purified using a QIAquick PCR purification kit (Qiagen catalog no. 28106) and analyzed by PCR (35 cycles) using the following primers: 5′-GTTTCCTCTGAACACGATTG-3′ and 5′-CTAGAACATTCGAGCTGCTT-3′ (which amplify a 215-bp region of the hsp-16.1 promoter) and 5′-CTTATATACCCGCATTCTGC-3′ and 5′-ACATTCGGTACATGGAAAAG-3′ (which amplify a 153-bp region of the hsp-16.2 promoter). To determine the specific promoter region bound by PBAF, we carried out PCR analysis (30 cycles) to amplify the following regions: a 119-bp region upstream of the ESRE sites of hsp-16.1 (5′-TCTTGAAGTTTAGAGAATGAACAGTAA-3′ and 5′-CTAGGACCTTCTAGAACATTCTAA-3′); 76 bp upstream of the ESRE sites of hsp-16.2 (5′-GCACTAGAACAAAGCGTG-3′ and 5′-CATTCTGTAAGGCTGCAGA-3′); 99 bp spanning the ESRE sites of hsp-16.1 (5′-GCATTCATTTCAAAATACACCCCA-3′ and 5′-ATTCAGAACATTGAGAAATAGTGTG-3′); 92 bp spanning the ESRE sites of hsp-16.2 (5′-TTCGTTTGAAAATACTCCCGG-3′ and 5′-GGTACATGGAAAAGTAGTGTACAC-3′); 119 bp downstream of the ESRE sites of hsp-16.1 (5′-TCGCCCTCCTTTTGCAAG-3′ and 5′-GTAGTTTGAAGATTTCACAATTAGAGTG-3′); and a 130-bp region downstream of the ESRE sites of hsp-16.2 (5′-TCGCCCTCCTTTTGCAAC-3′ and 5′-CATGATTATAGTTTGAAGATTTCTAATTTC-3′). The following primers were used to amplify mutated ESRE sites of hsp-16.1 promoter: 5′-CCATAGGTGCAAGCATGCC-3′ and 5′-TGTTTGGTTCGGTTTTGTCACTG-3′.
Longevity and survival assays.
L1-stage worms were seeded onto NGM or RNAi plates and incubated at 20°C for two generations. For longevity assays, worms were scored for survival by their response to mechanical stimuli, and live animals were transferred to new plates every other day. Life-span was defined as the length of time from when animals were put onto the plates at the L1 larval stage until they were scored as dead. For stress-induced survival assays, young adults were exposed to 37°C heat shock and scored for survival based on their response to mechanical stimuli every 1 to 2 h for a total of 12 h.
Microscopy.
GFP fluorescence images were collected using a Nikon Eclipse epifluorescence microscope and OpenLab software, and quantification was carried out using ImageJ. Mean fluorescence was determined for each embryo or adult, and isovolumetric background fluorescence was subtracted from each image. Fluorescence intensities provided in the figures are in arbitrary units. Confocal images were obtained on an Olympus IX-71 inverted microscope using MetaMorph software (Molecular Devices).
Statistical analysis.
P values for comparisons of means were calculated using the two-tailed Student t test. The statistical significance of effects on survival and life-span was estimated using the log-rank test, performed using software available at http://bioinf.wehi.edu.au/software/russell/logrank/.
RESULTS
The ESRE motif recruits an endogenous sequence-specific DNA-binding activity.
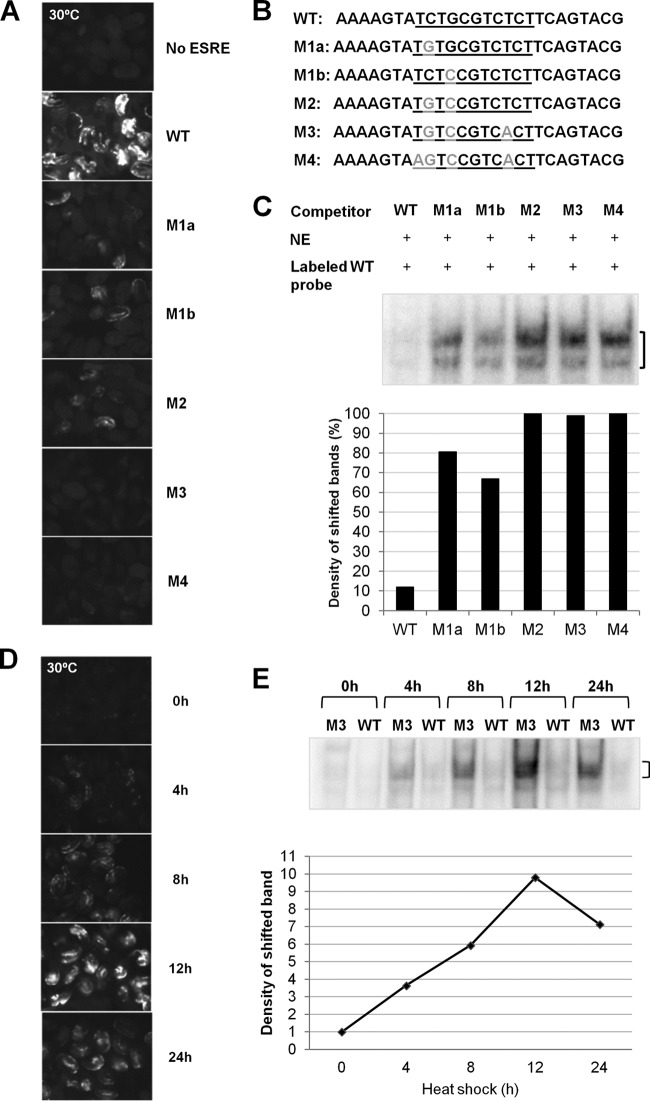
Although the ESRE network represents a functionally important and evolutionarily conserved stress pathway in C. elegans, its regulation remains largely uncharacterized. The pathway acts through a DNA element, the ESRE, which consists of a GC-rich core sequence of 11 bp (TCTGCGTCTCT) (34–36, 41). Importantly, the ESRE is sufficient to confer stress-inducible reporter expression within the context of an otherwise inactive minimal promoter in C. elegans (Fig. 1A) (34). C. elegans strains containing either extrachromosomal or integrated multicopy arrays composed of a 3×ESRE::GFP reporter and a rol-6(gf) coinjection marker showed weak basal expression in neurons and body wall muscles of both embryos and L1 larvae at 20°C (see Fig. S1A in the supplemental material). This expression was strongly enhanced within these tissues following heat shock at 30°C (Fig. 1A). Although expression of the 3×ESRE::GFP reporter was observed only in embryos and L1 larvae, the importance of the ESRE network in the stress response at later developmental stages has been well established (34–36).
FIG 1.
Analysis of ESRE sequence requirements for binding and transcription. (A) Fluorescence images of wild-type embryos containing arrays with either a 3×ESRE::GFP reporter (WT), a control GFP reporter with no ESRE, or ESRE::GFP mutational variants (M1a, M1b, M2, M3, and M4) exposed to 30°C heat shock for 20 h. Expression constructs contained three tandem repeats of the corresponding underlined sequences shown in panel B. All strains were marked with a rol-6(gf) plasmid, and exposure times for each image were identical. (B) Sequences of the “top” DNA strands of the competitors used for EMSAs. Competitors included both a core ESRE sequence (underlined), as well as flanking sequences. The WT (specific) competitor contained the wild-type ESRE motif, whereas the mutant competitors contained the altered nucleotides marked in gray. (C) ESRE-binding activity detected by EMSA using nuclear extracts (NE) from wild-type worms exposed to 30°C heat shock for 12 h. The bracket indicates the location of the shifted band(s). The graph represents quantification of the observed shift intensities. The results are normalized against the M4 shift, arbitrarily set at 100%. (D) Time course of wild-type embryos containing a 3×ESRE::GFP array exposed to 30°C heat shock. All images used identical exposure times. (E) ESRE-binding activity observed by EMSA using nuclear extracts from wild-type worms exposed to heat shock for the indicated times. Competitors (M3 or WT) are indicated for each lane. The bracket indicates the location of the shifted band(s). Also shown is a quantification of the specific shifted bands. Band intensity at 0 h was arbitrarily set to 1.
To identify nucleotide residues that are critical for ESRE function, we first carried out a mutational analysis of the 3×ESRE::GFP reporter. The ESRE sequence was altered at four highly conserved positions (Fig. 1B), and strains containing variant 3×ESRE::GFP reporters were generated. Whereas single and double point mutations partially abrogated reporter expression, three or more mutations rendered the motif nonfunctional and prevented GFP expression at both 20 and 30°C (Fig. 1A; see Fig. S1A in the supplemental material). These mutational variants similarly affected strains containing extrachromosomal arrays comprised of 3×ESRE::GFP reporters along with an unc-119 marker (see Fig. S1B and C in the supplemental material). Our mutational analysis specifically implicated base pairs in 5′ positions 2, 4, and 9 as critical for ESRE recognition by endogenous activators.
To detect endogenous ESRE-binding factors and to further validate our mutational analysis, we carried out EMSAs on mixed-stage worm populations exposed to a 30°C heat shock for 12 h. Nonspecific competitors used in our EMSAs corresponded to ESRE variants examined in the 3×ESRE::GFP mutational analysis (Fig. 1B). Whereas single variants could still associate weakly with endogenous ESRE-binding factors, variants containing two or more mutations failed to interact (Fig. 1C; see Fig. S2A in the supplemental material; additional controls for EMSAs are shown in Fig. S2B to D). These results are highly consistent with those obtained using the 3×ESRE::GFP reporter (Fig. 1A) and confirm that the ESRE is able to bind a sequence-specific DNA-binding protein(s) that is present in worm nuclear extract after stress.
ESRE expression and binding activity increase after exposure to stress.
To further characterize regulation of the ESRE, we measured both 3×ESRE::GFP expression and ESRE-binding activity at different time points during heat shock. Binding to the ESRE, which was barely detectable under nonstressed conditions, showed strong enhancement during the first hours of exposure, increasing by ∼4-fold after 4 h of heat shock (Fig. 1E; see Fig. S2E in the supplemental material). Continuous exposure to stress resulted in a steady increase in the intensity of the shifted ESRE band, which showed a maximal increase of ∼10-fold after 12 h. By 24 h of stress exposure, binding activity began to subside, consistent with results from the 3×ESRE::GFP reporter (Fig. 1D), as well as previous studies of endogenous ESRE gene expression (34). These results indicate that the ESRE stress pathway mediates the response to stress by increasing the abundance or binding affinity of one or more endogenous ESRE-binding proteins.
SWSN-7/BAF200 is required for the activation of stress-inducible ESRE genes.
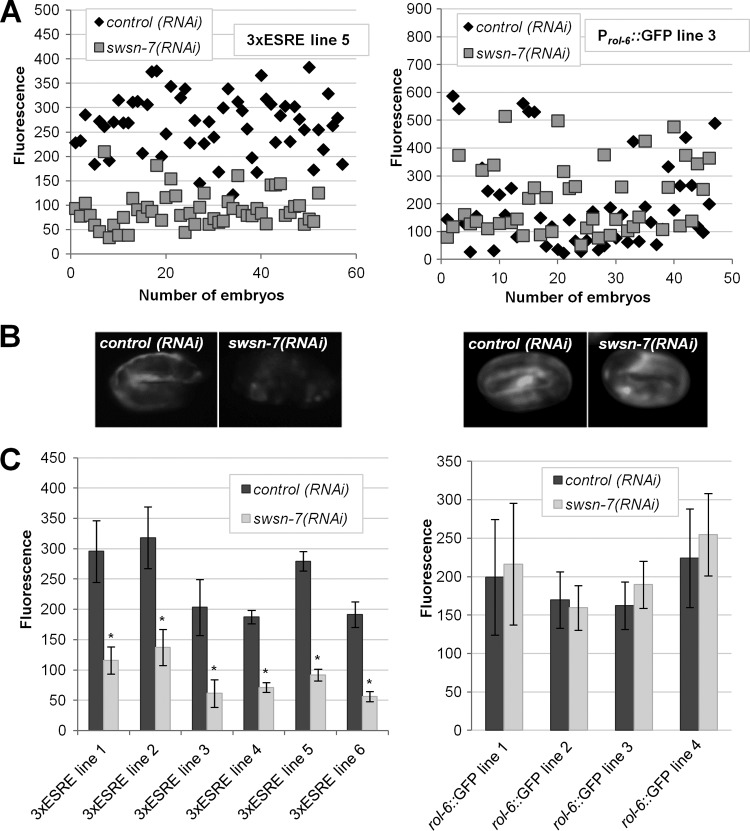
Previous studies had implicated SLR-2 and JMJC-1 as positive regulators of the ESRE response pathway (34). To identify additional positive regulators of the pathway, we performed an RNAi feeding screen on a subset of conserved transcriptional regulators in C. elegans (see Table S1 in the supplemental material). Positive RNAi clones would be expected to abolish or attenuate 3×ESRE::GFP expression following exposure to high temperatures. One of the tested clones, corresponding to swsn-7/C08B11.3, reduced 3×ESRE::GFP expression by ∼3-fold (Fig. 2A and B). This finding was consistent among multiple independent 3×ESRE::GFP expression lines tested (Fig. 2C). Effects of swsn-7(RNAi) on 3×ESRE::GFP induction were not due to the influence of SWSN-7 on non-ESRE elements within the transgene arrays, as similar results were observed for 3×ESRE::GFP strains that were marked with either unc-119 or rol-6 (see Fig. S3 in the supplemental material). Furthermore, swsn-7(RNAi) failed to inhibit either Prol-6::GFP or unc-119::GFP expression (Fig. 2; see Fig. S3 in the supplemental material), indicating that swsn-7 acts specifically through the ESRE and not through regulatory elements present in either the rol-6 or unc-119 genes.
FIG 2.
Knockdown of swsn-7 attenuates ESRE reporter expression. (A) Fluorescence intensities of individual embryos from representative strains carrying arrays with a rol-6(gf)-marked 3×ESRE::GFP (left) or a Prol-6::GFP (right) construct. Strains were pretreated with either control RNAi or swsn-7(RNAi). (B) Images of representative embryos from each strain in panel A. (C) Quantification of GFP fluorescence intensities in wild-type embryos from independent 3×ESRE::GFP and Prol-6::GFP lines pretreated with control RNAi or swsn-7(RNAi). Error bars indicate 95% confidence intervals (CIs). Statistical analysis was done using the Student t test; asterisks indicate statistical significance (P < 0.001) relative to the control (n = 40 to 60).
swsn-7 encodes an ortholog of human BAF200, a component of the mammalian SWI/SNF chromatin-remodeling complex (42). Furthermore, large-scale RNAi screens have shown swsn-7 to be essential for embryonic development in C. elegans (43, 44). To rule out the possibility that swsn-7(RNAi) led to a decrease in 3×ESRE::GFP expression because of reduced embryonic fitness or viability, we carried out a dilution series on swsn-7(RNAi) feeding plates. Stepwise dilutions resulted in a gradual decrease in embryonic lethality, which was completely eliminated by an 8-fold dilution of swsn-7 with control RNAi (see Fig. S4A in the supplemental material). Nevertheless, an 8-fold dilution of swsn-7(RNAi) still resulted in a significant (∼2-fold) decrease in 3×ESRE::GFP expression after heat shock (see Fig. S4B and C in the supplemental material), indicating that the effect of swsn-7(RNAi) on ESRE gene expression is separable from its role in embryogenesis (also see below).
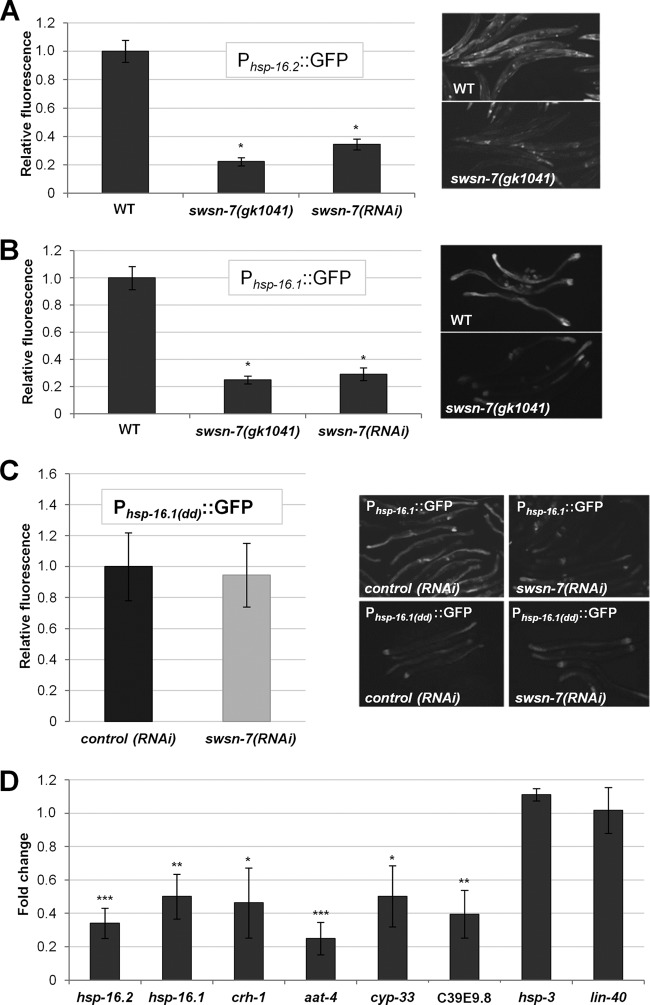
We next tested the requirement for SWSN-7 on ESRE-related gene expression within the context of a full-length promoter. The heat shock protein genes hsp-16.1 and hsp-16.2 are both upregulated after stress, and both genes contain two functional ESREs within their 5′-proximal promoter regions (35). Knockdown of swsn-7 by RNAi reduced the expression of Phsp-16.1::GFP and Phsp-16.2::GFP reporters in adult worms by 3.4- and 2.9-fold, respectively (Fig. 3A and B), a finding consistent with results obtained in embryos using 3×ESRE::GFP reporter strains (Fig. 2; see Fig. S3 in the supplemental material). Furthermore, using a null deletion mutation for swsn-7, gk1041, the expression of Phsp-16.1::GFP and Phsp-16.2::GFP was reduced by nearly 5-fold following heat shock (Fig. 3A and B). These findings indicate that SWSN-7 can regulate ESRE-mediated gene expression within the context of a full-length promoter. These results also demonstrate that the observed effects of swsn-7(RNAi) are not due to off-target RNAi silencing effects and show that SWSN-7 is required for ESRE-dependent expression in adults as well as embryos. In addition, swsn-7(gk1041) mutants were strongly compromised for induction of Phsp-16.1::GFP following hypoxia, demonstrating that SWSN-7 is required for ESRE gene expression under multiple stress conditions (see Fig. S5 in the supplemental material) (63).
FIG 3.
Knockdown of swsn-7 attenuates expression of endogenous ESRE-regulated genes. (A and B) Quantification of fluorescence intensities of Phsp-16.2::GFP (A) and Phsp-16.1::GFP (B) in swsn-7(RNAi)–treated worms or in the swsn-7(gk1041) deletion mutant background after exposure to heat shock (30°C) for 22 h. Integers on y axes indicate average levels of GFP fluorescence in swsn-7(RNAi) or gk1041 worms relative to those in control RNAi worms or worms grown on OP50, respectively (arbitrarily set to 1.0; n = 25 to 50). Also shown are representative GFP photomicrographs of heat-shocked animals. (C) Quantification of Phsp-16.1(dd)::GFP (the Phsp-16.1::GFP reporter with deleted ESRE sites) fluorescence intensities in heat-shocked wild-type adults pretreated with control RNAi or swsn-7(RNAi). Fluorescence is expressed relative to that in control RNAi worms. Error bars indicate 95% confidence intervals (CIs). Statistical analysis was performed using the Student t test (P = 0.716, n = 30). Also shown are micrographs of heat shock-induced expression of Phsp-16.1::GFP and Phsp-16.1(dd)::GFP pretreated with control RNAi or swsn-7(RNAi). (D) mRNA levels of endogenous ESRE-regulated genes were determined by qPCR for control RNAi and swsn-7(RNAi) strains exposed to 30°C heat shock. Fold changes indicate the expression levels of individual genes in worms treated with swsn-7(RNAi) relative to control RNAi. The data are the means of three biological replicates; error bars represent 95% CIs. Statistical analysis was done using the Student t test (***, P < 0.001; **, P < 0.01; *, P < 0.05).
hsp-16.1 and hsp-16.2 are both regulated by multiple DNA elements. In addition to ESRE sites, both genes are under the control of HSE, DBE (DAF-16 binding element), and hypoxia-responsive regulatory elements (11, 45). Thus, it was possible that the observed reduction in Phsp-16.1::GFP and Phsp-16.2::GFP expression following swsn-7 inhibition reflects a role for SWSN-7 in regulating an element that is distinct from the ESRE. We therefore made use of a Phsp-16.1::GFP reporter in which the two tandem ESRE sites were replaced with the binding site for SphI and SalI, respectively (GCATGC, GTCGAC; Phsp-16.1(dd)::GFP). After heat shock, expression of Phsp-16.1(dd)::GFP was greatly reduced relative to wild-type Phsp-16.1::GFP but was not entirely eliminated, presumably because of the activity of other enhancer elements (Fig. 3C). Notably, swsn-7(RNAi) failed to further reduce Phsp-16.1(dd)::GFP expression following heat shock relative to control RNAi (Fig. 3C), indicating that SWSN-7 regulates hsp-16.1 expression specifically through the ESRE pathway.
To assess the role of SWSN-7 on endogenous ESRE gene expression, we first measured endogenous mRNA levels of hsp-16.1 and hsp-16.2 in control RNAi and swsn-7(RNAi) L3-stage hermaphrodites following heat shock. Consistent with reporter strains, levels of hsp-16.1 and hsp-16.2 mRNA were downregulated ∼2- to 3-fold in swsn-7(RNAi) animals relative to controls (Fig. 3D). We next analyzed the expression of six putative ESRE-regulated genes (crh-1, aat-4, cyp-33C8, hsp-3, lin-40, and C39E9.8), which are stress inducible and are downregulated in slr-2 and jmjc-1 mutants (34). We observed four of the putative ESRE target genes (crh-1, aat-4, cyp-33, and C39E9.8) to be downregulated ∼2- to 5-fold after heat shock in swsn-7(RNAi) strains versus the control strain (Fig. 3D). Taken together, our findings strongly support a broad role for SWSN-7 in the positive regulation of stress-inducible ESRE genes in C. elegans.
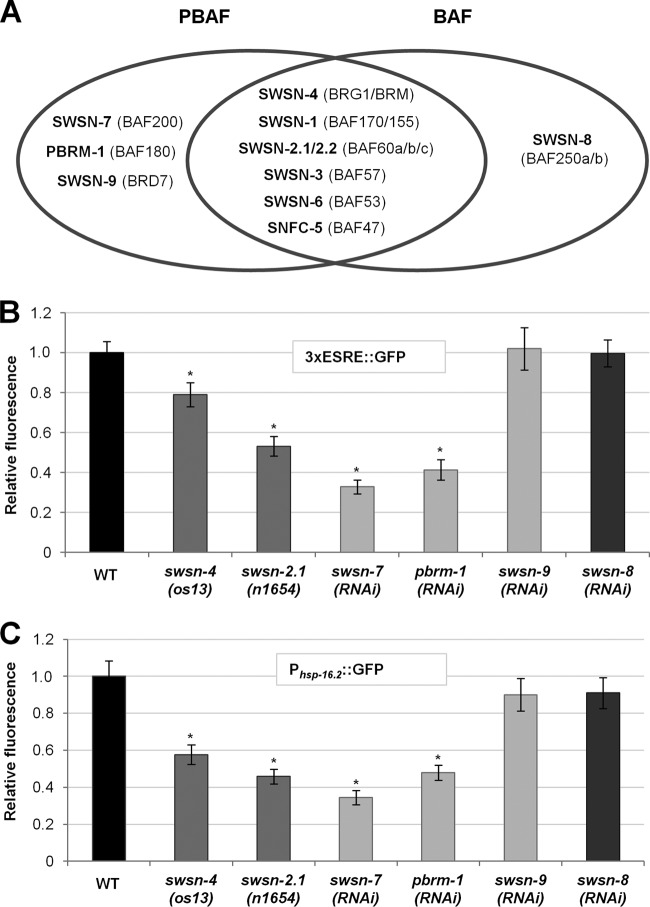
PBAF, but not BAF, promotes stress resistance.
SWSN-7/BAF200 is a conserved member of the mammalian PBAF subfamily of SWI/SNF nucleosome-remodeling complexes (42). PBAF, along with the closely related BAF complex, regulates transcription in a variety of contexts (17). Mammalian BAF and PBAF share eight common subunits, six of which are conserved in C. elegans (Fig. 4A). In addition, BAF and PBAF contain members that are unique to each complex, which are responsible for orchestrating complex-specific functions (Fig. 4A) (46). Unlike BAF, which contains only one highly conserved unique subunit, SWSN-8/BAF250, PBAF contains three specific components—SWSN-7/BAF200, PBRM-1/BAF180, and SWSN-9/BRD7—which combinatorially control the expression of distinct sets of genes. Whereas PBRM-1/BAF180 and SWSN-9/BRD7 are conserved in worms, mammals, and flies, SWSN-7/BAF200 is present in humans but not in Drosophila. To assess whether the PBAF complex requires all three PBAF-specific subunits to promote ESRE gene expression, we examined stress-induced expression of 3×ESRE::GFP and Phsp-16.2::GFP reporters after swsn-7, swsn-9, and pbrm-1 RNAi knockdown. In addition, we tested two of the shared subunits, SWSN-4/BRG1/BRM and SWSN-2.1/BAF60, and the BAF-specific subunit SWSN-8/BAF250. Notably, loss of the PBAF-specific subunits SWSN-7 and PBRM-1, as well as the common subunits (SWSN-2.1 and SWSN-4), led to a clear reduction in ESRE reporter expression after heat stress (Fig. 4B and C). In contrast, loss of the BAF-specific subunit, SWSN-8, as well as the PBAF subunit SWSN-9, failed to show effects (Fig. 4B and C). RNAi efficiency of the swsn-8 and swsn-9 clones was supported by our observation of embryonic lethality and slow-growth, respectively (see Fig. S6 in the supplemental material; also data not shown), consistent with previous reports (43, 47, 48). Our findings specifically implicate the PBAF complex in the regulation of ESRE gene induction but suggest that only a subset of the PBAF-specific members are required to carry out this function.
FIG 4.
PBAF is required for ESRE gene induction after heat stress. (A) Diagram of shared and specific subunits of BAF and PBAF in C. elegans, with their mammalian counterparts in parentheses. (B and C) Quantification of 3×ESRE::GFP (B) or Phsp-16.2::GFP (C) relative fluorescence intensities in RNAi-treated or mutant worms. WT and control RNAi levels were both arbitrarily set to 1.0. Relative levels were determined by comparison to wild-type strains grown on OP50 (os13 and n1654) or control RNAi plates (all others). Error bars indicate 95% CIs. Statistical analysis was done using the Student t test; asterisks indicate statistical significance (P < 0.001) in comparisons with either wild-type (os13 and n1654) or control RNAi (n = 40 to 60) worms.
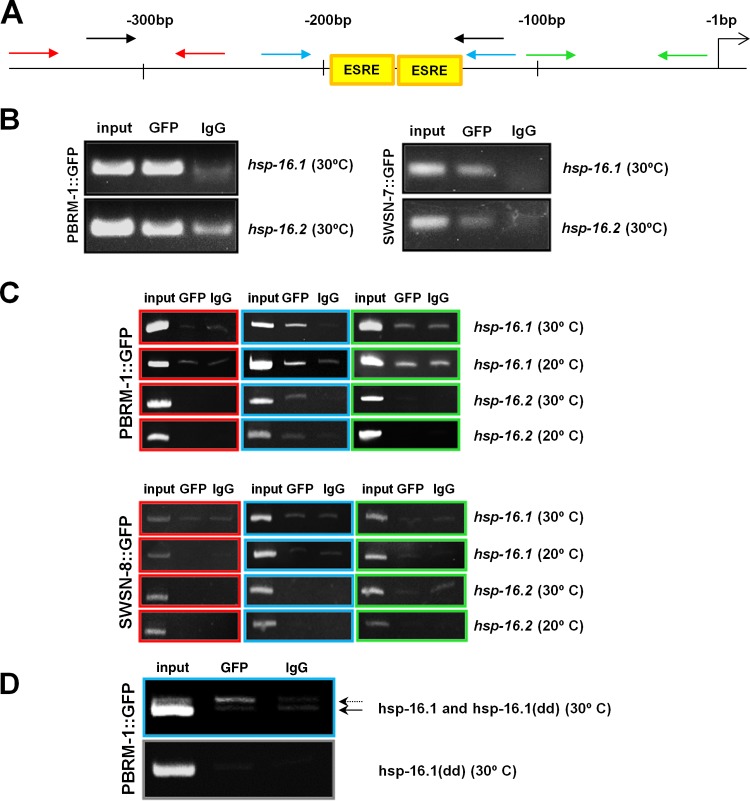
Using chromatin immunoprecipitation (ChIP) and deep-sequencing methods, it was recently shown that two subunits that are common to both BAF and PBAF (SWSN-1/BAF170/155 and SWSN-4/BRG1/BRM) are enriched at ESRE sites within the C. elegans genome (49). This observation, together with the results described above, suggested that the PBAF complex may be directly recruited to target genes containing an ESRE. To test this, we carried out ChIP studies using functional GFP-tagged SWSN-7 and PBRM-1 transgenes (see below) and assayed for their presence within an ∼200-bp region of the hsp-16.1 and hsp-16.2 promoters, each of which contains two ESRE sites. Notably, ChIP indicated that both PBRM-1 and SWSN-7 bind to the promoter regions of hsp-16.1 and hsp-16.2 in heat-shocked animals (Fig. 5A and B).
FIG 5.
ChIP analysis of hsp-16.1 and hsp-16.2 promoters. (A) Diagram of the hsp-16.1 and hsp-16.2 proximal promoter regions showing the positions of the start codon (right-most arrow), ESRE sites and PCR primer pairs used in the ChIP analysis. Colored primers correspond to gels shown in panels B to D. Numbers indicate nucleotide positions relative to start codon. (B to D) ChIP analysis was performed on strains expressing full-length functional PBRM-1::GFP, SWSN-7::GFP, or SWSN-8::GFP fusion proteins. Cross-linked DNA-protein complexes were immunoprecipitated with an anti-GFP antibody (GFP) or anti-IgG antibodies (IgG), and the DNA elutes were PCR amplified. (B) ChIP analysis (black primer set) showing association of both PBRM-1 and SWSN-7 with hsp-16.1 and hsp-16.2 proximal promoter regions at 30°C. (C) ChIP analysis of three promoter subregions (red, blue, and green primer sets) at either 20 or 30°C with PBRM-1::GFP or SWSN-8::GFP. Note that association with genomic DNA binding is specific to PBRM-1 and the ESRE-containing region (blue primer set). (D) ChIP analysis using a strain that contains both the wild-type endogenous hsp-16.1 promoter (dotted arrow) and an extrachromosomal array that contains copies of a mutated hsp-16.1 promoter (Phsp-16.1(dd)::GFP) in which the ESRE sites have been replaced, resulting in a region that is shorter by 15 bp (solid arrow). Note that association of PBRM-1 with DNA is specific to the endogenous hsp-16.1 locus containing wild-type ESRE sites.
To assess whether PBAF binds specifically to ESREs, ChIP analysis was carried out on the hsp-16.1 and hsp-16.2 promoters using the PBRM-1::GFP transgene. Enrichment was observed for primer sets that amplified a 100-bp region encompassing the ESRE sites but not for regions immediately upstream or downstream of the ESREs (Fig. 5A and C). Furthermore, no detectable binding was observed for the BAF-specific subunit, SWSN-8, indicating that binding to the ESRE sites was PBAF specific (Fig. 5A and C). To further confirm that PBAF specifically recognizes the ESRE motif, we performed ChIP analysis on a strain that contains wild-type ESRE sites within the endogenous hsp-16.1 promoter but that carries an extrachromosomal array containing mutated ESRE sites (Phsp-16.1(dd)::GFP). Notably, we observed clear enrichment of PBRM-1 binding at endogenous hsp-16.1 ESRE sites but not at mutated ESRE sites (Fig. 5D). These results strongly indicate that PBAF, but not BAF, is recruited directly to ESRE sites within the genome to control ESRE gene expression.
To determine whether PBAF associates with the ESRE sites in the absence of stress, we performed ChIP analysis on GFP-tagged PBRM-1 strain under nonstress conditions (20°C). Interestingly, PBRM-1 was clearly enriched at ESRE sites of both hsp-16.1 and hsp-16.2 promoters even in the absence of stress (Fig. 5A and C). Similar findings are known in yeast where the heat shock transcription factor (HSF) binds to promoters of heat shock protein (hsp) genes in the absence of stress (50).
swsn-7 and pbrm-1 are stress inducible and are required for viability after stress.
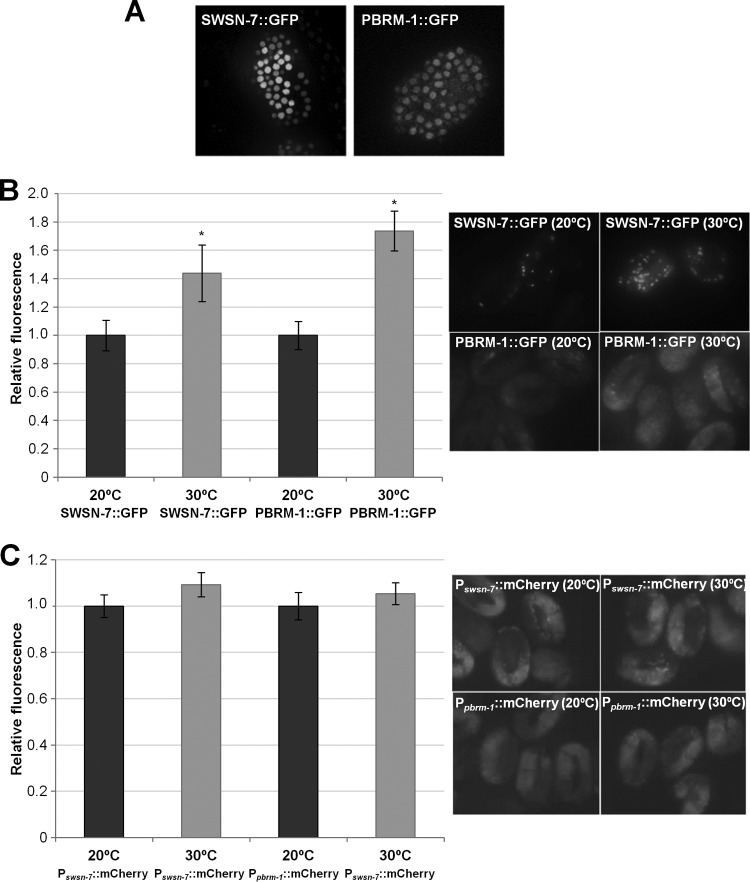
Given the importance of PBAF in the robust induction of the stress-inducible ESRE genes, we hypothesized that swsn-7 and pbrm-1 might themselves be stress inducible. We constructed a full-length SWSN-7::GFP reporter containing ∼900 bp of upstream genomic regulatory sequences. Biological activity of the reporter was verified by the rescue of the maternal-effect lethality of swsn-7(gk1041) mutants. In addition, we made use of a functional full-length PBRM-1::GFP, Pswsn-7::mCherry, and Ppbrm-1::mCherry reporters (46). Both SWSN-7::GFP and PBRM-1::GFP were expressed ubiquitously and localized to nuclei throughout development, as would be expected for members of a chromatin-remodeling complex (Fig. 6A). Moreover, both SWSN-7::GFP and PBRM-1::GFP were upregulated by ∼1.4- and 1.7-fold, respectively, after heat shock, indicating that both genes are stress inducible (Fig. 6B). Surprisingly, swsn-7 and pbrm-1 promoter fusions showed no change in expression following stress (Fig. 6C). These results suggest that upregulation of SWSN-7 and PBRM-1, following stress exposure, may be regulated at the level of translation or stability.
FIG 6.
SWSN-7 and PBRM-1 are stress-inducible nuclear proteins. (A) Embryonic expression pattern of full-length functional SWSN-7::GFP (left) and PBRM-1::GFP (right). Both reporters showed nuclear localization and early ubiquitous embryonic expression beginning at the ∼30-cell stage. (B) Quantification of GFP fluorescence intensities of SWSN-7::GFP and PBRM-1::GFP reporter strains at 20°C and after an 8-h exposure to a 30°C heat shock. Also shown are representative GFP photomicrographs of SWSN-7::GFP and PBRM-1::GFP reporter strains at 20 and 30°C. (C) Quantification of mCherry fluorescence intensities of Pswsn-7::mCherry and Ppbrm-1::mCherry reporter strains at 20°C and after an 8-h exposure to a 30°C heat shock. Representative fluorescent images of Pswsn-7::mCherry and Ppbrm-1::mCherry reporter strains at 20 and 30°C are shown on the right. Fluorescence intensities at 20°C were arbitrarily set to one for all reporters. Error bars indicate 95% CIs. Statistical analysis was done using the Student t test; asterisks indicate statistical significance (P < 0.001) of strains at 20°C versus 30°C (n = 50).
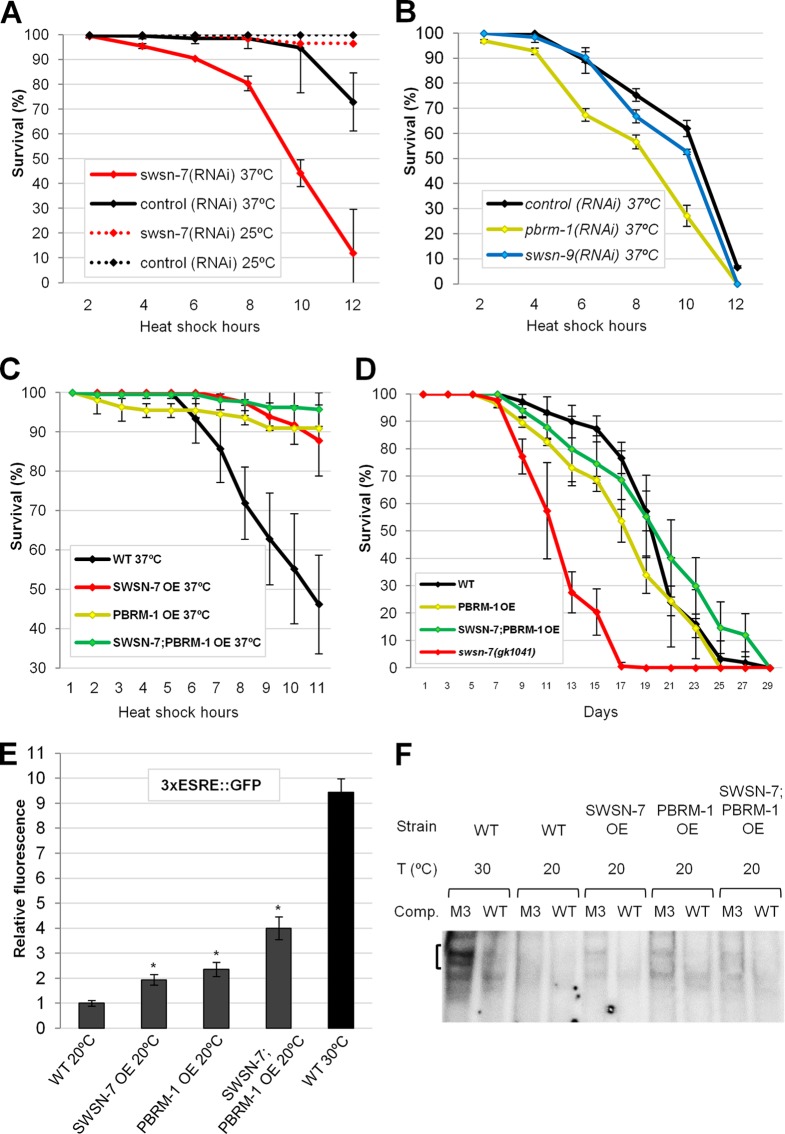
We next examined the functional consequences of inhibiting swsn-7 and pbrm-1 following stress. Whereas swsn-7(RNAi) had no effect on the viability of adult worms at 25°C, animals incubated at 37°C were much less robust than controls (Fig. 7A). Likewise, pbrm-1(RNAi) adults showed reduced survival after stress relative to controls, although effects were less dramatic than those observed for swsn-7(RNAi) (Fig. 7B). Consistent with results using 3×ESRE::GFP and Phsp-16.2::GFP reporters, swsn-9(RNAi) did not lead to a reduction in viability (Fig. 7B). Thus, PBAF is essential for normal survival following stress, although only a subset of the PBAF-specific subunits are required for this process.
FIG 7.
PBAF subunits SWSN-7 and PBRM-1 promote survival under stress conditions and regulate ESRE-binding activity. (A and B) Knockdown of swsn-7 (A) and pbrm-1 (B) by RNAi significantly reduced survival after heat shock at 37°C (P < 0.001 and P = 0.002, respectively). Note that depletion of swsn-9, a third PBAF-specific subunit, showed no effect on survival (P = 0.474), nor did swsn-7(RNAi) appreciably decrease survival under nonstressed conditions (25°C). (C) Overexpression (OE) of SWSN-7 and PBRM-1 subunits, either separately or together, increased resistance to thermal stress (37°C) compared to the wild type (P < 0.001). (D) Longevity assays showing that overexpression (OE) of SWSN-7 and PBRM-1 did not increase life-span (P = 0.183), whereas swsn-7(gk1041) mutants did have a reduced life-span (P < 0.001). Depicted results are averages from three independent experiments. Error bars indicate 95% CIs. Statistical analysis was done using a log-rank test. (E) Quantification of 3×ESRE::GFP fluorescence in worms that overexpressed SWSN-7 and PBRM-1 either separately or together. Controls included wild-type worms carrying a 3×ESRE::GFP at 20 and 30°C. Fluorescence intensity of the control worms at 20°C was arbitrarily set to one. Error bars indicate the 95% CIs. Statistical analysis was done using the Student t test; asterisks indicate statistical significance (P < 0.001; n = 50). (F) ESRE-binding activity observed using EMSA was carried out with nuclear extracts prepared from unstressed (at 20°C) and stressed (30°C) wild-type worms and worms overexpressing SWSN-7 and PBRM-1 (20°C only).
Overexpression of SWSN-7 and PBRM-1 increases resistance to stress and leads to the upregulation of ESRE-binding activity.
Given that the loss of the PBAF subunits SWSN-7 and PBRM-1 led to a reduction in ESRE reporter expression and decreased thermotolerance, we hypothesized that the overexpression of these proteins might be sufficient to enhance survival under stress conditions. To test this, we overexpressed SWSN-7 and PBRM-1, either separately or together, in strains containing the 3×ESRE::GFP reporter. Overexpression of each subunit alone at 20°C increased GFP expression by ∼2-fold, whereas extra copies of both of the genes resulted in an even greater enhancement (∼4-fold) (Fig. 7E). Although substantial, upregulation in these strains at 20°C was nevertheless less than that observed for wild type at 30°C, which is upregulated ∼10-fold (Fig. 7E), indicating that other factors are required for full induction of ESRE genes after stress.
Because extra copies of swsn-7 and pbrm-1 efficiently induced ESRE-mediated transcription, we next tested whether overexpression of PBAF subunits could increase ESRE-binding activity in worm extracts. EMSA was carried out using nuclear extracts derived from each of the overexpressing strains grown at 20°C. Notably, modest ESRE-binding activity was observed for each of these strains under nonstressed conditions (Fig. 7F; see Fig. S2F in the supplemental material), when ESRE binding is normally at or below levels of detection. These findings suggest that overexpression of PBAF can increase the abundance or affinity of an endogenous ESRE-binding protein(s).
To investigate whether increased levels of SWSN-7 and PBRM-1 are beneficial under stress conditions, we exposed overexpressing strains to heat shock (37°C) for 11 h and scored for survival. Notably, all PBAF-overexpressing strains that were tested exhibited significantly enhanced survival compared to the wild type (Fig. 7C). Because longevity and stress resistance are often coupled (2, 8, 10, 51–54), we next examined whether SWSN-7 and PBRM-1 levels affected the life-span of animals. Whereas swsn-7-null mutants lived significantly shorter in comparison with wild type, extra copies of swsn-7 and pbrm-1 did not detectably affect life-span (Fig. 7D). Taken together, these findings indicate that although overexpression of PBAF does not enhance longevity, the PBAF complex is nevertheless required for normal life-span as well as the adaptation to heat stress.
PBAF may cooperate with histone demethylase activity to modulate ESRE gene expression.
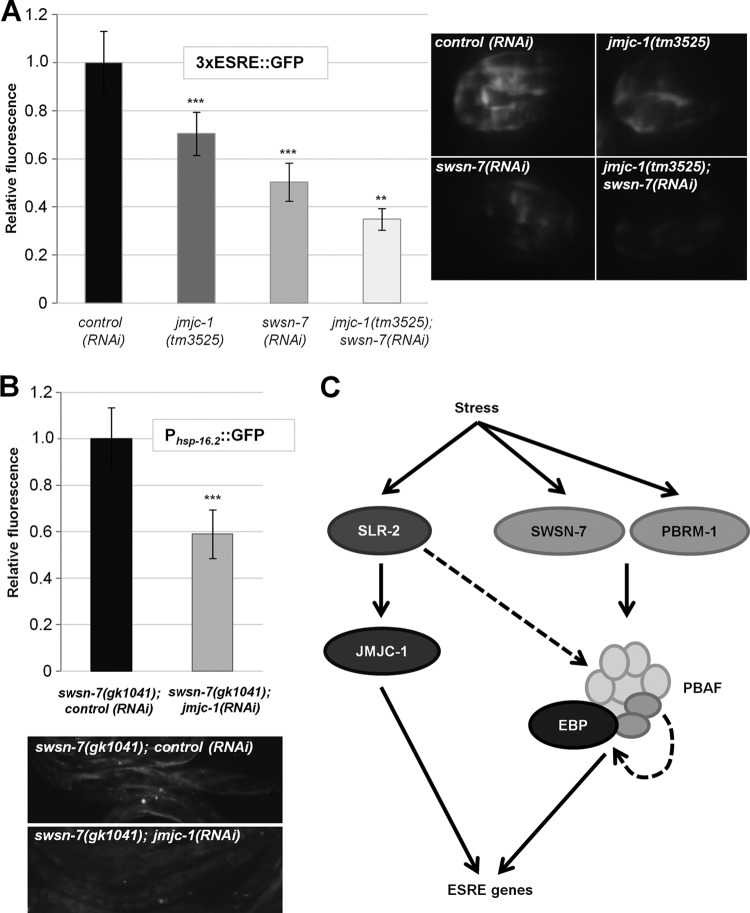
Our previous studies demonstrated that a conserved histone demethylase, JMJC-1/NO66, functions as a transcriptional activator of the ESRE genes in worms, flies, and mammals (34). To assess whether JMJC-1 and PBAF cooperate in regulating ESRE gene expression, we examined expression of the 3×ESRE::GFP reporter in heat-shocked embryos where either jmjc-1 or swsn-7 or both were depleted. Whereas depletion of each gene alone decreased 3×ESRE::GFP expression following stress, loss of both of the genes resulted in even greater reduction (Fig. 8A). In addition, we examined expression of the Phsp-16.2::GFP reporter in swsn-7 mutants exposed to control or jmjc-1(RNAi). Notably, RNAi of jmjc-1 significantly reduced expression of Phsp-16.2::GFP relative to the control (Fig. 8B). These results suggest that PBAF and JMJC-1 cooperate to promote ESRE gene expression.
FIG 8.
ESRE gene expression is cooperatively regulated by SWSN-7 and JMJC-1. (A) Quantification of fluorescence intensities of 3×ESRE::GFP in the wild type, in jmjc-1(tm3525) deletion mutants, in swsn-7(RNAi)-treated worms, and in jmjc-1(tm3525); swsn-7(RNAi) embryos after exposure to heat shock (30°C) for 12 h. Integers on y axes indicate average levels of GFP fluorescence relative to control RNAi (arbitrarily set to 1.0; n = 30 to 35). Also shown are representative GFP photomicrographs of heat-shocked embryos. (B) Quantification of Phsp-16.2::GFP relative fluorescence intensities in swsn-7(gk1041) mutant adults treated with control RNAi or jmjc-1(RNAi). Control RNAi levels were arbitrarily set to 1.0. Error bars indicate 95% CIs. Statistical analysis was done using the Student t test (***, P < 0.001; **, P < 0.01; n = 30). (C) Model of transcriptional regulation of ESRE genes. See the text for details.
DISCUSSION
We have shown that PBAF, a SWI/SNF family nucleosome-remodeling complex, is a critical regulator of the ESRE pathway, a master stress response network in C. elegans. Although the two main subclasses of SWI/SNF complexes, PBAF and BAF complex, are mostly identical at the level of protein composition, our data specifically implicate PBAF in the regulation of ESRE gene expression. Moreover, of the three PBAF-specific subunits, only two, SWSN-7 and PBRM-1, were required in the ESRE response. To our knowledge, this is the first report demonstrating direct regulation of a stress response network by the PBAF complex in vivo in a multicellular organism.
Recently, it was shown that the BAF complex regulates targets of IIS signaling in C. elegans (49). Specifically, BAF interacts directly with DAF-16/FOXO to mediate target gene expression. PBAF was not, however, implicated in the regulation of DAF-16/FOXO targets, supporting a model in which distinct nucleosome remodeling complexes are recruited by different stress response factors. Depletion of certain nucleosome-remodeling complexes (SWI/SNF, ISWI, and CHD) results in hypersensitivity to stress in yeast (22–24, 55), although their stress-related functions in multicellular organisms are less well documented. Other classes of chromatin regulators that have been implicated in stress-gene activation include modifiers of histones (12, 56–59). For example, the stress-responsive activator of p300, a heat shock-inducible transcription cofactor that facilitates HSF-1 binding, stimulates transcription of hsp70 and hsp90 genes in mammalian cells by increasing chromatin acetylation (59). An emerging view is that regional chromatin reorganization may often be necessary to achieve robust expression of genes following stress. Consistent with this, our previous studies demonstrated that a conserved histone demethylase, JMJC-1/NO66, functions as a transcriptional activator of the ESRE genes in worms, flies, and mammals (34). We showed here that SWSN-7 (PBAF) acts cooperatively with JMJC-1 to promote the expression of ESRE genes following stress (Fig. 8). In some cell types, both PBAF and JMJC-1 may associate with the promoters of ESRE genes through recruitment by one or more DNA-binding proteins. Alternatively, PBAF and JMJC-1 may have more specific functions or relative impact within distinct tissues. Our studies collectively show that neither histone modification nor nucleosome remodeling alone are sufficient to fully induce genes within the ESRE network, but that both activities are required for optimal gene expression following stress. We also note that SLR-2 transcriptional targets include several BAF/PBAF subunits, which are downregulated in slr-2 mutants (34). Thus, SLR-2 may contribute to the stress response and ESRE gene expression through both JMJC-1 and the PBAF pathways.
Based on our findings, we propose the model shown in Fig. 8C for the regulation of ESRE gene expression. An ESRE-binding protein (EBP) recruits the PBAF complex to target genes containing the motif. This complex may be targeted to some ESRE sites even in the absence of stress, based on findings for hsp-16.1 and hsp-16.2 (Fig. 5). This may permit genes to be rapidly activated following stress as has been demonstrated for HSF in yeast (50). Also consistent with this, DAF-16/FOXO, which regulates the expression of many stress-related genes, recruits the BAF complex to promoters under nonstress conditions (49). Direct regulation by the PBAF complex is supported by our studies with ESRE reporters (Fig. 3A and B, Fig. 4B and C), endogenous ESRE gene expression (Fig. 3D), and by ChIP assays (Fig. 5). Consistent with this, two subunits common to BAF and PBAF (SWSN-4/BRG1/BRM and SWSN-1/BAF170/BAF155) show enriched binding to ESRE sites in the C. elegans genome (49). A role for PBAF in the stress response is also supported by data showing that loss of PBAF function led to increased sensitivity to stress (Fig. 7A and B), whereas increased expression of PBAF led to increased thermotolerance (Fig. 7C). Moreover, the PBAF subunits SWSN-7 and PBRM-1 were themselves stress inducible (Fig. 6B). We speculate that preassembled PBAF complexes may be available to promote a rapid transcriptional response but that stress-inducible expression of PBAF subunits ensures that sufficient PBAF is available for the duration of the stress response. Our studies also suggest that the elevated expression of PBAF-specific subunits may be sufficient to promote increased formation of the PBAF complex leading to greater stress tolerance (Fig. 7C). This could be explained if the subunits that are common to both BAF and PBAF are normally present in excess of PBAF-specific subunits and thus PBAF-specific subunits are rate-limiting. Consistent with this, shared subunits are present at higher levels in mammalian and yeast cells than unique subunits (42, 60, 61). In addition, nuclear ESRE-binding activity also increased after stress exposure (Fig. 1D and E; see also Fig. S2E in the supplemental material), implying that a mechanism likely exists to promote EBP expression or DNA-binding activity. Furthermore, overexpression of PBAF leads to a small but detectable increase in ESRE-binding activity (Fig. 7F), suggesting that PBAF may positively regulate expression of the EBP. This result could be explained by the existence of a positive-feedback loop whereby the EBP, in conjunction with PBAF, activates its own expression.
Although our EMSAs clearly confirmed the presence of an endogenous EBP, its identity remains unknown. Although SLR-2 contains a recognizable DNA-binding domain, evidence suggests that neither SLR-2 nor JMJC-1 binds directly to the ESRE. First, whereas the ESRE is conserved across species, SLR-2 does not appear to have orthologs in non nematode species (34). Second, ESRE genes are induced following stress in slr-2- and jmjc-1-null mutants, albeit to lower levels than those observed in the wild type (34). Third, we observed wild-type levels of ESRE binding by endogenous factors in extracts prepared from slr-2- and jmjc-1-null mutants (see Fig. S7A in the supplemental material). Fourth, a SLR-2 recombinant protein prepared from E. coli failed to interact with the ESRE using EMSA (see Fig. S7B in the supplemental material). Finally, although JMJC-1 is an evolutionarily conserved histone demethylase, it lacks any recognizable DNA-binding domains.
Overexpression of several stress response factors, including DAF-16, SKN-1, and PHA-4, not only increases stress resistance but also leads to longer life-spans (8, 10, 62). In contrast, overexpression of the PBAF complex led to increased stress resistance (Fig. 7C) but not enhanced longevity, although PBAF appeared to be necessary for a normal life-span (Fig. 7D). Furthermore, overexpression of SLR-2 or JMJC-1 leads to increased stress resistance but only weakly affects longevity (34). Thus, factors that regulate the ESRE stress network appear to have relatively little or no impact on life-span underscoring that stress resistance and life-span are not always coupled. Future studies to identify the EBP, as well as other key ESRE-pathway regulators, will provide a more complete understanding of the complex cellular networks that coordinate the response to stress in multicellular organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Amy Fluet for editing, Harry Jarrett for technical advice and experimental assistance, and Junho Lee for reagents. We thank the Caenorhabditis elegans Genetics Center and the North American and Japanese (National BioResource Project) gene knockout consortiums for strains. We also thank Valerie Reinke and Michelle Kudron for advice on ChIP analysis.
This study was supported by National Institutes of Health grant R01 GM066868-06 and by INBRE (P20 GM103432).
Footnotes
Published ahead of print 13 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01502-13.
REFERENCES
- 1.Fulda S, Gorman AM, Hori O, Samali A. 2010. Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010:214074. 10.1155/2010/214074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kourtis N, Tavernarakis N. 2011. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 30:2520–2531. 10.1038/emboj.2011.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter K, Haslbeck M, Buchner J. 2010. The heat shock response: life on the verge of death. Mol. Cell 40:253–266. 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 4.An JH, Blackwell TK. 2003. SKN-1 links Caenorhabditis elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17:1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumeister R, Schaffitzel E, Hertweck M. 2006. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J. Endocrinol. 190:191–202. 10.1677/joe.1.06856 [DOI] [PubMed] [Google Scholar]
- 6.de Nadal E, Posas F. 2010. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 29:4–13. 10.1038/emboj.2009.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161:1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson ST, Johnson TE. 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11:1975–1980. 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- 9.Kell A, Ventura N, Kahn N, Johnson TE. 2007. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic. Biol. Med. 43:1560–1566. 10.1016/j.freeradbiomed.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. 2007. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447:550–555. 10.1038/nature05837 [DOI] [PubMed] [Google Scholar]
- 11.Hsu AL, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300:1142–1145. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- 12.Guertin MJ, Lis JT. 2010. Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6:e1001114. 10.1371/journal.pgen.1001114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shivaswamy S, Iyer VR. 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 28:2221–2234. 10.1128/MCB.01659-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiner A, Chen HV, Liu CL, Rahat A, Klien A, Soares L, Gudipati M, Pfeffner J, Regev A, Buratowski S, Pleiss JA, Friedman N, Rando OJ. 2012. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 10:e1001369. 10.1371/journal.pbio.1001369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgio G, La Rocca G, Sala A, Arancio W, Di Gesu D, Collesano M, Sperling AS, Armstrong JA, van Heeringen SJ, Logie C, Tamkun JW, Corona DF. 2008. Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet. 4:e1000089. 10.1371/journal.pgen.1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. 2011. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 39:8378–8391. 10.1093/nar/gkr535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 18.Ho L, Crabtree GR. 2010. Chromatin remodeling during development. Nature 463:474–484. 10.1038/nature08911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodeling complex. Nat. Rev. Cancer 11:588–596. 10.1038/nrc3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodelers and cancer. Nat. Rev. Cancer 11:481–492. 10.1038/nrc3068 [DOI] [PubMed] [Google Scholar]
- 21.Wu JI. 2012. Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim. Biophys. Sin (Shanghai) 44:54–69. 10.1093/abbs/gmr099 [DOI] [PubMed] [Google Scholar]
- 22.Mas G, de Nadal E, Dechant R, Rodriguez de la Concepcion ML, Logie C, Jimeno-Gonzalez S, Chavez S, Ammerer G, Posas F. 2009. Recruitment of a chromatin remodeling complex by the Hog1 MAP kinase to stress genes. EMBO J. 28:326–336. 10.1038/emboj.2008.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686–697. 10.1101/gad.13.6.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson B, Erdjument-Bromage H, Tempst P, Cairns BR. 2006. The RSC chromatin remodeling complex bears an essential fungal-specific protein module with broad functional roles. Genetics 172:795–809. 10.1534/genetics.105.047589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lant B, Storey KB. 2010. An overview of stress response and hypometabolic strategies in Caenorhabditis elegans: conserved and contrasting signals with the mammalian system. Int. J. Biol. Sci. 6:9–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez M, Snoek LB, De Bono M, Kammenga JE. 2013. Worms under stress: Caenorhabditis elegans stress response and its relevance to complex human disease and aging. Trends Genet. 29:367–374. 10.1016/j.tig.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 27.Mukhopadhyay A, Oh SW, Tissenbaum HA. 2006. Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 41:928–934. 10.1016/j.exger.2006.05.020 [DOI] [PubMed] [Google Scholar]
- 28.Murphy CT. 2006. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp. Gerontol. 41:910–921. 10.1016/j.exger.2006.06.040 [DOI] [PubMed] [Google Scholar]
- 29.Singh V, Aballay A. 2006. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U. S. A. 103:13092–13097. 10.1073/pnas.0604050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barsyte D, Lovejoy DA, Lithgow GJ. 2001. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 15:627–634. 10.1096/fj.99-0966com [DOI] [PubMed] [Google Scholar]
- 31.Murakami S, Johnson TE. 1996. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143:1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamitina ST, Strange K. 2005. Transcriptional targets of DAF-16 insulin signaling pathway protect Caenorhabditis elegans from extreme hypertonic stress. Am. J. Physiol. Cell Physiol. 288:C467–C474. 10.1152/ajpcell.00451.2004 [DOI] [PubMed] [Google Scholar]
- 33.Shivers RP, Youngman MJ, Kim DH. 2008. Transcriptional responses to pathogens in Caenorhabditis elegans. Curr. Opin. Microbiol. 11:251–256. 10.1016/j.mib.2008.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirienko NV, Fay DS. 2010. SLR-2 and JMJC-1 regulate an evolutionarily conserved stress-response network. EMBO J. 29:727–739. 10.1038/emboj.2009.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong M, Kwon JY, Shim J, Lee J. 2004. Differential hypoxia response of hsp-16 genes in the nematode. J. Mol. Biol. 344:369–381. 10.1016/j.jmb.2004.09.077 [DOI] [PubMed] [Google Scholar]
- 36.Kwon JY, Hong M, Choi MS, Kang S, Duke K, Kim S, Lee S, Lee J. 2004. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics 83:600–614. 10.1016/j.ygeno.2003.10.008 [DOI] [PubMed] [Google Scholar]
- 37.Stiernagle T. 11 February 2006. Maintenance of Caenorhabditis elegans. In The C. elegans Research Community, WormBook (ed), WormBook. 10.1895/wormbook.1.101.1 [DOI] [Google Scholar]
- 38.Ahringer J. 6 April 2006. Reverse genetics: Caenorhabditis elegans. In The C. elegans Research Community, WormBook (ed), WormBook. 10.1895/wormbook.1.47.1 [DOI] [Google Scholar]
- 39.Timmons L, Court DL, Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263:103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- 40.Stroeher VL, Kennedy BP, Millen KJ, Schroeder DF, Hawkins MG, Goszczynski B, McGhee JD. 1994. DNA-protein interactions in the Caenorhabditis elegans embryo: oocyte and embryonic factors that bind to the promoter of the gut-specific ges-1 gene. Dev. Biol. 163:367–380. 10.1006/dbio.1994.1155 [DOI] [PubMed] [Google Scholar]
- 41.GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, Lithgow G, Kim S, Link CD. 2002. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 12:701–712. 10.1101/gr.228902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. 2005. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19:1662–1667. 10.1101/gad.1323805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- 44.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434:462–469. 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- 45.Murphy CT. 2005. A review of genes that act downstream of the DAF-16 FOXO transcription factor to influence the life span of Caenorhabditis elegans. In Carey JR, Robine JM, Michel JP, Christen Y. (ed), Longevity and frailty. Springer-Verlag, Berlin, Germany [Google Scholar]
- 46.Shibata Y, Uchida M, Takeshita H, Nishiwaki K, Sawa H. 2012. Multiple functions of PBRM-1/Polybromo- and LET-526/Osa-containing chromatin remodeling complexes in Caenorhabditis elegans development. Dev. Biol. 361:349–357. 10.1016/j.ydbio.2011.10.035 [DOI] [PubMed] [Google Scholar]
- 47.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. 2000. Functional genomic analysis of Caenorhabditis elegans chromosome I by systematic RNA interference. Nature 408:325–330. 10.1038/35042517 [DOI] [PubMed] [Google Scholar]
- 48.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38:896–903. 10.1038/ng1844 [DOI] [PubMed] [Google Scholar]
- 49.Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, Ruvkun G. 2013. DAF-16 employs the chromatin remodeler SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15:491–501. 10.1038/ncb2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T, Parker CS. 2002. Dynamic association of transcriptional activation domains and regulatory regions in Saccharomyces cerevisiae heat shock factor. Proc. Natl. Acad. Sci. U. S. A. 99:1200–1205. 10.1073/pnas.032681299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin K, Dorman JB, Rodan A, Kenyon C. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278:1319–1322. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- 52.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- 53.Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. 2009. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8:524–541. 10.1111/j.1474-9726.2009.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. 2010. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 6:e1001048. 10.1371/journal.pgen.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erkina TY, Zou Y, Freeling S, Vorobyev VI, Erkine AM. 2010. Functional interplay between chromatin remodeling complexes RSC, SWI/SNF and ISWI in regulation of yeast heat shock genes. Nucleic Acids Res. 38:1441–1449. 10.1093/nar/gkp1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chervona Y, Costa M. 2012. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic. Biol. Med. 53:1041–1047. 10.1016/j.freeradbiomed.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta J, Tikoo K. 2012. Involvement of insulin-induced reversible chromatin remodeling in altering the expression of oxidative stress-responsive genes under hyperglycemia in 3T3-L1 preadipocytes. Gene 504:181–191. 10.1016/j.gene.2012.05.027 [DOI] [PubMed] [Google Scholar]
- 58.Nguyen MP, Lee S, Lee YM. 2013. Epigenetic regulation of hypoxia inducible factor in diseases and therapeutics. Arch. Pharm. Res. 36:252–263. 10.1007/s12272-013-0058-x [DOI] [PubMed] [Google Scholar]
- 59.Xu D, Zalmas LP, La Thangue NB. 2008. A transcription cofactor required for the heat-shock response. EMBO Rep. 9:662–669. 10.1038/embor.2008.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muchardt C, Yaniv M. 2001. When the SWI/SNF complex remodels the cell cycle. Oncogene 20:3067–3075. 10.1038/sj.onc.1204331 [DOI] [PubMed] [Google Scholar]
- 61.Nie Z, Yan Z, Chen EH, Sechi S, Ling C, Zhou S, Xue Y, Yang D, Murray D, Kanakubo E, Cleary ML, Wang W. 2003. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 23:2942–2952. 10.1128/MCB.23.8.2942-2952.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. 2008. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in Caenorhabditis elegans. Cell 132:1025–1038. 10.1016/j.cell.2008.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J. 2013. Hypoxia-inducible factor-1 (HIF-1)-independent hypoxia response of the small heat shock protein hsp-16.1 gene regulated by chromatin-remodeling factors in the nematode Caenorhabditis elegans. J. Biol. Chem. 288:1582–1589. 10.1074/jbc.M112.401554 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.