Abstract
HIV infiltrates the brain at early times postinfection and remains latent within astrocytes and macrophages. Because astrocytes are the most abundant cell type in the brain, we evaluated epigenetic regulation of HIV latency in astrocytes. We have shown that class I histone deacetylases (HDACs) and a lysine-specific histone methyltransferase, SU(VAR)3-9, play a significant role in silencing of HIV transcription in astrocytes. Our studies add to a growing body of evidence demonstrating that astrocytes are a reservoir for HIV.
TEXT
HIV establishes a state of latency, defined as period of nonproductive replication, as a means to evade immune responses and/or to ensure evolutionary coexistence with a host. Latency in HIV is best defined within resting CD4+ T cells, where reverse-transcribed HIV cDNA integrates into the host genome, and although the provirus is replication competent, its expression is silenced. Antiretroviral drug intensification has not been able to alter the size of the latent HIV pool (1, 2), representing a major obstacle toward eradication of HIV.
Much attention is focused on understanding mechanisms of HIV latency in CD4+ resting T cells. However, other cellular reservoirs and sanctuary sites for HIV remain, including the central nervous system (CNS). HIV invades the brain within weeks of infection, persists in the CNS at a steady state despite combination antiretroviral therapy (cART), and undergoes compartmentalization, as indicated by the evolution of HIV genetic sequences in the CNS that are distinct from those in plasma and lymphoid tissue (3–6). Further, studies of HIV genotyping from patients under cART with undetectable viremia indicate that these patients often experience “blips” in HIV replication and this reactivated virus is not derived from lymphoid/myeloid cells (7, 8), suggesting that additional sites for HIV latency and reactivation exist. The brain, among other sanctuary sites, is a source for latent HIV.
Astrocytes are latently infected by HIV.
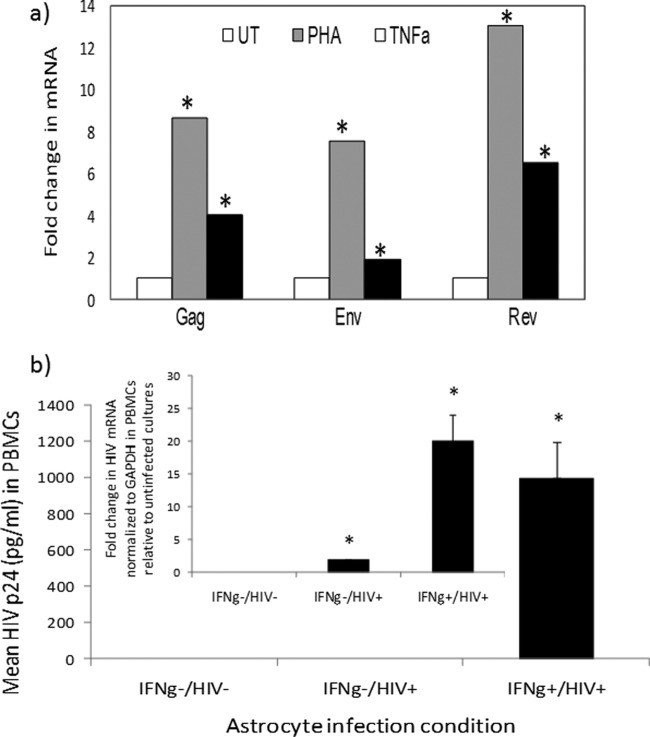
Astrocytes are the most abundant cell type in the brain. These cells perform vital functions to maintain brain homeostasis. A number of groups using postmortem tissue have detected integrated HIV DNA within astrocytes (9, 10). The frequency of HIV DNA within astrocytes ranges from 3 to 19%, with higher levels of HIV DNA within astrocytes associated with HIV encephalitis and a closer proximity of astrocytes to perivascular macrophages. HIV p24 in HIV-positive (HIV+) postmortem brain astrocytes is rarely detected, if at all. In vitro, astrocytes support low-level HIV replication, which is inducible in response to proinflammatory signals (11–13). Through a series of publications, we also demonstrated that robust/productive HIV replication in astrocytes is restricted but can be supported under inflammatory signals, such as gamma interferon (IFN-γ) (13–17). Priming astrocytes with IFN-γ, in particular, facilitates HIV replication in astrocytes (13–17). We used primary human progenitor-derived astrocytes (PDAs) and two astrocytoma cell lines (U87MG and U138) in this study to define the role of astrocytes in HIV latency. PDAs express hallmark features of astrocytes, including >90% positivity for glial fibrillary acidic protein (GFAP), positivity for glutamate transporter 1/EAAT2 and glutamine synthetase, and capability for glutamate uptake (18). They are also positive for s100b and aldehyde dehydrogenase (data not shown). We detected extremely low levels of early (Rev) and late (Gag and Env) mRNAs, which were closer to the lower limit of real-time PCR detection (threshold cycle [CT] ≥ 35), from PDAs infected with HIVBal (10 ng/106 cells/ml). Treatment with phytohemagglutinin (PHA) (a mitogen) or tumor necrosis factor alpha (TNF-α) induced HIV mRNA expression by 8- to 12-fold or 2- to 6-fold, respectively (Fig. 1a). The virions released from infected activated astrocytes are infectious, since they infected activated peripheral blood mononuclear cells (PBMCs) (Fig. 1b). Interestingly, while no detectable p24 in lymphocytes was measured in supernatant of infected/non-IFN-γ-primed astrocytes, probably due to poor sensitivity of the p24 enzyme-linked immunosorbent assay (ELISA) (200 to 40,000 pg/ml; AIDS Reagent Program), a low level of HIV is transmitted to lymphocytes, as measured by detection of HIV Env mRNA in lymphocytes exposed to supernatant from infected astrocytes (Fig. 1b). These data demonstrate that HIV from astrocytes is inducible and infectious.
FIG 1.
(a) Reactivation of HIV mRNAs in astrocytes. PDAs were infected with HIVBal (10 ng/ml/106 cells) and left untreated (UT) or treated with PHA (4 μg/ml) or TNF-α (TNFa) (100 U/ml), and HIV mRNAs were measured by qRT-PCR. Data are normalized to GAPDH mRNA and expressed as fold change relative to results for untreated cells. Asterisks denote P < 0.05 between results for treated and untreated cultures. (b) Inducible and low level of HIV replication in astrocytes is transmitted to lymphocytes. Astrocytes were primed with IFN-γ (75 U/ml) or left untreated and then infected with HIVBal (10 ng/ml/106 cells) and cultured with or without IFN-γ (IFNg). The initial virus inoculum was removed by mild pronase treatment and washing; the supernatant was harvested from astrocytes at day 7 postinfection and then exposed to anti-CD3/anti-CD28-costimulated PBMCs. HIV p24 from PBMC supernatant was measured by ELISA on day 6. HIV mRNA (Env) from PBMCs was quantified by real-time PCR on day 6, normalized to GAPDH, and presented as expression relative to that for uninfected cultures. HIV p24 from IFN-γ−/HIV− or HIV+ cultures was undetectable. *, P < 0.05 (Student's t test) between results for control and treated samples.
To assess mechanisms driving HIV latency in astrocytes, we established two key tools. (i) We generated latently infected primary astrocytes and astrocytic cell lines. PDAs and the U138 astrocytoma cell line were infected with HIVBal at 10 ng/ml of HIV p24 per 1 × 106 cells. The infected cells were propagated for several passages and subjected to PCR to detect HIV DNA. To determine the percentage of integrated provirus in infected PDAs and U138 cells, we mixed U1 cells (harboring 2 copies of HIV DNA/cell) and the parent uninfected cell line (U937) to mathematically create a pool of cellular DNA whereby 0 to 50% of DNA is from integrated HIV DNA (e.g., for 50% HIV DNA, the DNA is isolated from 25 U1 cells mixed with 75 U937 cells, etc.). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, and the HIV long terminal repeat (LTR) was amplified using Alu-PCR. A representative standard curve representing delta CT versus percent HIV-infected cells is shown in Fig. 2. Based on this analysis, we extrapolate that approximately 3% of U138 and PDAs harbor HIV DNA (Fig. 2), while 32% of PBMCs were infected by HIV. This value is an estimate, because some cells may harbor multiple DNA copies while others may not have some at all, and it is also a reflection of the specific experimental setting, where 10 ng/ml HIV p24/106 cells was used to infect the cells. (ii) We generated astrocytic cell lines that stably harbor the HIV LTR-Luc construct. We generated two astrocytic cell lines (U87MG and U251MG) expressing chromosomally integrated HIV LTR luciferase and selected by neomycin resistance, as previously described (16).
FIG 2.

Estimation of frequency of HIV infection in astrocytes: a standard curve was generated to determine the percentage of infected cells based on use of a mixture of DNA from U1 (contains 2 proviral copies/cell) and U937 (U1 parental cell line). HIVBal (10 ng/ml/106 cells) was used to infect stimulated PBMCs, primary astrocytes (PDA), or an astrocytic cell line (U138). HIV DNA was amplified using Alu-PCR followed by real-time PCR. Linear regression list dotted line and standard curve are shown.
Epigenetics regulates HIV latency in a number of cell models (19–22). Specifically, DNA CpG methylation and histone modifications (methylation and deacetylation) of the HIV promoter are hallmarks of HIV latency (20, 23–25). Histone deacetylases (HDACs) catalyze the removal of acetyl groups from the lysine ε-amino group of histones and other nonhistone substrates. HDACs are recruited to the HIV LTR, inducing LTR chromatin compaction and as a consequence viral latency (19, 26, 27). Several transcription factors and corepressor complexes are shown to recruit class I HDACs to the initiator and enhancer regions of the HIV-1 LTR (28–34). HDAC inhibitors reactivate HIV in cell culture models and in resting CD4+ T cells from HIV+ patients (19–22). Methylation of DNA by DNA methyltransferases (DMTs) and/or methylation of histones by histone methyltransferases (HMTs) can also induce HIV promoter silencing (23–25, 35, 36).
Class 1 HDACs regulate HIV LTR promoter activity in astrocytes.
To assess the impact of HDACs on HIV promoter activity in astrocytes, we first evaluated the expression of HDACs in PDAs and U87MG cells. There are 18 known human HDACs, which are divided into four classes: class I consists of HDAC1, -2, -3, and -8; class II consists of HDAC4, -5, -6, -7, -9, and -10; class III consists of the NAD+-dependent Sir2-like deacetylases or sirtuins; and class IV consists of HDAC11. Class I, II, and IV HDACs are zinc-dependent hydrolases (37–39). To determine which HDACs are driving HIV promoter silencing in astrocytes, we first determined mRNA and protein expression of HDACs in PDAs and U87MG. We calculated levels of mRNA expression for HDAC1 to -11 relative to that for cyclin D1 (CCDN1) in PDAs by real-time PCR (Fig. 3a). We show that HDAC2 had the highest mRNA expression, with a ∼9-fold increase, while HDAC11 mRNA expression was barely detectable (Fig. 3a). Expression levels of all other HDACs were slightly lower than that of CCDN1, except for HDAC8, which was similar to CCDN1 (Fig. 3a). To detect HDAC protein expression, we performed Western blotting on cell lysates from U87MG and PDAs. We used an HDAC antibody sample kit which has antibodies specific for HDAC1 to -4, -6, and -7. We detected HDAC1, -2, and -4 in both PDAs and U87MG, while HDAC3, -6, and -7 were undetectable (Fig. 3b and c). These data indicate that HDAC2 is expressed at the highest levels compared to HDAC1 and HDAC4 in both cell types (Fig. 3b and c).
FIG 3.

Transcript and protein expression profiles of HDACs in astrocytes. (a) RNA extracts (1 μg) from PDAs were subjected to real-time PCR to detect and quantify all of the 11 HDACs and CCDN1. GAPDH was used as an internal control, and data are presented as fold change relative to results for CCDN1. (b and c) Protein extracts (20 μg each) from PDAs (b) or U87MG (c) were used to perform Western blot analysis using antibodies specific to HDAC1, -2, -3, and -4 from an HDAC antibody sampling kit (Cell Signaling) or GAPDH. *, P < 0.05 (Student's t test) between results for CCDN1 and HDAC2.
In order to determine which class of HDACs is involved in silencing HIV LTR activity in astrocytic cell lines, we treated U87MG HIV LTR-Luc-stably expressing cells with specific classes of HDAC inhibitors. The list of these inhibitors and their target is shown in Table 1. Concentrations of inhibitors were selected based on their 50% inhibitory concentration (IC50) values reported for other cell types. Based on the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay, >90% of cells were viable at all drug concentrations tested. Suberoylanilide hydroxamic acid (SAHA) (a potent HDAC inhibitor), apicidin (class 1 and class II HDAC inhibitor), and trichostatin A (TSA) (an HDAC inhibitor) all potently induced HIV promoter activity (Fig. 4a to c). TSA-mediated induction of HIV LTR promoter activity was lower (5- to 14-fold) than that of SAHA and apicidin, which induced the HIV LTR by 100- to 150-fold. This is likely because TSA is a reversible inhibitor of HDACs, while SAHA and apicidin are potent nonreversible inhibitors. VAHA, a more potent inhibitor of class II and much less active with class I HDACs, exhibited no effect on LTR activity (Fig. 4d). Similarly, MC1568, another selective class II HDAC inhibitor, had no effect on LTR activity (Fig. 4e). These results suggested that class II HDACs are not involved in silencing of the HIV LTR in U87MG astrocytic cells. Salermide and splitomicin, the two well-known class III HDAC inhibitors, also had moderate to no effect on the HIV LTR (Fig. 4f and g), indicating that sirtuins do not play a role in epigenetic silencing of the LTR. Furthermore, an HDAC8 (class I)-specific inhibitor, 1-naphthohydroxamic acid (40), had no effect on stable LTR activity in U87MG cells at various concentrations tested (Fig. 4h). We could not test for class IV HDAC (HDAC11) because there are no specific inhibitors available. However, our expression analysis indicated that HDAC11 is expressed at very low levels. Similar results were observed using another astrocytoma cell line (U251MG) with a stable HIV LTR Luc construct (data not shown). Lastly, the induction of HIV promoter activity through inhibition of HDACs was minimally observed in HIV LTR-Luc-transiently transfected cells (2- to 3-fold induction; data not shown), underscoring the relevance of chromatin modification of integrated DNA rather than episomally expressed DNA constructs. Taken together, we conclude that HDACs play a significant role in epigenetic silencing of the HIV LTR in astrocytic cell lines, that class II, class III, and HDAC8 of class I do not contribute to HIV LTR silencing in astrocytes, and therefore that class I HDACs are specifically involved in transcriptional silencing of the integrated HIV-1 LTR in astrocytic cell lines.
TABLE 1.
Specificities of inhibitors of chromatin-modifying proteins
| Compound | Property |
|---|---|
| Trichostatin A | Reversible potent HDAC inhibitor |
| Apicidin | Potent irreversible class I and class II HDAC inhibitor |
| SAHA | Potent irreversible HDAC inhibitor (gold standard) |
| VAHA | HDAC inhibitor; more active on class II and less active on class I |
| MC1568 | Selective class II HDAC inhibitor |
| 1-Naphthohydroxamic acid | HDAC8 (class I HDAC) inhibitor |
| 5-AZA | DNA methyltransferase (DNMT) inhibitor |
| UNC0638 | Histone methyltransferase (HMT) inhibitor |
| Salermide | Class III HDCA inhibitor; Sirt1 (lower conc) and sirt2 (higher conc) |
| Splitomicin | Class III HDCA inhibitor |
| Garcinol | Potent HAT inhibitor (inhibits p300 and PCAF) |
| Bix 01294 | Selective inhibitor of G9a (HMT) |
| Chaetocin | Selective inhibitor of SU(VAR)3-9, an H3K9 trimethyltransferase (HMT) |
FIG 4.
HDAC class 1 inhibitors significantly induce HIV LTR activity in astrocytes: U87MG cells with stably integrated HIV LTR-Luc were treated with various concentrations of HDAC inhibitors having selective specificity as indicated in Table 1. The agents were SAHA (a), apicidin (b),TSA (c), VAHA (d), MC1568 (e), salermide (f), splitomicin (g), or 1-naphthohydroxamic acid (h). Approximately 24 h posttreatment, cells were lysed and measured for luciferase activity. The data are normalized to total protein content and presented as fold change with respect to results for DMSO-treated cells ± SD. *, P < 0.05 (Student's t test) between results for DMSO-treated and drug-treated samples.
Chaetocin, a potent HMT inhibitor, significantly induces HIV LTR activity in astrocytes.
Along with histone deacetylation, DNA methylation and histone methylation play significant roles in epigenetic silencing of the HIV-1 LTR (23–25, 35, 36). We evaluated the role of DNA methylation and histone methylation in HIV LTR activity in astrocytic cell lines. 5-Aza-2′-deoxycytidine (5-Aza) (a DNA methyltransferases inhibitor), garcinol (an inhibitor of P300 and pCAF), BIX01294 (an inhibitor of G9a, a dimethyltransferase that methylates histone H3 at lysine 9 [H3K9]), and UNC0638 (an inhibitor of both G9a [EHMT2] and GLP [EHMT1], an H3K9 monomethyltransferase), had no effect on HIV LTR activity in these cells (Fig. 5a to d). Chaetocin, a potent HMT inhibitor (41), dramatically induced HIV promoter activity (Fig. 5e). Chaetocin at lower concentrations (IC50 = 0.8 μM) specifically inhibits SU(VAR)3-9, an H3K9 trimethyltransferase (41). Collectively, these data indicate that in addition to HDACs, epigenetic silencing of the HIV-1 LTR in astrocytic cell lines is also modulated by HMTs, specifically by SU(VAR)3-9. It is possible that this protein may be recruited to the HIV-1 LTR in astrocytes, causing methylation on lysine 9 of histone H3, consequently compacting the LTR and diminishing its transcriptional ability.
FIG 5.
Induction of stable LTR activity is specific to trimethylation of histone H3 at Lys9 (H3K9me3). LTR-Luc-stably integrated U87MG cells were treated with AZA, a DNMT inhibitor (a), garcinol, a potent HAT inhibitor (b), UNC0638, GLP (H3K9 monomethyltransferase) and G9a (H3K9 dimethyltransferase) inhibitor (c), BIX0124, a specific G9a inhibitor (d), or chaetocin, a specific inhibitor of SU(VAR)3–9, an H3K9 trimethyltransferase (e), at the concentrations indicated. Approximately 24 h posttreatment, the cells were lysed and assayed for luciferase activity, normalized to total protein content. Data are presented as fold change with respect to results for DMSO-treated cells ± SD. *, P < 0.05 (Student's t test) between DMSO-treated and drug-treated samples.
Potent HDAC inhibitors (SAHA and apicidin) and a potent HMT inhibitor (chaetocin) induce HIV transcription in astrocytes.
To assess the inhibition capacity of class I HDACs and HMTs for HIV-1 proviral DNA, we generated long-term-infected PDAs and stably infected U138MG cells. Alu-PCR indicated ≈3% of viral DNA integration in these cells (Fig. 1). We treated these cells with SAHA, apicidin, or chaetocin for 24 h and quantified HIV-1 transcripts (Gag, Env, and Rev) by real-time PCR. SAHA, apicidin, and chaetocin potently induced HIV transcripts in comparison to results for the control (DMSO treated), albeit the magnitude of induction was noticeably lower in apicidin-treated cells than with SAHA and chaetocin treatment (Fig. 6a to c). Further, when supernatant from these infected and treated (with SAHA or chaetocin) or control (DMSO-treated) PDAs was incubated with activated PBMCs, we observed 12- to 20-fold induction for the HIV env transcript in PBMCs, indicating that HIV within PDAs is reactivated and is infectious (Fig. 6d). These data demonstrate that class 1 HDACs and HMT silence the HIV provirus in astrocytes, confirming our data using HIV LTR reporter cells.
FIG 6.
SAHA, apicidin, and chaetocin strongly induce HIV transcripts and infectious virus from PDAs and U138MG astrocytes. PDAs (a) or U138MG cells (b) were infected with HIVBal and treated with SAHA (10 μM), apicidin (1 μg/ml), chaetocin (200 nm), or vehicle control. RNA was isolated 24 h posttreatment, 1 μg reverse transcribed to cDNA, and subjected to real-time PCR to quantify HIV Gag, Env, and Rev (c) transcripts. Data are normalized to results for GAPDH and are represented as fold change with respect to results for control (DMSO)-treated cells ± SD. (d) Supernatants from SAHA-, chaetocin-, or DMSO-treated HIVBal-infected PDAs were added to anti-CD3/anti-CD28-activated PBMCs. At day 6, HIV transcript (Env) from PBMCs was quantified by real-time PCR. Data are normalized to results for GAPDH and are represented as fold change with respect to results for control-treated samples. *, P < 0.05 (Student's t test) between results for control (DMSO)-treated and inhibitor-treated samples.
Our studies and those of others build on growing evidence to indicate that astrocytes are reservoirs for HIV. They fulfill classical criteria defining “HIV reservoirs,” which include the following. (i) They have the ability to harbor provirus: considerable evidence from in vitro and in vivo studies indicates that astrocytes harbor integrated HIV DNA (provirus) (9, 10, 42). In fact, 1 to 3% of astrocytes from HIV-infected brains harbor HIV provirus (10). The size of this pool is impressive considering that the size of latent HIV in periphery is estimated to be 1 replication-competent virus/million cells in blood and lymph nodes of HIV+ patients (8, 43–46) and astrocytes are the most abundant cells in the brain. Astrocytes in essence could constitute a significant source of latent HIV. (ii) The provirus can be reactivated and is infectious: HIV from infected astrocytes is induced/reactivated by inflammatory signals (11–13). (iii) Low-level HIV transcripts without protein expression are detected: HIV-infected astrocytes express very low levels of early and late transcripts, and no detectable protein can be measured. (iv) The HIV LTR in astrocytes is subjected to classical pathways for gene silencing, including HDACs and HMTs. We provide evidence here to show that inhibition of class 1 HDACs and a specific HMT potently induce HIV promoter activity, indicating that HIV promoter activity in astrocytes is silenced by a mechanism similar to that reported to induce LTR repression in T cells (28–34). These collective criteria indicate that astrocytes are reservoirs for HIV.
Both HDACs and HMTs are linked to promoting HIV latency in a number of model systems, including resting CD4+ T cells, T cell lines, HeLa cells, and microglia. In astrocytes, these two pathways appear to be highly relevant to inducing a state of HIV latency in astrocytes. However, other regulators reported to be relevant in T cells do not seem to play a significant role in astrocytes. For example, BIX01294 induced HIV transcription in latently infected Jurkat (ACH-2) and promyelocyte (OM10.1) cell lines (35). In astrocytes, this specific inhibitor of a monomethyltransferase did not augment LTR activity, but specific inhibition of SU(VAR)3-9, a well-known H3K9 trimethyltransferase, by chaetocin caused a dramatic increase in LTR activity as well as proviral transcription. It is possible that SU(VAR)3-9 and G9a have distinct roles in chromatin modification by having distinct molecular partners (47). Also, DNA methylation does not seem to drive HIV latency in astrocytes, while it was reported to play a significant role in lymphocytes (23). In astrocytes, trimethylation of H3K9 in addition to antideacetylation is required to transform the LTR to a transcriptionally active euchromatin status and drive HIV out of latency.
Understanding mechanisms of HIV latency in the CNS by focusing on key cellular reservoirs, such as astrocytes and microglia/macrophages, is critical to design strategies to eliminate HIV from the CNS. Current strategies of reactivation of virus, using HDAC inhibitors and other approaches, do not take into consideration the presence of HIV in the CNS and most importantly the concern that reactivation of virus in the brain is likely to lead to episodes of encephalitis, which can set the stage for damage in the brain, and thus, while it may be transient and controlled by steroids, may have consequences that will not be fully appreciated until years later.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R01 MH100628 from the National Institute of Mental Health and grant R01 NS060632 from the National Institute of Neurologic Disease and Stroke. The studies were also supported by the Chicago Developmental Center for AIDS Research (D-CFAR) (P30AI082151), supported by NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
Footnotes
Published ahead of print 18 December 2013
REFERENCES
- 1.Le Douce V, Janossy A, Hallay H, Ali S, Riclet R, Rohr O, Schwartz C. 2012. Achieving a cure for HIV infection: do we have reasons to be optimistic? J. Antimicrob. Chemother. 67:1063–1074. 10.1093/jac/dkr599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O'Shea A, Callender M, Spivak A, Brennan T, Kearney MF, Proschan MA, Mican JM, Rehm CA, Coffin JM, Mellors JW, Siliciano RF, Maldarelli F. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 106:9403–9408. 10.1073/pnas.0903107106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnell G, Price RW, Swanstrom R, Spudich S. 2010. Compartmentalization and clonal amplification of HIV-1 variants in the cerebrospinal fluid during primary infection. J. Virol. 84:2395–2407. 10.1128/JVI.01863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 7:e1002286. 10.1371/journal.ppat.1002286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnell G, Spudich S, Harrington P, Price RW, Swanstrom R. 2009. Compartmentalized human immunodeficiency virus type 1 originates from long-lived cells in some subjects with HIV-1-associated dementia. PLoS Pathog. 5:e1000395. 10.1371/journal.ppat.1000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holman AG, Mefford ME, O'Connor N, Gabuzda D. 2010. HIVBrainSeqDB: a database of annotated HIV envelope sequences from brain and other anatomical sites. AIDS Res. Ther. 7:43. 10.1186/1742-6405-7-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO. 2009. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J. Virol. 83:8470–8481. 10.1128/JVI.02568-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun TW, Davey RT, Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, Fauci AS. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757–761. 10.1038/77481 [DOI] [PubMed] [Google Scholar]
- 9.Trillo-Pazos G, Diamanturos A, Rislove L, Menza T, Chao W, Belem P, Sadiq S, Morgello S, Sharer L, Volsky DJ. 2003. Detection of HIV-1 DNA in microglia/macrophages, astrocytes and neurons isolated from brain tissue with HIV-1 encephalitis by laser capture microdissection. Brain Pathol. 13:144–154. 10.1111/j.1750-3639.2003.tb00014.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churchill MJ, Wesselingh SL, Cowley D, Pardo CA, McArthur JC, Brew BJ, Gorry PR. 2009. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann. Neurol. 66:253–258. 10.1002/ana.21697 [DOI] [PubMed] [Google Scholar]
- 11.Atwood WJ, Tornatore CS, Traub R, Conant K, Drew PD, Major EO. 1994. Stimulation of HIV type 1 gene expression and induction of NF-kappa B (p50/p65)-binding activity in tumor necrosis factor alpha-treated human fetal glial cells. AIDS Res. Hum. Retroviruses 10:1207–1211. 10.1089/aid.1994.10.1207 [DOI] [PubMed] [Google Scholar]
- 12.Tornatore C, Nath A, Amemiya K, Major EO. 1991. Persistent human immunodeficiency virus type 1 infection in human fetal glial cells reactivated by T-cell factor(s) or by the cytokines tumor necrosis factor alpha and interleukin-1 beta. J. Virol. 65:6094–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll-Anzinger D, Al-Harthi L. 2006. Gamma interferon primes productive human immunodeficiency virus infection in astrocytes. J. Virol. 80:541–544. 10.1128/JVI.80.1.541-544.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll-Anzinger D, Kumar A, Adarichev V, Kashanchi F, Al-Harthi L. 2007. Human immunodeficiency virus-restricted replication in astrocytes and the ability of gamma interferon to modulate this restriction are regulated by a downstream effector of the Wnt signaling pathway. J. Virol. 81:5864–5871. 10.1128/JVI.02234-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Henderson LJ, Major EO, Al-Harthi L. 2011. IFN-gamma mediates enhancement of HIV replication in astrocytes by inducing an antagonist of the beta-catenin pathway (DKK1) in a STAT 3-dependent manner. J. Immunol. 186:6771–6778. 10.4049/jimmunol.1100099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narasipura SD, Henderson LJ, Fu SW, Chen L, Kashanchi F, Al-Harthi L. 2012. Role of beta-catenin and TCF/LEF family members in transcriptional activity of HIV in astrocytes. J. Virol. 86:1911–1921. 10.1128/JVI.06266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson LJ, Al-Harthi L. 2011. Role of beta-catenin/TCF-4 signaling in HIV replication and pathogenesis: insights to informing novel anti-HIV molecular therapeutics. J. Neuroimmune Pharmacol. 6:247–259. 10.1007/s11481-011-9266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson LJ, Sharma A, Monaco MC, Major EO, Al-Harthi L. 2012. Human immunodeficiency virus type 1 (HIV-1) transactivator of transcription through its intact core and cysteine-rich domains inhibits Wnt/beta-catenin signaling in astrocytes: relevance to HIV neuropathogenesis. J. Neurosci. 32:16306–16313. 10.1523/JNEUROSCI.3145-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. 2009. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS 23:1799–1806. 10.1097/QAD.0b013e32832ec1dc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18:1101–1108. 10.1097/00002030-200405210-00003 [DOI] [PubMed] [Google Scholar]
- 21.Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. 2009. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 284:6782–6789. 10.1074/jbc.M807898200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. 2009. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res. Hum. Retroviruses 25:207–212. 10.1089/aid.2008.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. 10.1371/journal.ppat.1000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. 10.1371/journal.ppat.1000495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, Bertrand E, Marcello A, Emiliani S, Kiernan R, Benkirane M. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26:424–435. 10.1038/sj.emboj.7601517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolis DM. 2011. Histone deacetylase inhibitors and HIV latency. Curr. Opin. HIV AIDS 6:25–29. 10.1097/COH.0b013e328341242d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Lint C, Emiliani S, Ott M, Verdin E. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112–1120 [PMC free article] [PubMed] [Google Scholar]
- 28.Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790–6799. 10.1128/JVI.74.15.6790-6799.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai K, Okamoto T. 2006. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J. Biol. Chem. 281:12495–12505. 10.1074/jbc.M511773200 [DOI] [PubMed] [Google Scholar]
- 30.Jiang G, Espeseth A, Hazuda DJ, Margolis DM. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81:10914–10923. 10.1128/JVI.01208-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malcolm T, Chen J, Chang C, Sadowski I. 2007. Induction of chromosomally integrated HIV-1 LTR requires RBF-2 (USF/TFII-I) and Ras/MAPK signaling. Virus Genes 35:215–223. 10.1007/s11262-007-0109-9 [DOI] [PubMed] [Google Scholar]
- 32.Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. 2007. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26:412–423. 10.1038/sj.emboj.7601516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyagi M, Karn J. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995. 10.1038/sj.emboj.7601928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25:139–149. 10.1038/sj.emboj.7600900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai K, Togami H, Okamoto T. 2010. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 285:16538–16545. 10.1074/jbc.M110.103531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavez L, Kauder S, Verdin E. 2011. In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods 53:47–53. 10.1016/j.ymeth.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregoretti IV, Lee YM, Goodson HV. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338:17–31. 10.1016/j.jmb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 38.Trapp J, Jung M. 2006. The role of NAD+ dependent histone deacetylases (sirtuins) in ageing. Curr. Drug Targets 7:1553–1560. 10.2174/1389450110607011553 [DOI] [PubMed] [Google Scholar]
- 39.Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. 2007. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 17:195–211. 10.1038/sj.cr.7310149 [DOI] [PubMed] [Google Scholar]
- 40.Krennhrubec K, Marshall BL, Hedglin M, Verdin E, Ulrich SM. 2007. Design and evaluation of ‘Linkerless' hydroxamic acids as selective HDAC8 inhibitors. Bioorg. Med. Chem. Lett. 17:2874–2878. 10.1016/j.bmcl.2007.02.064 [DOI] [PubMed] [Google Scholar]
- 41.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. 2005. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat. Chem. Biol. 1:143–145. 10.1038/nchembio721 [DOI] [PubMed] [Google Scholar]
- 42.Dewhurst S, Sakai K, Bresser J, Stevenson M, Evinger-Hodges MJ, Volsky DJ. 1987. Persistent productive infection of human glial cells by human immunodeficiency virus (HIV) and by infectious molecular clones of HIV. J. Virol. 61:3774–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chun TW, Justement JS, Pandya P, Hallahan CW, McLaughlin M, Liu S, Ehler LA, Kovacs C, Fauci AS. 2002. Relationship between the size of the human immunodeficiency virus type 1 (HIV-1) reservoir in peripheral blood CD4+ T cells and CD4+:CD8+ T cell ratios in aviremic HIV-1-infected individuals receiving long-term highly active antiretroviral therapy. J. Infect. Dis. 185:1672–1676. 10.1086/340521 [DOI] [PubMed] [Google Scholar]
- 44.Chun TW, Justement JS, Lempicki RA, Yang J, Dennis G, Jr, Hallahan CW, Sanford C, Pandya P, Liu S, McLaughlin M, Ehler LA, Moir S, Fauci AS. 2003. Gene expression and viral production in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc. Natl. Acad. Sci. U. S. A. 100:1908–1913. 10.1073/pnas.0437640100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chun TW, Justement JS, Moir S, Hallahan CW, Maenza J, Mullins JI, Collier AC, Corey L, Fauci AS. 2007. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195:1762–1764. 10.1086/518250 [DOI] [PubMed] [Google Scholar]
- 46.Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. 2011. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 204:135–138. 10.1093/infdis/jir208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee DY, Teyssier C, Strahl BD, Stallcup MR. 2005. Role of protein methylation in regulation of transcription. Endocr. Rev. 26:147–170 http://press.endocrine.org/doi/full/10.1210/er.2004-0008 [DOI] [PubMed] [Google Scholar]