Abstract
Viruses, in particular DNA viruses, generate microRNAs (miRNAs) to control the expression of host and viral genes. Due to their essential roles in virus-host interactions, viral miRNAs have attracted extensive investigations in recent years. To date, however, most studies on viral miRNAs have been conducted in cell lines. In this study, the viral miRNAs from white spot syndrome virus (WSSV) were characterized in shrimp in vivo. On the basis of our previous study and small RNA sequencing in this study, a total of 89 putative WSSV miRNAs were identified. As revealed by miRNA microarray analysis and Northern blotting, the expression of viral miRNAs was tissue specific in vivo. The results indicated that the viral miRNA WSSV-miR-N24 could target the shrimp caspase 8 gene, and this miRNA further repressed the apoptosis of shrimp hemocytes in vivo. As a result, the number of WSSV copies in shrimp in vivo was significantly increased compared with the control level (WSSV only). Therefore, our study presents the first report on the in vivo molecular events of viral miRNA in antiviral apoptosis.
INTRODUCTION
MicroRNAs (miRNAs) represent a class of small RNA regulators that are encoded by a diverse range of eukaryotic organisms (1, 2) and have important roles in many biological processes, including development, cell differentiation, apoptosis, oncogenesis, and immune defense (3–5). In recent years, since the first discovery of Epstein-Barr virus (EBV)-encoded miRNAs, a growing number of viral miRNAs have been demonstrated (6). To date, a total of 406 miRNAs have been discovered in various viruses (miRNA Registry, release 18.0; November 2011), such as EBV, bovine herpesvirus 1, herpesvirus of turkeys, human cytomegalovirus, and simian virus 40 (SV40). As reported, the majority of identified viral miRNAs are limited to mammalian DNA viruses, mainly from the herpesvirus family, suggesting that Drosha and DGCR8 proteins, which localize to the nucleus, are required for the initial pre-miRNA excision event. It seems that RNA viruses, which replicate exclusively in the cytoplasm, would not have ready access to this machinery. However, several recent reports showed that two RNA viruses, HIV-1 and West Nile virus (WNV), could also encode viral miRNAs (7, 8). Nevertheless, the relevance of these reports remains controversial. To date, very little information is available about the miRNAs of invertebrate viruses.
Many studies have revealed that viral miRNAs can target viral mRNAs and trigger the degradation of viral mRNAs to fine-tune patterns of viral gene expression. For example, two viral miRNAs (miR-S1-5p and miR-S1-3p) from the same miRNA precursor in the genome of the polyomavirus SV40 have been shown to be antisense relative to the viral T-antigen transcripts and to direct the cleavage of these early transcripts in virus infection (9). The downregulation of T antigen by these viral miRNAs is significant for virus replication, reducing cellular visibility of SV40-specific cytotoxic T lymphocytes (CTLs) (9). It has been reported that the viral miRNA miR-UL112-1, from human cytomegalovirus, plays an important role in establishing the latent viral infection state via inhibiting the expression of the viral gene IE1 (10, 11). Taken together, the data indicate that viruses have evolved to take advantage of the miRNA pathway to modulate expression of their own genes for successful infection.
In addition to “autoregulation” of viral targets, several virus-encoded miRNAs have been shown to target cellular or host genes. However, their functions are poorly understood. The well-characterized cellular gene targeted by Kaposi's sarcoma-associated herpesvirus (KSHV) miRNAs is the thrombospondin 1 gene (THBS1), which has functions in inhibiting angiogenesis and cell growth by activating transforming growth factor beta (TGF-β) (12). The inhibition of THBS1 expression by KSHV miRNAs promotes KSHV-infected cell survival and proliferation (12). The human cytomegalovirus miRNA miR-UL112-1 can target viral genes (10, 11) and host genes (13). Through binding with the 3′-untranslated region (3′ UTR) of the mRNA of major histocompatibility complex class 1-related chain B (MICB), viral miR-UL112-1 downregulates the expression of MICB and further reduces the susceptibility of virus-infected cells to killing by natural killer cells (13). EBV can encode miR-BHRF1-3 to inhibit the expression of CXC-chemokine ligand 11 (CXCL11), which is an interferon (IFN)-inducible T-cell chemoattractant and plays a major role in host defenses against EBV (14). Suppressing CXCL11 may aid infected cells in avoiding T-cell recognition (14). In recent years, viral miRNAs have attracted intensive study. So far, however, many viral miRNAs have been characterized in cell lines. As is well known, the roles of viral miRNAs in cell lines may be very different from those in hosts in vivo.
In this study, the viral miRNAs of white spot syndrome virus (WSSV) were characterized in shrimp in vivo. On the basis of our previous study (15) and high-throughput sequencing of small RNAs in this study, a total of 89 putative WSSV miRNAs were identified. It was found that the expression profiles of WSSV miRNAs were tissue specific. The results revealed that WSSV-miR-N24 was a key suppressor of host antiviral apoptosis, through downregulating the expression of shrimp caspase 8. Our findings highlight the key roles of viral miRNAs in the interactions between virus and host in vivo.
MATERIALS AND METHODS
Shrimp culture and WSSV infection.
Marsupenaeus japonicus shrimp of approximately 15 g each were reared in groups of 20 individuals in tanks filled with air-pumped circulating seawater at 25°C. The shrimp were kept temporarily for 2 to 3 days and acclimatized prior to the experiments. To ensure that the shrimp were WSSV-free before the experiments, three shrimp were randomly selected for WSSV detection by use of WSSV-specific primers (5′-TTGGTTTCATGCCCGAGATT-3′ and 5′-CCTTGGTCAGCCCCTTGA-3′). The virus-free shrimp were challenged with WSSV as described previously (16). Briefly, the tissues of WSSV-infected shrimp were homogenized in TN buffer (20 mM Tris-HCl, 400 mM NaCl, pH 7.4) at 0.1 g/ml. After centrifugation at 2,000 × g for 15 min, the supernatant was diluted with 0.85% NaCl to 1:100 and filtered using a 0.45-μm filter. Subsequently, the virus-free shrimp were infected with 0.1 ml of filtrate (105 virus copies/ml) by intramuscular injection. At various times postinfection (0, 6, 24, 48, and 72 h), the lymphoid organs, gills, and hemocytes of shrimp were collected for later use. Shrimp assays were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Zhejiang Province. The protocol was approved by the Committee on the Ethics of Animal Experiments of the University of Zhejiang Univesity, China.
Sequencing and sequence analysis of small RNAs.
Total RNAs were isolated from the lymphoid organs of WSSV-free and WSSV-infected shrimp at various time points (0, 6, 24, and 48 h) by use of a mirVana miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. The quality and integrity of total RNAs were evaluated by electrophoresis on 1.2% agarose gels. About 200 μg of total RNA was separated in a 15% polyacrylamide-8 M urea gel, and small RNAs of 16 to 30 nucleotides (nt) were recovered. Single libraries were sequenced on a GA-I genome analyzer (Illumina, San Diego, CA) according to the manufacturer's protocols. The small RNA sequencing reads were subjected to a search for putative WSSV-encoded miRNA sequences by using the ACGT V3.1 program, developed by LC Sciences (Houston, TX). After the removal of adaptor sequences and noncoding RNA sequences matching mRNA, rRNA, tRNA, snRNA, snoRNA, repeat, and other noncoding RNA sequences available in Rfam (http://www.sanger.ac.uk/software/Rfam), all the remaining high-quality sequences were searched against the WSSV genome (GenBank accession no. AF332093.1). The small RNA sequences (total counts of ≥5) of putative WSSV miRNAs were analyzed by a BLASTN search against the WSSV genome, allowing one or two mismatches between each pair of sequences. To analyze the potential precursor structures of 49 WSSV miRNA candidates, each sequence, including a fragment of 60 to 70 bases flanking the sequence, was subjected to miRNA secondary structure prediction using mFold online software (http://frontend.bioinfo.rpi.edu/applications/mfold/) with default parameters.
Northern blotting.
Total RNAs were isolated using a mirVanaP miRNA isolation kit according to the manufacturer's instructions (Ambion). Subsequently, the RNAs were treated with RNase-free DNase I (TaKaRa, Japan) at 37°C for 30 min. The concentration of the extracted RNAs was quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies). The total RNAs were electrophoresed in a denaturing 15% polyacrylamide gel containing 8 M urea and transferred to a Hybond-N+ membrane (Amersham Biosciences, Buckinghamshire, United Kingdom). After cross-linking with UV, the membrane was prehybridized in DIG Easy Hyb granule buffer (Roche, Basel, Switzerland) for 0.5 h and then hybridized with a digoxigenin (DIG)-labeled DNA probe complementary to a specific miRNA sequence for 20 h at 45°C. Signal detection was performed following the instructions in the manual for a DIG High Prime DNA labeling and detection starter kit II (Roche). RNA oligonucleotides of 20 nt, 30 nt, 100 nt, and 200 nt were synthesized and labeled with DIG at the 5′ end to serve as markers of small RNA.
Detection of primary miRNAs.
Total RNAs were extracted from various shrimp tissues or organs by using a mirVanaP RNA isolation kit according to the manufacturer's instructions (Ambion). The RNA extracts were treated with RNase-free DNase I (TaKaRa, Japan) at 37°C for 30 min. The first-strand cDNA synthesis was conducted using 1 μg of total RNA following the manufacturer's guidelines for a PrimeScript 1st strand cDNA synthesis kit (TaKaRa). The primary miRNAs were amplified by PCR with sequence-specific primers (5′-GTTCTGCTTCTTCTTCTCATTG-3′ and 5′-AGTCTTCTTGACATTTTGAGTA-3′ for pri-WSSV-miR-N24, 5′-TGGCCCGGCAAATTGGCCTAGA-3′ and 5′-AGCAAACCTTAAAGACTCAAAC-3′ for pri-WSSV-miR-N30, 5′-TATCTTCTTCATTTTCAAATAC-3′ and 5′-CACTGACGACCATACGAGATGG-3′ for pri-WSSV-miR-82, 5′-GACAATCTTGAAGTTGTTTTCT-3′ and 5′-ATTGTCTTGAAGCTCATAAG-3′ for pri-WSSV-miR-129, and 5′-GTTGTTGTTGTTTGAGGTAGTTG-3′ and 5′-AAACGAAAGAGATCTTCTGTTAAAAG-3′ for pri-WSSV-miR-162). The PCR protocol used was 94°C for 5 min followed by 40 cycles of 94°C for 40 s, 46°C for 45 s, and 72°C for 45 s, with a final elongation at 72°C for 10 min.
Prediction of genes targeted by miRNAs.
To predict the genes targeted by WSSV miRNAs, TargetScan 5.1 and miRanda algorithms were used to predict the target sites of miRNAs in the 3′ UTRs of shrimp, as described before (17).
miRNA microarray.
A WSSV miRNA microarray was prepared by LC Sciences (Houston, TX), using viral miRNAs revealed in our previous study (15) and this study. Total RNAs were extracted from shrimp by use of a mirVanaP miRNA isolation kit (Ambion) following the manufacturer's protocol and were subsequently labeled with Cy3. A group of control probes was included on each chip to evaluate microarray quality. Hybridization signals were detected using an Axon GenePix 4000B microarray scanner (Molecular Devices). Data were processed by subtracting the background and normalizing the signals by using a LOWESS filter. The ratios of the control and sample detected signals (log transformed) were generally defined as true when the P value of the t test was <0.01. The differentially expressed miRNAs were selected for cluster analysis.
Plasmid construction.
To examine whether the viral miRNA WSSV-miR-N24 could target the caspase 8 gene of M. japonicus shrimp, the 3′ UTRs of the shrimp caspase 8 gene and the enhanced green fluorescent protein (EGFP) gene were cloned into the pIZ/V5-His vector (Invitrogen). The EGFP gene was amplified and cloned into the pIZ vector to generate a control EGFP construct as described previously (18). The caspase 8 3′ UTR was cloned into the pIZ/V5-His vector downstream of EGFP by using primers 5′-ACTCTAGACACAATCAGTGTGTTTGAATCTTTAC-3′ and 5′-AACCCGCGGGAAAAGCTTTCTTTATTTAAGTGTATAATC-3′ to generate the EGFP-caspase 8 construct. As a control, the sequence of the caspase 8 3′ UTR complementary to the WSSV-miR-N24 seed sequence was mutated to yield the EGFP-Δcaspase 8 construct by using a QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). All recombinant plasmids were confirmed by sequencing.
Cell culture, transfection, and fluorescence assays.
Insect High Five cells (Invitrogen) were cultured at 27°C in Express Five SFM medium (Invitrogen) supplemented with l-glutamine (Invitrogen) as described previously (18). Briefly, 2 μg of EGFP, EGFP-caspase 8, or EGFP-Δcaspase 8 construct was transfected into High Five cells when the cells were about 70% confluent. At the same time, the cells were transfected with 100 pmol of either a synthesized WSSV-miR-N24 mimic (antisense strand, 5′-GUAUGAGUAGUGAUGAAGAAUCA-3′; and sense strand, 5′-UGAUUCUUCAUCAAUACUCAUAC-3′) or a synthesized control miRNA mimic (antisense strand, 5′-UGAGAUAGUAGAUAGAGAUCAUG-3′; and sense strand, 5′-CAUGAUCUCUAUCUACUAUCUCA-3′). All the miRNA mimics were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
To inhibit the activity of WSSV-miR-N24, the plasmid (EGFP, EGFP-caspase 8, or EGFP-Δcaspase 8)-transfected cells were simultaneously transfected with the synthesized WSSV-miR-N24 mimic (100 pmol) and a locked nucleic acid (LNA)-modified anti-WSSV-miR-N24 oligonucleotide (AMO-WSSV-miR-N24) (100 pmol) or with WSSV-miR-N24 (100 pmol) and an LNA-modified anti-control miRNA oligonucleotide (AMO-control) (100 pmol). The AMO constructs were obtained from Exiqon A/S (Vedbaek, Denmark). For AMO-WSSV-miR-N24 (5′-TGATTCTTCATCACTACTCATAC-3′), the sequence was modified at the 3rd, 6th, and 10th nucleotides with LNA, at the 13th and 16th nucleotides with 2′-O-methoxyethyl (MOE), and at the remaining nucleotides with phosphorothioate. For AMO-control (5′-AGTGAATGATGAGTATGTTGTTG-3′), the sequence was also modified as described above. All transfections were conducted in triplicate with Cellfectin transfection reagent (Invitrogen) according to the manufacturer's protocol. Twelve hours after transfection, the transfected cells were seeded in 96-well plates at a concentration of 2.0 × 104 cells per well. One day later, the EGFP fluorescence of the cells was recorded with a Flex Station II microplate reader (Molecular Devices) at 490/510 nm (excitation/emission). The fluorescence values were corrected by subtracting the autofluorescence of cells not expressing EGFP. The experiments were biologically repeated three times.
Quantitative real-time PCR.
To analyze the expression of the shrimp caspase 8 gene, quantitative real-time PCR was conducted as described previously (16). Shrimp β-actin was used as a control. The forward and reverse primers for amplification of the caspase 8 gene were 5′-TTCCGGAAAACCCTCAAAC-3′ and 5′-GGTTTGTGTGCTTTCGGAA-3′, respectively. The sequence of the TaqMan probe was 5′-6-carboxyfluorescein (FAM)-CGGCGTGTGTTTGAATCTTTACTTCGCCG-6-carboxytetramethylrhodamine (TAMRA)-3′. The primers for β-actin were 5′-CGAGCACGGCATCGTTACTA-3′ and 5′-TTGTAGAAAGTGTGATGCCAGATCT-3′, and the sequence of the TaqMan probe was 5′-FAM-CTGGGACGACATGGA-TAMRA-3′. The reactions were performed in a total volume of 25 μl consisting of 12.5 μl of Ex Taq premix (TaKaRa, Japan), 100 ng of cDNA template, 0.5 μl of 10 μM (each) primers, and 0.5 μl of 10 μM TaqMan fluorogenic probe. PCR was carried out at 95°C for 1 min followed by 40 cycles of 95°C for 15 s, 52°C for 45 s, and 72°C for 45 s.
To quantify WSSV virions in shrimp, the WSSV genome was extracted from virus-infected shrimp hemocytes by use of an SQ Tissue DNA kit (Omega Bio-Tek) according to the manufacturer's protocols and subsequently subjected to real-time PCR. The primers (forward, 5′-CCACCAATTCTACTCATGTACCAAA-3′; and reverse, 5′-TCCTTGCAATGGGCAAAATC-3′) were used to amplify a region from positions 260075 to 260138 of the WSSV genome (GenBank accession number AF332093.1), as described previously (15). The sequence of the TaqMan probe was 5′-FAM-CTGGGTTACGAGTCTAA-TAMRA-3′. A linearized plasmid containing a 1,400-bp DNA fragment from the WSSV genome was quantified and serially diluted 10-fold as an internal standard for real-time PCR (16). The real-time PCR mixture (25 μl) contained 12.5 μl Ex Taq premix (TaKaRa, Japan), 1 μl of the extracted DNA template or the internal standard plasmid, 0.5 μl of 10 μM (each) primers, and 0.5 μl of 10 μM TaqMan fluorogenic probe. The real-time PCR conditions were 95°C for 1 min followed by 45 cycles of 30 s at 95°C, 30 s at 52°C, and 30 s at 72°C.
Western blotting.
Proteins were analyzed in a 12% SDS-PAGE gel and transferred onto a nitrocellulose membrane (Bio-Rad). The membrane was immersed in blocking buffer (3% bovine serum albumin [BSA] in phosphate-buffered saline [PBS], pH 7.2) at 4°C overnight, followed by incubation with a specific antibody (anti-caspase 8 IgG or anti-β-actin IgG). The antibody against the caspase 8 protein of M. japonicus shrimp was prepared in our previous study (19). The membrane was then incubated with alkaline phosphatase (AP)-conjugated goat anti-mouse IgG (Sigma) for 1 h and detected with Nitro Blue Tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) solutions (BBI, Canada).
Annexin V assay of apoptosis.
Apoptosis assay of shrimp hemocytes by use of annexin V (Invitrogen) was performed according to the manufacturer's protocol. Briefly, shrimp hemocytes were collected with an equal volume of heparin sodium (40 mg/ml). After being washed twice in cold PBS, the cells were resuspended in 1× annexin binding buffer at ∼1 × 106 cells/ml, followed by the addition of 5 μl of Alexa Fluor 488-annexin V and 1 μl of 100 μg/ml propidium iodide (PI). The cells were incubated for 15 min in the dark, and then 400 μl of 1× annexin binding buffer was added to the sample. The sample was analyzed by flow cytometry (BD Bioscience, San Jose, CA) at 530 nm and 575 nm, using a 488-nm excitation wavelength.
TUNEL assay.
Apoptosis was detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay according to the manufacturer's instructions (Promega). Briefly, shrimp hemocytes were collected and mounted onto a poly-l-lysine-coated glass slide (Sigma), followed by incubation for 10 min at 4°C. The hemocytes were fixed with 4% paraformaldehyde for 25 min at 4°C. Subsequently, the hemocytes were washed twice with cold PBS and permeabilized with 0.2% Triton X-100 for 5 min. After being washed with PBS twice, the hemocytes were equilibrated with 100 μl of equilibration buffer at room temperature for 10 min. The hemocytes were incubated with rTdT mix containing green fluorescein-12–dUTP for 60 min at 37°C in a humidified chamber and then counterstained with PI. The reactions were stopped by immersing the slide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min. The slide was washed three times and mounted with antifade solution (Invitrogen). The number of TUNEL-positive hemocytes was determined.
Detection of caspase 3/7 activity.
The caspase activity of shrimp hemocytes was detected using a Caspase-Glo 3/7 assay (Promega) according to the manufacturer's instructions. Briefly, 100 μl of Caspase-Glo 3/7 reagent was mixed with shrimp hemocytes and incubated at room temperature for 2 h. Subsequently, the mixture was subjected to measurement using a Synergy 2 Multi-Mode microplate reader (BioTek).
Synthesis of siRNAs.
Small interfering RNAs (siRNAs) for RNA interference (RNAi) assays of shrimp were synthesized using an in vitro T7 transcription kit for siRNA synthesis (TaKaRa, Japan) according to the manufacturer's instructions, as described previously (18). The sequence-specific caspase 8-siRNA (5′-CUGUAGCACUUCAUAACUU-3′) was synthesized to silence the expression of the shrimp caspase 8 gene. As a control, one nucleotide of the caspase 8-siRNA sequence was mutated at random, yielding the corresponding control siRNA (5′-CUGUACCACUUCAUAACUU-3′). The synthesized siRNAs were dissolved in siRNA solution (50 mM Tris-HCl, 100 mM NaCl, pH 7.5) and quantified by spectrophotometry.
RNAi assays of shrimp in vivo.
Shrimp were simultaneously injected with WSSV virions (104 genome copies/shrimp) and 30 μg of a synthesized siRNA (caspase 8-siRNA or control siRNA), with WSSV virions (104 genome copies/shrimp) and 30 μg of a miRNA mimic (WSSV-miR-N24 mimic or control miRNA mimic), or with WSSV virions (104 genome copies/shrimp) and 15 μg of an LNA-modified anti-miRNA oligonucleotide (AMO-WSSV-miR-N24 or AMO-control). At the same time, WSSV only (104 genome copies/shrimp), WSSV-miR-N24 mimic only, AMO-WSSV-miR-N24 only, and PBS only were included in injections as controls. For each treatment, 20 individuals were used. At different time points after injection (0, 24, 48, and 72 h), the hemocytes of four randomly selected shrimp from each treatment were collected and subjected to real-time PCR to quantify the number of WSSV genome copies. Forty-eight hours after infection with WSSV, the collected hemocytes were also subjected to real-time PCR or Western blotting to quantify caspase 8 gene expression, to Northern blot analysis to examine WSSV-miR-N24 expression, and to apoptosis assays with annexin V, TUNEL, and caspase 3/7 activity detection to evaluate the apoptotic activity of shrimp. The above-described assays were biologically repeated three times.
Statistical analysis.
The numerical data were analyzed by one-way analysis of variance to calculate the means and standard deviations for triplicate assays. Statistically significant differences between treatments were determined using Student's t test, with significance defined as having a P value of <0.05 or <0.01.
Microarray data accession number.
The microarray data are available at Gene Expression Omnibus (GEO) under accession number GSE53900.
RESULTS
Identification of viral miRNAs in WSSV-infected shrimp in vivo.
Our previous study showed that WSSV possessed the capacity to encode 40 distinct viral miRNAs, suggesting that WSSV miRNAs might play important roles in virus infection (15). To characterize the WSSV miRNAs in shrimp in vivo, the miRNAs of lymphoid organs from WSSV-infected shrimp collected at different times (0, 6, 24, and 48 h) postinfection were subjected to high-throughput small RNA sequencing. The sequencing generated 1.2 to 3.8 million reads for each treatment. In total, the high-throughput sequencing identified 81 distinct miRNAs that mapped to stem-loop structures found within the WSSV genome. According to the criteria for miRNA generation (20), among them, 49 putative WSSV miRNAs were not identified previously (Table 1; see Fig. S1 in the supplemental material). It was found that the 49 novel miRNAs could map to the 5′ or 3′ arm of the predicted pre-miRNA hairpins, bearing signature 2-nt 3′ overhangs, and were abundant in read counts (Table 1; see Fig. S1). The results indicated that these putative miRNAs were generated from RNAi machinery processing. Taking the findings of our previous and present studies together, a total of 89 putative WSSV miRNAs were revealed. The 89 putative WSSV miRNAs were distributed across the viral genome, on both the positive and negative strands.
TABLE 1.
Sequences and genome locations of WSSV miRNAs
| miRNA | Sequence (5′–3′) | Length (nt) | Genome localization |
No. of reads after WSSVinfection of shrimp |
5′ or 3′ arm | Presence of overhanga | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Strand | Start position | End position | 0 h | 6 h | 24 h | 48 h | |||||
| WSSV-miR-N1 | CUAGGUAGAAGGAGAGAGAGUGCU | 24 | − | 41689 | 41666 | 0 | 0 | 0 | 8 | 5′ | Y |
| WSSV-miR-N2 | AAGUGCCUCGUAGAUUGGA | 19 | − | 102167 | 102149 | 0 | 8 | 9 | 8 | 3′ | Y |
| WSSV-miR-N3 | GGGAGAAACAUAUGAGCUGUU | 21 | − | 89871 | 89852 | 0 | 7 | 0 | 10 | 3′ | Y |
| WSSV-miR-N4 | GGAGCAACAGUCGGUUCUGG | 20 | − | 92004 | 91985 | 0 | 0 | 10 | 8 | 3′ | Y |
| WSSV-miR-N5 | CACUUGGCGGUCAUCGUUUUGAA | 23 | − | 19096 | 19074 | 0 | 0 | 8 | 10 | 3′ | Y |
| WSSV-miR-N6 | GAAACGACUGUUGCUCCAGAAAC | 23 | + | 92241 | 92263 | 0 | 0 | 9 | 9 | 5′ | Y |
| WSSV-miR-N7 | GAUGAUGGAGAAGAAGCGACU | 21 | + | 167461 | 167481 | 0 | 10 | 0 | 8 | 5′ | Y |
| WSSV-miR-N8 | GUAGUUGUAGAAUUAACAUCAUCCU | 25 | + | 200605 | 200629 | 0 | 0 | 0 | 9 | 5′ | N |
| WSSV-miR-N9 | GAAGAAUUAACCUGCUUUUCACUG | 24 | + | 189330 | 189353 | 0 | 0 | 9 | 0 | 3′ | Y |
| WSSV-miR-N10 | AUUGGGAGAUAACUUGGUC | 19 | − | 95954 | 95935 | 0 | 0 | 8 | 8 | 5′ | Y |
| WSSV-miR-N11 | AUCAAUUUCUUCCUCUUCUUCCU | 23 | − | 80360 | 80338 | 0 | 8 | 0 | 9 | 5′ | Y |
| WSSV-miR-N12 | UUUUAUCGAGAGAAUGAGAAUA | 22 | − | 126197 | 126173 | 0 | 0 | 9 | 10 | 5′ | Y |
| WSSV-miR-N13 | ACAUGAUUGACGAUAGGUUGACU | 23 | − | 294940 | 294918 | 0 | 0 | 0 | 10 | 3′ | Y |
| WSSV-miR-N14 | GAGGAGGAGGAGGAUGAAGAA | 21 | + | 152189 | 152209 | 0 | 10 | 8 | 7 | 3′ | Y |
| WSSV-miR-N15 | CUUGUUGGAACGUUUUGUCG | 20 | + | 299863 | 299882 | 0 | 0 | 9 | 0 | 5′ | Y |
| WSSV-miR-N16 | AUCCGAUGAGUAUGAUUUUGAUGAU | 25 | + | 216476 | 216500 | 0 | 0 | 71 | 331 | 3′ | Y |
| WSSV-miR-N17 | CCUGAAAUGACUGCAGAUUUAUUG | 24 | + | 294338 | 294361 | 0 | 7 | 0 | 10 | 3′ | Y |
| WSSV-miR-N18 | GAAGAAGAAUGGUCUCUAAGCA | 22 | − | 147378 | 147357 | 0 | 8 | 45 | 83 | 3′ | Y |
| WSSV-miR-N19 | GUGGGAGAAUCAUGUGUAUGGA | 22 | − | 171625 | 171604 | 0 | 0 | 10 | 10 | 5′ | Y |
| WSSV-miR-N20 | GUUGUCAAUCAUUGUAUUUUGUC | 23 | + | 298272 | 298294 | 0 | 0 | 6 | 87 | 3′ | Y |
| WSSV-miR-N21 | CGCUGGGUCGGCCUGAUGUCA | 21 | + | 197504 | 197524 | 0 | 10 | 0 | 9 | 3′ | Y |
| WSSV-miR-N22 | GGGGCGUAAAAAGACUGUAGG | 21 | + | 212144 | 212164 | 0 | 0 | 47 | 67 | 3′ | Y |
| WSSV-miR-N23 | GUGGUCUUAACGAAGGGCAUU | 21 | + | 197462 | 197482 | 0 | 10 | 10 | 8 | 5′ | Y |
| WSSV-miR-N24 | GUAUGAGUAGUGAUGAAGAAUCA | 23 | + | 45037 | 45059 | 0 | 0 | 84 | 817 | 5′ | Y |
| WSSV-miR-N25 | GAAUGGCUCUGAAAAGUUA | 19 | − | 33373 | 33355 | 0 | 8 | 0 | 9 | 5′ | Y |
| WSSV-miR-N26 | UAUUUGUCUUGGAAGUAACUU | 21 | + | 10462 | 10482 | 0 | 6 | 6 | 8 | 5′ | Y |
| WSSV-miR-N27 | UUCUUAGUAGGGUGUGGGUUU | 21 | − | 290828 | 290808 | 0 | 0 | 0 | 10 | 5′ | Y |
| WSSV-miR-N28 | UUCGAGUGUCCGAAUAUUCGCGUC | 24 | + | 56226 | 56249 | 0 | 0 | 9 | 8 | 5′ | Y |
| WSSV-miR-N29 | AAGAGGACAAAAACACAGGGU | 21 | + | 197055 | 197075 | 0 | 10 | 0 | 9 | 3′ | Y |
| WSSV-miR-N30 | AUUGGCCUAGAUGACUCUGUAGAUU | 25 | + | 298346 | 298371 | 0 | 8 | 25 | 54 | 5′ | Y |
| WSSV-miR-N31 | CUCUUGUACAAGCACUCUGAG | 21 | + | 110522 | 110542 | 0 | 0 | 0 | 8 | 5′ | Y |
| WSSV-miR-N32 | AGGCGGGGGAAGGUUUGGUG | 22 | − | 180879 | 180859 | 0 | 0 | 10 | 0 | 5′ | Y |
| WSSV-miR-N33 | GACGUGCGAUAUUUCUGCCUU | 21 | − | 250278 | 250258 | 0 | 10 | 93 | 87 | 5′ | Y |
| WSSV-miR-N34 | CACCGACGGCUUUUUUAAUGCA | 22 | + | 151899 | 151920 | 0 | 0 | 9 | 8 | 3′ | N |
| WSSV-miR-N35 | AAGGUCAGAUAUUGUAUUGGGAAA | 24 | − | 102253 | 102220 | 0 | 0 | 0 | 9 | 3′ | Y |
| WSSV-miR-N36 | AGUUUCUGUAUUGACAGAUA | 20 | + | 52831 | 52850 | 0 | 0 | 8 | 45 | 5′ | Y |
| WSSV-miR-N37 | UUUAGAGCAAUUUCUCGCUCAG | 22 | − | 73250 | 73229 | 0 | 0 | 10 | 10 | 5′ | Y |
| WSSV-miR-N38 | UUAGUCGGUAUCGGAAUCAGUG | 22 | − | 97399 | 97378 | 0 | 0 | 8 | 8 | 3′ | Y |
| WSSV-miR-N39 | UGAGGAUAGUGGACAUGUUGAA | 22 | + | 145804 | 145825 | 0 | 10 | 8 | 9 | 3′ | Y |
| WSSV-miR-N40 | AUCGAGGAUGAACAUGCAAGACA | 23 | + | 18553 | 18575 | 0 | 0 | 9 | 9 | 3′ | Y |
| WSSV-miR-N41 | GAAGAAAUUUGGGGUAGGCAUC | 22 | − | 13783 | 13762 | 0 | 10 | 8 | 9 | 3′ | Y |
| WSSV-miR-N42 | UGCUGCAGCUGUUGCAACCUCG | 22 | − | 29203 | 29182 | 0 | 0 | 0 | 10 | 5′ | Y |
| WSSV-miR-N43 | CGAGGAGGAACAUGGGCAGGUA | 22 | − | 9125 | 9104 | 0 | 6 | 0 | 10 | 5′ | Y |
| WSSV-miR-N44 | CGACGACGGAUCUUCUACAUC | 21 | + | 24075 | 24095 | 0 | 9 | 7 | 61 | 3′ | Y |
| WSSV-miR-N45 | GAGGACUUUCUAAGCAUGAGAAA | 23 | + | 212053 | 212075 | 0 | 10 | 48 | 80 | 5′ | Y |
| WSSV-miR-N46 | AGUGCCAAGAUAACGGUUGAAG | 22 | + | 5292 | 5313 | 0 | 0 | 9 | 8 | 5′ | Y |
| WSSV-miR-N47 | CUGGUGAUAUGUCUGGAGCU | 20 | + | 99894 | 99913 | 0 | 10 | 0 | 0 | 5′ | Y |
| WSSV-miR-N48 | ACGAGGAGAUGGUUGGGGACU | 21 | + | 109295 | 109315 | 0 | 0 | 0 | 75 | 3′ | Y |
| WSSV-miR-N49 | GUGAGAUUUGGUUUCAUGCCC | 21 | + | 226509 | 226529 | 0 | 0 | 0 | 49 | 3′ | Y |
Y, yes; N, no.
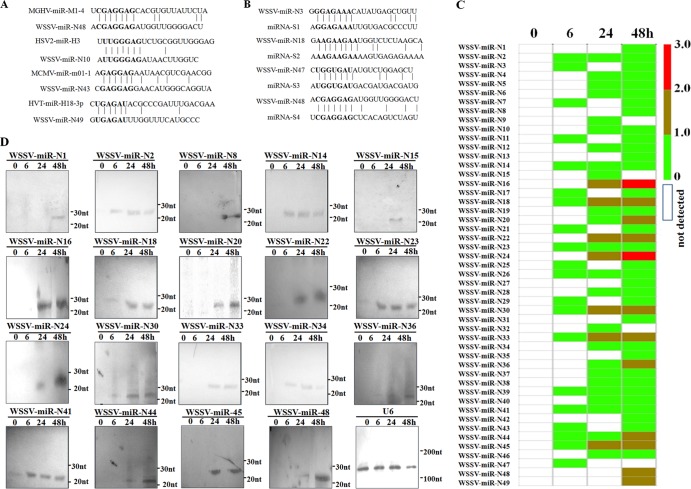
Based on homology analysis of the 49 putative WSSV miRNAs, the results indicated that WSSV-miR-N10, -N43, -N48, and -N49 shared identical miRNA seed regions with miRNAs of mouse gammaherpesvirus, herpes simplex virus 2, mouse cytomegalovirus, and herpesvirus of turkey, respectively (Fig. 1A), suggesting that these viral miRNAs might regulate conserved cellular transcripts. The homology analysis showed that WSSV-miR-N3, -N18, -N47, and -N48 had the same seed regions as those of shrimp miRNAs (Fig. 1B), indicating that the viral miRNAs might have evolved to imitate host miRNAs and hijack existing gene regulatory networks.
FIG 1.
Characterization of WSSV miRNAs. (A) Sequence alignments of WSSV miRNAs and known viral miRNAs. MGHV, mouse gammaherpesvirus; HSV2, herpes simplex virus 2; MCMV, mouse cytomegalovirus; HVT, herpesvirus of turkey. (B) Sequence alignments of WSSV miRNAs and shrimp miRNAs. (C) Time course expression patterns of WSSV miRNAs in WSSV-infected shrimp at different times postinfection. The lane headings show the time points postinfection. The numbers on the right indicate the log numbers of miRNA copies. (D) Northern blots of WSSV miRNAs. Total RNAs isolated from lymphoid organs of virus-free and WSSV-infected shrimp at different times postinfection were blotted with DIG-labeled oligodeoxynucleotide probes. The RNA sizes are shown to the right of the blots. U6 was used as a loading control. Lane headings show the time points postinfection.
The time course expression profiles of the 49 putative WSSV miRNAs showed that 20 viral miRNAs were expressed at 6 h postinfection, while no viral miRNA was detected in virus-free shrimp (Fig. 1C), indicating that the 20 putative miRNAs may be early viral miRNAs. The results revealed that the expression levels of WSSV-miR-N16 and WSSV-miR-N24 were the highest at 24 and 48 h postinfection (Fig. 1C), suggesting that these two miRNAs might play important roles during WSSV infection. To confirm the expression of the 49 putative WSSV miRNAs revealed in this study, 19 of them were selected at random for Northern blotting. The results showed that the expression profiles of viral miRNAs were similar to those obtained by small RNA sequencing (Fig. 1D).
Tissue-specific expression of viral miRNAs in shrimp in vivo.
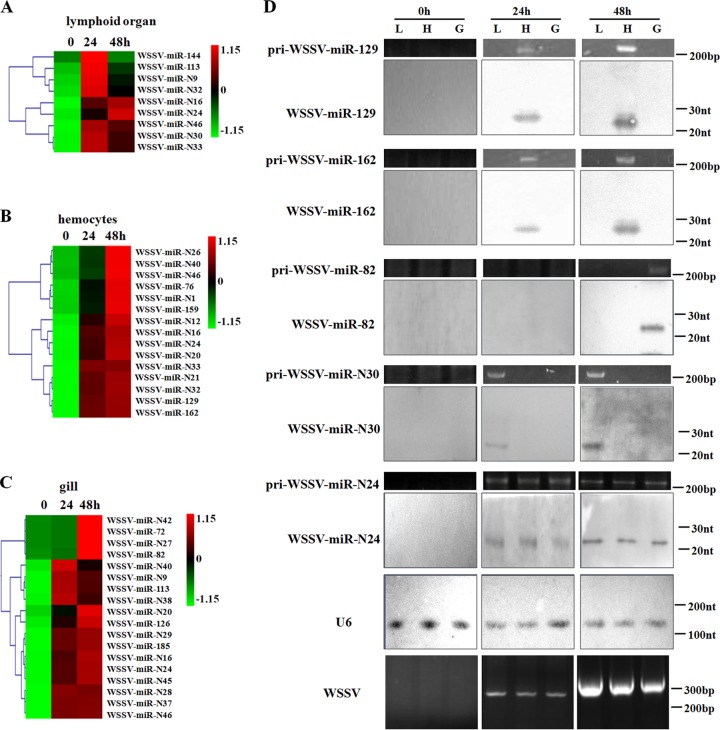
To evaluate the tissue-specific expression of viral miRNAs, the 89 putative WSSV miRNAs, including the 49 novel miRNAs revealed in this study and the other 40 miRNAs identified in the previous study (15), were used to prepare a virus-encoded miRNA microarray. The quantitative detection of WSSV virions by real-time PCR revealed that WSSV could infect lymphoid organs, hemocytes, and gills of shrimp (data not shown). The miRNA microarray results showed that the expression of viral miRNAs was tissue specific (Fig. 2A to C). Of the 89 WSSV miRNAs, 9 miRNAs were detected in the lymphoid organs of shrimp (Fig. 2A). WSSV-miR-144 and WSSV-miR-N30 were uniquely detected in the lymphoid organs (Fig. 2A). In the hemocytes of shrimp, 15 viral miRNAs were expressed (Fig. 2B). The results revealed that 8 viral miRNAs (WSSV-miR-N1, -N12, -N21, -N26, -76, -129, -159, and -162) were specifically present in shrimp hemocytes (Fig. 2B). As shown in Fig. 2C, 18 viral miRNAs were found to be present in WSSV-infected gills. Among them, 11 WSSV miRNAs were detected only in shrimp gills (Fig. 2C). These findings revealed that the expression of viral miRNAs in vivo is tissue specific.
FIG 2.
Tissue-specific expression of viral miRNAs in shrimp. Based on the results of WSSV miRNA microarray analysis, the normalized hybridization signals of viral miRNAs from shrimp lymphoid organs (A), hemocytes (B), and gills (C) at different times postinfection are shown. Lane headings show the time points postinfection. (D) Detection of selected viral miRNAs. Zero, 24, and 48 h after infection with WSSV, the lymphoid organs (L), gills (G), and hemocytes (H) of shrimp were collected and subjected to Northern blotting. At the same time, the collected samples were used for primary miRNA detection and WSSV detection by PCR, followed by agarose gel electrophoresis. The probes for Northern blotting are shown on the left, and the RNA sizes are indicated on the right. U6 was used as a loading control.
To confirm the microarray data, Northern blot analysis and the detection of primary miRNAs were performed. The results indicated that WSSV could be detected in shrimp lymphoid organs, hemocytes, and gills (Fig. 2D). Northern blots demonstrated that the expression of WSSV-miR-N30, WSSV-miR-82, WSSV-miR-129, and WSSV-miR-162 was tissue specific, whereas WSSV-miR-N24 was detected in all shrimp organs and tissues examined (Fig. 2D). At the same time, it was revealed that the primary miRNAs shared the same expression profiles as their corresponding viral miRNAs (Fig. 2D). These results were consistent with the data from the miRNA microarray analysis. Taken together, these findings reveal that the expression of viral miRNAs in vivo is tissue specific.
Interaction between viral miRNA and host gene.
Increasing evidence has indicated that viral miRNAs have important roles in host-virus interactions. As revealed above, among the WSSV miRNAs, WSSV-miR-N24 was upregulated in the shrimp tissues examined, suggesting that WSSV-miR-N24 might have great effects on WSSV infection. In this context, WSSV-miR-N24 was characterized further.
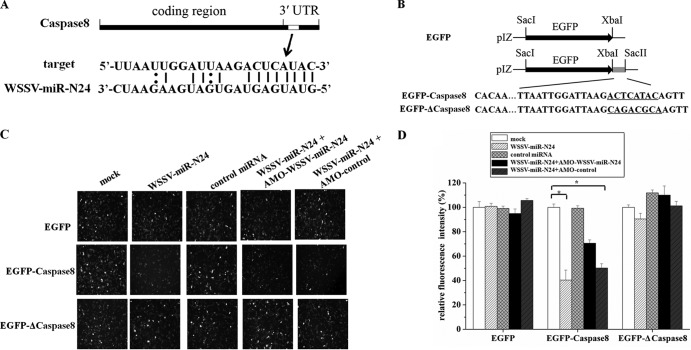
The predictions by miRanda and TargetScan showed that the host caspase 8 gene, a gene required for shrimp apoptosis (19), was a target gene of WSSV-miR-N24 (Fig. 3A), indicating that WSSV-miR-N24 may be involve in the regulation of apoptosis in shrimp.
FIG 3.
Interaction between WSSV-miR-N24 and M. japonicus shrimp caspase 8 gene. (A) Region of the host caspase 8 3′ UTR sequence targeted by WSSV-miR-N24. (B) Constructs with the wild-type and mutated 3′ UTRs of the host caspase 8 gene. The sequences complementary to the seed region of WSSV-miR-N24 are underlined. (C) Interaction between WSSV-miR-N24 and the host caspase 8 gene in insect cells. Insect High Five cells were simultaneously transfected with the WSSV-miR-N24 mimic and different reporter plasmids (EGFP, EGFP-caspase 8, or EGFP-Δcaspase 8). As a control, the control miRNA mimic was included in the transfections. The plasmid (EGFP, EGFP-caspase 8, or EGFP-Δcaspase 8)-transfected cells were cotransfected with the WSSV-miR-N24 mimic, the control miRNA, AMO-WSSV-miR-24, or AMO-control. Fluorescent images were obtained 48 h after transfection. The miRNAs and AMO-modified anti-miRNA oligonucleotides that were used for transfections are indicated at the top. The plasmids are shown on the left. (D) Summary of the data shown in panel C. The effects of the miRNAs and/or AMO constructs on gene expression of different plasmids (EGFP, EGFP-caspase 8, or EGFP-Δcaspase 8) were determined by comparing fluorescence intensities in the absence of miRNAs and/or AMO constructs with that of mock transfection. Statistically significant differences between treatments are indicated by asterisks (P < 0.05).
To evaluate the interaction between WSSV-miR-N24 and the host caspase 8 gene, we constructed the EGFP-caspase 8 plasmid, containing EGFP and the caspase 8 3′ UTR of M. japonicus shrimp (Fig. 3B). The synthesized WSSV-miR-N24 mimic and the EGFP-caspase 8 plasmid were cotransfected into insect High Five cells. The results showed that the fluorescence intensity in the cotransfected cells was significantly reduced compared with the intensity in the EGFP-caspase 8-transfected cells (Fig. 3C and D), indicating that the synthesized WSSV-miR-N24 mimic repressed the expression of the caspase 8 gene through targeting of its 3′ UTR. However, the control miRNA mimic had a negligible effect on the expression of EGFP-caspase 8. When the activity of the synthesized WSSV-miR-N24 mimic was inhibited by LNA-modified AMO-WSSV-miR-N24, the fluorescence intensity in the synthesized WSSV-miR-N24 mimic-, AMO-WSSV-miR-N24-, and EGFP-caspase 8-cotransfected cells was recovered (Fig. 3C and D).
To investigate the specificity of WSSV-miR-N24-mediated inhibition of caspase 8, the sequence of the caspase 8 3′ UTR complementary to the seed region of WSSV-miR-N24 was mutated, resulting in the pIZ/EGFP-Δcaspase 8 construct (Fig. 3B). It was revealed that mutation of predicted WSSV-miR-N24 recognition sites within the 3′ UTR of the caspase 8 gene completely disrupted inhibition by the WSSV-miR-N24 mimic (Fig. 3C and D), suggesting that the WSSV-miR-N24 seed sequence was essential for targeting of the caspase 8 3′ UTR.
Taken together, the findings indicated that WSSV-miR-N24 could target the host caspase 8 3′ UTR and might play important roles in apoptosis.
In vivo apoptotic regulation of host hemocytes mediated by viral miRNA.
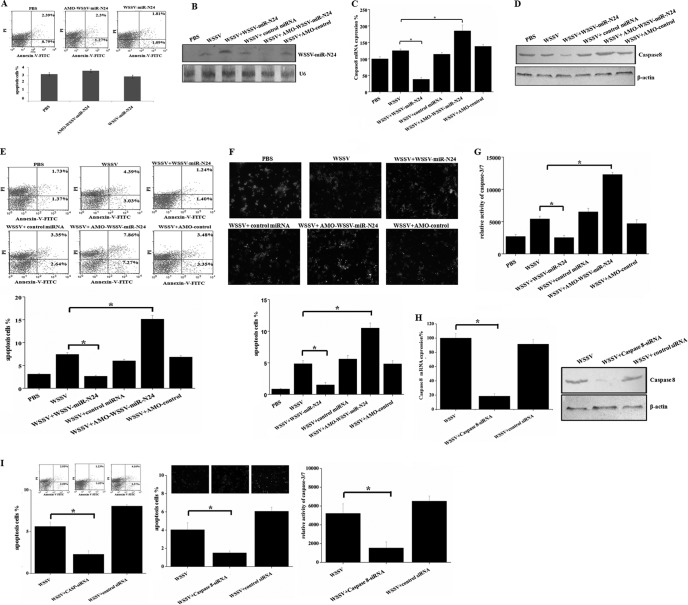
In an attempt to reveal the in vivo apoptotic regulation by viral miRNA, the apoptosis of shrimp hemocytes was assayed. To characterize the in vivo interaction between WSSV-miR-N24 and host caspase 8 mRNA, the synthesized WSSV-miR-N24 mimic was injected into shrimp. The results indicated that the WSSV-miR-N24 mimic had no effect on apoptosis of shrimp hemocytes (Fig. 4A), suggesting that WSSV-miR-N24 was not cytotoxic for shrimp. Northern blots showed that WSSV-miR-N24 was upregulated in hemocytes of WSSV-infected shrimp 48 h after injection of the WSSV-miR-N24 mimic compared with the control level (WSSV only) (Fig. 4B). Meanwhile, real-time PCR was used to detect the amount of the caspase 8 transcript. It was found that the overexpression of WSSV-miR-N24 led to a significant decrease in the transcript level of the caspase 8 gene at 48 h postinfection (Fig. 4C). However, the control miRNA mimic had little effect on the expression of the caspase 8 gene (Fig. 4C). To inhibit the expression of WSSV-miR-N24, AMO-WSSV-miR-N24 was injected into WSSV-infected shrimp. It was revealed that AMO-WSSV-miR-N24 was not cytotoxic for shrimp (Fig. 4A), and the results indicated that the expression of WSSV-miR-N24 was significantly knocked down in shrimp (Fig. 4B). As a result, there was a significant upregulation of caspase 8 mRNA (Fig. 4C). Western blots yielded results similar to those obtained by real-time PCR (Fig. 4D). These findings reveal that the viral miRNA WSSV-miR-N24 interacts with the host caspase 8 mRNA in shrimp in vivo.
FIG 4.
In vivo apoptotic regulation of host hemocytes by viral miRNA. (A) Effects of the synthesized WSSV-miR-N24 mimic and AMO-WSSV-miR-N24 on apoptosis of shrimp hemocytes. Forty-eight hours after injection of the WSSV-miR-N24 mimic or AMO-WSSV-miR-N24, the apoptosis of shrimp hemocytes was assayed using annexin V analysis. Lane headings indicate the treatments. (B) Expression of WSSV-miR-N24 in shrimp. Either the synthesized WSSV-miR-N24 mimic or AMO-WSSV-miR-N24 was administered to WSSV-infected shrimp. The shrimp hemocytes were collected and subjected to Northern blotting with a WSSV-miR-N24-specific probe 48 h after injection. Shrimp U6 was used as a control. (C) Real-time PCR detection of caspase 8 mRNA in hemocytes of WSSV-infected shrimp treated with the synthesized WSSV-miR-N24 mimic or AMO-WSSV-miR-N24. The control miRNA and AMO-control construct were included in the experiments. Forty-eight hours after injection, the shrimp hemocytes were collected and subjected to real-time PCR detection for caspase 8 mRNA. Shrimp β-actin was used as a control. The statistically significant differences between treatments are indicated with asterisks (P < 0.05). (D) Effects of WSSV-miR-N24 on expression of host caspase 8 gene in Western blot. Western blotting was performed with AP-conjugated goat anti-mouse IgG, using shrimp hemocytes collected 48 h after injection. Shrimp β-actin was used as a control. The lane headings indicate the different treatments. The antibodies used are shown on the right. (E) Effects of WSSV-miR-N24 overexpression or silencing on early stage of apoptosis. WSSV-miR-N24 was overexpressed or knocked down in virus-infected shrimp. At 48 h postinfection, the apoptosis of shrimp hemocytes was assayed using annexin V analysis. Lane headings indicated the treatments. The statistically significant differences between treatments are indicated with asterisks (P < 0.05). (F) Influence of WSSV-miR-N24 overexpression or silencing on late stage of apoptosis. The expression of WSSV-miR-N24 was inhibited or overexpressed in WSSV-infected shrimp. At 48 h postinfection, the apoptosis of shrimp hemocytes was evaluated with TUNEL assays. Lane headings indicate the treatments. The statistically significant differences between treatments are indicated with asterisks (P < 0.05). (G) Detection of caspase 3/7 activities in hemocytes of virus-infected shrimp at 48 h postinfection. Data are the means for three independent experiments. The statistically significant differences between treatments are indicated with asterisks (P < 0.05). (H) Silencing of caspase 8 expression in shrimp in vivo. Shrimp were injected with WSSV and caspase 8-siRNA or with WSSV and caspase 8-mutation-siRNA. At 48 h postinfection, the shrimp hemocytes were collected and subjected to real-time PCR (left) and Western blotting (right). Shrimp β-actin was used as a control. (I) Evaluations of apoptosis of shrimp hemocytes. In the case of the silencing of caspase 8 expression, the apoptosis of hemocytes was assayed by annexin V (left), TUNEL (middle), and caspase-3/7 activity (right) detection. Data are the means for three independent experiments. *, P < 0.05.
The apoptosis of shrimp hemocytes was evaluated under conditions where WSSV-miR-N24 was overexpressed or knocked down. The results of annexin V analysis showed that the overexpression of WSSV-miR-N24 led to a significant decrease of the percentage of apoptotic hemocytes from virus-infected shrimp at 48 h postinfection (Fig. 4E), indicating that WSSV-miR-N24 repressed host apoptosis at an early stage. In contrast, silencing of WSSV-miR-N24 expression resulted in an increase (about 2-fold) of the percentage of annexin V-positive hemocytes in comparison to that for the control with WSSV only (Fig. 4E). The detection of late-stage apoptosis of shrimp hemocytes by TUNEL yielded results similar to those of annexin V detection (Fig. 4F). It was found that the caspase 3/7 activity of shrimp hemocytes was significantly decreased by treatment of the WSSV-infected shrimp with the WSSV-miR-N24 mimic 48 h after injection, whereas the inhibition of WSSV-miR-N24 expression promoted the apoptosis of shrimp hemocytes (Fig. 4G). The above-described data indicate that WSSV-miR-N24 plays a negative role in the regulation of apoptosis of shrimp hemocytes in vivo.
To further examine the influence of host caspase 8 on apoptosis, the expression of the caspase 8 gene was silenced by sequence-specific siRNA, followed by evaluation of hemocyte apoptosis. The real-time PCR and Western blot results showed that caspase 8 mRNA and protein were not detected 48 h after the injection of caspase 8-siRNA, while the control siRNA (caspase 8-mutation-siRNA) had no effect on the expression of the caspase 8 gene (Fig. 4H), indicating that the expression of the caspase 8 gene was knocked down by the sequence-specific siRNA. In this case, the apoptotic activity of shrimp hemocytes was determined. The data from apoptosis assays with annexin V, TUNEL, and caspase 3/7 activity analyses revealed that the silencing of caspase 8 expression led to statistically significant decreases of apoptosis activity compared with that of the controls (Fig. 4I). The findings demonstrate that caspase 8 plays an essential role in apoptosis in vivo.
Taken together, these findings show that WSSV-miR-N24 mediates the regulation of in vivo apoptosis by targeting host caspase 8 mRNA.
Role of viral miRNA in virus-host interaction.
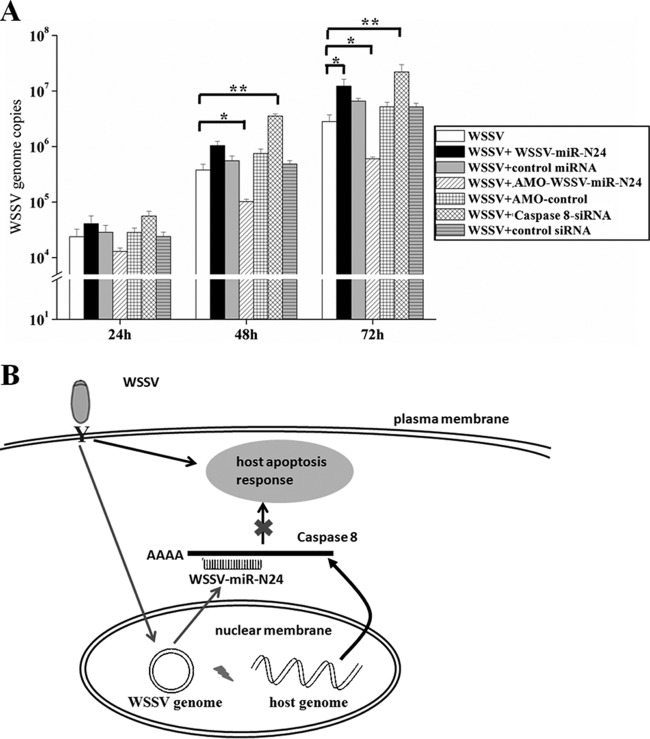
To assess the effects of WSSV-miR-N24 on WSSV replication, WSSV-miR-N24 in WSSV-infected shrimp was silenced by use of AMO-WSSV-miR-N24. The results showed that at 48 and 72 h postinfection, the number of WSSV copies in hemocytes of AMO-WSSV-miR-N24-treated shrimp was significantly decreased compared with that for the control with WSSV only, whereas the control inhibitor (AMO-control) had no effect on WSSV replication (Fig. 5A). On the other hand, injection of the synthesized WSSV-miR-N24 mimic into WSSV-infected shrimp led to a statistically significant increase in the number of WSSV copies in hemocytes at 72 h postinfection compared with that for WSSV only, while the control miRNA mimic had a negligible effect on WSSV infection (Fig. 5A). These results indicate that WSSV-miR-N24 plays important roles in WSSV infection.
FIG 5.
Role of viral miRNA in virus-host interactions. (A) Effect of WSSV-miR-N24 on virus infection. Shrimp were injected simultaneously with WSSV and the synthesized WSSV-miR-N24 mimic or AMO-WSSV-miR-N24. The treatments are indicated at the right. At various times postinfection, shrimp hemocytes were collected and subjected to real-time PCR to monitor WSSV replication. The numbers on the x axis indicate the time points postinfection. The statistically significant differences between treatments are represented with asterisks (*, P < 0.05; **, P < 0.01). (B) Proposed model for the role of WSSV-miR-N24 in virus-host interaction. WSSV-miR-N24 is generated from the viral genome and inhibits the expression of host caspase 8 by directly targeting the 3′ UTR of caspase 8 mRNA, further inhibiting host antiviral apoptosis.
It was revealed that the knockdown of caspase 8 gene expression resulted in a significant increase in the number of WSSV copies in hemocytes of virus-infected shrimp at 48 and 72 h postinfection compared with that for WSSV only (P < 0.01) (Fig. 5A). The control siRNA, however, had a negligible effect on virus infection (Fig. 5A). The findings indicate that WSSV-miR-N24 is involved in the virus-host interaction.
Based on the results described above, a model for the role of WSSV-miR-N24 in the virus-host interaction was proposed (Fig. 5B). In this model, WSSV-miR-N24 plays a key role in the regulation of in vivo apoptosis by targeting the host caspase 8 gene, which further leads to its involvement in the virus-host interaction.
DISCUSSION
Recently, it was revealed that hosts can use virus-derived siRNA or cellular miRNA to restrict viral propagation (18, 21–24). In turn, a number of viruses, in particular DNA viruses, seem to have evolved mechanisms to express their own miRNAs and employ miRNA-mediated regulations to counter host antiviral responses (25–27). Thus far, many viral miRNAs have been found to target the mRNAs of host or virus genes and to trigger the corresponding signaling pathways (25–27). To date, there have been some reports documenting the inhibition of host targets by viral miRNA species (25–27). However, many studies were completed in cultured cell lines, which may not be good models for characterizations of viral miRNAs in virus-host interactions. The in vitro events in cell lines may not be the same as molecular events in vivo. In the present study, viral miRNAs were investigated in vivo during virus infection. The results revealed that a total of 89 putative WSSV miRNAs were involved in virus-host interactions. Among them, the viral miRNA WSSV-miR-N24 could regulate host antiviral apoptosis by targeting the host caspase 8 gene. The results showed that WSSV-miR-N24 was employed by WSSV to regulate the expression of host caspase 8 to facilitate viral replication by inhibiting the host antiviral immune response. To our knowledge, this is the first study of the in vivo molecular events of a viral miRNA in antiviral apoptosis. Our study highlights a novel aspect of a viral miRNA in the virus-host interactions in vivo.
In recent years, EBV miRNAs were documented to have differential expression in distinct cultured cell types, similar to what has been observed in many studies of cellular miRNAs. It was found that the BamHI A rightward transcript (BART) miRNAs encoded by EBV were highly expressed in latently infected epithelial cells rather than in B cells (28). Some investigations revealed that 4 of 24 miRNAs expressed from the rhesus cytomegalovirus (RhCMV) genome were detected exclusively in infected fibroblasts, while 2 were specific for infected salivary glands (29, 30). The differential expression of viral miRNAs in different cell lines indicates that their targets may be different among cultured cells. Our study presented in vivo data showing that the expression profiles of viral miRNAs are tissue specific. Among 89 putative WSSV-encoded miRNAs, the findings showed that WSSV-miR-N30 and WSSV-miR-144 were uniquely detected in the lymphoid organs but not the hemocytes and gills of WSSV-infected shrimp. At the same time, 8 WSSV miRNAs were preferentially expressed in hemocytes, and 11 viral miRNAs were uniquely detected in gills of WSSV-infected shrimp. To date, the differential expression of viral miRNAs in various tissues in vivo remains unknown. Therefore, our study provided a clue to reveal the in vivo molecular events mediated by viral miRNAs in virus-host interactions. Due to the specificity of viral miRNA expression in different tissues, it could be inferred that the regulatory strategies and mechanisms mediated by viral miRNAs are tissue specific during virus infection. This issue merits further investigation.
It is believed that viral miRNAs play very important roles in the virus life cycle in vivo. Our previous study documented the existence of 40 WSSV miRNAs, all of which are expressed in hemocytes of virus-infected shrimp at the early stage of virus infection (15). This study extended the earlier data by documenting that WSSV, with its 305-kb double-stranded circular DNA genome, encodes at least 89 viral miRNAs. The ratio of viral miRNAs to the WSSV genome is about 800 times higher than that of human miRNAs to the human genome. One possibility raised by this high content of viral miRNAs is that viral miRNAs might greatly contribute to viral viability in response to selective pressures in the host environment. Thus, it appears that the use of viral miRNAs is an ideal and effective strategy for WSSV infection, with the tight space in the virus genome, to control distinct genes of shrimp and WSSV itself to establish a favorable environment during virus replication or to prevent infected cells and individuals from mounting antiviral immune responses.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Hi-Tech Research and Development Program of China (863 Program of China) (grant 2011AA10A216-3), the National Basic Research Program of China (grant 2012CB114403), and the Ministry of Agriculture, China (grant 201103034).
Footnotes
Published ahead of print 18 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03575-13.
REFERENCES
- 1.Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139. 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. 2004. The functions of animal microRNAs. Nature 431:350–355. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 5.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922. 10.1038/nature06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. 2004. Identification of virus-encoded microRNAs. Science 304:734–736. 10.1126/science.1096781 [DOI] [PubMed] [Google Scholar]
- 7.Chiang K, Rice AP. 2011. Mini ways to stop a virus: microRNAs and HIV-1 replication. Future Virol. 6:209–221. 10.2217/fvl.10.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, Khromykh AA, Asgari S. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 40:2210–2223. 10.1093/nar/gkr848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686. 10.1038/nature03576 [DOI] [PubMed] [Google Scholar]
- 10.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carrington JC, Nelson J. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095–12099. 10.1128/JVI.79.18.12095-12099.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U. S. A. 105:5453–5458. 10.1073/pnas.0711910105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. 10.1371/journal.ppat.0030065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381. 10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ., Jr 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442. 10.1158/0008-5472.CAN-07-5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Zhang X. 2012. Comprehensive characterization of viral miRNAs involved in white spot syndrome virus (WSSV) infection. RNA Biol. 9:1019–1029. 10.4161/rna.20741 [DOI] [PubMed] [Google Scholar]
- 16.Liu W, Han F, Zhang X. 2009. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. J. Proteome Res. 8:1198–1206. 10.1021/pr800840x [DOI] [PubMed] [Google Scholar]
- 17.Huang T, Xu D, Zhang X. 2012. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics 13:159. 10.1186/1471-2164-13-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T, Zhang X. 2012. Functional analysis of a crustacean microRNA in host-virus interactions. J. Virol. 86:12997–13004. 10.1128/JVI.01702-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Zhi B, Wu W, Zhang X. 2008. Requirement for shrimp caspase in apoptosis against virus infection. Dev. Comp. Immunol. 32:706–715. 10.1016/j.dci.2007.10.010 [DOI] [PubMed] [Google Scholar]
- 20.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. 2003. A uniform system for microRNA annotation. RNA 9:277–279. 10.1261/rna.2183803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang T, Zhang X. 2013. Host defense against DNA virus infection in shrimp is mediated by the siRNA pathway. Eur. J. Immunol. 43:137–146. 10.1002/eji.201242806 [DOI] [PubMed] [Google Scholar]
- 22.Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 84:8849–8860. 10.1128/JVI.00456-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86:13729–13734. 10.1128/JVI.02041-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronkhorst AW, van Cleef KW, Vodovar N, Ince IA, Blanc H, Vlak JM, Saleh MC, van Rij RP. 2012. The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. U. S. A. 109:E3604–E3613. 10.1073/pnas.1207213109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen BR. 2013. MicroRNAs as mediators of viral evasion of the immune system. Nat. Immunol. 14:205–210. 10.1038/ni.2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894. 10.1101/gad.17352611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164. 10.1101/gad.1793309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. 10.1371/journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer C, Grey F, Kreklywich CN, Andoh TF, Tirabassi RS, Orloff SL, Streblow DN. 2011. Cytomegalovirus microRNA expression is tissue specific and is associated with persistence. J. Virol. 85:378–389. 10.1128/JVI.01900-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancock MH, Tirabassi RS, Nelson JA. 2012. Rhesus cytomegalovirus encodes seventeen microRNAs that are differentially expressed in vitro and in vivo. Virology 425:133–142. 10.1016/j.virol.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.