ABSTRACT
We previously reported a proof-of-concept study for curing chronic hepatitis B virus (HBV) infection using a foreign-antigen recombinant HBV (rHBV) as a gene therapy vector. Targeted elimination of wild-type HBV (wtHBV)-infected cells could be achieved by functionally activating an in situ T-cell response against the foreign antigen. However, as chronic HBV infection spreads to all hepatocytes, specific targeting of virus-infected cells is thought to be less critical. It is also feared that rHBV may not induce active immunization in a setting resembling natural infection. For this immunotherapeutic approach to be practically viable, in the present study, we used a recombinant adenovirus (rAd) vector for rHBV delivery. The rAd vector allowed efficient transduction of wtHBV-producing HepG2 cells, with transferred rHBV undergoing dominant viral replication. Progeny rHBV virions proved to be infectious, as demonstrated in primary tupaia hepatocytes. These results greatly expanded the antiviral capacity of the replication-defective rAd/rHBV in wtHBV-infected liver tissue. With prior priming in the periphery, transduction with rAd/rHBV attracted a substantial influx of the foreign-antigen-specific T-effector cells into the liver. Despite the fully activated T-cell response, active expression of rHBV was observed for a prolonged time, which is essential for rHBV to achieve sustained expansion. In a mouse model of HBV persistence established by infection with a recombinant adeno-associated virus carrying the wtHBV genome, rAd/rHBV-based immunotherapy elicited a foreign-antigen-specific T-cell response that triggered effective viral clearance and subsequent seroconversion to HBV. It therefore represents an efficient strategy to overcome immune tolerance, thereby eliminating chronic HBV infection.
IMPORTANCE Adenovirus-delivered rHBV activated a foreign-antigen-specific T-cell response that abrogated HBV persistence in a mouse model. Our study provides further evidence of the potential of foreign-antigen-based immunotherapy for the treatment of chronic HBV infection.
INTRODUCTION
For effective immunotherapy against chronic hepatitis B virus (HBV) infection, antiviral immunity has to be reset in favor of a fully activated T-cell response. We previously described a novel strategy based on a T-cell response against a foreign antigen that specifically targets HBV-infected cells (1). In brief, the coding sequence for a cluster of immunodominant foreign epitopes was inserted into the HBV core gene to create a recombinant virus (rHBV). Disruption of the core gene rendered rHBV replication deficient except in cells already infected with wild-type HBV (wtHBV). Thus, targeted viral elimination was achieved through functional activation of an in situ T-cell response to a foreign antigen.
However, the rHBV vector-based immunotherapy approach has several challenges. (i) It is unclear whether rHBV can superinfect wtHBV-infected hepatocytes with high efficiency. (ii) All hepatocytes are infected with wtHBV during chronic disease (2–7), and therefore, targeted elimination of virus-infected cells appears less critical. (iii) Despite high immunogenicity of the viral antigens, HBV is believed to establish a stealthy infection without alerting the innate immune system (8). Virus-specific T-cell activation is reported to be significantly delayed even if self-limited acute hepatitis progresses (2, 3, 5). It is therefore important to establish whether the rHBV vector facilitates active immunization, albeit with high expression of the foreign polyepitope. In this regard, surrogate delivery of rHBV DNA via hydrodynamic injection, as used in our previous study (1), could not be applied in humans, although it triggered a potent T-cell response in the murine liver (9). (iv) For sustained expansion, rHBV vector has to survive the functionally activated T-cell response, rather than be largely suppressed in the liver, as suggested in our previous study (1).
To overcome these issues, in the present study, we used a recombinant adenovirus (rAd) vector to transduce the rHBV genome. The rAd vector is among the most widely studied gene transfer vehicles because of its high transduction efficiency in vitro and in vivo. Moreover, rAd vectors are predominantly sequestered by the liver after intravenous administration due to the architecture and vascular system of the organ (10). Also, rAd typically induces potent CD8+ T-cell responses specific to HBV transgenes (9, 11). Of particular significance, rAd vectors persist in vivo even after boosting the immune response (12–14), thus allowing the cargo gene (i.e., rHBV) to fulfill its function.
Liver transduction efficiency, rather than specificity for individual wtHBV-infected cells, is primarily emphasized in rAd/rHBV-based immunotherapy. In particular, the strategy exploits preexisting wtHBV in the host liver to expand the antiviral capacity of the replication-defective rAd vector. Ideally, administration of a small dose of rAd/rHBV would lead to the spread of rHBV in the liver, which would provoke a strong foreign-antigenic T-cell response that clears wtHBV via cytolytic or noncytolytic mechanisms (Fig. 1). In the present study, rAd/rHBV-based immunotherapy was validated in a mouse model of HBV persistence via infection with a recombinant adeno-associated virus type 8 (rAAV8) carrying the wtHBV genome (rAAV8-HBV) (15). This proved to be a valuable approach to overcome immune tolerance, thereby eliminating chronic HBV infection.
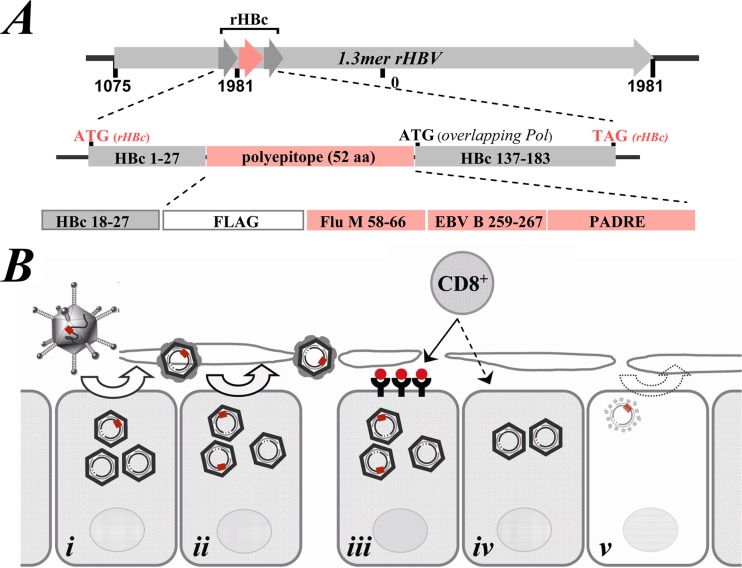
FIG 1.
Schematic illustration of rAd/rHBV-based immunotherapy strategy. (A) rHBV contains a sequence of a foreign polyepitope that is fused to the HBV core gene (rHBc). The polyepitope comprises a B-cell epitope (FLAG), two HLA-A2-restricted T-cell epitopes (influenza virus matrix, aa 58 to 66 [Flu M 58-66], and EBV BMLF-1, aa 259 to 267 [B 259-267]), and a CD4+ T-cell epitope (PADRE). HLA-A2-restricted HBc aa 18 to 27 is preserved at the N-terminal domain of rHBc. The start codon for an overlapping viral pol gene is also indicated. The expression of the pol gene is thought to be more efficient due to its reduced distance from the 5′ end of the pregenomic RNA. (B) During chronic HBV infection, most hepatocytes in the liver are infected. Administration of low-dose rAd/rHBV may result in a spread of rHBV, eliciting a potent intrahepatic T-cell response. (i) rHBV delivered to the liver through rAd transduction. (ii) Core-rescued rHBV displays dominant replication over wtHBV, with progeny virions invading neighboring cells. (iii) Following rHBV replication, foreign epitopes (red) are presented on liver cells as targets for specific T-effector cells. (iv) Neighboring cells during the bystander effect of in situ-activated T-cell response. Rather than direct destruction of cells, cytokine-mediated noncytolytic response plays a principal role in virus elimination. (v) rHBV stops replication after the natural HBV infection is cleared.
MATERIALS AND METHODS
DNA constructs.
The plasmid carrying rHBV, named prHBV1.3s, was constructed as previously described (1), except that an HLA-A2-restricted epitope from the HIV gag protein was removed from the polyepitope sequence due to its hyporesponsiveness (1) (Fig. 1A). prHBV1.3s-C carried an additional expression cassette of the HBV core protein, driven by the simian virus 40 (SV40) early promoter, downstream from the rHBV genome. The construct pCMV-rHBc contains the foreign-antigenic rHBc under cytomegalovirus (CMV) promoter control, as previously described (1). pIC-polyepitope was constructed on the basis of the pCI-neo vector (Promega) with the expression cassette under CMV promoter control. The fusion gene encoded a mouse li invariant-chain (82-amino-acid [aa]-encoding) gene (16) which was fused to a sequence of 55-aa foreign polyepitope at the 3′ terminus.
Viral vectors.
rAd/rHBV1.3 and rAd/rHBV1.1 were constructed commercially (E1-deleted human rAd vector of serotype 5; Shanghai SBO Medical Biotechnology Co., Ltd.) by recombining rAd with the shuttle vectors pAVsi1-ΔCMV-rHBV1.3s and pAVsil-rHBV1.1s, respectively, in cotransfected HEK293 cells. rAd/GFP carries a CMV promoter-driven green fluorescent protein (GFP) gene for control experiments. The rAAV8-HBV (containing 1.3 copies of the HBV genome, ayw3 subtype) vector was obtained from the Beijing FivePlus Molecular Medicine Institute (1.0 × 1012 genome equivalents [GEs]/ml).
Cells.
Hepatic cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. HepG2-4H5 cells stably producing rHBV virions were derived from prHBV1.3s-C-transfected HepG2 cells through G418 selection. Huh-7 cells constitutively expressing the HBV core antigen were generated by lentiviral vector transduction. Primary tupaia hepatocytes (PTHs) were isolated by a two-step perfusion protocol (17, 18). For viral infection, 1 × 106 freshly isolated PTHs were seeded in collagen-coated 6-well plates (BD Biosciences, USA); 1 day after seeding, the PTHs were inoculated with wtHBV isolated from the infectious sera of HBV-infected patients or rHBV virions concentrated from culture supernatants by polyethylene glycol 8000. The cultures were maintained for 10 to 12 days, with the medium replaced daily.
Mice.
HLA-A2 transgenic mice [C57BL/6-Tg (HLA-A2.1)1Enge/J; Jackson Laboratory] were purchased from the Model Animal Research Center of Nanjing University. DNA immunization was achieved by injecting 100 μg of plasmid DNA as described previously (16). For rAd/rHBV administration, 1 × 109 PFU of the vectors was intravenously administered (9, 19). To induce persistent HBV infection, 4- to 5-week-old male mice (HLA-A2) were infused with 7 × 1010 GEs of rAAV8-HBV vectors via the tail vein. All animal studies were approved by the Animal Ethics Committee of the Institut Pasteur of Shanghai (no. A2012008).
DNA and RNA hybridization.
rHBV/HBV-associated viral DNA was extracted from HepG2.2.15 or Huh-7 cells after transient transfection, as previously described (1). To detect secreted rHBV virions, the culture supernatants were harvested and ultracentrifuged on a 20% sucrose cushion at 25,000 rpm for 16 h. The pellets were resuspended in 10 mM Tris-HCl buffer (pH 7.5) containing 5.5 mM MgCl2. The samples were treated with RNase A and micrococcal nuclease for 1 h at 37°C. Viral DNA was ethanol precipitated after protease K (Merck) digestion in the presence of 1% SDS for 2 h at 55°C. RNA was extracted with TRIzol reagent (Invitrogen). Aliquots of total RNA (20 μg) were subjected to electrophoresis on a 1.0% formaldehyde agarose gel and blotted onto a nylon membrane. Hybridization was performed using a 32P-labeled probe of the full-length HBV genome (20). To discriminate rHBV from wtHBV DNA intermediates, the DNA encoding the polyepitope and the core gene fragment (nucleotides [nt] 1982 to 2308; genotype D [ayw3]; GenBank accession no. V01460) were used as probes, respectively. In experiments involving the reprobing of a single membrane, a digoxigenin-labeled HBV probe, prepared using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany), was used for the first round of hybridization.
ELISPOT and flow cytometry analysis.
The procedures of enzyme-linked immunosorbent spot assay (ELISPOT) and flow cytometry analysis were described previously (1).
Quantitative real-time PCR analysis.
The media of rAd/rHBV1.3-infected HepG2.2.15 cell cultures were replaced daily. Viral DNA was extracted using a QIAamp blood DNA minikit (Qiagen). The virus titers were quantified using a SYBR green Real-Time PCR Master Mix kit (Toyobo, Osaka, Japan). Two pairs of primers were used for the specific amplification of rHBV alone (Forward, 5′-GACGACAAGACCAAGGGCA-3′; Reverse, 5′-GAAGTGTTGATAGGATAGGGGC-3′) or with wtHBV and rHBV in total (Forward, 5′-GAGCCCCTATCCTATCAACACT-3′; Reverse, 5′-CACCTTATGAGTCCAAGGAATAC-3′). For absolute quantification, prHBV1.3s DNA was used to generate a standard curve.
HBsAg detection.
HBsAg was quantified using an architect platform (Abbott). An enzyme-linked immunosorbent assay (ELISA) kit (Shanghai Kehua Bio-Engineering Co., Ltd.) was used to detect HBsAg in the supernatants of rHBV-infected PTH cultures.
Statistics.
The data are expressed as means ± standard errors of the mean (SEM). Statistical analysis was performed with Student's t test or two-way analysis of variance (ANOVA) with Tukey's multiple-comparison test using GraphPad Prism 6.0.
RESULTS
rHBV replication is dependent on HBV core protein provided in trans.
Cytokine-mediated viral clearance occurs at many steps of HBV replication (21–25) and plays a major role in achieving high-efficiency immunotherapy (26–28). These steps are thought to be consistent with those of the core-rescued rHBV. We rescued rHBV replication with a core gene coexpression construct (prHBV1.3s-C). In transiently transfected Huh-7 cells, the expression of core antigen was detected mainly in the cytoplasm (29) (Fig. 2A), which was accompanied by viral replication and release of progeny virions (Fig. 2B and C). Transfection with rHBV (prHBV1.3s) alone failed to initiate the replicative cycle, despite the production of abundant viral RNA transcripts (Fig. 2D). Electron microscopy revealed intracellular recombinant virions as typical spherical particles in prHBV1.3s-C-transfected HepG2 cells and as 42-nm Dane particles in the culture medium (Fig. 2E, inset). Secreted rHBV virions, which were purified by CsCl density gradient centrifugation, exhibited characteristics similar to those of wtHBV, as revealed by dot blot hybridization (Fig. 2F) (30). These data indicated that core-rescued rHBV was virologically similar to wtHBV, through which the foreign antigen-activated immune clearance could eventually act on wtHBV.
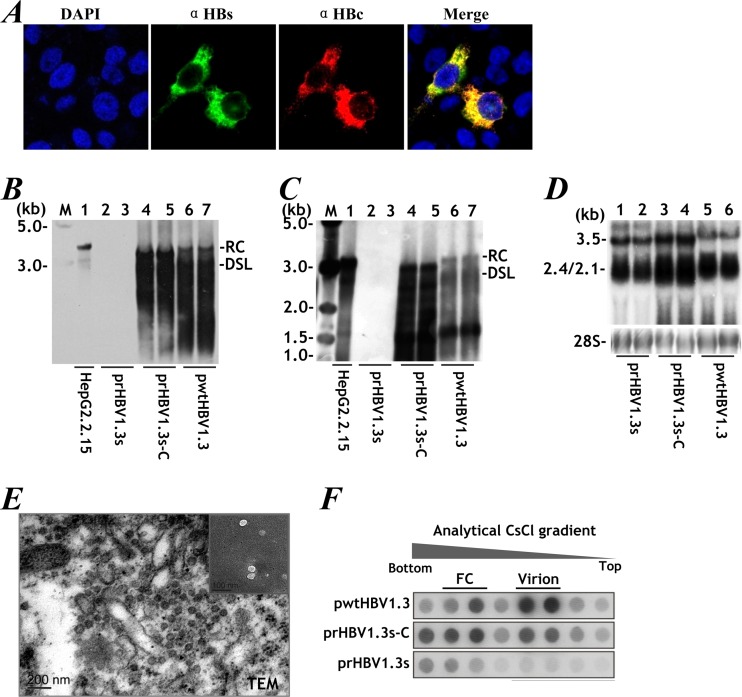
FIG 2.
rHBV is intrinsically similar to wtHBV. (A) HBsAg (green) and HBcAg (red) expression revealed by immunofluorescence staining of Huh-7 cells transfected with prHBV1.3s-C. DAPI, 4′,6-diamidino-2-phenylindole. (B) Intracellular viral replication detected by Southern blotting of Huh-7 cells transfected with prHBV1.3s, prHBV1.3s-C, or pwtHBV1.3. wtHBV DNA extracted from HepG2.2.15 cells served as a control. The positions of relaxed circular (RC) and double-stranded linear (DSL) forms are indicated. (C) Extracellular rHBV virions assayed as described for panel B. (D) Viral transcription by Northern blotting. (E) Transmission electron microscopy (TEM) for rHBV particles in HepG2-4H5 cells stably expressing prHBV1.3s-C. (Inset) Secreted rHBV virions. (F) rHBV virions identified by HBV DNA-specific dot blotting after CsCl density gradient centrifugation. FC, nonenveloped nucleocapsids. M, 1-kb DNA ladder (NEB).
rHBV virions generated by transcomplementation can infect tupaia hepatocytes.
The spread of core-rescued rHBV to neighboring cells would increase the antiviral potential, especially considering that all hepatocytes are infected during chronic disease. This issue was addressed in PTHs, one of the few currently available in vitro models of HBV infection. Cells were inoculated with rHBV with and without wtHBV coinfection. Newly synthesized HBsAg was detected in culture supernatants 8 to 10 days later (Fig. 3A) (18). In line with this finding, immunofluorescent staining on day 10 postinfection revealed the foreign-antigenic rHBc, as well as HBsAg, in the cytoplasm. The replication-defective rHBV did not need the aid of wtHBV to infect hepatocytes (Fig. 3B, middle). De novo rHBV expression was also detected in PTHs during coinfection with wtHBV, although the immunostaining did not identify the two viruses in a single cell (Fig. 3B, bottom). Collectively, these data indicated that the replication-defective rHBV virions were infectious in PTHs.
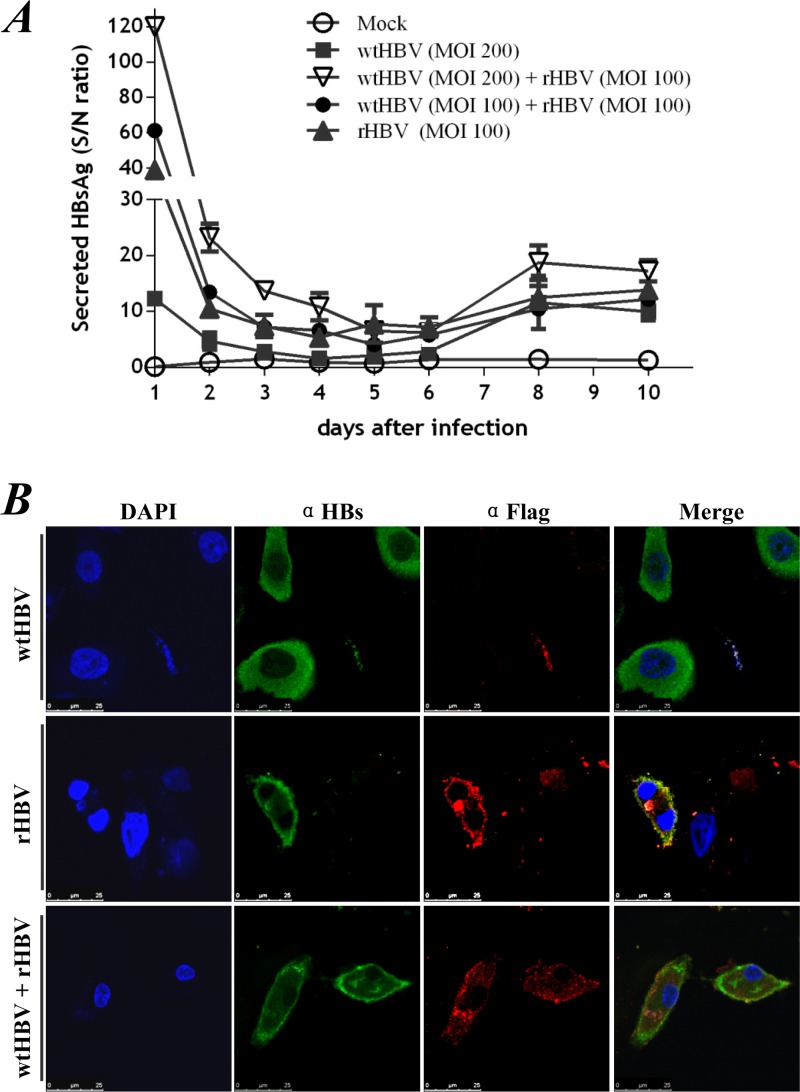
FIG 3.
rHBV is infectious in PTHs. PTHs were cultured with concentrated preparations of rHBV virions isolated from prHBV1.3s-C-transfected HepG2-4H5 cells in the presence or absence of wtHBV from the infectious sera of HBV-infected patients. (A) Secreted HBsAg was measured at the indicated time points by ELISA. Relative titers of HBsAg were calculated as signal/noise (S/N) ratios, where the sample absorbance (signal) was divided by the average negative-control absorbance (noise). HBsAg recurrence was defined as double the baseline ratio for the same curve. PBS-treated (Mock) PTHs were used as a control. (B) Immunostaining of HBsAg (α-HBs) and foreign-antigenic rHBc (α-Flag) in PTHs 10 days after inoculation with wtHBV (top), rHBV (middle), or mixed wtHBV and rHBV at a 2:1 ratio (bottom).
rAd vector enables efficient rHBV transduction in vitro.
The rAd constructs contained either the 1.3-copy overlength rHBV genome (rAd/rHBV1.3) or a 1.1-copy genome driven by the CMV promoter (rAd/rHBV1.1) (Fig. 4A). In our study, immunological analysis used rAd/rHBV1.1 because it abundantly expressed the antigenic rHBc (Fig. 4B), while rAd/rHBV1.3 encoding rHBV under the control of its endogenous promoter was used for virological assessments. In cells expressing HBV core antigen, rAd/rHBV1.3 transduction led to intracellular replication of rHBV (Fig. 4C), with progeny virions detected in the culture medium (data not shown).
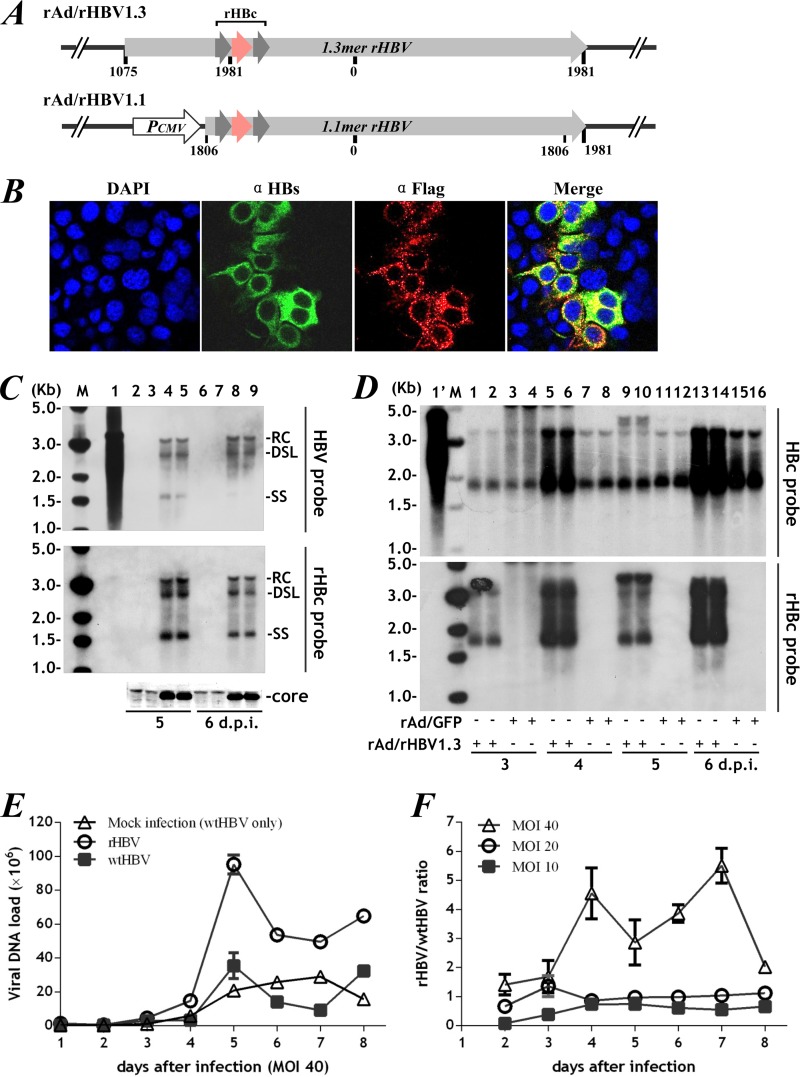
FIG 4.
Efficient rHBV transduction via rAd vectors in vitro. (A) Schematic representation of the rAd/rHBV1.3 and rAd/rHBV1.1 constructs used in this study. (B) Immunofluorescence for HBsAg (green) and rHBc (α-Flag; red) 7 days after rAd/rHBV1.1 transduction of Huh-7 cells. (C) Following rAd/rHBV1.3 transduction, intracellular rHBV was assayed in Huh-7 cells (lanes 2 and 3, 6 and 7) and in cells constitutively expressing the HBV core protein (lanes 4 and 5, 8 and 9) by Southern blotting with HBV (full-length genome)-specific or rHBc (polyepitope only)-specific probes. wtHBV DNA from HepG2.2.15 cells was used as a control (lane 1). The positions of relaxed circular (RC), double-stranded linear (DL), and single-stranded (SS) forms are indicated. HBV core protein expression was verified by Western blotting. The smaller bright bands are specific signals (bottom). (D) Intracellular replicative wtHBV and rHBV in the HepG2.2.15 cell line 3 to 6 days after transduction with rAd/rHBV1.3 (MOI, 40) (lanes 1 and 2, 5 and 6, 9 and 10, and 13 and 14), as determined by hybridization with rHBc probe (bottom) or HBc probe (specific to the core gene fragment missing in rHBV) (top) on a single blot. Mock infection with rAd/GFP was used as a control (lanes 3 and 4, 7 and 8, 11 and 12, and 15 and 16). Lane 1′, wtHBV DNA extract from HepG2.2.15 cells without rAd coinfection. (E) rHBV and wtHBV virions in culture supernatants (medium replaced daily) were quantified by specific quantitative PCR (qPCR) on days 2 to 8 following transduction with rAd/rHBV1.3 at an MOI of 40. rAd/GFP mock-infected HepG2.2.15 cells producing wtHBV only were used as a control. (F) rHBV/wtHBV ratios at the indicated time points after transduction at MOIs of 10, 20, and 40. The error bars indicate SEM.
The rAd vectors allowed efficient transduction of HepG2.2.15 cells, which produce a high yield of wtHBV. When rAd/rHBV1.3 was inoculated at a multiplicity of infection (MOI) of 40, more than 90% of cells were transduced. Both rHBV and wtHBV displayed robust viral replication, which, however, fluctuated over time (Fig. 4D), probably reflecting limited core protein production by HepG2.2.15 cells during rHBV coinfection. Dose-dependent accumulation of rHBV virions in culture medium was observed. At an MOI of 10 to 20, when only 60 to 80% of cells were transduced, the rHBV/wtHBV ratio remained steady throughout the experiment. Albeit with apparent saturation, the rHBV production was largely increased by rAd/rHBV1.3 transduction at an MOI of 40, with 5-fold more than wtHBV virions on day 7 (Fig. 4E and F). Considering multiple integrations of the wtHBV transgene (four 5′-3′ tandem copies of the viral genome) (20), as well as the established episomal covalently closed circular DNA (cccDNA) pool in HepG2.2.15 cells, these data suggested that rHBV replication was dominant over that of wtHBV.
Intravenous delivery of rAd/rHBV alone leads to a compromised immune response.
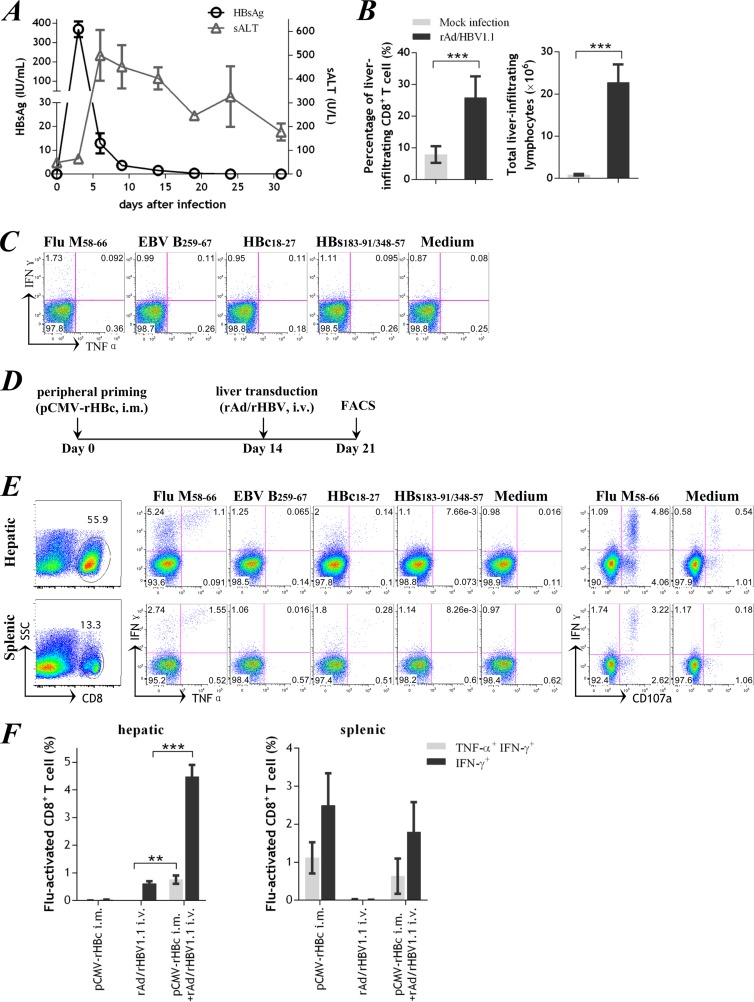
rAd/rHBV1.1 was administered intravenously to HLA-A2 transgenic mice (Fig. 5A). A high level of HBsAg antigenemia was observed at day 3 after infection and quickly declined thereafter. Serum alanine transaminase (sALT) activity peaked 6 days after infection and remained mildly elevated over 31 days (177.0 ± 36.0 U/liter; day 31). Both the frequency of CD8+ T cells and the absolute number of lymphocytes were significantly increased in the liver, as determined at day 10 (19, 31) following rAd infection (Fig. 5B). After ex vivo stimulation with the foreign polyepitope-derived peptides, however, few gamma interferon-positive (IFN-γ+) T-effector cells were detected, and they did not produce tumor necrosis factor alpha (TNF-α) (Fig. 5C). Similarly, no IFN-γ+ T-effector cells were found in the spleen despite substantial antigenemia. The T-cell response was also measured at days 14 to 16, which showed similar results (data not shown). These data were consistent with previous findings that direct administration of rAd vectors via the intravenous route might compromise the immune response (9, 32).
FIG 5.
rAd/rHBV-based active immunization in HLA-A2 transgenic mice. (A) Kinetics of serum HBsAg and sALT after intravenous rAd/rHBV1.1 administration (1 × 109 PFU) in HLA-A2 mice. (B) Proportions of CD8+ T cells (left) and absolute numbers of liver-infiltrating lymphocytes (right) from mice mock infected with PBS or from mice 10 days after injection of rAd/rHBV1.1. (C) Hepatic CD8+ T cells from a representative mouse given rAd/rHBV1.1 as in panel B, gated for intracellular IFN-γ and TNF-α staining after stimulation with the peptide influenza virus M58-66, EBV B259-67, HBc18-27, or HBsAg183-191/348-357. (D) Protocol used for rAd/rHBV-based active immunization. FACS, fluorescence-activated cell sorting. (E) At 7 days after rAd/rHBV-based active immunization, CD8+ T cells from mouse liver and spleen were analyzed by intracellular IFN-γ and TNF-α staining or CD107a surface staining after stimulation with influenza virus (Flu) M58-66, EBV B259-67, HBc18-27, or HBsAg183-191/348-357. (F) Percentages of influenza virus-activated IFN-γ-positive or IFN-γ/TNF-α-double-positive CD8+ T cells in mice immunized with pCMV-rHBc alone (n = 4; tested on day 14), rAd/rHBV1.1 infection alone (n = 4; tested on day 10), or rAd/rHBV1.1 infection with prior pCMV-rHBc priming (n = 4; tested on day 7 after rAd infection). These time points corresponded to the peak T-cell responses upon the different treatments and were based on previous studies for rAd/wtHBV-based immunization in mice (19, 31). **, P < 0.01; ***, P < 0.001. i.m., intramuscular injection; i.v., intravenous injection. The error bars indicate SEM.
A prime-boost strategy retargeted the functional T-cell response to the liver.
A prime-boost protocol for rAd/rHBV-based active immunization was used to retarget a functional T-cell response to the mouse liver (Fig. 5D). First, a foreign-antigen-specific T-cell response was activated by intramuscular vaccination with pCMV-rHBc. When the peripheral response peaked at day 14, mice were infused intravenously with rAd/rHBV1.1, which was sequestered in the liver, thereby boosting a functional in situ T-cell response.
Following the immunization protocol, a vigorous T-cell response was mounted in mice, with a large percentage of CD8+ T cells accumulating in the liver (52.7% ± 1.1%) and fewer in the spleen (16.4% ± 0.9%; P < 0.0001) (Fig. 5E, left, 7 days after infection). Strikingly, the total number of infiltrating lymphocytes amounted to 65.3 ± 6.8 million per mouse liver, compared to an average 1 to 2 million in PBS-treated mice (P < 0.01).The foreign-antigenic influenza virus M58-66 (Fig. 1A) was the most immunodominant epitope (1). The peptide stimulation resulted in substantial IFN-γ and modest TNF-α production (33) in the liver-infiltrating CD8+ T cells (Fig. 5E, middle, and Fig. 5F). Most influenza virus-specific IFN-γ+ T cells expressed surface CD107a, which was a surrogate marker for functional cytolytic activity (Fig. 5E, right). The CD8+ T cells specific to HBc18-27 and Epstein-Barr virus (EBV) B259-267 (two other HLA-A2-restricted epitopes from rHBc [Fig. 1A]) were only slightly activated, possibly reflecting competition among HLA-A2-restricted epitopes. Collectively, the foreign-antigen-specific CD8+ T cells (i.e., influenza virus M58-66) represented a dominant and mostly functional population in the liver-infiltrating lymphocytes following rAd/rHBV-based active immunization.
Sustained rHBV gene expression despite fully activated intrahepatic T-cell response.
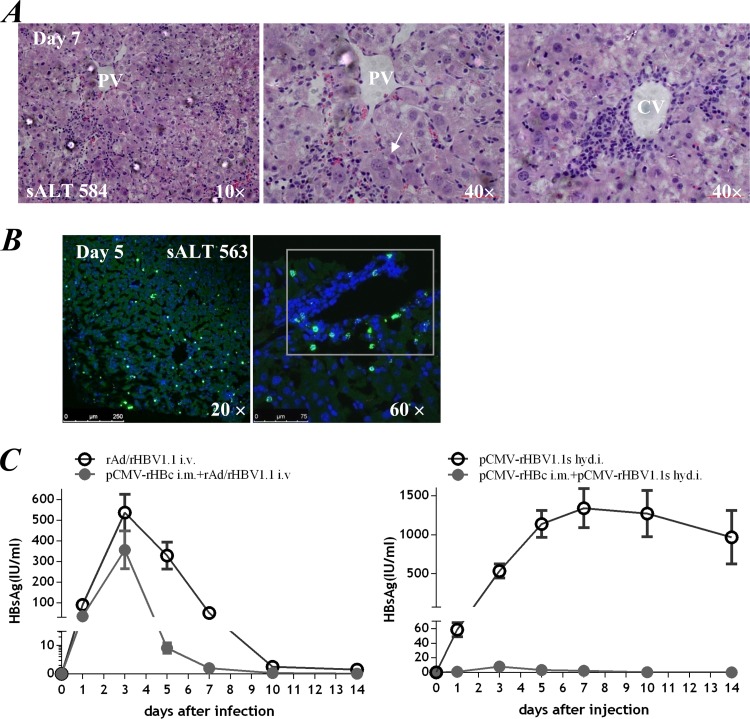
Histological staining revealed a rapidly developing and localized immune response in mouse liver following a rAd/rHBV-based active immunization protocol. Infiltrating lymphocytes were widely distributed or aggregated into inflammatory foci, mostly in the central vein or portal area (Fig. 6A), reminiscent of acute hepatitis. Large areas of necrosis were not observed in liver sections, consistent with the significant but not massive increase in sALT levels (584 ± 39.0 U/liter).
FIG 6.
Active rHBV expression in the presence of fully activated intrahepatic immune response. With prior pCMV-rHBc priming in the periphery, intravenous transduction of rAd/rHBV1.1 (1 × 109 PFU) triggered a vigorous immune response in the livers of HLA-A2 transgenic mice. (A) Hematoxylin-eosin staining of mouse liver sections on day 7 after rAd infection. Arrow, hepatocyte undergoing cytoplasmic condensation and regeneration. sALT activity (mean value) and magnification are indicated. PV, portal vein; CV, central vein. (B) Immunofluorescence for HBsAg (green) in frozen liver sections on day 5 after rAd infection. rHBV expression was distributed throughout the hepatic lobule (left) or within the inflammatory focus (right, boxed). (C) Serum HBsAg kinetics in mice given rAd/rHBV1.1 with (n = 4) or without (n = 4) prior pCMV-rHBc priming (left). rHBV delivered through hydrodynamic injection (hyd.i.) of pCMV-rHBV1.1s (10 μg), which encoded a 1.1-copy rHBV genome driven by a CMV promoter, was used as a control assay (right). The error bars indicate SEM.
Interestingly, active rHBV expression was observed despite the fully activated T-cell response. In frozen liver sections taken 5 days after rAd infection (with prior pCMV-rHBc priming), HBsAg staining was distributed throughout the liver lobules and was also present in inflammatory foci (Fig. 6B), indicating a noncytolytic immune response is involved. Foreign antigen was also detected (data not shown), although it was not completely colocalized with HBsAg-positive cells, possibly due to CMV promoter-induced expression in nonparenchymal cells. Consistent with the findings in liver samples, significant HBsAg antigenemia was observed and remained detectable for at least 10 days after rAd/rHBV1.1 infection. The mice displayed a small reduction in the HBsAg kinetics, compared to that of mice receiving rAd/rHBV1.1 infection alone (Fig. 6C, left). In contrast, when rHBV was delivered through hydrodynamic injection (with prior pCMV-rHBc priming), the HBsAg expression was scarcely detected in serum (Fig. 6C, right) or in the liver (data not shown). This result indicated that the rAd vector persisted in vivo with active transcription, even after the immune response was boosted (12–14), which allowed the rHBV cargo gene to undergo sustained expansion in chronically wtHBV-infected liver tissue.
rAd/rHBV abrogated HBV persistence in rAAV8-HBV-infected mice.
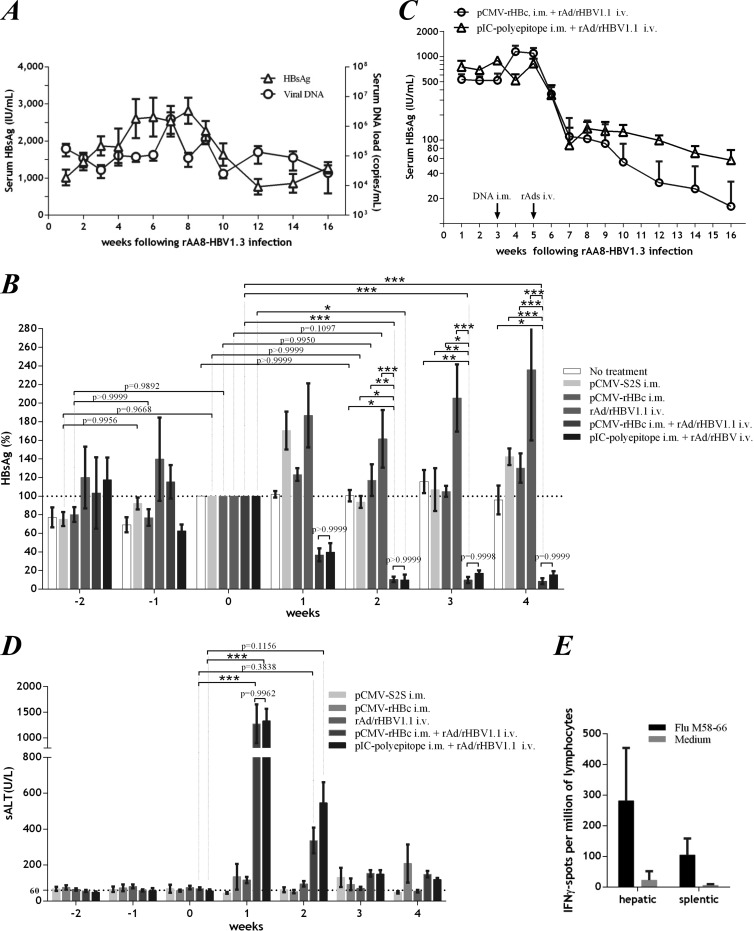
We administered rAAV8-HBV to HLA-A2 transgenic mice as a model of HBV persistence. Following intravenous inoculation of rAAV8-HBV, the mice developed substantial HBsAg antigenemia that plateaued within 3 to 9 weeks (Fig. 7A). Although it declined from week 10, the circulating HBsAg persisted at lower levels for the duration of the experiment (>20 weeks). The HBV DNA load in serum displayed similar kinetics. It is noteworthy that DNA vaccination with pCMV-S2S, a plasmid encoding the middle and small envelope proteins of HBV (34, 35), did not affect HBV persistence (Fig. 7B), indicating a prominent immune tolerance developed in the mouse liver (36).
FIG 7.
rAd/rHBV-based active immunotherapy abolished persistent viral infection in the rAAV8-HBV mouse model. (A) HBsAg and viral DNA persistence in mouse (HLA-A2 transgenic) sera after intravenous administration of 7 × 1010 GEs of rAAV8-HBV (n = 4). (B) Serum HBsAg at the indicated time points in mice with rAAV8-HBV persistence. The mice were not treated (n = 4), immunized with pCMV-S2S (n = 4) or pCMV-rHBc (n = 4) alone (at week −2), infected with rAd/rHBV1.1 (1 × 109 PFU) alone (n = 4), or given rAd/rHBV1.1 (1 × 109 PFU) with prior pCMV-rHBc (n = 8) or pIC-polyepitope (n = 4) priming in the periphery. The results are percentages of the HBsAg level when rAd/rHBV was administered (week 0; arbitrarily defined as 100%). (C) Follow-up of serum HBsAg concentration in rAAV8-HBV mice that underwent rAd/rHBV-based active immunization (1 × 109 PFU of rAd/rHBV1.1) with prior DNA priming by either pCMV-rHBc (n = 4) or pIC-polyepitope (n = 4). Arrows, times at which DNA priming and rAd/rHBV1.1 infection were performed. (D) sALT activities at the indicated time points in groups of mice (as depicted in panel B) that were immunized with pCMV-S2S or pCMV-rHBc alone, infected with rAd/rHBV1.1 (1 × 109 PFU) alone, or received rAd/rHBV1.1 with prior DNA priming by either pCMV-rHBc or pIC-polyepitope. (E) Splenic and hepatic lymphocytes from mice (n = 4) by ex vivo ELISPOT assay. Four weeks after rAd/rHBV1.1 injection (with prior pCMV-rHBc priming), IFN-γ-specific spots were developed after stimulation with influenza virus M58-66. Two-way ANOVA with Tukey's multiple-comparison test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The error bars indicate SEM.
rAd/rHBV-based active immunotherapy was then evaluated in this rAAV8-HBV mouse model. With prior pCMV-rHBc priming, infection with rAd/rHBV1.1 (1 × 109 PFU) led to rapid HBV clearance. A 90% reduction in HBsAg titers was achieved in the mice within 2 weeks (Fig. 7B). Sustained viral clearance was observed in a 16-week follow-up study of these mice (Fig. 7C). In contrast, pCMV-rHBc immunization alone did not abrogate HBV persistence, while rAd infection alone actually increased HBsAg antigenemia over several weeks (Fig. 7B). HBV clearance was also achieved in mice when a pIC-polyepitope plasmid was used for periphery priming before rAd infection (Fig. 7B and C). Unlike pCMV-rHBc, the pIC-polyepitope encoded a foreign polyepitope fusion that was unrelated to HBV. These results demonstrated that the immune response against foreign polyepitope was mainly responsible for viral clearance. A decline in the serum HBV DNA load was also observed in these mice (data not shown).
In conjunction with viral clearance, large increases in sALT activity (1,273 ± 377.5 U/liter, 1 week after rAd infection) (Fig. 7D) were observed in the mice following rAd/rHBV-based active immunization, suggesting a major cytolytic T lymphocyte (CTL) response. Indeed, a T-cell response against the foreign polyepitope (i.e., influenza virus M58-66) was significantly induced in both the liver and spleen (Fig. 7E), but HBV antigen-specific T-cell reactivation was rarely observed, as tested at 4 weeks after rAd infection (data not shown). Nevertheless, anti-HBeAg was identified in all mice at long-term follow-up (as shown in Fig. 7C) after rAd/rHBV-based active immunization, with three out of eight mice positive for anti-HBsAg at 13 weeks after rAd administration. The seroconversion clearly indicated that rAd/rHBV-based active immunization could overcome immune tolerance and eliminate chronic HBV infection in the mouse model.
DISCUSSION
In our strategy, we use preexisting wtHBV in the host liver to expand the antiviral capacity of a replication-defective rAd vector. Ideally, a small dose of rAd/rHBV would be administered for the immunotherapy to achieve high efficiency. However, the T-cell response induced by rAd vector is critically influenced by the viral dose and route of immunization (32). Direct rAd/rHBV infusion, even at 1 × 109 PFU, only marginally activated intrahepatic T-effector cells (Fig. 5C). This result was consistent with a recent report that significant levels of circulating HBV antigens, but no virus-specific CTLs, were seen in mice treated with low doses of rAd vectors carrying wtHBV (9).
To resolve this issue, we used a prime-boost vaccination regimen for active immunization. A characteristic of rAd-based vaccines is that they induce T-cell responses with a protracted contraction phase that is associated with the persistence of low levels of transcriptionally active rAd vector at the site of inoculation and in liver or lymphatic tissues, even after boosted immunization (12–14). In our study, with prior DNA priming, intravenously administered rAd/rHBV attracted a substantial influx of functional T-effector cells to the liver. In the presence of this vigorous T-cell response, active rHBV expression was still observed in the liver and in circulation for at least 1 week (Fig. 6B and C). Similar results have been reported for intravenous administration of rAd/wtHBV following HBsAg-specific DNA vaccination (19). Thus, the rAd vector might allow sustained expansion of an rHBV cargo gene despite a fully activated T-cell response.
rHBV replicates only in wtHBV-infected hepatocytes, which was thought to confer distinct specificity for rHBV-based immunotherapy. However, since the chronic HBV infection spreads to nearly all hepatocytes, targeted elimination of HBV-infected cells might be less critical. In contrast, the simplistic notion of HBV clearance through CTL-activated cell destruction has been replaced with a new paradigm emphasizing cytokine-mediated HBV clearance (26, 28). Virus-specific CTLs do not merely kill target cells; they also release antiviral cytokines that can purge viruses noncytopathically from large numbers of additional infected cells. The foreign-antigen-activated T-cell response has been shown to restrict rHBV expression in a mouse model with limited liver injury (1). This noncytolytic mechanism for rHBV clearance would act on wtHBV because of the intrinsic similarity between rHBV and wtHBV, whether it occurs by epigenetic modulation or by preventing any step of the viral replication.
This updated rationale does not alter the specificity of rAd/rHBV-based immunotherapy. The rAd vectors are predominantly sequestered by the liver after intravenous administration, while foreign-antigen polyepitope would be productively amplified only in the livers of chronically HBV-infected patients. We found that rHBV displayed significant dominance over wtHBV in viral replication (Fig. 4). Thus, rAd-delivered rHBV likely persisted continuously until wtHBV was cleared. Using the in vitro PTH model, we also confirmed the infectivity of rHBV for hepatocytes (Fig. 3). This infectivity is required for rHBV progeny to spread the antiviral activity. In this respect, productive replication of core-rescued rHBV remains essential for the accumulation of sufficient antigen for active immunization.
A major obstacle to in vivo studies of chronic HBV infection is the lack of suitable animal models. We used a recently developed mouse model of rAAV8-HBV persistence in which prominent immune tolerance was established by a defect in the HBV-specific functional T-cell response and a high number of regulatory T cells in the liver (36). This tolerance was fundamentally different from that of HBV transgenic mice, which produce HBV proteins as self-antigens in the liver from birth. DNA vaccination with pCMV-S2S, which has been used in clinical trials, did not abolish HBV persistence in this model (Fig. 7B). It must be noted that the persistent infection may reflect an attribute of the rAAV vector that differs from natural HBV infection. Also, murine hepatocytes were not susceptible to HBV or rHBV infection, and no intracellular viral cycle was established due to the absence of cccDNA. Nevertheless, HBV tolerance was clearly developed in the mouse liver, validating the rAd/rHBV-based active immunization.
The rAd/rHBV-based active immunization effectively eliminated viral persistence in the rAAV-HBV mouse model. Once immune tolerance was overcome, sALT activity was sharply increased, probably due to a synergistic release of rAAV8-HBV-niched CTLs that were previously hyporesponsive in the chronically infected liver. The massive influx of liver-infiltrating lymphocytes, including non-HBV-specific effector T cells, suggests that a bystander effect could also be important in this model of viral clearance.
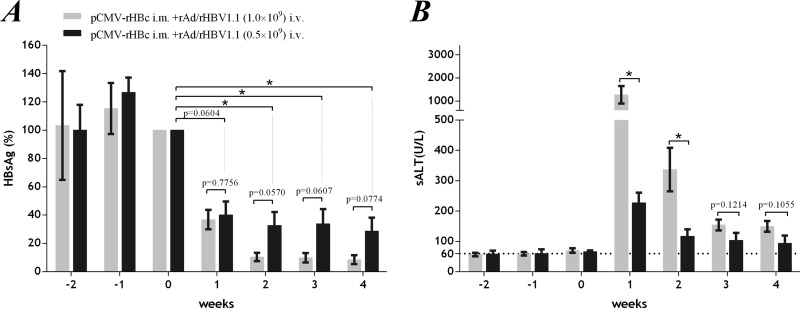
For immunotherapy to be safe, the hepatic inflammation must be managed to avoid severe liver injury. An intrahepatic T-cell response could be induced moderately by adjusting the rAd/rHBV dose and timing of administration rather than administering the virus when the peripherally primed T-cell response is fully activated. In our study, an rAd vector dose of 1 × 109 PFU was routinely used for inoculations, which appeared to target more than 90% of hepatocytes in the mouse liver (9). Ameliorated hepatic injury (sALT; 228 ± 32.9 U/liter; week 1 after rAd infection) was observed in mice given a low dose of rAd/rHBV (0.5 × 109 PFU, with prior pCMV-rHBc priming), which, however, compromised the viral clearance (only 60 to 70% reduction in HBsAg titers within 2 weeks) in the mouse model (Fig. 8). In this regard, an expansion of rHBV is expected to accumulate sufficient antiviral response in chronically wtHBV-infected liver tissue, although this study was limited by use of a murine model not susceptible to HBV infection.
FIG 8.
Ameliorated hepatic injury following rAd/rHBV-based active immunization at a reduced rAd dose in the rAAV8-HBV mouse model. With prior pCMV-rHBc priming in the periphery (week −2), rAAV8-HBV mice were infected with rAd/rHBV1.1 (week 0) at 1 × 109 PFU (n = 8) or a reduced dose of 0.5 × 109 PFU (n = 7). (A) Relative concentrations of serum HBsAg at the indicated time points. The data are expressed as percentages of the HBsAg level when rAd/rHBV was administered (week 0; arbitrarily defined as 100%). (B) sALT activities in mice at the indicated time points. *, P < 0.05. Statistical analysis was performed by Student's t test or two-way ANOVA with Tukey's multiple-comparison test. The error bars indicate SEM.
In summary, the foreign-antigen-activated immunization efficiently transformed chronic liver tolerance to functional anti-HBV immunity in an rAAV8-HBV mouse model. This provided further evidence of the potential of rAd/rHBV-based immunotherapy for the treatment of chronic HBV infection.
ACKNOWLEDGMENTS
This work was supported by grants from the National Science and Technology Major Projects (2008ZX10002007), Natural Science Foundation (30900065 and 31270976), and National Key Basic Research Program of China (2012CB519000). We also thank Roche China for the basic research grant.
We thank Yi Ni for the dot blot assay, Chunhui Yang for the FACS analysis, Demin Yu and Jiehong Jiang for serum HBsAg and viral DNA assays, and Qibing Leng and Xia Jin for constructive discussions.
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Deng Q, Mancini-Bourgine M, Zhang X, Cumont MC, Zhu R, Lone YC, Michel ML. 2009. Hepatitis B virus as a gene delivery vector activating foreign antigenic T cell response that abrogates viral expression in mouse models. Hepatology 50:1380–1391. 10.1002/hep.23150 [DOI] [PubMed] [Google Scholar]
- 2.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, Chisari FV. 2009. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J. Virol. 83:9652–9662. 10.1128/JVI.00867-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825–829. 10.1126/science.284.5415.825 [DOI] [PubMed] [Google Scholar]
- 4.Guo JT, Zhou H, Liu C, Aldrich C, Saputelli J, Whitaker T, Barrasa MI, Mason WS, Seeger C. 2000. Apoptosis and regeneration of hepatocytes during recovery from transient hepadnavirus infections. J. Virol. 74:1495–1505. 10.1128/JVI.74.3.1495-1505.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. 2003. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77:68–76. 10.1128/JVI.77.1.68-76.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y, Yamamoto T, Cullen J, Saputelli J, Aldrich CE, Miller DS, Litwin S, Furman PA, Jilbert AR, Mason WS. 2001. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J. Virol. 75:311–322. 10.1128/JVI.75.1.311-322.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nuriya H, Inoue K, Tanaka T, Hayashi Y, Hishima T, Funata N, Kaji K, Hayashi S, Kaneko S, Kohara M. 2010. Detection of hepatitis B and C viruses in almost all hepatocytes by modified PCR-based in situ hybridization. J. Clin. Microbiol. 48:3843–3851. 10.1128/JCM.00415-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wieland SF, Chisari FV. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 79:9369–9380. 10.1128/JVI.79.15.9369-9380.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang LR, Gabel YA, Graf S, Arzberger S, Kurts C, Heikenwalder M, Knolle PA, Protzer U. 2012. Transfer of HBV genomes using low doses of adenovirus vectors leads to persistent infection in immune competent mice. Gastroenterology 142:1447–1450. 10.1053/j.gastro.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 10.Shayakhmetov DM, Li ZY, Ni S, Lieber A. 2004. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J. Virol. 78:5368–5381. 10.1128/JVI.78.10.5368-5381.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godfrey DI, Uldrich AP, Baxter AG. 2012. NKT cells—an early warning system for HBV infection. Nat. Med. 18:1014–1016. 10.1038/nm.2853 [DOI] [PubMed] [Google Scholar]
- 12.Tatsis N, Fitzgerald JC, Reyes-Sandoval A, Harris-McCoy KC, Hensley SE, Zhou D, Lin SW, Bian A, Xiang ZQ, Iparraguirre A, Lopez-Camacho C, Wherry EJ, Ertl HC. 2007. Adenoviral vectors persist in vivo and maintain activated CD8+ T cells: implications for their use as vaccines. Blood 110:1916–1923. 10.1182/blood-2007-02-062117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finn JD, Bassett J, Millar JB, Grinshtein N, Yang TC, Parsons R, Evelegh C, Wan Y, Parks RJ, Bramson JL. 2009. Persistence of transgene expression influences CD8+ T-cell expansion and maintenance following immunization with recombinant adenovirus. J. Virol. 83:12027–12036. 10.1128/JVI.00593-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett JD, Swift SL, Bramson JL. 2011. Optimizing vaccine-induced CD8(+) T-cell immunity: focus on recombinant adenovirus vectors. Expert Rev. Vaccines 10:1307–1319. 10.1586/erv.11.88 [DOI] [PubMed] [Google Scholar]
- 15.Dong XY, Yu CJ, Wang G, Tian WH, Lu Y, Zhang FW, Wang W, Wang Y, Tan WJ, Wu XB. 2010. Establishment of hepatitis B virus (HBV) chronic infection mouse model by in vivo transduction with a recombinant adeno-associated virus 8 carrying 1.3 copies of HBV genome (rAAN8-1.3HBV). Bing Du Xue Bao 26:425–431 (In Chinese.) [PubMed] [Google Scholar]
- 16.Bayard F, Malmassari S, Deng Q, Lone YC, Michel ML. 2010. Hepatitis B virus (HBV)-derived DRB1*0101-restricted CD4 T-cell epitopes help in the development of HBV-specific CD8+ T cells in vivo. Vaccine 28:3818–3826. 10.1016/j.vaccine.2010.03.026 [DOI] [PubMed] [Google Scholar]
- 17.Walter E, Keist R, Niederost B, Pult I, Blum HE. 1996. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology 24:1–5. 10.1002/hep.510240101 [DOI] [PubMed] [Google Scholar]
- 18.Hong R, Bai W, Zhai J, Liu W, Li X, Zhang J, Cui X, Zhao X, Ye X, Deng Q, Tiollais P, Wen Y, Liu J, Xie Y. 2013. Novel recombinant hepatitis B virus vectors efficiently deliver protein and RNA encoding genes into primary hepatocytes. J. Virol. 87:6615–6624. 10.1128/JVI.03328-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isogawa M, Kakimi K, Kamamoto H, Protzer U, Chisari FV. 2005. Differential dynamics of the peripheral and intrahepatic cytotoxic T lymphocyte response to hepatitis B surface antigen. Virology 333:293–300. 10.1016/j.virol.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 20.Sells MA, Zelent AZ, Shvartsman M, Acs G. 1988. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 62:2836–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uprichard SL, Wieland SF, Althage A, Chisari FV. 2003. Transcriptional and posttranscriptional control of hepatitis B virus gene expression. Proc. Natl. Acad. Sci. U. S. A. 100:1310–1315. 10.1073/pnas.252773599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9913–9917. 10.1073/pnas.0504273102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, Guo H, Pan XB, Mao R, Yu W, Xu X, Wei L, Chang J, Block TM, Guo JT. 2010. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J. Virol. 84:9332–9340. 10.1128/JVI.00918-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puro R, Schneider RJ. 2007. Tumor necrosis factor activates a conserved innate antiviral response to hepatitis B virus that destabilizes nucleocapsids and reduces nuclear viral DNA. J. Virol. 81:7351–7362. 10.1128/JVI.00554-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. 2012. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 122:529–537. 10.1172/JCI58847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guidotti LG, Chisari FV. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65–91. 10.1146/annurev.immunol.19.1.65 [DOI] [PubMed] [Google Scholar]
- 27.Chisari FV. 2000. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am. J. Pathol. 156:1117–1132 10.1016/S0002-9440(10)64980-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feld J, Lee JY, Locarnini S. 2003. New targets and possible new therapeutic approaches in the chemotherapy of chronic hepatitis B. Hepatology 38:545–553. 10.1053/jhep.2003.50389 [DOI] [PubMed] [Google Scholar]
- 29.Melegari M, Wolf SK, Schneider RJ. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J. Virol. 79:9810–9820. 10.1128/JVI.79.15.9810-9820.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni Y, Sonnabend J, Seitz S, Urban S. 2010. The pre-s2 domain of the hepatitis B virus is dispensable for infectivity but serves a spacer function for L-protein-connected virus assembly. J. Virol. 84:3879–3888. 10.1128/JVI.02528-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Freyend MJ, Untergasser A, Arzberger S, Oberwinkler H, Drebber U, Schirmacher P, Protzer U. 2011. Sequential control of hepatitis B virus in a mouse model of acute, self-resolving hepatitis B. J. Viral Hepat. 18:216–226. 10.1111/j.1365-2893.2010.01302.x [DOI] [PubMed] [Google Scholar]
- 32.Holst PJ, Orskov C, Thomsen AR, Christensen JP. 2010. Quality of the transgene-specific CD8+ T cell response induced by adenoviral vector immunization is critically influenced by virus dose and route of vaccination. J. Immunol. 184:4431–4439. 10.4049/jimmunol.0900537 [DOI] [PubMed] [Google Scholar]
- 33.Wherry EJ, Ahmed R. 2004. Memory CD8 T-cell differentiation during viral infection. J. Virol. 78:5535–5545. 10.1128/JVI.78.11.5535-5545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mancini-Bourgine M, Fontaine H, Scott-Algara D, Pol S, Brechot C, Michel ML. 2004. Induction or expansion of T-cell responses by a hepatitis B DNA vaccine administered to chronic HBV carriers. Hepatology 40:874–882. 10.1002/hep.20408 [DOI] [PubMed] [Google Scholar]
- 35.Michel ML, Deng Q, Mancini-Bourgine M. 2011. Therapeutic vaccines and immune-based therapies for the treatment of chronic hepatitis B: perspectives and challenges. J. Hepatol 54:1286–1296. 10.1016/j.jhep.2010.12.031 [DOI] [PubMed] [Google Scholar]
- 36.Dion S, Bourgine M, Godon O, Levillayer F, Michel ML. 2013. Adeno-associated virus-mediated gene transfer leads to persistent hepatitis B virus replication in mice expressing HLA-A2 and HLA-DR1 molecules. J. Virol. 87:5554–5563. 10.1128/JVI.03134-12 [DOI] [PMC free article] [PubMed] [Google Scholar]