ABSTRACT
Within the polyprotein encoded by hepatitis C virus (HCV), the minimum components required for viral RNA replication lie in the NS3-5B region, while virion assembly requires expression of all virus components. Here, we have employed complementation systems to examine the role that HCV polyprotein precursors play in RNA replication and virion assembly. In a trans-complementation assay, an HCV NS3-5A polyprotein precursor was required to facilitate efficient complementation of a replication-defective mutation in NS5A. However, this requirement for precursor expression was partially alleviated when a second functional copy of NS5A was expressed from an additional upstream cistron within the RNA to be rescued. In contrast, rescue of a virion assembly mutation in NS5A was more limited but exhibited little or no requirement for expression of functional NS5A as a precursor, even when produced in the context of a second replicating helper RNA. Furthermore, expression of NS5A alone from an additional cistron within a replicon construct gave greater rescue of virion assembly in cis than in trans. Combined with the findings of confocal microscope analysis examining the extent to which the two copies of NS5A from the various expression systems colocalize, the results point to NS3-5A playing a role in facilitating the integration of nonstructural (NS) proteins into viral membrane-associated foci, with this representing an early stage in the steps leading to replication complex formation. The data further imply that HCV employs a minor virion assembly pathway that is independent of replication.
IMPORTANCE In hepatitis C virus-infected cells, replication is generally considered an absolute prerequisite for virus particle formation. Here we investigated the role that the viral protein NS5A has in both replication and particle assembly using complementation assays and microscopy. We found that efficient rescue of replication required NS5A to be expressed as part of a larger polyprotein, and this correlated with detection of NS5A at sites where replication occurred. In contrast, rescue of particle assembly did not require expression of NS5A within the context of a polyprotein. Interestingly, although only partial restoration of particle assembly was possible by complementation, that proportion that could be rescued benefitted from expressing NS5A from the same RNA being packaged. Collectively, these findings provide new insight into aspects of polyprotein function. They also support the existence of a minor virion assembly pathway that bypasses replication.
INTRODUCTION
Hepatitis C virus (HCV) infects an estimated 170 million individuals worldwide and is one of the leading causes of liver failure and hepatocellular carcinoma. It is an enveloped positive-strand RNA virus with a 9.6-kb genome that encodes a single open reading frame (ORF) flanked by both 5′ and 3′ untranslated regions (UTRs). An internal ribosome entry site (IRES) contained within the 5′ UTR promotes translation of the viral polyprotein (1), the first third of which gives rise to both structural proteins (core, E1, and E2) and a viroporin (p7), while the last two-thirds of the polyprotein encodes nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B (2, 3). Release of the structural proteins from the polyprotein is through the action of host signal peptidase and signal peptide peptidase proteases (3–5). In contrast, maturation of the nonstructural proteins requires two virus-encoded proteases, NS2/3 and NS3 (6–8). NS2/3 is a zinc metalloprotease that operates in cis to cleave at the NS2-NS3 boundary, whereas NS3 is a serine protease that cuts at the remaining boundaries between the NS proteins.
Replication of the viral RNA genome is dependent on both cis-acting RNA elements present within the 5′ and 3′ ends (9–11) and the NS proteins NS3, NS4A, NS4B, NS5A, and NS5B (12). RNA synthesis occurs at discrete endoplasmic reticulum (ER)-derived membrane sites within infected cells that contain high concentrations of NS proteins (13). At resolutions achievable by light microscopy, these assemblies appear as membrane-associated foci (MAF) and under an electron microscope are identified as a collection of membranous structures that are vesicular in nature and that possess either a single membrane or both single and double membranes (13, 14). How these different vesicular structures relate to the site where replication occurs has yet to be elucidated, although more recent data implicate the double membrane vesicles as being the structural component of the replication complex (RC) (14, 15). Knowledge regarding the activities of the NS proteins within the MAF is also incomplete, despite considerable efforts to decipher what these may be. Nonetheless, certain key functions have been ascribed to the different NS proteins and almost certainly relate to their contribution to viral RNA replication within the MAF. As well as containing a protease domain, NS3 possesses a helicase domain critical for replicative function (10). NS4A associates with and helps anchor NS3 to the ER-derived membrane as well as acts as a cofactor for NS3 protease activity (8, 16, 17). Polymerase activity is provided by NS5B (18), and the final two proteins, NS4B and NS5A, are RNA-binding proteins (19, 20) which are thought to play a more structural role in replication complex formation, as their expression as individual proteins induces membrane alterations similar to those seen in cells expressing the entire HCV polyprotein (14, 21). While it is unclear how they orchestrate membrane remodeling, NS4B and NS5A possess amphipathic helices (22–25) and undergo oligomerization/dimerization (26–28), properties implicated in the membrane curvature activities of other proteins (29).
A considerable number of studies support the notion that communication between the RC and sites of virion assembly occurs in much the same way that it does for other positive-strand RNA viruses, and this notion is consistent with a view that nascent RNA generated from the RC can be directed into the maturing virus particle. Cell imaging shows that the RC lies proximal to sites where virus assembly probably occurs, since double-stranded RNA and MAF colocalize with envelope proteins and core-coated lipid droplets (30–32); the latter structures are thought to represent both a repository and a transitionary step in the maturation of core from a monomeric protein to a capsid shell (33, 34). In addition, it has been conclusively demonstrated that mutations in the NS proteins can affect the efficiency of virus particle production independently of any effect that these changes may have on RNA replication (30, 35–48). Such data illustrate the tight link between viral RNA replication and virion assembly.
Arguably one of the most studied RC proteins involved in virus particle assembly is NS5A. This protein associates with membranes as a result of its aforementioned amphipathic helix and contains three domains, the first two of which are necessary for replication (49–51), while the third one is necessary for virus particle formation (35, 41, 47). Numerous activities have been ascribed to NS5A, and these fit with putative functional roles in virus particle assembly. First, it interacts with and coassociates with core on lipid droplets in virus-infected cells, an interaction that appears to be necessary for virus particle formation (30, 35, 41). It also binds to NS2 (52), a nonstructural protein considered to be an essential scaffold for bringing components of the RC together with the HCV envelope proteins and p7 (31, 43, 52). Interestingly, the use of cyclophilin inhibitors that target the NS5A function(s) has suggested a link between viral RNA binding, colocalization to core-coated lipid droplets, and its ability to facilitate virus production (53). Thus, there is the distinct possibility that rather than simply acting as a virus assembly scaffold, NS5A may be involved in shuttling viral RNA during steps leading up to virus particle formation.
Genetic analysis can provide important insight into aspects of replication and virus assembly. Reverse genetic studies have shown that replication of the HCV genome requires NS proteins to be expressed in cis and that most NS protein functions cannot be provided by a helper virus in trans (54, 55). There is also good evidence to suggest that physical linkage between individual NS proteins within the NS3-5B cassette, prior to proteolytic processing, is necessary for certain aspects of replicative function (56–58). However, a select number of replication and virion assembly defects in the NS3-5B-coding region can be trans-complemented. Early studies using genotype 1b replicons demonstrated trans-complementation of certain NS5A mutations, with most but not all concluding that this required expression of NS5A as an NS3-5A polyprotein (54, 59, 60). Consistent with such reports, hyperphosphorylation of NS5A, a posttranslational modification associated with replication competency, also requires expression of NS5A as an NS3-5A precursor (61, 62). The advent of more robust replication systems based on the JFH-1 genotype 2a virus has allowed a reappraisal of the situation. Consequently, other mutations in both NS5A and NS4B that can be trans-complemented using helper virus have been identified (38, 63, 64). Similarly, a virion assembly defect resulting from the deletion of domain 3 in NS5A can be rescued (35).
Given the newly emerging information regarding complementation of NS functions, we wished to examine the role that the polyprotein might play in licensing NS4B and NS5A for replication. We chose to use a combined approach of looking at the ability of different polyprotein constructs to complement defective mutations and in parallel examine the subcellular distribution of NS proteins expressed from the same polyprotein precursors. We also extended our studies to determine the polyprotein requirement for complementation of virion assembly defects in NS5A to gain insight into the nature of the association between replication and virion assembly. Our results are consistent with the NS3-5A precursor being able to facilitate integration of NS proteins into MAF formed during the process of replication. Unexpectedly, they also point to the existence of a minor virion assembly pathway that bypasses the need for RNA replication.
MATERIALS AND METHODS
Cells and viruses.
HepG2, Huh7, and Huh7.5 cells (obtained from M. Harris [University of Leeds, United Kingdom], R. Bartenschlager [University of Heidelberg, Germany], and C. Rice [Rockefeller University, USA], respectively) were maintained in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum (FCS), 1 × nonessential amino acids, 50 U/ml penicillin, and 50 μg/ml streptomycin. Sf9 cells were maintained in TC100 medium (Invitrogen) supplemented with 10% FCS, 50 U/ml penicillin, and 50 μg/ml streptomycin. Recombinant baculoviruses were produced using Invitrogen's Bac-to-Bac system as described previously (65).
DNA constructs.
To generate a bicistronic JFH-1 replicon containing a FLAG epitope in NS5A, PCR using primer pairs JFH(6439-59)–FLAG-JFH(rev) and FLAG-JFH(fwd)–JFH(9477-57) (see Table 1 for primer sequences) were used to amplify two products from the pSGRJFH1luc template (66) (referred to as pSGR-wt for the remainder of this report) that were combined in a second-round PCR and ligated back into pSGR-wt via RsrII and SfiI restriction sites to create pSGR-FLAG. The equivalent green fluorescent protein (GFP)-containing vector was created by amplifying the GFP-coding region from pEGFP-N1 (Clontech) using primers GFP(fwd-AsiSI) and GFP(rev-AsiSI), with the AsiSI sites flanking the FLAG tag sequence in pSGR-FLAG used to facilitate an exchange of NS5A tags, creating pSGR-GFP. To remove the JFH-1-coding region encompassing residues 2419 to 2433, two primer pairs, JFH(7449-70)–Δ2419-2433(rev) and JFH(7803-84)–Δ2419-2433(fwd), were used in a two-step PCR to amplify and combine DNA fragments, with the resultant single DNA being cloned into pSGR-wt via RsrII and BsrGI sites to create pSGR(Δ2419-2433). To introduce the G1911A and S2208I mutations into pSGR-FLAG, NsiI-RsrII DNA fragments containing these mutations were transferred from NS5A GFP-tagged replicons, the construction of which has already been described (38). To generate pSGR(ΔGDD) and pSGR-GND, the RsrII-XbaI fragments from pSGRJFH1neo(ΔGDD) and pSGRJFH1neo(GND), respectively (67), were introduced into pSGR-wt cleaved with RsrII and XbaI.
TABLE 1.
Primers used to generate viral constructs
| Primer name | Sequence (5′–3′)a |
|---|---|
| NS3(fwd) | GCTGCGGCCGCGCCACCATGGCTCCCATCACTGCTTATGCCCAGCA |
| NS4A(fwd) | GCTGCGGCCGCGCCACCATGAGCACGTGGGTCCTAGCTGGAG |
| NS4B(fwd) | GCTGCGGCCGCGCCACCATGGCCTCTAGGGCGGCTCTCATCG |
| NS5A(fwd) | GCTGCGGCCGCGCCACCATGTCCGGATCCTGGCTCCGC |
| NS3(rev) | GCTGCGGCCGCTCAGGTCATGACCTCAAGGTCAGC |
| NS4A(rev) | GCTGCGGCCGCTCAGCATTCCTCCATCTCATCAAAAG |
| NS4B(rev) | GCTGCGGCCGCTCAGCATGGGATGGGGCAGTCC |
| NS5A(rev) | GCTGCGGCCGCCTAGCAGCACACGGTGGTATCGTCCTCCTC |
| NS5B(rev) | GCTGCGGCCGCATCCTACCGAGCGGGGAGTAGGAAGAGG |
| FLAG-JFH(fwd) | GATTACAAGGATGACGATGACAAAGCGATCGCAGGATCTTCATCTGATCAGGTAGAGCTTCAACCTC |
| FLAG-JFH(rev) | CTTTGTCATCGTCATCCTTGTAATCTGCGATCGCGCTACTTCCCTCCAGGTCCGGATCTCCAGGCTCC |
| JFH(6439-59) | CCCTTGCGGCGCCAACATCTC |
| JFH(7449-70) | CCGAATCCGGCGGTCCGACGTC |
| JFH(7803-84) | GAGGCGCTCTTTGATGTTGT |
| JFH(9477-57) | CAGTTAGCTATGGAGTGTACC |
| GFP(fwd-AsiSI) | GGGCGATCGCAATGGTGAGCAAGGGCGAGGAG |
| GFP(rev-AsiSI) | GGGCGATCGCTTGTACAGCTCGTCCATGCC |
| Δ2419-2433(fwd) | CCAGGGGGGGGAGGAGGACGATACCACCGTG |
| Δ2419-2433(rev) | CGTCCTCCTCCCCCCCCTGGGGGGGAGGTTGAAGC |
| Ren(fwd) | GAAGATCTATGACTTCGAAAGTTTATG |
| 2A_NS5A(fwd) | CCCCGGTCCGTCCGGATCCTGGCTCCGCG |
| 2A_NS5A(rev) | CAGGATCCGGACGGACCGGGGTTGGACTCG |
| NS5A(term)_PmeI | GTGTGTTTAAACTCAGCAGCACACGGTGGTATCG |
| NS4B(DVR_fwd) | GAGAGACGGTCCGGCCAGCAGAGCCGCTCTGATC |
| NS4B(DVR_rev) | GAGAGAGTTTAAACCTAGCAGGGGATTGGGCAATCCTCG |
| NS5A(DVR_fwd) | GAGAGACGGTCCGAGCGGCTCTTGGCTGAGAGATG |
Restriction sites used to facilitate cloning are underlined.
For creation of baculovirus mammalian expression constructs, pSGR-wt and, where appropriate, pSGR-GFP were used as the templates for amplification of open reading frames encoding various NS proteins and NS polyproteins using primers NS3(fwd), NS4A(fwd), NS4B(fwd) NS5A(fwd), NS3(rev), NS4A(rev), NS4B(rev), NS5A(rev), and NS5B(rev). These were subsequently cloned into pFBM (68) via NotI, with the resulting vectors being employed in the Bac-to-Bac system to make recombinant baculovirus (see above). The construction of baculoviruses FBMJFH1 containing the core to NS2 region [FBMJFH1(C-NS2)] and AcCAlacZ has already been described (68, 69).
Because of potential stability issues surrounding the duplication of sequence within a single plasmid vector, all replicon construct cDNAs carrying duplicated NS-coding regions were maintained in a low-copy-number plasmid vector, pFK-CMV. This was created by first cloning an AseI-MluI-cut and Pfu-polished cassette from pEGFP-N1 into an SpeI-HindIII-cut and Pfu-polished pFK5.1 backbone (70), followed by removing the GFP-coding region with NheI, polishing with Pfu, and religating the vector. In the first instance, replicon cDNAs from pSGR-wt, pSGR(Δ2419-2433), and pSGR(ΔGDD) were transferred to pFK-CMV using EcoRI and XbaI, creating pFKSGR-wt, pFKSGR(Δ3D5A), and pFKSGR(ΔGDD), respectively. To introduce the G1911A and S2208I mutations in combination with FLAG-tagged NS5A into the second cistron of these vectors, their NsiI-SfiI fragments were exchanged with the equivalent NsiI-SfiI fragment from the relevant pSGR-FLAG vector. To create the initial Renilla-foot and mouth disease virus 2A (FMDV2A)-NS5A cassettes, two primer pairs, Ren(fwd)–2A_NS5A(rev) and 2A_NS5A(fwd)–NS5A(term)_PmeI, were used to PCR amplify DNA fragments from pTNTRen2AJVFF (71) and either pSGR-wt or pSGR(Δ3D5A), respectively. The fragments were combined using a second round of PCR and cloned into pCR-Blunt (Invitrogen), creating pCRB-Ren-2A-NS5A and pCRB-Ren-2A-NS5A(Δ2419-2433). The cassettes from these constructs were then cloned into pFKSGRJFH1luc, pFKSGRJFH1luc(Δ2419-2433), and pFKSGRJFH1luc(ΔGDD) via BglII and PmeI sites to create pRep_R2NS5A/NS3-5B, pRep_R2NS5A(Δ2419-2433)/NS3-5B, pRep-R2NS5A/NS3-5B(Δ2419-2433), pRep_R2NS5A(Δ2419-2433)/NS3-5B(Δ2419-2433), and pRep_R2NS5A/NS3-5B(ΔGDD) (the transcripts from these plasmids are referred to as wt/wt, Δ/wt, wt/Δ, Δ/Δ, and wt/wt(ΔGDD), respectively, throughout the report). To expand the NS protein-coding region within the first cistron of these pRep vectors, a synthetic DNA (Invitrogen) which encoded the NS3-5A polyprotein of JFH-1 with a V5 epitope tag at the end of NS5A was purchased. This synthetic DNA had a range of rare codon usage similar to that of JFH-1 but incorporated a level of degeneracy so as to be 67.5% identical to the original JFH-1 NS3-5A-coding region. To introduce it into the Renilla-FMDV2A cassette system, the Ren-2A-NS5A fragment from pCRB-Ren-2A-NS5A first had to be excised and cloned into LITMUS28 (NEB) via EcoRI sites to create LIT_Ren-2A-NS5A. Exchange of the original NS5A-coding region with that of the synthetic NS3-5AV5-coding region could then be facilitated by RsrII and PmeI sites. Introduction of the synthetic NS4B- and NS5AV5-coding regions into LIT_Ren-2A-NS5A involved PCR amplification of these two fragments from the original vector carrying the NS3-5AV5 sequence using primer pairs NS4B(DVR_fwd)–NS4B(DVR_rev) and NS5A(DVR_fwd)–M13(rev) and cloning of these fragments via RsrII and PmeI sites. Exchange of the Ren-2A-NS cassettes from their respective LITMUS-based vectors with the firefly luciferase ORF within the pFKSGR-FLAG vectors was facilitated by BglII and PmeI sites to generate the pRep_R2NS4B/NS3-5BFLAG, pRep_R2NS5AV5/NS3-5BFLAG, and pRep_R2NS3-5AV5/NS3-5BFLAG series of vectors.
All plasmid vectors were cloned and propagated in either the DH5α or the Stbl2 strain of Escherichia coli grown at 30°C, with the latter strain being used for those constructs containing duplicated sequences.
RNA transcription and electroporation of cells.
Five micrograms of plasmid DNA was linearized with XbaI (Fermentas), polished with mung bean nuclease (NEB), purified by phenol-chloroform extraction, ethanol precipitated, and treated with the RNAsecure reagent (Ambion) according to the manufacturer's recommendations. The DNA was used in a 50-μl reaction mixture containing 40 units T7 polymerase plus associated buffer (Fermentas), 50 units RiboLock RNase inhibitor (Fermentas), and 8 mM recombinant nucleoside triphosphates (Promega). After incubation at 30°C for ≥6 h, 2.5 units RQ1 DNase was added and the reaction mixture was left at 37°C for a further 30 min. RNAs were recovered using RNA Clean & Concentrator-25 spin columns (Zymo Research), and transcript integrity was assessed by gel electrophoresis. Huh7 or Huh7.5 cells were detached by trypsin, washed twice in ice-cold diethyl pyrocarbonate (DEPC)-treated phosphate-buffered saline (PBS), and resuspended at a final density of 1 × 107 cells/ml in DEPC-treated PBS. Four hundred microliters of cells was typically mixed with 2 μg of RNA transcript, transferred to a 0.4-cm-gap electroporation cuvette (VWR), and pulsed using a Bio-Rad Gene Pulser apparatus set at 270 V and 960 μF. However, 0.7 pmol RNAs (equivalent to 2 μg of SGR-wt) were used in the experiments whose results are described in Fig. 5 to compensate for differences in transcript length.
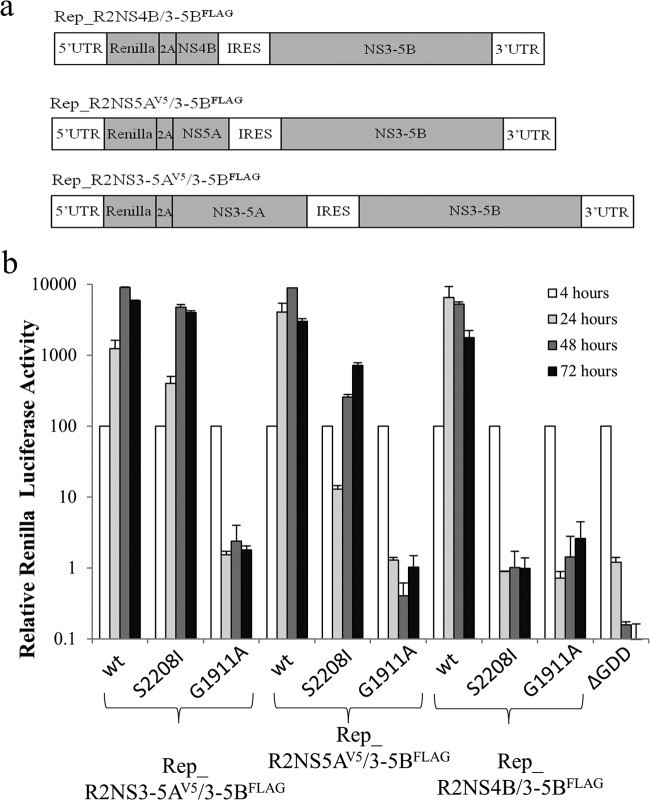
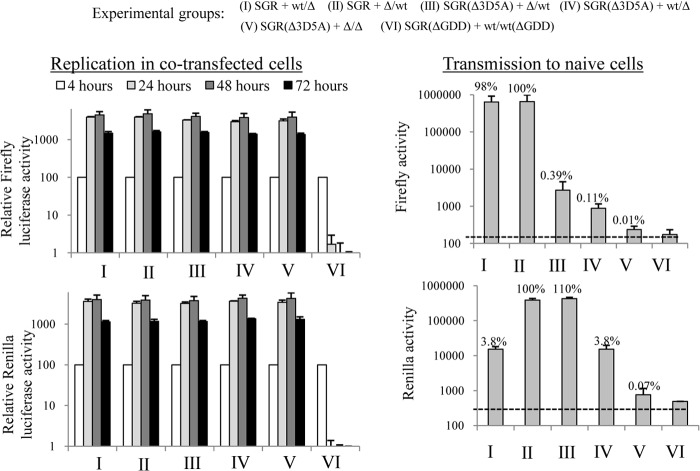
FIG 5.
Intragenomic complementation of defective HCV RNA replication. (a) Schematic representation of bicistronic replicon constructs with the first cistron encoding a Renilla luciferase-FMDV2A reporter fusion protein linked to NS4B (Rep_R2NS4B/3-5BFLAG), NS5A (Rep_R2NS5AV5/3-5BFLAG), and NS3-NS5A (Rep_R2NS3-5AV5/3-5BFLAG). (b) Transient replication assays in Huh7.5 cells. wt, S2208I, and G1911A refer to the NS3-5B polyprotein expressed in the second cistron for each construct; wt denotes wt versions of NS5A and NS4B, S2208I indicates the mutation in NS5A, and G1911A denotes the mutation in NS4B. The ΔGDD construct expresses a nontagged version of NS5A in the first cistron and NS3-5B containing a defective HCV RNA polymerase in the second cistron. Luciferase activity was monitored over 72 h and normalized to that at the 4-h time point (mean ± SD; n = 2).
Transient replication assays.
To assess viral RNA replication, Huh7.5 cells were electroporated with RNA transcripts and then sampled over a 72-h time period using either a luciferase assay system (Promega), a Renilla luciferase assay kit (Biotium), or a dual-luciferase reporter assay system (Promega). In the case of trans-complementation experiments assessing replication, cells were transduced with 5 × 107 PFU/ml of the relevant baculovirus for 4 h and allowed to recover for a further 2 h prior to electroporation.
trans-Encapsidation assays.
Huh7.5 cells were electroporated with RNA transcripts and allowed to recover over a period of 24 h before being transduced with baculovirus. For those experiments where more than one baculovirus was required in any experimental group, cells were transduced with 2 × 107 PFU/ml of each viral construct, such that the total amount of virus applied to the cells was 4 × 107 PFU/ml, with titers being made up where necessary with the control β-galactosidase-expressing baculovirus, AcCAlacZ. For those experiments where only a single baculovirus construct was required per experimental group, transduction was performed with 2 × 107 PFU/ml virus. After 6 h transduction, virus was removed and the cells were allowed to recover for 48 h in fresh growth medium supplemented with 25 mM HEPES (Invitrogen). Supernatants were subsequently collected, clarified by centrifugation at 7,000 × g for 6 min, and transferred onto naive subconfluent Huh7.5 cells, which were then left for a further 48 h before assaying luciferase activity.
Western blot analysis.
Except when phosphatase treatment was required, cells were lysed in radioimmunoprecipitation assay buffer (0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris [pH 8.0]) supplemented with 2× Complete protease inhibitor (Roche), 10 mM NaF, and 1 mM Na3VO4. When phosphatase treatment was required, cells were washed in Tris-buffered saline (TBS; 150 mM NaCl, 50 mM Tris [pH 7.5]) and lysed in TBS supplemented with 1% Triton X-100, and the clarified lysate was supplemented with 0.16 units/μl calf intestinal phosphatase (Fermentas) and incubated at 37°C for 60 min. All lysates were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were blocked with 5% (wt/vol) low-fat dried milk, 0.1% Tween 20 (Merck) in Tris-buffered saline. Antibodies used included sheep polyclonal anti-NS3 and anti-NS5A (a gift from M. Harris), goat polyclonal anti-GFP (Serotec), rabbit polyclonal anti-NS4B, mouse monoclonal anti-NS3 (BioFront) and anti-V5 (a gift from R. Randall, University of St. Andrews), and rat monoclonal anti-FLAG (Biolegend).
Indirect immunofluorescence.
Huh7.5 cells transduced with a relevant baculovirus constructs(s) at 5 × 107 PFU/ml for 4 h were allowed to recover for a further 4 h before electroporation with RNA transcripts and seeding onto glass coverslips. Cells were fixed at 24 h postelectroporation with 4% paraformaldehyde, washed in PBS, permeabilized in saponin buffer (0.1% saponin, 10% FCS, 0.1% sodium azide) for 1 h at 4°C, labeled with rat anti-FLAG primary antibody (Biolegend), and detected with anti-rat-Alexa 568 secondary antibody (Invitrogen). Antibody incubations were performed for 1 h at room temperature, and the incubation mixtures were diluted in saponin buffer, with washes in saponin buffer between steps. Coverslips were counterstained with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Fisher Scientific), washed a final time in PBS, and mounted onto slides with ProLong Gold antifade reagent (Invitrogen). Images were captured using a Leica TCP SP5 confocal microscope. For analysis of NS5A localization from the pRep_R2NS5AV5/NS3-5BFLAG and pRep_R2NS3-5AV5/NS3-5BFLAG replicons, cells were fixed with methanol at −20°C for 30 min. Cells were rehydrated with PBS, followed by blocking with PBS containing 2% FCS (PBS-FCS) for 10 min. Cells were then probed with anti-V5 and anti-FLAG antibodies for 1.5 h at room temperature and then washed extensively with PBS-FCS and finally incubated with anti-mouse-Alexa 594 and anti-rat-Alexa 488 for 1 h at room temperature. All antibody incubations were carried out in PBS-FCS. Cells were washed with PBS-FCS, followed by PBS, rinsed with H2O, and then mounted with Vectashield (Vector Laboratories) that contained DAPI to stain the nuclei. Images were captured with a Zeiss LSM 710 confocal microscope. Up to 30 images, captured as 12-bit data, were taken for each z-stack. Images were processed by blinded deconvolution using Autoquant software and then analyzed by Imaris Bitplane software for scatterplot analysis as well as derivation of Pearson's correlation coefficients (PCC).
Nucleotide sequence accession number.
The synthetic DNA (Invitrogen) which encoded the NS3-5A polyprotein of JFH-1 with a V5 epitope tag at the end of NS5A is available through GenBank accession number KF797814.
RESULTS
Insertion of different tags into NS5A does not disrupt HCV RNA replication.
To both examine the need for NS polyprotein precursors to complement replication defects and determine their subcellular location(s), a system to distinguish between polyproteins expressed from two different RNAs was required. We made use of the ability of NS5A to accommodate insertions (72) and introduced a FLAG tag and a GFP-coding region into a firefly luciferase bicistronic replicon, SGR-wt, creating SGR-FLAG and SGR-GFP, respectively (Fig. 1a). In vitro-synthesized RNA from these constructs as well as from SGR-wt and an NS5B polymerase-defective construct (SGR-GND) was electroporated into Huh7 cells, and luciferase activity was monitored over time to gauge viral RNA replication. Both tagged replicons yielded luciferase values by 48 and 72 h that were indistinguishable from those for SGR-wt (Fig. 1b), indicative of efficient replication. At this time point, both NS5A-GFP and NS5A-FLAG were also readily detected by immunofluorescence (data not shown).
FIG 1.
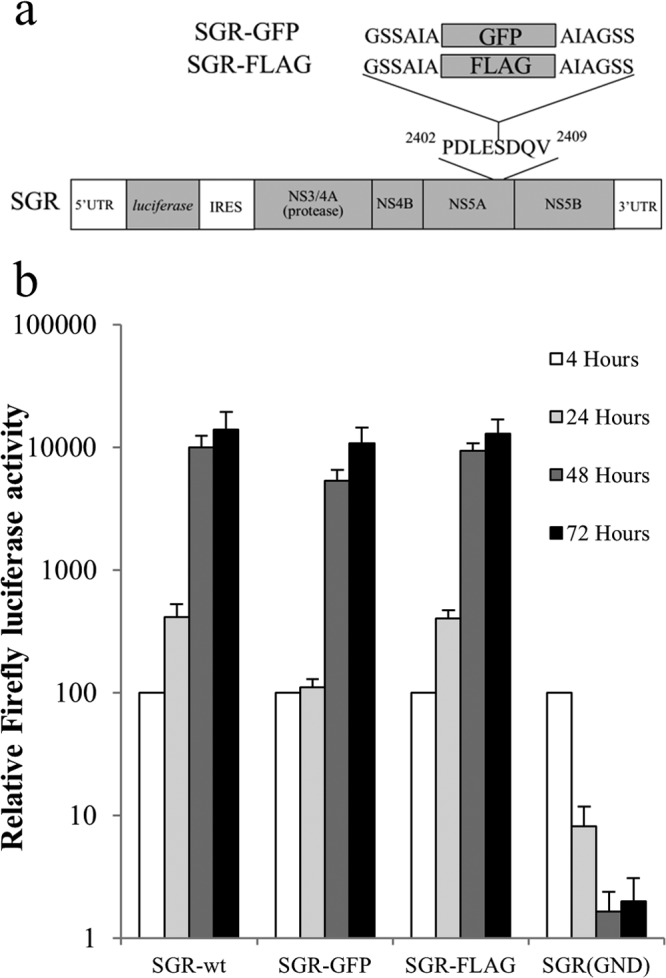
Replication of FLAG- and GFP-tagged bicistronic replicons. (a) Schematic representation of the firefly luciferase bicistronic replicon construct used in this study, showing the locations of the FLAG tag and GFP-coding-region inserts within NS5A. (b) Huh7 cells were electroporated with the indicated SGR RNAs, and luciferase activities were monitored over 72 h (mean ± SD; n = 2).
trans-Complementation of defects in NS4B and NS5A that block replication.
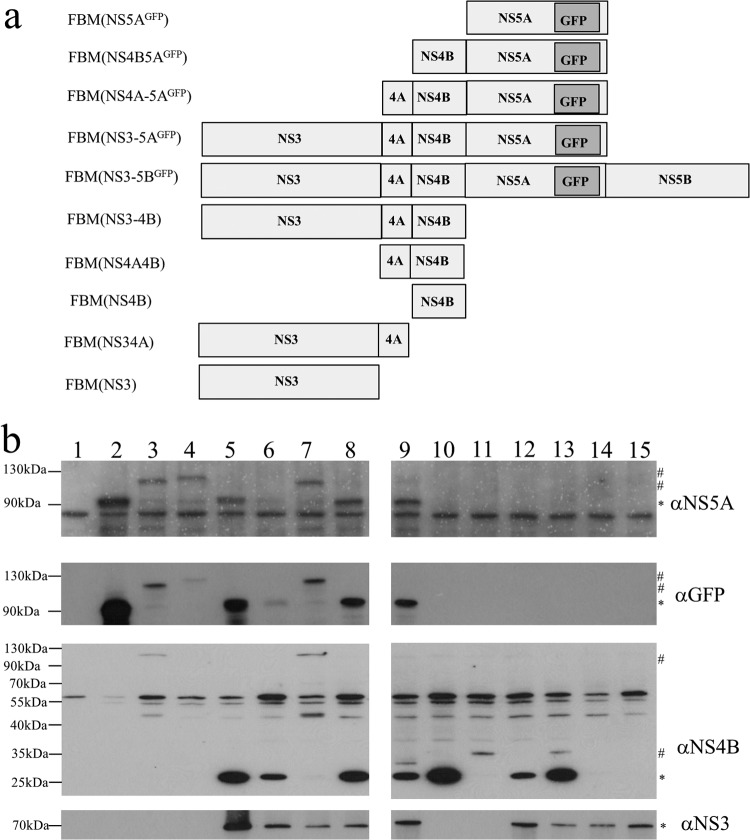
As insertions into NS5A did not disrupt viral RNA replication, SGR-GFP was used as a template to produce a series of mammalian baculovirus constructs expressing a variety of polyprotein precursor open reading frames (ORFs). Since the plan was to study replication-defective mutations in NS4B and NS5A, these combinations included all possible NS4B- and NS5A-containing permutations that could be derived from a NS3-5A precursor as well as NS3-5B (Fig. 2a). Western blot analysis of Huh7.5 cells transduced with these baculoviruses revealed detectable NS5A-GFP expression with the appropriate constructs (Fig. 2b, top, lanes 2 to 9). Typically, constructs expressing larger open reading frames yielded lower levels of expression, such that detecting NS5A-GFP was difficult with construct FBM(NS3-5BGFP), which expressed the largest precursor. Such low-level expression complicated the detection of other NS proteins, where only immunological reagents with lower avidity were available. Therefore, the ability of each baculovirus to express an NS protein(s) was verified by Western blotting of transduced cells of the HepG2 cell line (Fig. 2b, bottom three panels), a cell line that supports high levels of transduction by this virus. Western blot analysis detected NS3, NS4B, and NS5A-GFP, as well as the respective uncleaved polyprotein precursors, in those experimental groups that lacked the viral NS3 protease. As reported previously, cleavage of the NS4B-5A boundary required expression of both NS3 and the NS4A cofactor (Fig. 2b, lanes 7 and 8) (16, 73).
FIG 2.
Expression of the HCV NS proteins by baculovirus vectors. (a) Schematic of the various baculovirus constructs generated. (b) Western blot analysis of NS proteins after transduction of Huh7.5 (top) and HepG2 (bottom three panels) with the baculovirus constructs. Cells were transduced with the following baculovirus vectors: FBM(NS5AGFP) (lane 2), FBM(NS4B-5AGFP) (lane 3), FBM(NS4A-5AGFP) (lane 4), FBM(NS3-5AGFP) (lane 5), FBM(NS3-5BGFP) (lane 6), FBM(NS4B-5AGFP) plus FBM(NS3) (lane 7), FBM(NS4B-5AGFP) plus FBM(NS3-4A) (lane 8), FBM(NS4A-5AGFP) plus FBM(NS3) (lane 9), FBM(NS4B) (lane 10), FBM(NS4A-4B) (lane 11), FBM(NS3-4B) (lane 12), FBM(NS4A-4B) plus FBM(NS3) (lane 13), FBM(NS3) (lane 14), and FBM(NS3-4A) (lane 15). For comparison, cells were transduced with a baculovirus that expressed lacZ (lane 1). *, bands corresponding to the expected size of mature HCV proteins; #, bands corresponding to the size of uncleaved polyprotein products.
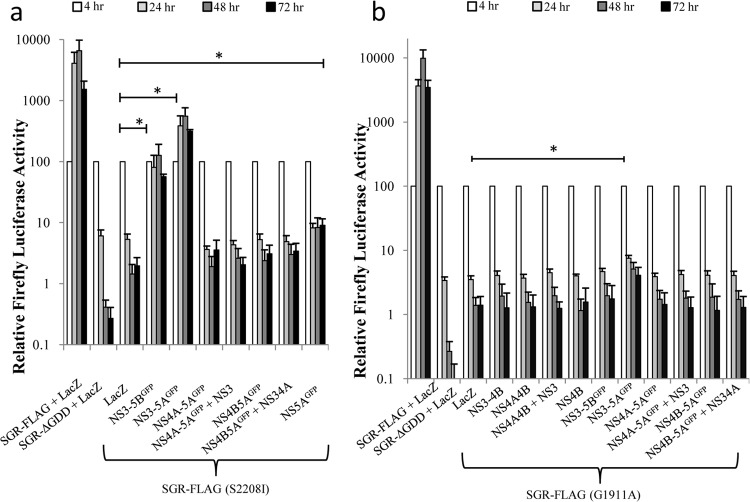
The baculovirus constructs were then assessed for their ability to support replication of SGR-FLAG, which had been modified to contain a lethal mutation in either NS4B (G1911A) or NS5A (S2208I) (38). For the NS5A mutant, efficient trans-complementation was observed using FBM(NS3-5AGFP) and FBM(NS3-5BGFP) (Fig. 3a). No other baculovirus construct gave detectable trans-complementation, except for FBM(NS5AGFP), which produced a slight but statistically significant increase in luciferase activity compared to that of the lacZ-expressing baculovirus (Fig. 3a) from 24 h onwards. Compared to the efficient trans-complementation of the NS5A defect, the ability to trans-complement the NS4B mutation was much reduced, and only FBM(NS3-5AGFP) gave a significant increase in luciferase activity above that seen using the control lacZ-expressing baculovirus (Fig. 3b). We could not exclude the possibility that certain NS4B-containing precursors might have also complemented the G1911A mutation, as it was not possible to achieve NS4B expression levels comparable to the level seen for FBM(NS3-5AGFP) in all transduced groups. However, at least in the case of FBM(NS4B), the level of NS4B expression did match that of FBM(NS3-5AGFP), demonstrating that trans-complementation of the G1911A mutation by NS4B required the protein to be expressed as part of a larger precursor.
FIG 3.
trans-Complementation of defective HCV RNA. Huh7.5 cells were transduced with baculoviruses expressing the indicated HCV NS proteins or LacZ for 4 h, incubated for a further 2 h, and then transfected with SGR-FLAG (S2208I) (a) and SGR-FLAG (G1911A) (b) RNAs. Luciferase activities were monitored over 72 h (mean ± SEM). *, significant differences (P < 0.05) in luciferase activity at 72 h posttransfection (n ≥ 3; 2-tailed t test).
Localization of baculovirus-expressed NS5A within replicon cells.
Taking advantage of the different NS5A tags in the constructs, baculoviruses expressing NS5A-GFP were used to transduce Huh7.5 cells 6 h prior to transfection with replication-competent SGR-FLAG, such that the onset of expression from the two constructs would be similar. The extent to which baculovirus- and replicon-derived NS5A colocalized was then examined at 24 h posttransfection. Expression of NS5A alone or as a component of an NS4B-5A or NS4A-5A polyprotein with or without coexpression of NS3-4A and NS3, respectively, resulted in very limited colocalization with replicon-derived NS5A, with the GFP distribution being quite diffuse in nature, in contrast to the punctate signal given by FLAG-NS5A. On the other hand, expression of NS3-5A resulted in the formation of GFP foci that were frequently coincident with the NS5A foci expressed from the replicon (Fig. 4). This pattern of the NS5A-GFP distribution was also similar to that seen in cells lacking the replicon (data not shown).
FIG 4.
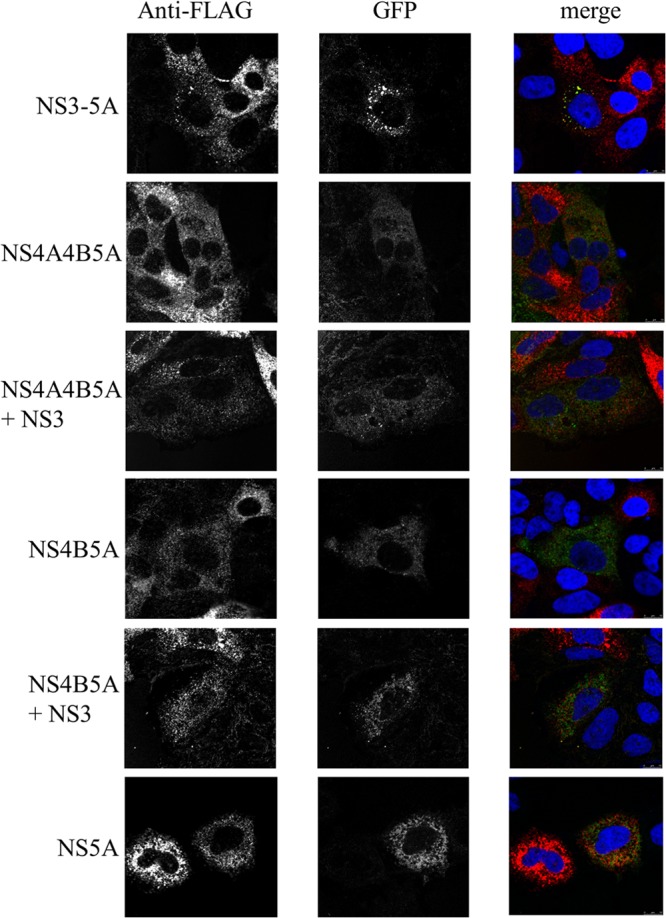
Colocalization of NS5A species expressed from SGR-FLAG RNA and baculovirus vectors. Huh7.5 cells were transduced with baculoviruses expressing the indicated NS proteins for 4 h, incubated for a further 2 h, and then transfected with SGR-FLAG RNA. Cells were fixed at 24 h posttransfection, labeled with anti-FLAG (red), and analyzed by confocal microscopy; the panels labeled as GFP represent NS5A-GFP expressed from the various baculovirus vectors.
Intragenomic complementation of replication.
The trans-complementation and colocalization data were consistent with a model in which NS3-5A allowed NS proteins to be integrated into viral MAF. As translationally active viral RNA has been reported to be closely associated with MAF (74), we were interested to determine whether replicative functions could be complemented by a second NS protein-coding cistron within the RNA that was required to be rescued. Therefore, the first cistron in the SGR-FLAG series of replicons was modified to encode a Renilla luciferase-FMDV2A fusion protein linked to NS4B, NS5A, or NS3-5A, creating a series of replicons referred to as Rep_R2NS4B/3-5BFLAG, Rep_R2NS5AV5/3-5BFLAG, and Rep_R2NS3-5AV5/3-5BFLAG, respectively (Fig. 5a). For all constructs lacking replication-defective mutations in the second cistron, replication was readily detected, as evidenced by increased levels of luciferase activity over 72 h (Fig. 5b). However, on the basis of the initial increase in luciferase activity over the first 24 h, there appeared to be a spectrum of replication efficiency (Rep_R2NS4B/3-5BFLAG > Rep_R2NS5AV5/3-5BFLAG > Rep_R2NS3-5AV5/3-5BFLAG) that reflected a correlation between an increase in the length of the sequence in the first cistron and a lower replication competence. For those constructs carrying replication-defective mutations in the second cistron, a variety of replicative profiles was seen. Rep_R2NS3-5AV5/3-5BFLAG(S2208I) had a replication efficiency almost identical to that of Rep_R2NS3-5AV5/3-5BFLAG, indicating that complementation of the S2208I mutation in NS5A was readily mediated by NS3-5A, as described above. Rep_R2NS5AV5/3-5BFLAG(S2208I) also exhibited quite robust levels of replication. Although the luciferase values were lower than those seen for Rep_R2NS3-5AV5/3-5BFLAG(S2208I), luciferase values at 72 h were no more than 6-fold less than those seen for any of the other replication-competent constructs. In contrast, complementation of the G1911A mutation could not be detected by expressing NS4B either as a single protein or as part of an NS3-5A polyprotein. These data indicate that, unlike the situation with NS5A, there was no added benefit of expressing a second functional copy of NS4B within the RNA to be rescued.
Localization of NS5A expressed from replicons encoding two copies of NS5A.
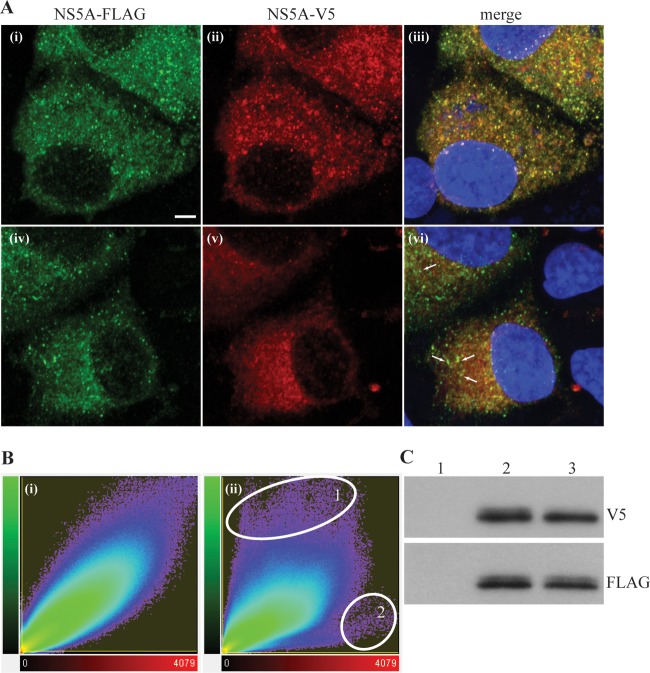
Transduction of Huh7.5 cells is relatively inefficient, and thus, demonstrating colocalization of NS5A expressed from baculoviruses with that produced by HCV replicons is relatively inefficient; our results consistently yielded low numbers of cells that coexpressed NS5A from both expression systems (typically, 2 to 3% of the cell population). Moreover, the relative abundance of NS5A produced from baculoviruses and replicons in the same cell was often dissimilar, which made any quantitative assessment of colocalization difficult. Expression of differentially tagged NS5A from Rep_R2NS5AV5/3-5BFLAG and Rep_R2NS3-5AV5/3-5BFLAG overcame such problems and provided an opportunity to quantitatively evaluate the extent of colocalization of NS5A expressed either as an individual polypeptide or as a component of the NS3-5A polyprotein. First, Western blot analysis of Huh7.5 cells electroporated with RNAs from Rep_R2NS5AV5/3-5BFLAG and Rep_R2NS3-5AV5/3-5BFLAG revealed that both replicon RNAs generated equivalent amounts of hyperphosphorylated and basally phosphorylated FLAG-tagged NS5A from the NS3-5B replicase unit in each RNA (Fig. 6C, bottom, lanes 2 and 3). The NS3-5A polyprotein in Rep_R2NS3-5AV5/3-5BFLAG also produced the two phosphorylated forms of NS5A, as detected by V5 antibody (Fig. 6C, top, lane 2). In contrast, Rep_R2NS5AV5/3-5BFLAG RNA expressed only basally phosphorylated V5-tagged NS5A (Fig. 6C, top, lane 3); this is consistent with the need to express NS5A as part of an NS3-5A precursor to generate the hyperphosphorylated form of the protein. However, there were comparable levels of NS5A produced from the first cistron in Rep_R2NS5AV5/3-5BFLAG and Rep_R2NS3-5AV5/3-5BFLAG. Cells electroporated with either Rep_R2NS5AV5/3-5BFLAG or Rep_R2NS3-5AV5/3-5BFLAG RNA were then examined by confocal microscopy using anti-V5 and anti-FLAG antibodies. A series of z-stacks was collected from cells expressing both tagged forms of NS5A and processed by deconvolution, followed by image reconstruction (Fig. 6A). Visual inspection of the images showed V5- and FLAG-tagged NS5A proteins from Rep_R2NS3-5AV5/3-5BFLAG at foci, which putatively represent MAF (Fig. 6A, panels i to iii). FLAG-tagged NS5A from Rep_R2NS5AV5/3-5BFLAG was also found at foci (Fig. 6A, panel iv). In contrast, V5-tagged NS5A from this construct gave only a reticular pattern, consistent with a distribution at the ER membrane (Fig. 6A, panel v). To provide an unbiased, statistical approach for assessing colocalization, scatterplot analysis was applied to the z-stacks taken from the cells shown in Fig. 6A and other cells electroporated with either Rep_R2NS5AV5/3-5BFLAG or Rep_R2NS3-5AV5/3-5BFLAG RNA (Fig. 6B). This approach revealed that, for Rep_R2NS3-5AV5/3-5BFLAG, fluorescent intensities for V5- and FLAG-tagged NS5A followed a linear relationship and gave a PCC value of 0.93. This indicates almost complete colocalization between NS5A expressed from NS3-5A and NS3-5B polyprotein precursors, even at high fluorescent intensities that are typically found at MAF. Similar analysis of the two forms of NS5A expressed by Rep_R2NS5AV5/3-5BFLAG showed a different scatterplot profile, in which there was evidence of colocalization, but to a lesser extent for the high fluorescent intensities produced by FLAG-tagged NS5A. Here, there was a shift in the fluorescence toward the y axis (circle 1 in Fig. 6B, panel ii), indicating that FLAG-tagged NS5A at foci colocalizes to a lesser extent with V5-tagged NS5A. A reduction in the extent of colocalization was also reflected in a lower PCC value of 0.85. From these data, we conclude that NS5A expressed from a polyprotein can integrate more efficiently with MAF than NS5A expressed as an individual component.
FIG 6.
Colocalization of NS5A expressed from the intragenomic replicons. Huh7.5 cells were electroporated with intragenomic replicon RNAs and examined at 72 h by confocal microscopy (A, B) or Western blot analysis (C). (A) Cells were fixed with methanol and then probed with antibodies that recognize the V5 and FLAG tags. Cells were electroporated with Rep_R2NS3-5AV5/3-5BFLAG (i to iii) and Rep_R2NS5AV5/3-5BFLAG RNAs (iv to vi). Images represent reconstructions of z-stacks, each consisting of 30 slices. White arrows in panel vi, foci containing FLAG-tagged but not V5-tagged NS5A. Bar, 20 μm. (B) Scatterplot analysis of fluorescent intensity for FLAG-tagged (green) and V5-tagged (red) NS5A. Histogram plots are shown for the cells in panel A electroporated with Rep_R2NS3-5AV5/3-5BFLAG (i) and Rep_R2NS5AV5/3-5BFLAG (ii). Circle 1 in panel ii, high-intensity voxels for FLAG-tagged NS5A, represented by foci that show colocalization only with low- to medium-intensity voxels for V5-tagged NS5A; circle 2 in panel ii, background staining by the V5 antibody in the lower-left region of panels v and vi from panel A. (C) Western blot analysis of cell lysates electroporated with Rep_R2NS3-5AV5/3-5BFLAG RNA (lane 2) and Rep_R2NS5AV5/3-5BFLAG RNA (lane 3); the cell lysate in lane 1 is from mock-electroporated cells. Lysates were probed with V5 (top) and FLAG (bottom) antibodies.
trans-Complementation of virion assembly.
NS5A plays a critical role in both replication and virion assembly and has been suggested to facilitate the interaction between replication complexes and virion assembly factories (30, 35, 41). Furthermore, the RNA-binding properties of NS5A (20, 75) have led to the suggestion that it may act to chaperone newly synthesized RNA between sites of replication and sites of virion assembly. Given that NS5A expressed from a polyprotein can target MAF and complement replication defects, we wished to examine whether complementation of virion assembly defects in NS5A showed a similar dependency on polyprotein expression. To address this question, a further series of baculovirus constructs expressing nontagged versions of NS3-5A and NS5A was created since the location of the tag interfered with virion assembly (data not shown). Huh7.5 cells were electroporated with SGR-wt or SGR(Δ3D5A), a modified version of SGR-wt lacking residues 2419 to 2433 within NS5A, which are necessary for virion assembly (47). We assessed the ability of NS5A to complement virion assembly using a trans-encapsidation assay (Fig. 7c). As expected, transfer of trans-encapsidated SGR-wt to naive recipient Huh7.5 cells occurred when donor cells were transduced with FBMJFH1(C-NS2), a baculovirus that provided the structural and nonstructural proteins necessary for virion assembly; in contrast, trans-encapsidation was not detected for donor cells transduced with a lacZ-expressing baculovirus. Similarly, transfer of SGR(Δ3D5A) from donor cells was not evident following transduction with FBMJFH1(C-NS2) alone but was detected by cotransduction with another baculovirus expressing NS5A. Unlike the results obtained with trans-complementation of replication by NS5A expressed from different constructs, complementation of packaging was similar for NS5A expressed either as an individual polyprotein or as part of an NS3-5A precursor. Even allowing for the differences in NS5A expression levels between the two baculovirus constructs (Fig. 7a), it was apparent that trans-encapsidation was less dependent on expression of NS5A as part of a polyprotein than complementation of defective NS5A in replication assays. A further notable feature was the limited extent to which virion assembly could be complemented, as transfer of SGR(Δ3D5A) to recipient cells was ∼0.5% of that seen for SGR-wt. This reduced transfer of infectivity was not due to replication differences between the two replicons, as both exhibited comparable levels of replication (Fig. 7b).
FIG 7.
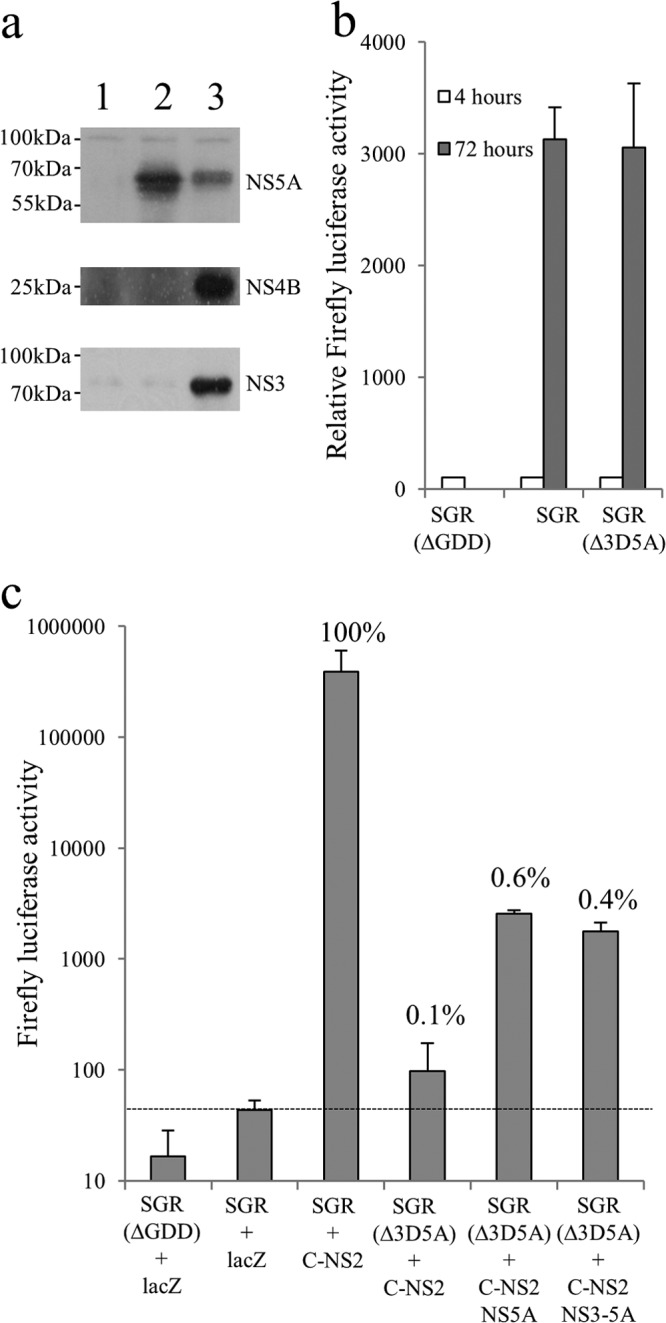
trans-Complementation of NS5A virion assembly functions. (a) Expression of NS proteins in Huh7.5 cells at 24 h postransduction with baculoviruses encoding LacZ (lane 1), NS5A (lane 2), and NS3-5A (lane 3). (b) Transient replication of SGR, SGR(Δ3D5A), and SGR(ΔGDD) following transfection into Huh7.5 cells. Luciferase activity was measured at 4 and 72 h (mean ± SD; n = 2). (c) Transmission of replicons from donor to recipient Huh7.5 cells by trans-encapsidation assay. Cells were electroporated with the indicated SGR RNAs and then transduced after 24 h with baculovirus expressing the HCV core to NS2 region (C-NS2) alone or with baculoviruses expressing NS5A or NS3-5A for a further 6 h. At 48 h postransduction, supernatants were harvested and used to inoculate naive Huh7.5 cells. Luciferase activities were assayed at 48 h after inoculation of cells (mean ± SD; n = 2). Baculovirus expressing LacZ was used as a negative transduction control. For the purpose of comparison, values show the percent transmission compared to that observed for SGR-wt plus C-NS2.
Intragenomic complementation of virion assembly defects.
Given that expression of NS5A alone was able to support replication when produced from the same RNA being rescued, it was possible that a similar phenomenon might occur for packaging. Therefore, a series of replicons expressing a nontagged NS5A both in the first cistron as a Renilla-FMDV2A-NS5A fusion protein and in the second cistron as a functional NS3-5B polyprotein was constructed. Sequences encoding residues 2419 to 2433 in NS5A, which are necessary for efficient virion assembly, were removed from both cistrons (Δ/Δ), the first cistron (Δ/wt), the second cistron (wt/Δ), or neither cistron (wt/wt). Western blot analysis of cells transfected with these replicons revealed the presence of up to three distinct NS5A species (Fig. 8a). On the basis of the assumption that NS5A expressed from the first cistron would not be hyperphosphorylated, as was observed for Rep_R2NS5AV5/3-5BFLAG, it seemed likely that the middle species represented both basal phosphorylated wild-type (wt) NS5A and hyperphosphorylated NS5A(Δ2419-2433), while the upper and lower bands indicated hyperphosphorylated wt NS5A and basal phosphorylated NS5A(Δ2419-2433), respectively. Consistent with this notion, these species could be further resolved into 1 band (Δ/Δ and wt/wt) or 2 bands (Δ/wt and wt/Δ) upon prior phosphatase treatment of the cell lysate. The intensity of the two bands in the Δ/wt and wt/Δ lanes suggested only a small difference in the amount of NS5A expressed from the two cistrons.
FIG 8.
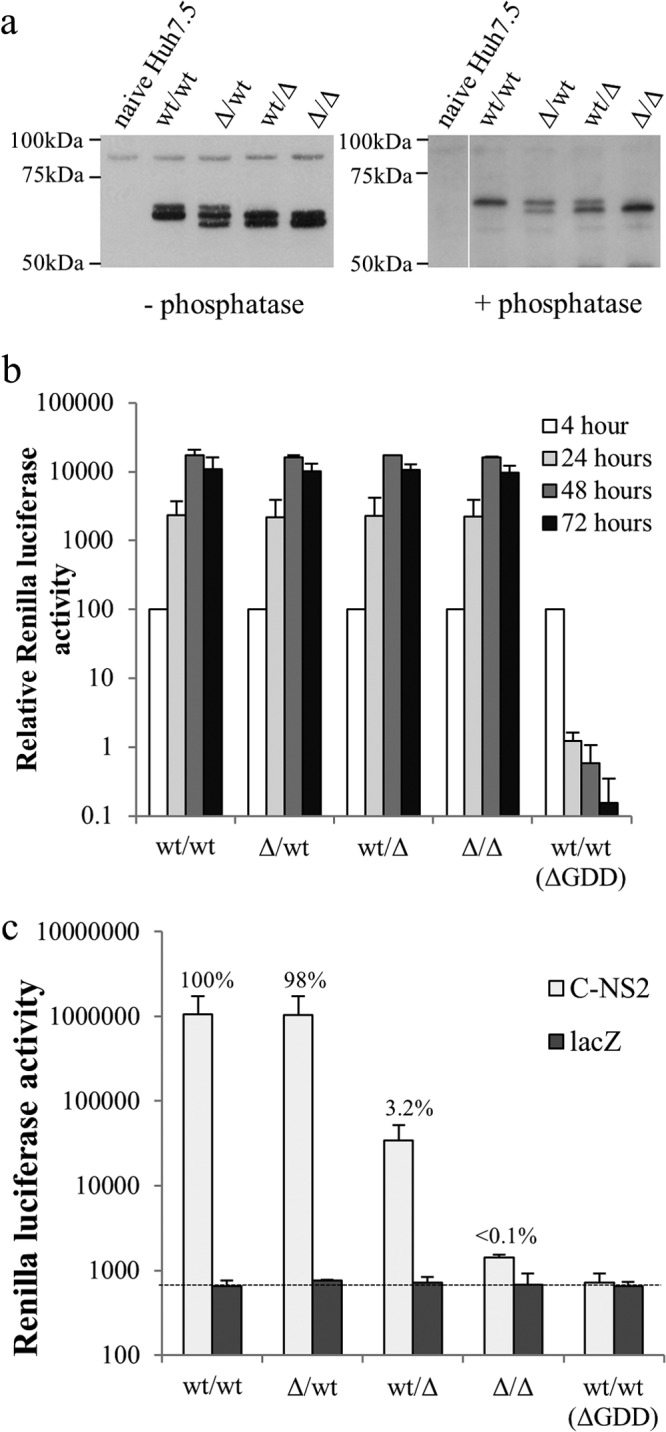
Intragenomic complementation of defective virion assembly. (a) Western blot analysis of lysates from Huh7.5 cells electroporated with intragenomic replicons expressing NS5A only in the first cistron and engineered to contain the deletion of residues 2419 to 2433 in both cistrons (Δ/Δ), the first cistron alone (Δ/wt), the second cistron alone (wt/Δ), or neither cistron (wt/wt). Cells were harvested at 48 h posttransfection. Lysates were either untreated (left) or treated with phosphatase (right) prior to probing with a sheep anti-NS5A polyclonal antibody. (b) Replication of RNA from intragenomic constructs following electroporation into Huh7.5 cells. Luciferase activity was monitored over 72 h, with the values normalized to those at the 4-h time point (mean ± SD; n = 2). (c) Transmission of intragenomic replicons from donor to recipient Huh7.5 cells by trans-encapsidation assay. Cells were electroporated with the indicated intragenomic RNAs and then transduced after 24 h with baculoviruses expressing either the HCV core to NS2 region (C-NS2) or LacZ for a further 6 h. At 48 h postransduction, supernatants were harvested and used to inoculate naive Huh7.5 cells. Luciferase activities were assayed at 48 h after inoculation of cells. Data shown are luciferase values assayed at 48 h after inoculation of cells (mean ± SD; n = 2). For the purpose of comparison, values showing the percent transmission compared to that observed for the wt/wt plus C-NS2 group are included.
Huh7.5 cells transfected with these constructs were then used in a trans-encapsidation assay to determine whether NS5A from the different cistrons supported virion assembly (Fig. 8c). The Δ/Δ construct resulted in levels of luciferase activity in the recipient cells comparable to that seen for the control wt/wt construct lacking polymerase activity [wt/wt(ΔGDD)], whereas luciferase activity was much higher in the other experimental groups, confirming that trans-encapsidation required functional NS5A. In the case of the wt/wt and Δ/wt replicons, virion assembly was almost identical, indicating that that the defective NS5A expressed from the first cistron did not inhibit packaging. In contrast, luciferase activities in recipient cells seen for the wt/Δ construct were only 3% of those seen for the wt/wt and Δ/wt constructs. These differences could not be attributed to replication competence, as the luciferase activities in the transfected donor cells were comparable for all constructs (Fig. 8b). We conclude that expression of NS5A alone within the RNA being rescued does facilitate virion assembly, but similar to the results of the trans-complementation experiments shown in Fig. 7c, the efficiency of rescue is low.
trans-Complementation of the virion assembly defect using a helper replicon.
The results obtained so far indicated that expression of NS5A alone allowed rescue of only a proportion of the virion assembly. More intriguingly, the trans-complementation data for packaging hinted that expression of NS5A within the context of an NS3-5A polyprotein did not extend its capacity to support virion assembly in the same manner as for replication. To further examine the extent to which the assembly defect in NS5A could be complemented, the SGR-wt and SGR(Δ3D5A) replicons were cotransfected with the Δ/Δ, wt/Δ, or Δ/wt replicon and a trans-encapsidation assay was performed to assess whether the latter constructs would rescue the former constructs (Fig. 9). Cotransfection of SGR(Δ3D5A) with the wt/Δ construct gave inefficient transmission of the former replicon, with firefly luciferase activities being 1,000-fold lower than those obtained by cotransfection of SGR-wt with the wt/Δ construct (i.e., when the SGR replicon expressed its own NS5A that was functional for packaging). Nonetheless, the firefly luciferase activity seen in recipient cells was greater than that seen for cells cotransfected with SGR(Δ3D5A) and the Δ/Δ construct. This confirmed that NS5A expressed alone from the wt/Δ construct could trans-complement SGR-based virion assembly. Surprisingly, cotransfection of SGR(Δ3D5A) with the Δ/wt construct rather than the wt/Δ construct gave only a 3- to 4-fold increase in transmission, with firefly luciferase levels in the recipient cells being 250-fold lower than those obtained by cotransfection of SGR-wt with the Δ/wt construct. Equally remarkable was the fact that transmission of Renilla activity from the wt/Δ construct remained approximately 3% of that from the Δ/wt construct in these experiments, which was considerably higher than the extent to which virion assembly of the Δ2419-2433 mutation in SGR(Δ3D5A) could be restored by trans-complementation with either the wt/Δ or Δ/wt construct. None of the observed differences in transmission between the experimental groups were due to differences in replication, as the Renilla and firefly luciferase activities in donor cells electroporated with replication-competent constructs were identical between the different experimental groups. The differences in transmission seen between the experimental groups were also not due to competition for limiting host factors involved in virion assembly, as neither the packaging-competent Renilla-based replicon (Δ/wt) nor the packaging-competent firefly-based replicon (SGR-wt) experienced a drop in transmission when competing with their packaging-defective and packaging-competent counterparts. We conclude that the Δ2419-2433 virion assembly defect is predominantly refractile to trans-complementation, even when the rescuing NS5A is expressed from a functional NS3-5B polyprotein that effectively supported replication and packaging of a helper RNA.
FIG 9.
trans-Complementation of defective virion assembly by cotransfection with replicon RNA. Huh7.5 cells were coelectroporated with either SGR-wt or SGR(Δ3D5A) in combination with one of the three intragenomic complementation replicons, Δ/wt, wt/Δ, or Δ/Δ. Cotransfection of cells with SGR(ΔGDD) and wt/wt(ΔGDD) was included as a negative control for both replication and transmission of replicon RNAs to recipient cells. (Left) RNA replication was assessed by measuring firefly and Renilla luciferase activities over 72 h; data were normalized to those at the 4-h time point (mean ± SD; n = 2). (Right) Transmission of infectivity was assessed by transducing all experimental groups with FBMJFH1(C-NS2) and performing a trans-encapsidation assay. Data represent luciferase values from recipient cells 48 h after inoculation with donor cell supernatants (mean ± SD; n = 2). For the purpose of comparison, values showing the percent transmission compared to that observed for the SGR-wt plus Δ/wt group are included.
DISCUSSION
It is well established that the HCV NS proteins are essential for both HCV RNA replication and virion assembly. In terms of replication, the NS3-5B region is sufficient to sustain viral RNA synthesis (12), and each of the proteins derived from this polyprotein is also known to contribute to virus production (30, 35–42, 44–48). Our previous studies have shown that the kinetics of cleavage at boundaries between the NS proteins plays a critical role in regulating replication (56). Therefore, the viral proteins do not operate solely as individual components that are capable of interacting with each other, the viral genome, and cellular factors; their ability to function is regulated at other levels, where the context of expression is a further key factor. Here, we have explored this question in more detail by examining how the NS proteins contribute to RNA replication and virion assembly either as components of a viral polyprotein or as individual polypeptides, using complementation as a model system. We have also developed a novel strategy for examining complementation by expressing wt and mutant forms of the NS proteins from the same RNA molecule; we refer to this approach as intragenomic complementation.
To examine trans-complementation of RNA replication, we tested previously identified mutations in NS4B (G1911A) and NS5A (S2208I) that abrogate genome synthesis (38). In a previous report, we demonstrated that the S2208I mutation could be readily trans-complemented by expressing a wt form of NS5A from the NS3-5B replicase unit in a second helper replicon. Restoration of replication for NS4B with the G1911A mutation was possible by use of the same helper replicon, although replication levels were considerably diminished compared to the recovery achieved for NS5A with the S2208I mutation (38). We have extended this analysis to determine whether the ability to trans-complement these defective mutations was possible by expressing wt NS4B and NS5A either as part of a polyprotein or as distinct polypeptides. We found that the G1911A mutation was rescued to a very limited extent by coexpressing wt NS3-5A from a recombinant baculovirus, but production of NS4B alone with this system failed to restore any detectable replication. By expressing wt NS4B either individually or as part of NS3-5A from the same RNA encoding an NS3-5B polyprotein carrying the G1911A mutation, we also failed to detect any restoration in replication. We concluded that compared to the baculovirus system, in which a small but significant level of replication could be recovered, insufficient wt NS4B was produced from the intragenomic system to enable any rescue of replication. It is possible that to reach the low level of trans-complementation detected for this mutation in NS4B, it is necessary to produce an excess of NS4B, e.g., from the baculovirus system, as shown in this study, or from a second helper replicon that replicates to high levels (see the work of Jones et al. [38]). However, there may be insufficient translation of wt NS4B from the intragenomic system, since the G1911A mutation is highly inhibitory to replication and only a low level of complementation is possible. Thus, the overall effect could be that there are never sufficient numbers of active RCs generated by expressing two copies of NS4B from the same RNA to achieve a threshold that enables detectable replication. In contrast to the low complementation achieved with the G1911A mutation, S2208I could be efficiently rescued by expressing wt NS5A from an NS3-5A polyprotein produced either from baculovirus or from the same RNA carrying the S2208I mutation in the NS3-5B polyprotein. Expressing wt NS5A alone from baculovirus gave low levels of rescue, but interestingly, producing the protein from the same RNA containing the S2208I mutation in NS3-5B did promote replication, although at an efficiency lower than that achieved by expressing NS5A as part of the NS3-5A polyprotein. This shows that it is possible to complement a mutation in NS5A by expressing the protein independently from the other components of the viral polyprotein.
Our imaging studies indicated that expressing NS5A from NS3-5A and NS3-5B polyproteins gives almost complete colocalization of both proteins, including at foci which are the presumed sites of RNA replication. Indeed, there was no evidence of foci that did not have both copies of NS5A. In contrast, NS5A expressed alone, even from the intragenomic system which did partially complement replication, was to a large extent excluded from foci containing NS5A produced from the NS3-5B polyprotein. Our results are consistent with the notion that pre-RC complexes formed from a polyprotein are capable of interaction with other similar complexes made on either a different or the same RNA molecule. The formation of such higher-order structures is likely to arise during or shortly after translation, ultimately giving rise to MAF where replication occurs. Thus, the initial complexes formed during polyprotein translation and maturation may influence the interacting partners, which are then capable of contributing to RNA replication. It is these early steps in replicase complex formation that may be one of the factors that determine the extent to which replication-defective mutations can be complemented. For the S2208I mutation, we did find that the intragenomic system enabled some rescue of replication by expressing only NS5A. Here, it is likely that the close proximity between translation of wt NS5A and NS3-5B carrying the S2208I mutation from the same RNA molecule would allow incorporation of some wt protein into these pre-RC complexes. The extent of incorporation may not be detected either by imaging studies or by Western blot analysis for detection of hyperphosphorylated NS5A, which is typically generated through production of the protein from a polyprotein. This may reflect events within the MAF that elude microscopic analysis due to resolution limits but are influential in facilitating replication, for instance, a preponderance of the viral genome associating with NS proteins derived from a single NS3-5B polyprotein encoded by an ORF adjacent to the 3′ UTR. Alternatively, subtle differences in the mobility of individual proteins within and outside the MAF could also influence complementation efficiency. In another report, we have shown that NS5A is mobile on ER membranes but relatively immobile when associated with MAF (76).
In contrast to the ability of NS5A to trans-complement defective replication, there is only one report describing the ability of NS5A to trans-complement defective virion assembly (35). The authors concluded that the lack of virion assembly as a consequence of a truncation in NS5A could be efficiently trans-complemented using a helper virus; the authors of this report also commented on their inability to achieve trans-complementation by expressing NS5A alone. Our complementation results on virion assembly were therefore unexpected. Such differences could arise from the distinct systems used to study virion assembly. Our experimental strategy employed a trans-encapsidation system (68), whereas the previous study used a helper virus to provide the structural proteins. However, virion assembly studies using trans-encapsidation systems have thus far recapitulated observations made using cell culture-derived infectious HCV (68, 77, 78), making this explanation unlikely. Instead, we propose that the differences arise from the characteristics of the NS5A deletions in the two studies. In the report by Appel et al. (35), the deletion selected to block virion assembly encompassed the entire domain 3 of NS5A (residues 2328 to 2435). The authors did not opt for a shorter truncation, as they found such deletions were less effective at disrupting virion assembly. Significantly, one of the smaller truncations tested (with deletion of residues 2404 to 2435) reduced the titer of secreted infectious virions by only 100-fold, whereas the decrease reported by others when removing an even shorter segment of domain 3 (residues 2419 to 2433) was >5 log units (47). Importantly, the truncation construct with the deletion of residues 2419 to 2433 was the one selected for use in our study. We therefore consider our inability to complement the bulk of virion assembly to be due to a strong cis-acting dominant negative effect elicited by removing residues 2419 to 2433 and acting within the context of the RC. Certainly, such a scenario fits with our finding that virion assembly exhibited little or no requirement for NS5A to be expressed as part of a polyprotein or from a helper virus, as residual virion assembly activities involving NS5A would therefore reside outside the RC and thus not require targeting to the MAF. These low levels of complementation could also be easily interpreted to be a lack of complementation under experimental conditions where complementation was operating efficiently, such as those encountered by Appel and colleagues (35). It is tempting to speculate that if MAF targeting of NS5A is not required to support low-level virion assembly, then NS5A likely facilitates particle formation through its association with lipid droplets, an interaction that is known to be important for virion assembly (30, 35, 41) and one that does not require expression of NS5A within the context of the polyprotein (79). However, the involvement of lipid droplets does not explain why virion assembly exhibits a preference for functional NS5A to be expressed in cis with the RNA to be rescued. To our minds, the most plausible explanation is that the link between replication and virion assembly is not absolute and that HCV RNA can transition directly between a translational state and a packaging-competent state without passing through the MAF and RC. This suggestion is not without precedent, as transfection of HCV genotype 1b transcripts encoding a dysfunctional NS5B polymerase results in the formation of virus-like particles, albeit ones that cannot be tested for infectivity (45). Further work is needed to establish whether alternative assembly pathways exist and whether the mechanism underlying such virion formation has requirements distinct from those of the major virion assembly pathway. Thus, as with transmission of infectivity by extracellular virus or cell-to-cell contact (80, 81), there may be different pathways for producing infectious virus.
In conclusion, a mechanistic explanation as to why NS3-5A facilitates the complementation of NS5A in replication has been identified. Unexpected insight into the possible existence of virion assembly pathways that operate independently of replication has also been gained. The novel complementation systems described in this report, where constructs expressed two copies of the same NS protein, were instrumental to both arms of the investigation. We anticipate that they will provide a valuable addition to the tools available to investigate both virion assembly and packaging.
ACKNOWLEDGMENTS
This work was supported by MRC grants G0701215 and 4050295596.
We are grateful to Mark Harris for the gift of anti-NS5A antisera, Rick Randall for the gift of the anti-V5 monoclonal antibody, and Takaji Wakita for the gift of JFH-1-based replicon constructs.
Footnotes
Published ahead of print 18 December 2013
REFERENCES
- 1.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin C, Lindenbach BD, Pragai BM, McCourt DW, Rice CM. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547–5551. 10.1073/pnas.88.13.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980–3988. 10.1093/emboj/cdf414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057. 10.1128/JVI.75.24.12047-12057.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046–2051. 10.1128/JVI.74.4.2046-2051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. U. S. A. 96:2291–2295. 10.1073/pnas.96.5.2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- 13.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492. 10.1128/JVI.77.9.5487-5492.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8:e1003056. 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87:10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Pragai BM, Grakoui A, Xu J, Rice CM. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12–22 [PMC free article] [PubMed] [Google Scholar]
- 19.Einav S, Gerber D, Bryson PD, Sklan EH, Elazar M, Maerkl SJ, Glenn JS, Quake SR. 2008. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nat. Biotechnol. 26:1019–1027. 10.1038/nbt.1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417–36428. 10.1074/jbc.M508175200 [DOI] [PubMed] [Google Scholar]
- 21.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984. 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brass V, Bieck E, Montserret R, Wolk B, Hellings JA, Blum HE, Penin F, Moradpour D. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130–8139. 10.1074/jbc.M111289200 [DOI] [PubMed] [Google Scholar]
- 23.Elazar M, Liu P, Rice CM, Glenn JS. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 78:11393–11400. 10.1128/JVI.78.20.11393-11400.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouttenoire J, Montserret R, Kennel A, Penin F, Moradpour D. 2009. An amphipathic alpha-helix at the C terminus of hepatitis C virus nonstructural protein 4B mediates membrane association. J. Virol. 83:11378–11384. 10.1128/JVI.01122-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83:6257–6268. 10.1128/JVI.02663-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouttenoire J, Roingeard P, Penin F, Moradpour D. 2010. Amphipathic alpha-helix AH2 is a major determinant for the oligomerization of hepatitis C virus nonstructural protein 4B. J. Virol. 84:12529–12537. 10.1128/JVI.01798-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang J, Huang L, Cordek DG, Vaughan R, Reynolds SL, Kihara G, Raney KD, Kao CC, Cameron CE. 2010. Hepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 84:12480–12491. 10.1128/JVI.01319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu GY, Lee KJ, Gao L, Lai MM. 2006. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80:6013–6023. 10.1128/JVI.00053-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- 30.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 31.Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Heliot L, Rouille Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and nonstructural proteins towards virus assembly. PLoS Pathog. 7:e1001278. 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195. 10.1128/JVI.01565-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Counihan NA, Rawlinson SM, Lindenbach BD. 2011. Trafficking of hepatitis C virus core protein during virus particle assembly. PLoS Pathog. 7:e1002302. 10.1371/journal.ppat.1002302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentzsch J, Brohm C, Steinmann E, Friesland M, Menzel N, Vieyres G, Perin PM, Frentzen A, Kaderali L, Pietschmann T. 2013. Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathog. 9:e1003355. 10.1371/journal.ppat.1003355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. 10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouklani H, Bull RA, Beyer C, Coulibaly F, Gowans EJ, Drummer HE, Netter HJ, White PA, Haqshenas G. 2012. Hepatitis C virus nonstructural protein 5B is involved in virus morphogenesis. J. Virol. 86:5080–5088. 10.1128/JVI.07089-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Q, Manna D, Belton K, Cole R, Konan KV. 2013. Modulation of hepatitis C virus genome encapsidation by nonstructural protein 4B. J. Virol. 87:7409–7422. 10.1128/JVI.03523-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177. 10.1128/JVI.01885-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DM, Atoom AM, Zhang X, Kottilil S, Russell RS. 2011. A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J. Virol. 85:12351–12361. 10.1128/JVI.05313-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639. 10.1128/JVI.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976. 10.1128/JVI.00826-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 85:6963–6976. 10.1128/JVI.00502-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phan T, Beran RK, Peters C, Lorenz IC, Lindenbach BD. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379–8395. 10.1128/JVI.00891-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phan T, Kohlway A, Dimberu P, Pyle AM, Lindenbach BD. 2011. The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly. J. Virol. 85:1193–1204. 10.1128/JVI.01889-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 5:e1000475. 10.1371/journal.ppat.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pokrovskii MV, Bush CO, Beran RK, Robinson MF, Cheng G, Tirunagari N, Fenaux M, Greenstein AE, Zhong W, Delaney WE, Paulson MS. 2011. Novel mutations in a tissue culture-adapted hepatitis C virus strain improve infectious-virus stability and markedly enhance infection kinetics. J. Virol. 85:3978–3985. 10.1128/JVI.01760-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. 10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638. 10.1128/JVI.01890-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross-Thriepland D, Amako Y, Harris M. 2013. The C terminus of NS5A domain II is a key determinant of hepatitis C virus genome replication, but is not required for virion assembly and release. J. Gen. Virol. 94:1009–1018. 10.1099/vir.0.050633-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587. 10.1074/jbc.M407787200 [DOI] [PubMed] [Google Scholar]
- 51.Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083. 10.1128/JVI.00328-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Y, Anantpadma M, Timpe JM, Shanmugam S, Singh SM, Lemon SM, Yi M. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86–97. 10.1128/JVI.01070-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nag A, Robotham JM, Tang H. 2012. Suppression of viral RNA binding and the assembly of infectious hepatitis C virus particles in vitro by cyclophilin inhibitors. J. Virol. 86:12616–12624. 10.1128/JVI.01351-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Appel N, Herian U, Bartenschlager R. 2005. Efficient rescue of hepatitis C virus RNA replication by trans-complementation with nonstructural protein 5A. J. Virol. 79:896–909. 10.1128/JVI.79.2.896-909.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans MJ, Rice CM, Goff SP. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 78:12085–12089. 10.1128/JVI.78.21.12085-12089.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herod MR, Jones DM, McLauchlan J, McCormick CJ. 2012. Increasing rate of cleavage at boundary between non-structural proteins 4B and 5A inhibits replication of hepatitis C virus. J. Biol. Chem. 287:568–580. 10.1074/jbc.M111.311407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 5:e1000546. 10.1371/journal.ppat.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Lahser FC, Yi M, Wright-Minogue J, Xia E, Weber PC, Lemon SM, Malcolm BA. 2004. Conserved C-terminal threonine of hepatitis C virus NS3 regulates autoproteolysis and prevents product inhibition. J. Virol. 78:700–709. 10.1128/JVI.78.2.700-709.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graziani R, Paonessa G. 2004. Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon. J. Gen. Virol. 85:1867–1875. 10.1099/vir.0.80006-0 [DOI] [PubMed] [Google Scholar]
- 60.Tong X, Malcolm BA. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res. 115:122–130. 10.1016/j.virusres.2005.07.012 [DOI] [PubMed] [Google Scholar]
- 61.Koch JO, Bartenschlager R. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neddermann P, Clementi A, De Francesco R. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984–9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fridell RA, Valera L, Qiu D, Kirk MJ, Wang C, Gao M. 2013. Intragenic complementation of hepatitis C virus NS5A RNA replication-defective alleles. J. Virol. 87:2320–2329. 10.1128/JVI.02861-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. 2013. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 9:e1003359. 10.1371/journal.ppat.1003359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormick CJ, Brown D, Griffin S, Challinor L, Rowlands DJ, Harris M. 2006. A link between translation of the hepatitis C virus polyprotein and polymerase function; possible consequences for hyperphosphorylation of NS5A. J. Gen. Virol. 87:93–102. 10.1099/vir.0.81180-0 [DOI] [PubMed] [Google Scholar]
- 66.Targett-Adams P, McLauchlan J. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 86:3075–3080. 10.1099/vir.0.81334-0 [DOI] [PubMed] [Google Scholar]
- 67.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. 10.1053/j.gastro.2003.09.023 [DOI] [PubMed] [Google Scholar]
- 68.Adair R, Patel AH, Corless L, Griffin S, Rowlands DJ, McCormick CJ. 2009. Expression of hepatitis C virus (HCV) structural proteins in trans facilitates encapsidation and transmission of HCV subgenomic RNA. J. Gen. Virol. 90:833–842. 10.1099/vir.2008.006049-0 [DOI] [PubMed] [Google Scholar]
- 69.Shoji I, Aizaki H, Tani H, Ishii K, Chiba T, Saito I, Miyamura T, Matsuura Y. 1997. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 78:2657–2664 [DOI] [PubMed] [Google Scholar]
- 70.Krieger N, Lohmann V, Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614–4624. 10.1128/JVI.75.10.4614-4624.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCormick CJ, Salim O, Lambden PR, Clarke IN. 2008. Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 82:8917–8921. 10.1128/JVI.02362-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409. 10.1128/JVI.78.14.7400-7409.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Failla C, Tomei L, De Francesco R. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu HM, Aizaki H, Machida K, Ou JH, Lai MM. 2012. Hepatitis C virus translation preferentially depends on active RNA replication. PLoS One 7:e43600. 10.1371/journal.pone.0043600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Foster TL, Belyaeva T, Stonehouse NJ, Pearson AR, Harris M. 2010. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84:9267–9277. 10.1128/JVI.00616-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones DM, Gretton SN, McLauchlan J, Targett-Adams P. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88:470–475. 10.1099/vir.0.82363-0 [DOI] [PubMed] [Google Scholar]
- 77.Ishii K, Murakami K, Hmwe SS, Zhang B, Li J, Shirakura M, Morikawa K, Suzuki R, Miyamura T, Wakita T, Suzuki T. 2008. Trans-encapsidation of hepatitis C virus subgenomic replicon RNA with viral structure proteins. Biochem. Biophys. Res. Commun. 371:446–450. 10.1016/j.bbrc.2008.04.110 [DOI] [PubMed] [Google Scholar]
- 78.Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 82:7034–7046. 10.1128/JVI.00118-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198–210. 10.1006/viro.2001.1225 [DOI] [PubMed] [Google Scholar]
- 80.Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, Wong-Staal F, Balfe P, McKeating JA. 2011. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J. Virol. 85:596–605. 10.1128/JVI.01592-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, Owsianka AM, Angus AG, Keck ZY, Foung SK, Pietschmann T, Rice CM, Patel AH. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48–58. 10.1099/vir.0.006700-0 [DOI] [PMC free article] [PubMed] [Google Scholar]