Abstract
Mumps is a highly contagious human disease, characterized by lateral or bilateral nonsuppurative swelling of the parotid glands and neurological complications that can result in aseptic meningitis or encephalitis. A mumps vaccination program implemented since the 1960s reduced mumps incidence by more than 99% and kept the mumps case numbers as low as hundreds of cases per year in the United States before 2006. However, a large mumps outbreak occurred in vaccinated populations in 2006 and again in 2009 in the United States, raising concerns about the efficacy of the vaccination program. Previously, we have shown that clinical isolate-based recombinant mumps viruses lacking expression of either the V protein (rMuVΔV) or the SH protein (rMuVΔSH) are attenuated in a neurovirulence test using newborn rat brains (P. Xu et al., Virology 417:126–136, 2011, http://dx.doi.org/10.1016/j.virol.2011.05.003; P. Xu et al., J. Virol. 86:1768–1776, 2012, http://dx.doi.org/10.1128/JVI.06019-11) and may be good candidates for vaccine development. In this study, we examined immunity induced by rMuVΔSH and rMuVΔV in mice. Furthermore, we generated recombinant mumps viruses lacking expression of both the V protein and the SH protein (rMuVΔSHΔV). Analysis of rMuVΔSHΔV indicated that it was stable in tissue culture cell lines. Importantly, rMuVΔSHΔV was immunogenic in mice, indicating that it is a promising candidate for mumps vaccine development.
INTRODUCTION
Mumps is a human infectious disease characterized by lateral or bilateral nonsuppurative swelling of the parotid glands. In severe cases, mumps can lead to orchitis in postpuberty male patients and damage to the central nervous system. In the prevaccine era, 90% of the population turned seropositive for mumps virus (MuV) by 14 to 15 years of age, reflecting its highly contagious nature. Mumps virus is neurotropic and was one of the most common causes of aseptic meningitis before the implementation of mass mumps vaccination programs.
At present, the Jeryl Lynn (JL) vaccine is the most commonly used mumps vaccine, administered as lyophilized live virus with measles and rubella vaccine components. The JL vaccine strain originated from an infectious isolate from a mumps patient in 1963 (1). The virus was attenuated through continuous passages in embryonic hen eggs and chicken embryos/chicken embryo cell cultures (1). The JL vaccine was licensed in the United States in 1967 and has been used for over 40 years. This vaccine has been efficacious and safe overall (2–6). However, several large mumps outbreaks have occurred recently in the United States and worldwide in populations that have been vaccinated with the JL vaccine (7–10). Major mumps outbreaks in the United States include the 2006 multistate mumps outbreak, reporting 6,584 suspected cases originating from the state of Iowa (11, 12) and the 2009–2010 New York and New Jersey mumps outbreaks with a total of 2,078 suspected cases reported in 2010 (13). Both of the outbreaks occurred among highly vaccinated populations, raising questions about the efficacy of the current vaccination program in the United States. One possible causality is the antigenic differences between the genotype A vaccine strain and the genotype G circulating wild-type mumps viruses.
In this study, we seek to develop a mumps vaccine candidate through genetic modification of a clinically isolated mumps virus. Mumps virus is a member of the family Paramyxoviridae, subfamily Paramyxovirinae, and genus Rubulavirus (6, 14). It is an enveloped virus enclosing a negative-sense, single-stranded, nonsegmented RNA genome of 15,384 nucleotides in length which encodes 9 viral proteins (15–17). Studies of the function of the Paramyxovirus SH protein reveal that it blocks tumor necrosis factor alpha (TNF-α) induction, signaling, caspase activation, and NF-κB nuclear translocation in transfected and virus-infected cells (18–23). The V protein is an accessory protein translated from the authentic transcript of the V/P gene (24, 25). Mumps V protein is an antagonist of antiviral innate immunity. It interferes with type I interferon (IFN) induction by disrupting the recognition of intracellular viral double-stranded RNA (dsRNA) by MDA5 (26–28). It also blocks IFN signaling by targeting STAT proteins for proteasome-mediated degradation (29–35). Recombinant mumps viruses with either the V protein deletion (rMuVΔV) or the SH protein deletion (rMuVΔSH) are attenuated in neurotoxicity in intracerebrally (IC) infected rats (21, 36). In this study, we tested the immunogenicity of rMuVΔV and rMuVΔSH in mice. Furthermore, we generated a recombinant MuV lacking expression of both the SH and V proteins (rMuVΔSHΔV) and examined antibody and cellular immune responses in mice.
MATERIALS AND METHODS
Plasmids, viruses, and cells.
The MuV strain was obtained from a patient during the 2005–2006 Midwest mumps outbreak in the United States. A full-length cDNA clone of the virus (pMuV) was constructed as previously described (21). Recombinant MuV lacking the V protein (rMuVΔV), recombinant MuV lacking the SH protein, and recombinant MuV expressing a Renilla luciferase protein have been described before (21). A plasmid containing the MuV genome but lacking both V and SH was constructed by combining the SH open reading frame (ORF) deletion with the plasmid encoding the rMuVΔV genome. Primer sequences, detailed cloning strategies, and entire cDNA sequences of MuV are available upon request. Jeryl Lynn (JL) vaccine, isolated from the measles, mumps, and rubella (MMR) vaccine, was a gift from Paul Rota at the CDC.
To rescue an infectious virus, plasmid pMuVΔSHΔV (5 μg), along with plasmids pCAGGS-L (1 μg), pCAGGS-NP (1.5 μg), and pCAGGS-P (200 ng), were transfected into BSRT-7 cells. Three days later, transfected BSRT-7 cells were mixed with Vero cells at a 1:1 ratio. Ten to 14 days later, when syncytium formation was observed, supernatants containing putative rMuVΔSHΔV were collected and plaque purified in Vero cells. Plaques (developing 4 to 7 days postinfection [dpi]) were amplified in Vero cells once (P0), and their genomes were sequenced. All recombinant viruses used for the following experiments were expanded once in Vero cells from the P0 amplification (P1). The rescue procedure was repeated to produce independent stocks of rMuVΔSHΔV, resulting in 6 isolates of independently rescued rMuVΔSHΔV viruses (PX64-1, PX64-4, PX64-61, PX64-67, and PX64-84).
All mumps viruses were grown in Vero cells and harvested at 4 to 7 dpi. Virus titers were measured in Vero cells by plaque assay as described previously (37, 38). JL virus was grown in Vero cells and concentrated to achieve a working titer. Harvested virus stock was cushioned onto 20% sucrose using ultracentrifugation at 37,500 rpm (Thermo Scientific Sorvall RC 6 plus centrifuge). Pelleted viruses were resuspended in 1% bovine serum albumin (BSA)–Dulbecco's modified Eagle medium (DMEM) and stored at −80°C. Concentrated JL virus was retitrated in Vero cells by plaque assay.
Vero cells were maintained in DMEM with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) (Mediatech Inc., Holu Hill, FL). BSRT-7 cells were maintained in DMEM supplemented with 10% FBS, 1% P/S, 10% tryptose phosphate broth (TPB), and 400 μg/ml Geneticin G418 antibiotic. Cells were cultured at 37°C with 5% CO2 and passed the day before infection or transfection at appropriate dilution factors to achieve 80 to 90% confluence the next day. For virus infection, cells were inoculated with viruses in DMEM plus 1% BSA at an multiplicity of infection (MOI) of 0.01, 3, or 5 and incubated for 1 to 2 h at 37°C with 5% CO2. The inocula were then replaced with DMEM supplemented with 2% FBS and 1% P/S. Cells were transfected with plasmids using Plus and Lipofectamine reagents (Invitrogen, Carlsbad, CA) by following the manufacturer-provided protocols.
Sequencing of viruses.
Viral RNA was extracted from cell culture supernatants using the QIAamp viral RNA extraction minikit (Qiagen Inc., Valencia, CA) by following the manufacturer's protocol. Isolated viral RNA was reverse transcribed into cDNA using Super Script III reverse transcriptase with random hexamers (Invitrogen). Synthesized cDNA then served as templates for PCR using mumps virus genome-specific primers and Taq polymerase (Invitrogen). Fifteen sets of primers, each containing a forward and reverse primer, were designed to divide the genome into 15 overlapping fragments. The primers were then used for the subsequent sequencing of the PCR products (39). Primer sequences are available upon request.
Immunoblotting.
Vero cells in 6-well plates at approximately 90% confluence were mock infected or infected with rMuV or rMuVΔSHΔV at an MOI of 0.5. Cells were lysed and collected at different time points postinfection in 0.5 ml WCEB buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 0.5% NP-40, 0.00076% EGTA, 0.2 mM EDTA, 10% glycerol) with a mixture of protease inhibitors as described previously (30, 31). Cell lysates were briefly centrifuged to remove cell debris and loaded onto a 10% or 17.5% polyacrylamide gel and subjected to SDS-PAGE. Proteins were transferred to an Immobilon-FL transfer membrane (Millipore, Billerica, MA), incubated with primary antibody (anti-MuV V, 1:500; anti-MuV NP, 1:5,000; anti-MuV P, 1:2,000; and anti-MuV SH, 1:200) (21) and corresponding secondary antibodies conjugated to horseradish peroxidase (1:1,000) (KPL, Inc.) and detected using an Amersham ECL Western blotting detection kit (GE Healthcare Bioscience, Piscataway, NJ).
Multicycle growth curve in Vero cells.
Vero cells in 6-cm plates or 6-well plates were mock infected or infected with rMuV, JL, rMuVΔV, rMuVΔSH, or rMuVΔSHΔV (multiple isolates) at an MOI of 0.01. One ml (6-cm plate) or 100 μl (6-well plate) of supernatant was collected at 1, 2, 3, 4, 5, and 6 dpi, supplemented with 1% BSA, and stored at −80°C. Virus titers were determined by plaque assay in triplicate using Vero cells in 6-well plates. After 1 to 2 h of incubation with the viruses, the growth medium was changed to DMEM with 2% FBS, 1% P/S, and 1% low-melting-point agarose. Four to 7 dpi, the Vero cells were stained with Giemsa stain and plaques were counted.
Immunization of mice.
BALB/c mice (female, 6 to 8 weeks old) were purchased from Charles River Laboratories (Frederick, MD). Mice were immunized with 1 × 106 PFU of rMuV, JL, rMuVΔV, rMuVΔSH, or rMuVΔSHΔV in a volume of 100 μl for intranasal (i.n.) vaccination. For intramuscular (i.m.) vaccination, mice were injected with 25 μl of inoculum into each side of the caudal thigh bilaterally (106 PFU). i.n.- or i.m.-vaccinated mice were boosted with the same amount of virus inocula as the primary vaccination on the 21st or 22nd day after primary vaccination. Blood samples were obtained from mock or recombinant MuV-vaccinated mice through tail vein puncture. At the termination of each experiment, mice were euthanized with 500 μl of Avertin (2,2,2-tribromoethanol) (Sigma-Aldrich) followed by cervical dislocation. Spleens were removed from the mice for splenocyte isolation and in vitro analysis. All mouse immunizations and studies with mumps viruses were performed in enhanced biosafety level 2 facilities with HEPA-filtered isolators and were conducted by following protocols reviewed and approved by the Institutional Animal Care and Use Committee of the University of Georgia.
The ELISPOT assay.
Splenocytes were isolated from mouse spleens at the time of euthanasia. Spleens were ground, filtered through cell strainers (BD Falcon), and washed once with 50 ml of Hanks' balanced salt solution (Life Technologies) per spleen. Washed splenocytes from each spleen were treated with 3 ml of Gey's solution (ammonium chloride, 8.29 g/liter; potassium bicarbonate, 1 g/liter) for 5 min at room temperature (RT) to lyse red blood cells. The residual splenocytes were washed once with 50 ml Hanks balanced salt solution (HBSS) per spleen and resuspended in 10 ml complete tumor medium (CTM) containing 0.75 g/liter d-glucose (Sigma), 7.5 ml/liter essential amino acids (50×) (Invitrogen), 14 ml/liter nonessential amino acids (100×) (Invitrogen), 10 ml/liter sodium pyruvate (100×) (Gibco), 10 ml/liter l-glutamine (100×) (Gibco), 0.85 g/liter sodium bicarbonate (Sigma), 1% gentamicin-penicillin G-streptomycin sulfate (Sigma), and 3.4 μl/liter 2-mercaptoethanol (Fisher) in minimum essential medium, Spinner modification (S-MEM; Sigma). Splenocytes were counted and reconstituted to a concentration of 3 × 106 cells/ml and 1.5 × 106 cells/ml in CTM. One hundred μl of splenocytes was plated onto prepared enzyme-linked immunosorbent spot (ELISPOT) plates (MultiScreen-IP without underdrain; 0.45 μm, white, sterile; Millipore). The ELISPOT plates were precoated with anti-mouse IFN-γ (AN-18; MABTECH) overnight, washed with sterile PBS five times, and incubated with CTM for 1 h at RT. One hundred μl of CTM containing either mock-infected or MuV-infected Vero cell lysates at 50 μg/ml was overlaid onto splenocytes as a stimulant. Vero cell lysates were prepared by rounds of sonication and several freeze-thaw cycles to inactivate any infectious viral particles. The mixture of splenocytes and viral antigens was incubated for 40 to 48 h at 37°C with 5% CO2. The plates were washed after incubation, blotted with biotinylated anti-mouse IFN-γ antibody (MAb R4-6A2; MABTECH) and streptavidin-alkaline phosphatase (MABTECH), and developed in 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (KPL).
ELISA.
Enzyme-linked immunosorbent assay (ELISA) was performed as previously described (40). Briefly, immulon 2 HB 96-well microtiter plates (ThermoLab Systems) were coated with MuV proteins at 2 μg/ml and incubated at 4°C overnight. Plates were then washed with KPL wash solution (KPL, Inc.), and each well was blocked with 200 μl KPL wash solution with 5% nonfat dry milk and 0.5% BSA (Blotto) for 1 h at RT. Serum samples were inactivated by heating at 56°C for 0.5 h and were serially diluted 2-fold or 4-fold in Blotto. One hundred μl of diluted serum samples was transferred to the coated plate and incubated for 1 h at RT. To detect anti-MuV specific antibodies, alkaline phosphatase (AP)-labeled, goat anti-mouse IgG (KPL, Inc.) was diluted in Blotto according to the manufacturer's instructions, added to each well, and incubated for 1 h at RT. Plates were washed and developed by adding 100 μl pNPP phosphatase substrate (KPL, Inc.) per well. Optical density (OD) was measured at 405 nm on a Bio-Tek Powerwave XS plate reader.
Luciferase activity-based neutralization assay.
Serum samples were serially diluted 2-fold starting from 1:10 or 1:40 up to 1:20,480. Recombinant virus expressing a Renilla luciferase protein (rMuV-Luc) was diluted to 2,000 PFU/ml in 1% BSA-DMEM. One portion of serum (40 μl) was mixed with an equal volume of rMuV-Luc virus (80 PFU/40 μl) into each well of a 96-well plate and incubated at 37°C with 5% CO2. Each 96-well plate contained five serum samples and one standard in duplicate. After 1 h of incubation, trypsinized Vero cells in 4% FBS, 2% P/S in DMEM were added to each well of the 96-well plates. At 48 to 72 h postinfection, infected Vero cells were lysed and analyzed for total luciferase activity per well using the Renilla Luciferase assay system (Promega) and a Veritas microplate luminometer (Promega). The neutralizing titer was calculated as the highest dilution level with luciferase readings exceeding that produced by 40 PFU of rMuV-Luc virus in standard control wells.
Statistics.
P values were calculated using Student's t test (two-tailed, type 2). Correlations of titers determined by luciferase activity-based neutralization assay to that determined by plaque reduction neutralization assay were calculated by R2.
RESULTS
Immunogenicity of recombinant mumps viruses lacking either the V protein or the SH protein in mice.
To analyze the immunogenicity of the current mumps vaccine, JL, and the clinical MuV isolate from the 2006 outbreak (referred to as MuV), mice were vaccinated with JL or MuV via the intranasal (i.n.) or intramuscular (i.m.) route and boosted at 22 days postprimary vaccination with the same virus, dose, and route as the primary vaccination. Serum samples were collected at 14 days postboost (dpb). As expected, JL generated higher neutralizing antibody titers against JL than MuV, and MuV generated higher anti-MuV titers than JL, regardless of the route of immunization (Fig. 1). This result is consistent with a previous report that sera from JL-vaccinated humans had higher anti-JL neutralizing titers than anti-MuV neutralizing titers (41).
FIG 1.
Cross-reactivity of JL and MuV. BALB/c mice were i.n. or i.m. immunized with PBS, MuV, or JL at 106 PFU/mouse and boosted at 22 days postvaccination with the same virus at 106 PFU/mouse. Serum samples were collected at 14 days postboost. Heat-inactivated serum samples of individual mice from the same group were pooled to perform the plaque reduction neutralization test (PRNT). Serum samples were 2-fold serial diluted from 1:30 to 1:3,840. A volume of 120 μl diluted serum was mixed with 120 μl diluted virus containing 80 PFU of either JL or rMuV virus and incubated at 37°C for 1 h. The count of residual unneutralized PFU per 100 μl was determined by plaque assay in 6-well plates of Vero cells. PRNT titer is determined as the first dilution level with residual PFU of more than half of the input per 100 μl.
Previous studies have shown that rMuVΔV (lacking V protein expression) or rMuVΔSH (lacking SH protein expression) are attenuated in a neurovirulency potency test in rat brains (21, 36), suggesting these viruses are good candidates for vaccine development. To investigate the immunogenicity and vaccine potential of rMuVΔV and rMuVΔSH in mice, BALB/c mice were mock vaccinated (PBS) or vaccinated with rMuV, JL, rMuVΔSH, or rMuVΔV and boosted at 22 days postprimary vaccination through i.m. injection with the same virus and dose as the primary vaccination. We chose the i.m. route because the trivalent MMR vaccine is usually administered by intramuscular (i.m.) or deep subcutaneous injection (42), but mostly via i.m. administration. Serum samples were collected at 14 dpb. The total IgG antibody titer against MuV was measured by ELISA using plates coated with purified MuV (Fig. 2). We found that all groups generated robust anti-MuV antibody responses.
FIG 2.
i.m. immunization with rMuVΔSH or rMuVΔV induced antibody responses in mice. BALB/c mice were i.m. vaccinated with PBS, rMuV, JL, rMuVΔSH, and rMuVΔV at 106 PFU and boosted 22 days postvaccination with 106 PFU. Serum samples were collected at 14 dpb, and total antibody titers in these samples were measured by ELISA coated with MuV viral proteins.
Mumps virus is a human respiratory virus transmitted via respiratory secretions such as saliva and nose and throat discharge (6). i.n. vaccination induces both local immunity in the respiratory tract and systemic immunity. Mucosal immunity provides direct and rapid protection against virus challenge. To examine the immunogenicity of rMuVΔV and rMuVΔSH compared to that of rMuV and JL, BALB/c mice were mock vaccinated (PBS) or vaccinated with rMuV, JL, rMuVΔSH, or rMuVΔV intranasally and boosted at 22 days postprimary vaccination with the same virus type and dose as the primary i.n. vaccination. Serum samples were collected at 14 dpb. Total IgG antibody titers against MuV were measured by ELISA (Fig. 3). All groups generated robust anti-MuV antibody responses.
FIG 3.
i.n. immunization with rMuVΔSH or rMuVΔV induced antibody responses in mice. BALB/c mice were i.n. vaccinated with PBS, rMuV, JL, rMuVΔSH, and rMuVΔV with 106 PFU and boosted 22 days postvaccination with 106 PFU. Serum samples were collected at 14 dpb, and total antibody titers of these samples were measured by ELISA coated with MuV viral proteins.
Rescue of recombinant viruses lacking both V and SH proteins.
To further enhance the safety of vaccine candidates, we constructed a recombinant virus lacking expression of both the V and SH proteins. The genome length of the newly synthesized cDNA (pMuVΔSHΔV) complied with the rule of six for Paramyxovirus (43). Infectious recombinant viruses (rMuVΔSHΔV) were rescued from BSRT-7 cells transfected with pMuVΔSHΔV and helper plasmids as described before (21). To confirm rescue of the virus, viral RNA was extracted from cell culture medium containing rescued viruses (Fig. 4B). The SH gene and the V/P gene region were amplified using reverse transcription-PCR and sequenced. As shown in Fig. 4, the SH ORF truncation as well as the V deletion was confirmed (Fig. 4C and D).
FIG 4.
Generation of recombinant MuV lacking V and SH proteins (rMuVΔSHΔV). (A) Schematics of pMuVΔSHΔV. A 156-bp section was removed from the SH gene of pMuVΔV, a cDNA genome of mumps virus lacking expression of V protein. (B) Reverse transcription-PCR confirmed the mutation in the SH ORF in rescued rMuVΔSHΔV. Recombinant viruses (rMuVΔSHΔV) were rescued from pMuVΔSHΔV through transfection of BSRT-7 cells with pMuVΔSHΔV, together with the helper plasmids (pCAGGS-L, pCAGGS-NP, and pCAGGS-P). RNA was extracted from rMuVΔSHΔV-infected Vero cells. Two primers, PX47F and PX48R (sequences are available upon request), were used to amplify the SH gene region. (C and D) Sequence confirmation of the mutated regions in the SH ORF and the V/P editing site. The reverse transcription-PCR product of the SH gene was sent for sequencing. Sequencing results confirmed the mutation was successfully introduced into rMuVΔSHΔV. (E) Western blot confirmation of the deletion of V and SH proteins in rMuVΔSHΔV viruses. Vero cells were mock infected or infected with rMuV or rMuVΔSHΔV at an MOI of 0.5. Cell lysates were collected at 48 hpi and were blotted against MuV NP, P, V, and SH proteins.
To confirm that genomic changes in rMuVΔSHΔV abolish V and SH expression, Vero cells were mock infected or infected with rMuV or rescued rMuVΔSHΔV (PX64-67 strain). Expression levels of MuV NP, P, V, and SH proteins were examined using Western blotting. While NP and P were detected in both rMuV- and rMuVΔSHΔV-infected cells, expression of V or SH protein was only detected in rMuV-infected Vero cells (Fig. 4E).
Analysis of rMuVΔSHΔV in tissue culture cells.
To select an rMuVΔSHΔV virus that replicates well for vaccine production purposes, the replication capability of rMuVΔSHΔV viruses from 6 individual rescues, designated PX64-N, were compared by multicycle growth kinetics in Vero cells. While most rMuVΔSHΔV viruses reached the peak virus titer within the first 72 h postinfection (hpi), PX64-4 grew slower than the others and peaked at 96 hpi. Three strains (PX64-4, PX64-61, and PX64-67) grew to a titer close to 107 PFU/ml, and the other three strains (PX64-1, PX64-2, and PX64-81) had titers ranging from 5.5 × 105 to 6 × 105, 1 to 1.5 logs lower than the former strains (Fig. 5A). PX64-67 had a growth pattern similar to that of full-length rMuV (virus titer peaking during first 48 hpi) and the highest virus titer among rMuVΔSHΔV viruses. Therefore, it was chosen and designated rMuVΔSHΔV for the following studies. Previously, when we obtained rMuVΔV, mutations in regions other than the designed V/P editing site always arose. The entire genome of the rescued rMuVΔSHΔV viruses were sequenced to determine whether genome-wide mutations occurred during virus rescue. PX64-67 contained an additional single-nucleotide change (C-T) in genomic position 1913 (termed 1913 C-T) (NP gene end) and 7894 T-A silent (HN ORF) mutations compared to rMuV. To compare the growth of rMuVΔSHΔV to rMuV and parental viruses (rMuVΔSH and rMuVΔV), Vero cells were infected with rMuV, rMuVΔSH, rMuVΔV, and rMuVΔSHΔV at an MOI of 0.01. While rMuVΔSH showed growth kinetics comparable to those of rMuV, consistent with previously published data (21), rMuVΔV and rMuVΔSHΔV were about a half log lower in virus titer. The virus titer of rMuVΔSHΔV decreased after 48 hpi, remaining about a half log lower than that of rMuVΔV and one log lower than that of rMuV or rMuVΔSH (Fig. 5B).
FIG 5.
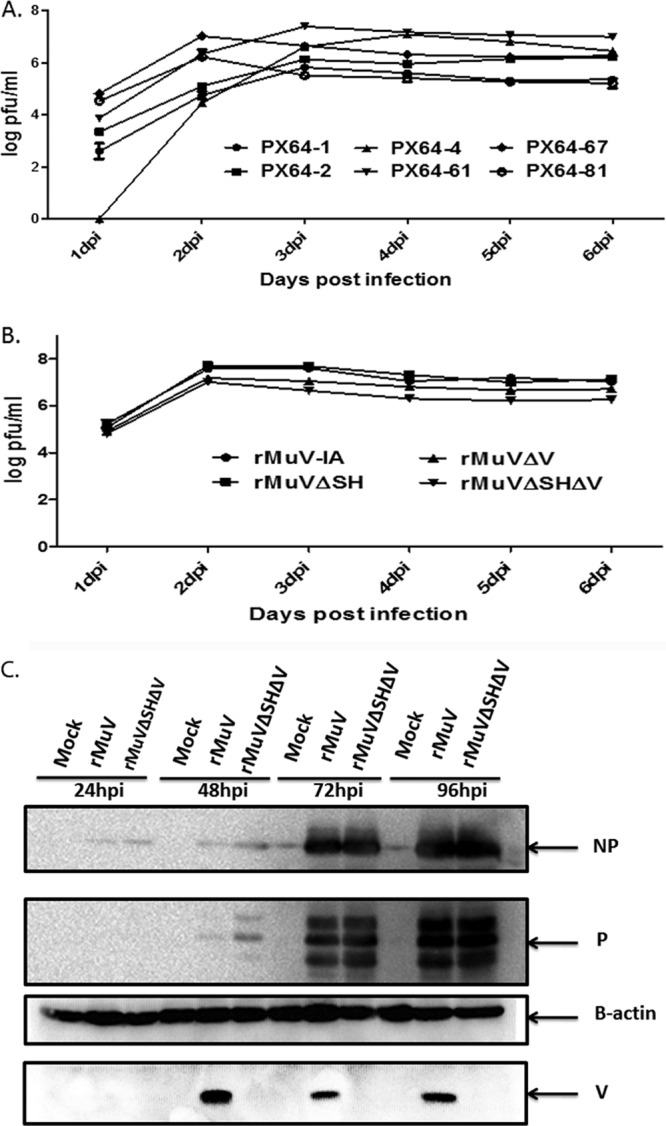
Analysis of rMuVΔSHΔV in tissue culture cells. (A) Multicycle growth rate of rMuVΔSHΔV. Vero cells were mock infected or infected with rMuVΔSHΔV viruses from different rescues (PX64-1, PX64-2, PX64-4, PX64-61, or PX64-67) at an MOI of 0.1. Supernatants collected from culture medium of infected cells at 1, 2, 3, 4, 5, and 6 dpi were plated onto Vero cells for plaque assay, from which virus titer was calculated. (B) Multicycle growth curve of rMuVΔSHΔV virus compared to parent viruses. PX64-67 was selected for subsequent animal experiments. In vitro growth of rMuVΔSHΔV (PX64-67) was compared to rMuV, JL, rMuVΔV, and rMuVΔSH in Vero cells as described for panel A. (C) Viral protein expression levels of rMuVΔSHΔV at different time points postinfection. NP, P, and V protein expression levels in rMuVΔSHΔV-infected Vero cells (MOI of 0.01) were examined from 1 to 4 dpi and compared to those of rMuV infection.
Intracellular viral protein expression of rMuVΔSHΔV was compared to that of rMuV. NP, P, and V protein expression levels were examined (Fig. 5C). Comparable NP and P protein levels were detected in Vero cells infected with rMuV or rMuVΔSHΔV. However, secretion of infectious viral particles of rMuVΔSHΔV-infected Vero cells was less than that of rMuV-infected Vero cells at all time points postinfection (Fig. 5C). At 48 hpi, a more intense P protein band was observed in rMuVΔSHΔV-infected cells than in rMuV-infected cells, consistent with a previous report of higher P expression at early time points in rMuVΔV viruses, a likely result of increased P transcription from the V/P gene due to deletion of the V mRNA transcript (36). Expression of the V protein was only detected in rMuV-infected cells.
Maintenance of V and SH protein deletion in rMuVΔSHΔV through 10 passages in Vero cells.
To examine the stability of rMuVΔSHΔV, it was passed in Vero cells continuously for 10 passages at a low MOI. At passage 10 (rMuVΔSHΔV P10), the culture medium from infected Vero cells was used for viral RNA extraction, followed by whole-genome sequencing to determine the consensus genome sequence. Sequencing results revealed 3 additional mutations: 1 silent mutation in the HN ORF, an R154K mutation in P, and an N2063H mutation in L. Interestingly, the G-A nucleotide mutation at position 2445 in the P ORF is the first nucleotide of the 6-guanine editing site (GGGGGG) of the V/P gene, which has been altered to GAGGAGGG in rMuVΔSHΔV and rMuVΔV viruses (36). Importantly, none of these mutations affected deletion of V or SH.
Furthermore, 10 single plaques (designated SP-1 to SP-10) were obtained from rMuVΔSHΔV P10, and the V/P and SH gene regions were sequenced (Table 1). The V protein and the SH ORF deletion were maintained in all 10 progeny strains from passage 10, including the 1913 C-T mutation in the NP gene end region. While 9 out of 10 strains contained the 2445 G-A mutation in the V/P gene editing site, 1 strain lost/failed to retain this mutation. To confirm that the 2445 G-A mutation had no effect on the V protein deletion, expression of the V protein in SP-1- to SP-10-infected Vero cells was examined by Western blotting (Fig. 6). No expression of V was detected, indicating that the mutation had no effect on V protein deletion.
TABLE 1.
V/P gene and SH gene sequences of rMuVΔSHΔV P10 single plaque-purified virusesa
| Virus | V protein deletion | NP GE or V/P GS mutation | V/P editing site mutation | SH ORF deletion |
|---|---|---|---|---|
| SP-1 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-2 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-3 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-4 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-5 | Yes | 1913 C-T | 1578 A-C | Yes |
| SP-6 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-7 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-8 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-9 | Yes | 1913 C-T | 2445 G-A | Yes |
| SP-10 | Yes | 1913 C-T | 2445 G-A | Yes |
rMuVΔSHΔV was passed continuously in Vero cells for 10 passages. Ten plaques were randomly obtained from rMuVΔSHΔV at passage 10 and grown in Vero cells (SP-1 to SP-10). The V/P and SH gene regions of SP-1 to SP-10 were sequenced. Mutations found in these regions are shown.
FIG 6.
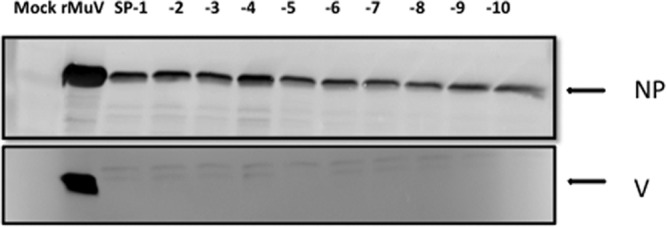
Lack of V expression in isolates from the 10th passage of rMuVΔSHΔV. Vero cells were mock infected or infected with rMuV or SP-1 to SP-10. One hundred μl of infected Vero cell lysates was subjected to Western blotting to detect NP and V protein expression levels.
Intramuscular immunization of BALB/c mice with rMuVΔSHΔV generated an antibody response against MuV.
BALB/c mice were i.m. vaccinated with rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV as described above. Serum samples were collected as described previously. Serum IgG antibody titers against MuV were measured using ELISA with plates coated with MuV viral proteins (lysed virions). Neutralizing antibody titers against MuV were measured by an rMuV-Luc-based neutralization assay as described in Materials and Methods (linear correlation of rMuV-Luc-based neutralization assay to traditional plaque reduction neutralization assay was confirmed by an R2 value of 0.9317 using ferret serum samples [data not shown]). Similar approaches have been used for adenovirus, measles virus, and respiratory syncytial virus (RSV) to substitute for the traditional plaque reduction neutralization test (PRNT) (44–47). We used rMuV-Luc, which was constructed based on the genetic background of MuV, as the targeting virus to compare the potentials of humoral responses induced by the vaccine candidates as well as the JL strain in mice to protect against the circulating mumps virus in the United States. No significant differences were detected among the groups for total antibody titers (Fig. 7A). However, the neutralizing antibody titers of serum samples showed some differences. The average neutralizing titer of JL-inoculated mice was significantly lower than that of rMuV-infected mice, which had the highest neutralizing titer. Mice inoculated with rMuVΔSH, rMuVΔV, or rMuVΔSHΔV had similar average titers (Fig. 7B).
FIG 7.
Evaluation of antibody responses in mice i.m. vaccinated with rMuVΔSHΔV. BALB/c mice were i.m. immunized with PBS, rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV at 106 PFU/mouse and boosted at 22 days postvaccination with the same virus at 106 PFU/mouse. (A) ELISA results measuring total antibody titer at 14 dpb. Serum samples were collected at 14 dpb. The total antibody titer against MuV was measured through ELISA. (B) Neutralizing antibody titer at 14 dpb. Neutralizing antibody titers in serum samples collected at 14 dpb were measured through an rMuV-Luc-based neutralization assay. P values of <0.05 are shown.
Intranasal immunization of BALB/c mice with rMuVΔSHΔV generated an antibody response against MuV.
BALB/c mice were i.n. inoculated with rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV at 106 PFU, and serum samples were collected for measurement of both total antibody titer and neutralizing titer against MuV. rMuVΔSH-inoculated mice developed the highest total antibody titer, and the rMuV group had a higher titer than the JL group. No significant differences were detected among the JL group, rMuVΔV group, and rMuVΔSHΔV groups (Fig. 8A). The rMuV group had the highest neutralizing titer, the JL group had the lowest neutralizing titer, and the other three groups (rMuVΔSH, rMuVΔV, and rMuVΔSHΔV) ranked between them (Fig. 8B). Although i.n.-immunized mice exhibited neutralizing antibody titer patterns like those observed in the i.m. groups, statistically significant differences were found between the rMuV and JL groups (P = 0.001), JL and rMuVΔSH groups (P = 0.001), JL and rMuVΔSHΔV groups (P = 0.038), and rMuVΔV and rMuVΔSH groups (P = 0.034).
FIG 8.
Evaluation of antibody responses in mice i.n. vaccinated with rMuVΔSHΔV. BALB/c mice were i.n. immunized with PBS, rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV at 106 PFU and boosted at 22 days postvaccination with the same virus at 106 PFU. (A) ELISA results measuring total antibody titer at 14 dpb. Serum samples were collected at 14 dpb. Total antibody titers against MuV were measured through ELISA. (B) Neutralizing antibody titer at 14 dpb. Neutralizing antibody titers in serum samples collected at 14 dpb were measured through an rMuV-Luc-based neutralization assay. P values for significantly different groups were found for rMuV and JL groups (0.001), rMuV and rMuVΔV groups (0.016), JL and rMuVΔSH groups (0.001), JL and rMuVΔSHΔV groups (0.038), and rMuVΔV and rMuVΔSH groups (0.034). For simplicity, P values comparing the rMuV and JL groups and JL and rMuVΔSHΔV groups are shown.
Adaptive T cell responses were induced in mice vaccinated with rMuVΔSHΔV.
To investigate the cellular immune responses induced by rMuVΔSHΔV, i.m.- or i.n.-inoculated mice were euthanized at 28 dpb and splenocytes were isolated for ELISPOT assay. In i.m.-inoculated mice, the JL group had the highest T cell response levels and rMuVΔSHΔV had the lowest T cell response levels, with no distinguishable differences among the rMuV, rMuVΔSH, and rMuVΔV groups (Fig. 9A). Differences were significant between JL and rMuVΔSH, JL and rMuVΔSHΔV, rMuVΔSH and rMuVΔSHΔV, and rMuVΔV and rMuVΔSHΔV. In i.n.-inoculated mouse groups, rMuV- and rMuVΔV-immunized mice had the lowest responding cell counts (Fig. 9B). Significant differences were observed between JL and rMuVΔV, rMuV and rMuVΔSH, rMuV and rMuVΔSHΔV, rMuVΔSH and rMuVΔV, and rMuVΔV and rMuVΔSHΔV groups.
FIG 9.
Cellular immune responses induced by rMuVΔSHΔV vaccination in mice. (A) Memory T cell responses in mumps virus i.m.-immunized mice. BALB/c mice were i.m. immunized with PBS, rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV at 106 PFU and boosted at 22 days postvaccination with the same virus at 106 PFU. Splenocytes were extracted from mouse spleens and used for ELISPOT assay. Splenocytes were stimulated with MuV-infected Vero cell lysates or with mock-infected Vero cell lysates at 50 μg/ml. P values of <0.05 are shown. (B) Memory T cell responses in mumps virus i.n.-immunized mice. BALB/c mice were i.m. immunized with PBS, rMuV, JL, rMuVΔSH, rMuVΔV, or rMuVΔSHΔV at 106 PFU and boosted at 22 days postvaccination with the same virus at 106 PFU. Splenocytes were extracted from mouse spleens and used for ELISPOT assay. Splenocytes were stimulated with MuV-infected Vero cell lysates or with mock-infected Vero cell lysates at 50 μg/ml. P values of <0.05 are shown.
DISCUSSION
The JL vaccine is one of the most successful vaccines developed during the third quarter of the last century. It was produced by the propagation of mumps virus in embryonated hen's eggs that resulted in attenuation (48–51). Introduction of the in vitro tissue/cell culture technique into vaccinology facilitated the development and production of the majority of currently licensed live-attenuated vaccines in the United States against viral infections (52–56). For mumps vaccine candidates, JL is the great success, but unfortunately there were many failures. Different passages of attenuated viruses were tested in animal models or in field trials in order to select a vaccine seed with the most reduced virulence and greatest immunogenicity (1). Selected vaccine candidates need to be biologically characterized in order to be distinguished from virulent strains. There are currently no standardized attenuation markers for mumps vaccines, partially due to the semirational and semiempirical nature of the traditional attenuation method (1, 57–60). The rate of aseptic meningitis following vaccination with JL (estimated one case per 1.8 million doses) is below background levels (61). However, other live attenuated mumps virus vaccines have had much higher incidences of vaccine-associated meningitis. The Urabe vaccine, which was widely distributed in Japan, Europe, and Canada, is estimated to cause one case of meningitis in every 1,000 to 11,000 doses distributed in the United Kingdom and one case of meningitis in every 62,000 doses distributed in Canada. The Urabe vaccine has been withdrawn due to safety concerns.
In this study, based on the establishment of reverse genetic technology of negative-sensed, nonsegmented RNA viruses (62–66), we examined the possibility of generating a new mumps vaccine candidate through deletion of V and SH protein expression from a clinical isolate from a 2006 Iowa mumps epidemic (MuV, genotype G). Deletion of either of the proteins has been previously reported to reduce mumps neurotoxicity in IC-infected rats. Importantly, since deletion of the V protein alone (from the MuV strain) is sufficient to reduce the neurovirulence potential of the recombinant virus to a level comparable to that of the JL vaccine (21, 36), the lack of V protein expression can be used as an attenuation marker for our vaccine candidates.
Attenuation based on targeted genetic modification has several advantages. The most commonly used mumps vaccine, JL, is a mixture of at least two well-distinguished components (67–69). Surveillance of the compositional balance between the two components during vaccine preparation and propagation has been proposed (68). However, cDNA-derived recombinant viruses have defined consensus sequences and higher homogeneity. They are plaque purified, whole genome sequenced, and passed in Vero cells only for the purpose of amplification. Vaccine candidates can be continuously rescued from the cDNA plasmids with a defined consensus sequence and clear genetic markers for attenuation. All processes are cell culture based, bypassing the necessity of high-quality pathogen-free chickens, chicken eggs, or any other animals used for in vivo adaption (70–72). Omission of the serial passages saves time for vaccine development and avoids potential adaption-induced antigen shifts of the vaccine strains; therefore, it retains the maximum amount of immunogenic epitopes.
Mumps viruses are classified into 12 genotypes based on genetic variability of the SH gene (73, 74). Different subtypes of mumps viruses exhibit distinguished geographic distribution worldwide. Although the driving force of such distribution remains unclear, emergence of new subclusters of circulating mumps viruses within a genotype (75, 76) indicates evolution of wild-type mumps viruses under various selection forces. Failure to detect genotype A wild-type mumps viruses in countries/regions immunized with genotype A vaccine in recent studies may be due to a vaccine-based selection pressure. This pressure may select for genotypes with increased virulence and heterogeneity compared to current vaccines (77–82). Decreased neutralization capabilities against heterogenotypes among subtypes of mumps viruses (83–86) and lack of cross protection between different subtypes (genotype D against genotype A) in human natural infection have been reported (87). It would be ideal to use a genotype-matched vaccine candidate (genotype G), which elicits more specific immune responses that effectively protect against the circulating mumps viruses in the United States (genotype G) (11, 13, 78, 79).
Although additional mutations occurred during rMuVΔSHΔV virus rescue and during passages of rMuVΔSHΔV in Vero cells, no regaining of the V protein or the SH protein was observed in any rMuVΔSHΔV viruses analyzed, indicating that rMuVΔSHΔV is stable in tissue culture cells. Interestingly, besides two silent mutations in the HN and L ORFs, rMuVΔSHΔV (PX64-67) possessed one nucleotide change (C-T) in genomic position 1913 in the V/P GS region, which has been previously seen in the course of rMuVΔV virus rescue (36). This mutation is believed to be important in regulating the transcription/translation level of P protein, emphasizing the significance of a proper ratio between NP and P protein during virus growth.
One challenge of developing a new mumps vaccine is the lack of correlation between protection and immune responses. While a neutralization titer is thought to be essential in protection against mumps infection (79), investigations of serum samples of patients versus nonpatients during recent mumps outbreaks revealed no defined cutoff neutralizing antibody titer against mumps virus, indicating a potential role for cellular immunity in effective protection against mumps challenge (41, 86). In this study, the investigation of immunogenicity of rMuVΔSHΔV in i.n.- and i.m.-vaccinated mice showed that rMuVΔSHΔV was able to induce a neutralizing titer comparable to those induced by rMuVΔSH and rMuVΔV and a higher titer than that induced by JL vaccine. Furthermore, rMuVΔSHΔV vaccination also stimulated T cell responses in mice, although the role of cell-mediated immunity in mumps disease protection remains to be demonstrated. We also observed that rMuVΔSH induced slightly higher total antibody titers than those induced by rMuV, and rMuVΔSHΔV induced higher antibody titers than those induced by rMuVΔV, suggesting that deletion of SH leads to better antigen presentation. Similar results have been reported for a closely related virus, parainfluenza virus 5 (PIV5), in which PIV5 lacking SH is more immunogenic than PIV5 (88). The mouse models have been widely used to test vaccine efficacy for various human viruses (89–92). However, it is not a good model for mumps virus infection. The efficacy of rMuVΔSHΔV in nonhuman primates, which is a good model for mumps virus infection (40), should be examined before testing this candidate in humans. In summary, rMuVΔSHΔV was able to elicit both antibody and cellular responses against MuV in i.n.- and i.m.-vaccinated mice, providing a safe and immunogenic mumps vaccine candidate.
ACKNOWLEDGMENTS
We appreciate the comments, suggestions, and technical help from members of the He laboratory. We thank Paul Rota for providing the JL vaccine strain. We are grateful to Kaori Sakamoto for carefully reading the manuscript prior to submission.
This work has been supported by a grant from the NIH (AI097368 to B.H.).
Footnotes
Published ahead of print 18 December 2013
REFERENCES
- 1.Buynak EB, Hilleman MR. 1966. Live attenuated mumps virus vaccine. 1. Vaccine development. Proc. Soc. Exp. Biol. Med. 123:768–775. 10.3181/00379727-123-31599 [DOI] [PubMed] [Google Scholar]
- 2.Pagano JS, Levine RH, Sugg WC, Finger JA. 1967. Clinical trial of new attenuated mumps virus vaccine (Jeryl Lynn strain): preliminary report. Prog. Immunobiol. Stand. 3:196–202 [PubMed] [Google Scholar]
- 3.Weibel RE, Stokes J, Jr, Buynak EB, Leagus MB, Hilleman MR. 1968. Jeryl Lynn strain live attenuated mumps virus vaccine. Durability of immunity following administration. JAMA 203:14–18 [PubMed] [Google Scholar]
- 4.Young ML, Dickstein B, Weibel RE, Stokes J, Jr, Buynak EB, Hilleman MR. 1967. Experiences with Jeryl Lynn strain live attenuated mumps virus vaccine in a pediatric outpatient clinic. Pediatrics 40:798–803 [PubMed] [Google Scholar]
- 5.Rubin SA, Afzal MA, Powell CL, Bentley ML, Auda GR, Taffs RE, Carbone KM. 2005. The rat-based neurovirulence safety test for the assessment of mumps virus neurovirulence in humans: an international collaborative study. J. Infect. Dis. 191:1123–1128. 10.1086/428098 [DOI] [PubMed] [Google Scholar]
- 6.Carbone KM, Wolinsky JS. 2001. Mumps virus, p 1381–1400 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4 ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 7.Smits G, Mollema L, Hahne S, de Melker H, Tcherniaeva I, Waaijenborg S, van Binnendijk R, van der Klis F, Berbers G. 2013. Seroprevalence of mumps in the Netherlands: dynamics over a decade with high vaccination coverage and recent outbreaks. PLoS One 8:e58234. 10.1371/journal.pone.0058234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahamud A, Fiebelkorn AP, Nelson G, Aguon A, McKenna J, Villarruel G, Gallagher K, Ortega-Sanchez IR. 2012. Economic impact of the 2009–2010 Guam mumps outbreak on the public health sector and affected families. Vaccine 30:6444–6448. 10.1016/j.vaccine.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 9.Rajcevic S, Seguljev Z, Petrovic V, Medic S, Nedelijkovic J, Milosevic V, Turo L, Ristic M. 2012. Ongoing mumps outbreak in Novi Sad, the autonomous province of Vojvodina, Serbia, January to April 2012. Euro Surveill. 17:20169. [PubMed] [Google Scholar]
- 10.Hassan J, Dean J, Moss E, Carr MJ, Hall WW, Connell J. 2012. Seroepidemiology of the recent mumps virus outbreaks in Ireland. J. Clin. Virol. 53:320–324. 10.1016/j.jcv.2011.12.022 [DOI] [PubMed] [Google Scholar]
- 11.Marin M, Quinlisk P, Shimabukuro T, Sawhney C, Brown C, Lebaron CW. 2008. Mumps vaccination coverage and vaccine effectiveness in a large outbreak among college students–Iowa, 2006. Vaccine 26:3601–3607. 10.1016/j.vaccine.2008.04.075 [DOI] [PubMed] [Google Scholar]
- 12.Barskey AE, Glasser JW, LeBaron CW. 2009. Mumps resurgences in the United States: a historical perspective on unexpected elements. Vaccine 27:6186–6195. 10.1016/j.vaccine.2009.06.109 [DOI] [PubMed] [Google Scholar]
- 13.Anonymous. 2009. Mumps outbreak–New York, New Jersey, Quebec, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:1270–1274 [PubMed] [Google Scholar]
- 14.Gordon JE, Kilham L. 1949. Ten years in the epidemiology of mumps. Am. J. Med. Sci. 218:338–359. 10.1097/00000441-194909000-00013 [DOI] [PubMed] [Google Scholar]
- 15.Rima BK, Roberts MW, McAdam WD, Martin SJ. 1980. Polypeptide synthesis in mumps virus-infected cells. J. Gen. Virol. 46:501–505. 10.1099/0022-1317-46-2-501 [DOI] [PubMed] [Google Scholar]
- 16.Elango N, Varsanyi TM, Kovamees J, Norrby E. 1988. Molecular cloning and characterization of six genes, determination of gene order and intergenic sequences and leader sequence of mumps virus. J. Gen. Virol. 69(Part 11):2893–2900. 10.1099/0022-1317-69-11-2893 [DOI] [PubMed] [Google Scholar]
- 17.Hosaka Y, Shimizu K. 1968. Lengths of the nucleocapsids of Newcastle disease and mumps viruses. J. Mol. Biol. 35:369–373. 10.1016/S0022-2836(68)80031-2 [DOI] [PubMed] [Google Scholar]
- 18.Fuentes S, Tran KC, Luthra P, Teng MN, He B. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81:8361–8366. 10.1128/JVI.02717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Bright AC, Rothermel TA, He B. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 77:3371–3383. 10.1128/JVI.77.6.3371-3383.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson RL, Fuentes SM, Wang P, Taddeo EC, Klatt A, Henderson AJ, He B. 2006. Function of small hydrophobic proteins of paramyxovirus. J. Virol. 80:1700–1709. 10.1128/JVI.80.4.1700-1709.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, Li Z, Sun D, Lin Y, Wu J, Rota PA, He B. 2011. Rescue of wild-type mumps virus from a strain associated with recent outbreaks helps to define the role of the SH ORF in the pathogenesis of mumps virus. Virology 417:126–136. 10.1016/j.virol.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Xu J, Patel J, Fuentes S, Lin Y, Anderson D, Sakamoto K, Wang LF, He B. 2011. Function of the small hydrophobic protein of J paramyxovirus. J. Virol. 85:32–42. 10.1128/JVI.01673-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in mdbk cells. J. Virol. 75:4068–4079. 10.1128/JVI.75.9.4068-4079.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott GD, Yeo RP, Afzal MA, Simpson EJ, Curran JA, Rima BK. 1990. Strain-variable editing during transcription of the P gene of mumps virus may lead to the generation of non-structural proteins NS1 (V) and NS2. J. Gen. Virol. 71(Part 7):1555–1560. 10.1099/0022-1317-71-7-1555 [DOI] [PubMed] [Google Scholar]
- 25.Paterson RG, Lamb RA. 1990. RNA editing by G-nucleotide insertion in mumps virus P-gene mRNA transcripts. J. Virol. 64:4137–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motz C, Schuhmann KM, Kirchhofer A, Moldt M, Witte G, Conzelmann KK, Hopfner KP. 2013. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 339:690–693. 10.1126/science.1230949 [DOI] [PubMed] [Google Scholar]
- 27.Parisien JP, Bamming D, Komuro A, Ramachandran A, Rodriguez JJ, Barber G, Wojahn RD, Horvath CM. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260. 10.1128/JVI.00153-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran A, Horvath CM. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163. 10.1128/JVI.01375-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishio M, Garcin D, Simonet V, Kolakofsky D. 2002. The carboxyl segment of the mumps virus V protein associates with stat proteins in vitro via a tryptophan-rich motif. Virology 300:92. 10.1006/viro.2002.1509 [DOI] [PubMed] [Google Scholar]
- 30.Poole E, He B, Lamb RA, Randall RE, Goodbourn S. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33–46. 10.1006/viro.2002.1737 [DOI] [PubMed] [Google Scholar]
- 31.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385–6393. 10.1128/JVI.77.11.6385-6393.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokosawa N, Yokota S, Kubota T, Fujii N. 2002. C-terminal region of STAT-1alpha is not necessary for its ubiquitination and degradation caused by mumps virus V protein. J. Virol. 76:12683–12690. 10.1128/JVI.76.24.12683-12690.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulane CM, Horvath CM. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160–166. 10.1006/viro.2002.1773 [DOI] [PubMed] [Google Scholar]
- 34.Andrejeva J, Poole E, Young DF, Goodbourn S, Randall RE. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379–11386. 10.1128/JVI.76.22.11379-11386.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parisien JP, Lau JF, Rodriguez JJ, Ulane CM, Horvath CM. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190–4198. 10.1128/JVI.76.9.4190-4198.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, Luthra P, Li Z, Fuentes S, D'Andrea JA, Wu J, Rubin S, Rota PA, He B. 2012. The V protein of mumps virus plays a critical role in pathogenesis. J. Virol. 86:1768–1776. 10.1128/JVI.06019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He B, Lamb RA. 1999. Effect of inserting paramyxovirus simian virus 5 gene junctions at the HN/L gene junction: analysis of accumulation of mRNAs transcribed from rescued viable viruses. J. Virol. 73:6228–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He B, Paterson RG, Ward CD, Lamb RA. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249–260. 10.1006/viro.1997.8801 [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Yu M, Zhang H, Magoffin DE, Jack PJ, Hyatt A, Wang HY, Wang LF. 2006. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology 346:219–228. 10.1016/j.virol.2005.10.039 [DOI] [PubMed] [Google Scholar]
- 40.Xu P, Huang Z, Gao X, Michel FJ, Hirsch G, Hogan RJ, Sakamoto K, Ho W, Wu J, He B. 2013. Infection of mice, ferrets, and rhesus macaques with a clinical mumps virus isolate. J. Virol. 87:8158–8168. 10.1128/JVI.01028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortese MM, Barskey AE, Tegtmeier GE, Zhang C, Ngo L, Kyaw MH, Baughman AL, Menitove JE, Hickman CJ, Bellini WJ, Dayan GH, Hansen GR, Rubin S. 2011. Mumps antibody levels among students before a mumps outbreak: in search of a correlate of immunity. J. Infect. Dis. 204:1413–1422. 10.1093/infdis/jir526 [DOI] [PubMed] [Google Scholar]
- 42.Carter H, Campbell H. 1993. Rational use of measles, mumps and rubella (MMR) vaccine. Drugs 45:677–683. 10.2165/00003495-199345050-00005 [DOI] [PubMed] [Google Scholar]
- 43.Calain P, Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822–4830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Chang JS, Nason M, Rangel D, Gall JG, Graham BS, Ledgerwood JE. 2010. A flow cytometry-based assay to assess RSV-specific neutralizing antibody is reproducible, efficient and accurate. J. Immunol. Methods 362:180–184. 10.1016/j.jim.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haralambieva IH, Ovsyannikova IG, Vierkant RA, Poland GA. 2008. Development of a novel efficient fluorescence-based plaque reduction microneutralization assay for measles virus immunity. Clin. Vaccine Immunol. 15:1054–1059. 10.1128/CVI.00008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprangers MC, Lakhai W, Koudstaal W, Verhoeven M, Koel BF, Vogels R, Goudsmit J, Havenga MJ, Kostense S. 2003. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J. Clin. Microbiol. 41:5046–5052. 10.1128/JCM.41.11.5046-5052.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuentes S, Crim RL, Beeler J, Teng MN, Golding H, Khurana S. 2013. Development of a simple, rapid, sensitive, high-throughput luciferase reporter based microneutralization test for measurement of virus neutralizing antibodies following respiratory syncytial virus vaccination and infection. Vaccine 31:3987–3994. 10.1016/j.vaccine.2013.05.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodpasture EW, Woodruff AM, Buddingh GJ. 1931. The cultivation of vaccine and other viruses in the chorioallantoic membrane of chick embryos. Science 74:371–372. 10.1126/science.74.1919.371 [DOI] [PubMed] [Google Scholar]
- 49.Habel K. 1946. Preparation of mumps vaccines and immunization of monkeys against experimental mumps infection. Public Health Rep. 61:1655–1664. 10.2307/4585906 [DOI] [PubMed] [Google Scholar]
- 50.Levens JH, Enders JF. 1945. The hemoagglutinative properties of amniotic fluid from embryonated eggs infected with mumps virus. Science 102:117–120. 10.1126/science.102.2640.117 [DOI] [PubMed] [Google Scholar]
- 51.Enders JF, Levens JH, et al. 1946. Attenuation of virulence with retention of antigenicity of mumps virus after passage in the embryonated egg. J. Immunol. 54:283–291 [PubMed] [Google Scholar]
- 52.Hilleman MR. 2002. Overview of the needs and realities for developing new and improved vaccines in the 21st century. Intervirology 45:199–211. 10.1159/000067911 [DOI] [PubMed] [Google Scholar]
- 53.Hilleman MR. 2002. Overview: past and future of immunologic intervention in the pathogenesis, prophylaxis and therapeusis of hepatitis B. J. Gastroenterol. Hepatol. 17(Suppl):S449–S451. 10.1046/j.1440-1746.17.s4.8.x [DOI] [PubMed] [Google Scholar]
- 54.U S. FDA. 2013. Complete list of vaccines licensed for immunization and distribution in the US. U.S. FDA, Washington, DC [Google Scholar]
- 55.Malik H, Khan FH, Ahsan H. 2013. Human papillomavirus: current status and issues of vaccination. Arch. Virol.[Epub ahead of print.] 10.1007/s00705-013-1827-z [DOI] [PubMed] [Google Scholar]
- 56.Doan HQ, Ung B, Ramirez-Fort MK, Khan F, Tyring SK. 2013. Zostavax: a subcutaneous vaccine for the prevention of herpes zoster. Expert Opin. Biol. Ther. 13:1467–1477. 10.1517/14712598.2013.830101 [DOI] [PubMed] [Google Scholar]
- 57.St Geme JW, Hawley LM, Davis WC. 1976. Comparative replication of natural, attenuated, and laboratory strains of mumps virus. Life Sci. 18:129–134. 10.1016/0024-3205(76)90283-6 [DOI] [PubMed] [Google Scholar]
- 58.Yamanishi K, Hosai H, Ueda S, Takahashi M, Okuno Y. 1970. Studies on live attenuated mumps virus vaccine. II. Biological characteristics of the strains adapted to the amniotic and chorioallantoic cavity of developing chick embryos. Biken J. 13:127–132 [PubMed] [Google Scholar]
- 59.Kaptsova TI, Alekseeva AK, Gordienko NM, Rozina EE, Ermakova MN. 1976. Results of a study of live mumps vaccine from strain L-3 produced by the Moscow research institute for viral preparations. Characteristics of the vaccine. Voprosy Virusologii 1976:674–685 [PubMed] [Google Scholar]
- 60.Gluck R, Hoskins JM, Wegmann A, Just M, Germanier R. 1986. Rubini, a new live attenuated mumps vaccine virus strain for human diploid cells. Dev. Biol. Stand. 65:29–35 [PubMed] [Google Scholar]
- 61.Nalin DR. 1989. Mumps vaccine complications: which strain? Lancet ii:1396. [DOI] [PubMed] [Google Scholar]
- 62.Schnell MJ, Mebatsion T, Conzelmann KK. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. U. S. A. 92:11563–11567. 10.1073/pnas.92.25.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350. 10.1073/pnas.96.16.9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Afzal MA, Pickford AR, Forsey T, Heath AB, Minor PD. 1993. The Jeryl Lynn vaccine strain of mumps virus is a mixture of two distinct isolates. J. Gen. Virol. 74(Part 5):917–920. 10.1099/0022-1317-74-5-917 [DOI] [PubMed] [Google Scholar]
- 68.Amexis G, Rubin S, Chizhikov V, Pelloquin F, Carbone K, Chumakov K. 2002. Sequence diversity of Jeryl Lynn strain of mumps virus: quantitative mutant analysis for vaccine quality control. Virology 300:171–179. 10.1006/viro.2002.1499 [DOI] [PubMed] [Google Scholar]
- 69.Chambers P, Rima BK, Duprex WP. 2009. Molecular differences between two Jeryl Lynn mumps virus vaccine component strains, JL5 and JL2. J. Gen. Virol. 90:2973–2981. 10.1099/vir.0.013946-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barrett PN, Mundt W, Kistner O, Howard MK. 2009. Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines. Expert Rev. Vaccines 8:607–618. 10.1586/erv.09.19 [DOI] [PubMed] [Google Scholar]
- 71.Clark HF, Offit PA, Plotkin SA, Heaton PM. 2006. The new pentavalent rotavirus vaccine composed of bovine (strain WC3)-human rotavirus reassortants. Pediatr. Infect. Dis. J. 25:577–583. 10.1097/01.inf.0000220283.58039.b6 [DOI] [PubMed] [Google Scholar]
- 72.Bernstein DI. 2006. Live attenuated human rotavirus vaccine, Rotarix. Semin. Pediatr. Infect. Dis. 17:188–194. 10.1053/j.spid.2006.08.006 [DOI] [PubMed] [Google Scholar]
- 73.Jin L, Rima B, Brown D, Orvell C, Tecle T, Afzal M, Uchida K, Nakayama T, Song JW, Kang C, Rota PA, Xu W, Featherstone D. 2005. Proposal for genetic characterisation of wild-type mumps strains: preliminary standardisation of the nomenclature. Arch. Virol. 150:1903–1909. 10.1007/s00705-005-0563-4 [DOI] [PubMed] [Google Scholar]
- 74.Santos CL, Ishida MA, Foster PG, Sallum MA, Benega MA, Borges DB, Correa KO, Constantino CR, Afzal MA, Paiva TM. 2008. Detection of a new mumps virus genotype during parotitis epidemic of 2006–2007 in the state of Sao Paulo, Brazil. J. Med. Virol. 80:323–329. 10.1002/jmv.21068 [DOI] [PubMed] [Google Scholar]
- 75.Tecle T, Mickiene A, Johansson B, Lindquist L, Orvell C. 2002. Molecular characterisation of two mumps virus genotypes circulating during an epidemic in Lithuania from 1998 to 2000. Arch. Virol. 147:243–253. 10.1007/s705-002-8317-y [DOI] [PubMed] [Google Scholar]
- 76.Utz S, Richard JL, Capaul S, Matter HC, Hrisoho MG, Muhlemann K. 2004. Phylogenetic analysis of clinical mumps virus isolates from vaccinated and non-vaccinated patients with mumps during an outbreak, Switzerland 1998–2000. J. Med. Virol. 73:91–96. 10.1002/jmv.20064 [DOI] [PubMed] [Google Scholar]
- 77.Greenland K, Whelan J, Fanoy E, Borgert M, Hulshof K, Yap KB, Swaan C, Donker T, van Binnendijk R, de Melker H, Hahne S. 2012. Mumps outbreak among vaccinated university students associated with a large party, the Netherlands, 2010. Vaccine 30:4676–4680. 10.1016/j.vaccine.2012.04.083 [DOI] [PubMed] [Google Scholar]
- 78.Anonymous. 2012. Mumps outbreak on a university campus–California, 2011. MMWR Morb. Mortal. Wkly. Rep. 61:986–989 [PubMed] [Google Scholar]
- 79.Hviid A, Rubin S, Muhlemann K. 2008. Mumps. Lancet 371:932–944. 10.1016/S0140-6736(08)60419-5 [DOI] [PubMed] [Google Scholar]
- 80.Echevarria JE, Castellanos A, Sanz JC, Perez C, Palacios G, Martinez de Aragon MV, Pena Rey I, Mosquera M, de Ory F, Royuela E. 2010. Circulation of mumps virus genotypes in Spain from 1996 to 2007. J. Clin. Microbiol. 48:1245–1254. 10.1128/JCM.02386-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Echevarria AC, Sanz JEJC, Martinez de Aragon MV, Pena Rey I, Mosquera M, Fde Ory Royuela E. 2010. Mumps virus genotyping: basis and known circulating genotypes. Open Vaccine J. 3:37–41. 10.2174/1875035401003020037 [DOI] [Google Scholar]
- 82.Muhlemann K. 2004. The molecular epidemiology of mumps virus. Infect. Genet. Evol. 4:215–219. 10.1016/j.meegid.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 83.Santak M, Lang-Balija M, Ivancic-Jelecki J, Kosutic-Gulija T, Ljubin-Sternak S, Forcic D. 2013. Antigenic differences between vaccine and circulating wild-type mumps viruses decreases neutralization capacity of vaccine-induced antibodies. Epidemiol. Infect. 141:1298–1309. 10.1017/S0950268812001896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orvell C, Tecle T, Johansson B, Saito H, Samuelson A. 2002. Antigenic relationships between six genotypes of the small hydrophobic protein gene of mumps virus. J. Gen. Virol. 83:2489–2496 [DOI] [PubMed] [Google Scholar]
- 85.Rubin SA, Qi L, Audet SA, Sullivan B, Carbone KM, Bellini WJ, Rota PA, Sirota L, Beeler J. 2008. Antibody induced by immunization with the Jeryl Lynn mumps vaccine strain effectively neutralizes a heterologous wild-type mumps virus associated with a large outbreak. J. Infect. Dis. 198:508–515. 10.1086/590115 [DOI] [PubMed] [Google Scholar]
- 86.Rubin SA, Link MA, Sauder CJ, Zhang C, Ngo L, Rima BK, Duprex WP. 2012. Recent mumps outbreaks in vaccinated populations: no evidence of immune escape. J. Virol. 86:615–620. 10.1128/JVI.06125-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nojd J, Tecle T, Samuelsson A, Orvell C. 2001. Mumps virus neutralizing antibodies do not protect against reinfection with a heterologous mumps virus genotype. Vaccine 19:1727–1731. 10.1016/S0264-410X(00)00392-3 [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Gabbard JD, Mooney A, Chen Z, Tompkins SM, He B. 2013. Efficacy of parainfluenza virus 5 mutants expressing hemagglutinin from H5N1 influenza A virus in mice. J. Virol. 87:9604–9609. 10.1128/JVI.01289-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boyoglu-Barnum S, Gaston KA, Todd SO, Boyoglu C, Chirkova T, Barnum TR, Jorquera P, Haynes LM, Tripp RA, Moore ML, Anderson LJ. 2013. A respiratory syncytial virus (RSV) anti-G protein F(ab′)2 monoclonal antibody suppresses mucous production and breathing effort in RSV rA2-line19F-infected Balb/c mice. J. Virol. 87:10955–10967. 10.1128/JVI.01164-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu L, Bao L, Deng W, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, Xu Y, Huang L, Li Y, Liu J, Yao Y, Yu P, Yong W, Wei Q, Zhang L, Qin C. 2013. The mouse and ferret models for studying the novel avian-origin human influenza A (H7N9) virus. Virol. J. 10:253. 10.1186/1743-422X-10-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Gabbard JD, Mooney A, Gao X, Chen Z, Place RJ, Tompkins SM, He B. 2013. Single-dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 virus provides broad immunity against influenza A viruses. J. Virol. 87:5985–5993. 10.1128/JVI.00120-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Z, Mooney AJ, Gabbard JD, Gao X, Xu P, Place RJ, Hogan RJ, Tompkins SM, He B. 2013. Recombinant parainfluenza virus 5 expressing hemagglutinin of influenza A virus H5N1 protected mice against lethal highly pathogenic avian influenza virus H5N1 challenge. J. Virol. 87:354–362. 10.1128/JVI.02321-12 [DOI] [PMC free article] [PubMed] [Google Scholar]