Abstract
In goats, several field studies have identified coding mutations of the gene encoding the prion protein (I/M142, N/D146, S/D146, R/Q211, and Q/K222) that are associated with a lower risk of developing classical scrapie. However, the data related to the levels of resistance to transmissible spongiform encephalopathies (TSEs) of these different PRNP gene mutations are still considered insufficient for developing large-scale genetic selection against scrapie in this species. In this study, we inoculated wild-type (WT) PRNP (I142R154R211Q222) goats and homozygous and/or heterozygous I/M142, R/H154, R/Q211, and Q/K222 goats with a goat natural scrapie isolate by either the oral or the intracerebral (i.c.) route. Our results indicate that the I/M142 PRNP polymorphism does not provide substantial resistance to scrapie infection following intracerebral or oral inoculation. They also demonstrate that H154, Q211, and K222 PRNP allele carriers are all resistant to scrapie infection following oral exposure. However, in comparison to WT animals, the H154 and Q211 allele carriers displayed only moderate increases in the incubation period following i.c. challenge. After i.c. challenge, heterozygous K222 and a small proportion of homozygous K222 goats also developed the disease, but with incubation periods that were 4 to 5 times longer than those in WT animals. These results support the contention that the K222 goat prion protein variant provides a strong but not absolutely protective effect against classical scrapie.
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal neurodegenerative disorders occurring in small ruminants (scrapie), cattle (bovine spongiform encephalopathy [BSE]), and humans (Creutzfeldt-Jakob disease [CJD]). The key event in TSEs is the conversion of a normal cellular protein (PrPC) into an abnormal isoform (PrPSc) which accumulates in tissues in infected individuals. According to the prion concept, abnormal PrP is the causative agent of TSEs (1).
In sheep, susceptibility to TSEs is strongly modulated by polymorphisms of the prion protein gene (PRNP) and the nature of the prion disease agent (strain) (2). The A136R154R171 allele is associated with a highly protective effect against natural or experimental infection with classical scrapie and BSE agents, while the V136R154Q171 allele and the wild-type (WT) A136R154Q171 allele are associated with susceptibility (3, 4). However, in sheep, the ARR allele does not provide any particular protection against atypical scrapie, whereas the R/H154 or L/F141 amino acid substitution is associated with an increased risk of occurrence of this TSE (5–8).
At the European level, the selection of ARR allele carriers was successfully applied for controlling and eradicating classical scrapie in infected sheep flocks (9). Large-scale selection programs were also implemented at the population level. They aimed to increase the frequency of the ARR allele in the general population, making it less favorable to TSE agent circulation and spreading. This “breeding for resistance” policy, in combination with other eradication measures, resulted in a significant reduction of the prevalence of classical scrapie in populations where it was comprehensively applied (10–12).
In goats, several field studies have identified coding mutations of the PRNP gene that are associated with a lower risk of developing classical scrapie, namely, the I/M142, N/D146 and S146, R/Q211, and Q/K222 alleles (13–18). However, the low frequencies of these alleles in the goat population limit the possibility of reaching an unequivocal conclusion about the resistance/susceptibility to infection associated with these different PRNP genotypes (2, 13, 18). In this context, experimental TSE inoculation of goats is a straightforward and robust approach to better assess the levels of resistance associated with certain PRNP polymorphisms in this species (19, 20).
In this study, we inoculated wild-type goats and homozygous and/or heterozygous I/M142, R/H154, R/Q211, and Q/K222 goats with a goat natural scrapie isolate, by either the intracerebral (i.c.) or the oral route, in order to characterize their relative resistances/susceptibilities to infection.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were performed in compliance with institutional and French national guidelines, in accordance with European Community Council Directive 86/609/EEC. The experimental protocol was approved by the INRA Toulouse/ENVT ethics committee.
Scrapie inoculum.
The inoculum was derived from a single natural scrapie field case (clinical) in a 3.5-year-old goat with the wild-type PRNP genotype. This animal was necropsied under TSE-sterile conditions, and its central nervous system (CNS; brain and spinal cord) was used to prepare a 10% tissue homogenate in 5% glucose. The stock homogenate was aliquoted and stored at −80°C.
Experimental animal production.
Goat kids intended to be used in the experiment were produced by direct mating of PRNP-sequenced Alpine and Saanen female goats and bucks. Parents were selected from three herds that are managed by the French National Agronomic Institute (INRA). Selection was based on the PRNP polymorphisms at codons 142 (I/M), 154 (R/H), 211 (R/Q), and 222 (Q/K), which were identified by previous studies as influencing susceptibility to natural scrapie (14, 18, 21). Animals were then naturally mated to produce the goats used for experimental inoculation. Exon 3 of the PRNP gene of each goat kid was sequenced as previously described (22).
Goat oral challenge experiments.
For the oral challenge experiments, gravid goats were relocated to ANSES Niort A2 facilities. Within 48 h after birth, each goat kid received 1.5 g brain-equivalent material through natural suckling (1% diluted stock inoculum in 5% glucose). A second inoculation (same material and route) was performed at the age of 30 days. Considering (i) the logistic constraints (housing of goats and goat kids) and (ii) the fact that the parent goats were heterozygous only for the alleles of interest, the oral inoculation of goat kids that would have been homozygous for the mutated PRNP alleles was not feasible in the framework of this experiment.
Two separate oral inoculation experiments were performed. The first one aimed at establishing the PrPSc dissemination scheme and kinetics in animals with the wild-type PRNP genotype. For that purpose, 3 or 4 animals were culled at 30, 90, 120, 360, 540, and 940 days postinoculation (dpi). A last group of animals (n = 4) was kept until the occurrence of clinical signs.
The second experiment aimed at establishing the relative susceptibilities of goats harboring various genotypes to scrapie following oral exposure. WT and heterozygous I/M142, R/Q211, and Q/K222 animals were orally challenged using the same isolate as in the first experiment and were culled at 120, 360, 760, and 1,040 dpi. Five animals of each genotype were killed at each of the different time points.
In addition, a group of animals from each of these genotypes and a group (n = 6) of heterozygous R/H154 animals that had also been challenged orally were kept alive for establishment of the incubation period. Because of space constraints in the animal facilities, it was not possible to challenge a sufficient number of R/H154 PRNP allele carriers to complete the time point experiment (see below).
Goat intracerebral challenge.
After weaning, goat kids selected by genotype were transported to UMR INRA ENVT A2 animal facilities for i.c. inoculation. When the animals were 6 months of age, they were anesthetized (ketamine plus diazepam [Valium]), and 400 μl of the stock inoculum was injected into the temporal cortex.
Clinical monitoring and sample collection.
Inoculated goats were clinically monitored on a daily basis. The animals that developed TSE were euthanized if they exhibited locomotor signs that impaired their feeding capacity. Animals that developed intercurrent health problems were treated by qualified veterinarians and euthanized if the condition was not curable.
Dead animals were systematically necropsied, and the CNS and a variety of lymphoid (mesenteric lymph nodes, tonsils, prescapular lymph nodes, and Peyer's patches) and nonlymphoid tissues were collected (Table 1). Half of the samples were formalin fixed, while the other half were stored frozen (−20°C).
TABLE 1.
PrPSc detection in tissues of WT PRNP goats orally challenged with scrapie agent and sequentially killed
| Organa | 30 dpi |
90 dpi |
180 dpi |
360 dpi |
540 dpi |
940 dpi |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | |
| Obex | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 0/4 | − | 4/4 | +/+++ |
| Spinal cord (cervical) | ND | ND | ND | 0/3 | − | 0/4 | − | 4/4 | +/++ | |||
| Spinal cord (thoracic) | ND | ND | ND | 0/3 | − | 0/4 | − | 4/4 | +/+++ | |||
| Spinal cord (lumbar) | ND | ND | ND | 0/3 | − | 0/4 | − | 4/4 | +/+++ | |||
| Tonsil | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | ++ | 2/4 (a, b) | +/+++ | 4/4 | +++ |
| Parotid LN | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | + | 3/4 (a to c) | ++/+++ | 4/4 | +++ |
| Retropharyngeal LN | 0/3 | − | 0/4 | − | 0/3 | − | 2/3 (a, b) | +/++ | 3/4 (a to c) | ++/+++ | 4/4 | ++++ |
| Spleen | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 1/4 (a) | + | 4/4 | +/+++ |
| Duodenal PP | 0/3 | − | 0/4 | − | 0/3 | − | 3/3 | +++ | 4/4 | +++ | 4/4 | +++ |
| Jejunal PP | 0/3 | − | 0/4 | − | 2/3 (a, b) | + | 3/3 | +++ | 4/4 | +++ | 4/4 | +++ |
| Ileal PP | 0/3 | − | 0/4 | − | 2/3 (a, b) | +/++ | 3/3 | +++ | 4/4 | +++ | 4/4 | +++ |
| Cecal PP | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | ++ | 4/4 | ++/+++ | 4/4 | ++/+++ |
| Jejunal MLN | 0/3 | − | 0/4 | − | 2/3 (a, b) | + | 3/3 | +/+++ | 4/4 | +++ | 4/4 | +++ |
| Ileal MLN | 0/3 | − | 0/4 | − | 1/3 (a) | + | 3/3 | +++ | 4/4 | ++++ | 4/4 | +++ |
| Mediastinal LN | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | + | 2/4 (a, b) | +/+++ | 4/4 | +++ |
| Prescapular LN | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | + | 0/4 | − | 4/4 | +/+++ |
| Retrohepatic LN | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | ++ | 4/4 | ++/+++ | 4/4 | ++/+++ |
| Duodenum (ENS) | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 4/4 | +/++ | 4/4 | +/++ |
| Jejunum (ENS) | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | + | 4/4 | +/++ | 4/4 | +/++ |
| Ileum (ENS) | 0/3 | − | 0/4 | − | 0/3 | − | 1/3 (a) | + | 4/4 | +/++ | 4/4 | +/++ |
| Cecum (ENS) | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 3/4 (a to c) | +/++ | 4/4 | +/++ |
| Colon (ENS) | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 3/4 (a to c) | +/++ | 4/4 | +/++ |
| Sciatic nerve | 0/3 | − | 0/4 | − | 0/3 | − | 0/3 | − | 0/4 | − | 4/4 | + |
| Brachial nerve | 0/3 | − | 0/3 | − | 0/3 | − | 0/3 | − | 0/4 | − | 4/4 | + |
| External ocular muscle | 0/3 | − | 0/3 | − | 0/3 | − | 0/3 | − | 0/4 | − | 4/4 | + |
For the spinal cord, cervical (C3-C4), thoracic (Th7-Th8), and lumbar (L3-L4) segments were analyzed. LN, lymph nodes; PP, Peyer's patches; MLN, mesenteric lymph nodes; ENS, enteric nervous system.
At each time point, the positive goats are identified arbitrarily by the letters a to d. ND, no data.
PrPSc labeling intensities are indicated as follows (as previously described [26]): −, negative; +, minimal to slight; ++, moderate; and +++, strong.
PrPSc IHC and PrPres enzyme-linked immunosorbent assay (ELISA) detection.
PrPSc immunohistochemistry (IHC) detection was performed as described by Lacroux et al., using the 8G8 antibody, raised against a human recombinant PrP protein and specifically recognizing amino acids 95 to 108 (SQWNKP) of the PrP protein (23).
WB of abnormal PrP.
Proteinase K (PK)-resistant abnormal PrP (PrPres) extraction and Western blotting (WB) were performed as previously described (24), using a commercial extraction kit (Bio-Rad, France). PrP immunodetection was performed using monoclonal antibody Sha31 (0.06 μg per ml) (YEDRYYRE epitope [amino acids 145 to 152]) or 12B2 (4 μg/ml) (WGQGG epitope [amino acids 93 to 97]) (25).
For glycoprofiling of PrPres, the signal volume and relative percentage associated with each band were established using Quantity One software (Bio-Rad) following immunoblotting. For each sample, three independent measures were determined for three different gels.
RESULTS
Oral challenge in goats.
Our first oral challenge experiment in goats was designed to establish the scheme and kinetics of PrPSc dissemination in the tissues of WT PRNP animals. For that purpose, goat kids obtained by natural mating of WT PRNP goats and bucks were orally challenged within the first 48 h following birth. Groups of these animals were culled at different time points after inoculation (Table 1). PrPSc accumulation was first observed in the gut-associated lymphoid tissue (Peyer's patches) of animals, at more than 180 dpi. As already described for sheep, PrPSc progressively spread to all lymphoid organs before becoming detectable (between 180 and 360 dpi) in the enteric nervous system (ENS) and, later (between 540 and 940 dpi), in the central nervous system (26).
On the basis of these results, a second oral challenge experiment was designed. The goal of this experiment was to characterize the impact of the investigated polymorphisms on the susceptibility and PrPSc dissemination in the tissues of orally exposed animals. Groups of wild-type animals and heterozygous I/M142, R/Q211, and Q/K222 animals were produced by natural mating and orally challenged using the same procedure and scrapie isolate as in the first experiment.
In the second experiment, the PrPSc dissemination scheme observed in WT animals was consistent with the results of the first experiment (Table 2). No PrPSc deposition was observed in the tissues collected from goats killed at 120 dpi. At 360 dpi, significant PrPSc deposition was observed in various lymphoid tissues (Peyer's patches, mesenteric lymph nodes, and tonsils) of some of the challenged individuals. PrPSc deposition was observed in ENS, CNS, and peripheral nervous tissues and skeletal muscles in four of the five animals culled at 760 dpi.
TABLE 2.
PrPSc detection in tissues of WT and I/M142 PRNP goats orally challenged with scrapie agent and sequentially killed
| Organa | 120 dpi |
360 dpi |
760 dpi |
1,040 dpi |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT |
I/M142 |
WT |
I/M142 |
WT |
I/M142 |
WT |
I/M142 |
|||||||||
| No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | No. of positive individuals/no. of animals testedb | PrPSc label intensityc | |
| Obex | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/+++ | 0/5 | − | 5/5 | +++ | 2/5 (a, b) | +/++ |
| Spinal cord (cervical) | ND | ND | 0/5 | − | 0/5 | − | 1/5 (a) | + | 0/5 | − | 4/5 (a to d) | +/+++ | 0/5 | − | ||
| Spinal cord (thoracic) | ND | ND | 0/5 | − | 0/5 | − | 1/5 (a) | ++ | 0/5 | − | 5/5 | ++/+++ | 0/5 | − | ||
| Spinal cord (lumbar) | ND | ND | 0/5 | − | 0/5 | − | 1/5 (a) | +/++ | 0/5 | − | 5/5 | ++/+++ | 0/5 | − | ||
| Tonsil | 0/5 | − | 0/5 | − | 1/5 (a) | + | 0/5 | − | 4/5 (a to d) | +/+++ | 5/5 | 5/5 | 3/5 (a to c) | +/+++ | ||
| Parotid LN | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/++ | 3/5 (a to c) | +/+++ | 5/5 | 3/5 (a to c) | +/+++ | |
| Retropharyngeal LN | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/+++ | 5/5 | +/+++ | 5/5 | 3/5 (a to c) | +/+++ | |
| Spleen | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/++ | 2/5 (a, b) | + | 5/5 | +/+++ | 2/5 (a, b) | + |
| Duodenal PP | 0/5 | − | 0/5 | − | 1/5 (a) | ++ | 0/5 | − | 5/5 | +++ | 5/5 | ++/+++ | 5/5 | ++/+++ | 4/5 (a to d) | ++/+++ |
| Jejunal PP | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | +++ | 5/5 | ++/+++ | 5/5 | +/+++ | 4/5 (a to d) | ++/+++ |
| Ileal PP | 0/5 | − | 0/5 | − | 2/5 (a, b) | ++ | 1/5 (a) | + | 5/5 | +++ | 5/5 | ++/+++ | 5/5 | +++ | 4/5 (a to d) | ++/+++ |
| Cecal PP | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | ++/+++ | 4/5 (a to d) | +/+++ | 5/5 | ++/+++ | 4/5 (a to d) | +/+++ |
| Jejunal MLN | 0/5 | − | 0/5 | − | 1/5 (a) | + | 1/5 (a) | + | 5/5 | ++ | 5/5 | ++ | 5/5 | +++ | 4/5 (a to d) | +/+++ |
| Ileal MLN | 0/5 | − | 0/5 | − | 2/5 (a, b) | ++ | 0/5 | − | 5/5 | +++ | 5/5 | ++ | 5/5 | +++ | 4/5 (a to d) | +++ |
| Mediastinal LN | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/++ | 4/5 (a to d) | + | 5/5 | ++/+++ | 2/5 (a, b) | ++/+++ |
| Prescapular LN | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/++ | 0/5 | − | 5/5 | ++/+++ | 2/5 (a, b) | +/++ |
| Retrohepatic LN | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | ++/+++ | 3/5(a to c) | +/++ | 5/5 | ++/+++ | 4/5 (a to d) | ++/+++ |
| Duodenum (ENS) | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | +/++ | 4/5 (a to d) | +/++ | 5/5 | +/++ | 4/5 (a to d) | +/++ |
| Jejunum (ENS) | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | +/++ | 5/5 | +/++ | 5/5 | +/++ | 4/5 (a to d) | +/++ |
| Ileum (ENS) | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | +/++ | 5/5 | +/++ | 5/5 | +/++ | 4/5 (a to d) | +/++ |
| Cecum (ENS) | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 5/5 | +/++ | 5/5 | +/++ | 5/5 | +/++ | 4/5 (a to d) | +/++ |
| Colon (ENS) | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 4/5 (a to d) | +/++ | 4/5 (a to d) | +/++ | 5/5 | +/++ | 4/5 (a to d) | +/++ |
| Sciatic nerve | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 1/5 (a) | + | 0/5 | − | 5/5 | + | 0/5 | − |
| Brachial nerve | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 1/5 (a) | + | 0/5 | − | 5/5 | + | 0/5 | − |
| External ocular muscle | 0/5 | − | 0/5 | − | 0/5 | − | 0/5 | − | 1/5 (a) | + | 0/5 | − | 5/5 | + | 0/5 | − |
For the spinal cord, cervical (C3-C4), thoracic (Th7-Th8), and lumbar (L3-L4) segments were analyzed. LN, lymph nodes; PP, Peyer's patches; MLN, mesenteric lymph nodes; ENS, enteric nervous system.
At each time point, the positive goats are identified arbitrarily by the letters a to d. ND, no data.
PrPSc labeling intensities are indicated as follows (as previously described [26]): −, negative; +, minimal to slight; ++, moderate; and +++, strong.
In the orally challenged heterozygous I/M142 animals, a similar but slightly delayed PrPSc accumulation scheme was observed; PrPSc was first detected in the gut-associated lymphoid tissue (Peyer's patches) at 360 dpi, but it was only detected at 1,040 dpi in the CNS (Table 2).
No PrPSc was observed in any of the tissues collected from the heterozygous R/Q211 and Q/K222 animals that had been orally challenged and killed at the different time points (5 animals per genotype per time point), and none of the animals bearing these genotypes had developed a clinical TSE at >2,500 dpi.
In both oral challenge experiments, groups of animals harboring the different PRNP genotypes (I/M142, R/Q211, and Q/K222) were kept alive and clinically monitored for TSE development (Table 3). Similarly, a group of heterozygous R/H154 animals (n = 6) that had been orally challenged with the same inoculum was also monitored.
TABLE 3.
Scrapie incubation periods and PrPSc deposition in central nervous system and lymphoid tissues in goats inoculated by the oral route, shown according to genotypes at codons 142, 154, 211, 222, and 240 of the PRNP gene
| Genotype | No. of scrapie-affected animals/total no. of animals | Scrapie incubation period (dpi) (mean ± SD) | Death from intercurrent disease |
PrPSc accumulation (no. of goats/total no. examined) |
||
|---|---|---|---|---|---|---|
| No. of goats/total no. in groupa | Time to death (dpi) | CNS | Lymphoid tissuesb | |||
| IRRQS/IRRQS (wild type) | 9/9 | 1,141 ± 93 | 9/9 | 9/9 | ||
| M142RQP240/IRRQS | 4/4 | 1,490 ± 126 | 4/4 | 4/4 | ||
| IH154RQS/IRRQS | 0/6 | 3/6 | 966, 1,002, 1,853 | 0/3 | 0/3 | |
| IRQ211QS/IRRQS | 0/5 | 2/5 | 1,234, 1,678 | 0/2 | 0/2 | |
| IRRK222S/IRRQS | 0/5 | 1/5 | 1,815 | 0/1 | 0/1 | |
Animals that were still alive were at <2,500 days postinoculation at the time of writing.
Tonsils, prescapular lymph nodes, ileal/jejunal Peyer's patches, and mesenteric lymph nodes.
In WT goats, incubation periods in the first (n = 4) and second (n = 5) experiments were not different. All the challenged I/M142 goats (n = 4) also developed clinical TSEs, but with a slightly longer incubation period (1,490 ± 126 dpi) than that in WT animals (1,141 ± 93 dpi). For both genotypes, affected animals showed PrPSc deposition in CNS and lymphoid tissues (Table 2).
After more than 2,500 days of incubation, none of the orally inoculated heterozygous R/H154, R/Q211, and Q/K222 animals had developed a clinical TSE. Some of the heterozygous R/H154 (n = 3) or Q/K222 (n = 2) goats and one homozygous K/K222 animal died from intercurrent disease (Table 3). In these orally challenged animals, none of the investigated tissues (lymphoid organs and CNS) displayed any detectable PrPSc deposition.
Together, these findings support the contention that R/H154, R/Q211, and Q/K222 PrP mutant alleles have a strong protective effect against scrapie infection following oral exposure.
Intracerebral challenge in goats.
To further assess the resistance to scrapie associated with the I/M142, R/H154, R/Q211, and Q/K222 PrP alleles, groups of heterozygous and homozygous animals were inoculated intracerebrally with the same isolate as that used for oral challenge (Table 4). As expected, all the intracerebrally inoculated WT goats developed clinical TSEs. In those animals, PrPSc deposits were observed in both the central nervous system and lymphoid tissues.
TABLE 4.
Scrapie incubation periods and PrPSc deposition in central nervous system and lymphoid tissues in intracerebrally inoculated goats, shown according to genotypes at codons 142, 154, 211, 222, and 240 of the PRNP genea
| Genotype | No. of clinically TSE-affected animals/total no. of animals | Scrapie incubation period (dpi) (mean ± SD) | Death from intercurrent disease |
PrPSc accumulation (no. of goats/total no. examined) |
||
|---|---|---|---|---|---|---|
| No. of goats/total no. in group | Time to death (dpi) | CNS | Lymphoid tissuesb | |||
| I142R154R211Q222/IRRQ (wild type) | 5/5 | 486 ± 21 | 5/5 | 5/5 | ||
| M142RQ/IRRQ | 5/5 | 788 ± 99 | 5/5 | 5/5 | ||
| IH154RQ/IRRQ | 5/5 | 624 ± 148 | 5/5 | 0/5 | ||
| IRQ211Q/IRRQ | 5/5 | 1,291 ± 325 | 5/5 | 5/5 | ||
| IRQ211Q/IRRQ211Q | 10/10 | 770 ± 139 | 10/10 | 1/10 | ||
| IRRK222/IRRQ | 2/5 | 1,900, 2,174 | 3/5 | 568, 898, 1,062 | 2/5c | 0/5 |
| IRRK222/IRRK222 | 1/5 | 2,101 | 1/1 | 0/1 | ||
Groups of five goats were intracerebrally challenged in the temporal cortex with the same classical scrapie isolate used for oral challenge. Animals that were still alive were at >2,400 days postinoculation at the time of writing.
Tonsils, prescapular lymph nodes, ileal/jejunal Peyer's patches, and mesenteric lymph nodes.
The two PrPSc-positive animals were clinically affected.
In contrast to the results of the oral challenge experiment, the heterozygous I/M142, R/H154, and R/Q211 animals, and also homozygous Q/Q211 animals, developed clinical TSEs. Strikingly, the heterozygous R/Q211 individuals displayed a longer incubation period than the homozygous Q/Q211 animals. PrPSc deposition was observed in the lymphoid tissues of the heterozygous I/M142 and homozygous Q/Q211 animals. No or limited PrPSc accumulation was observed in lymphoid tissues from R/H154 and R/Q211 animals. With the Sha31 antibody, the PrPres WB patterns observed for the brains of all the scrapie-affected R/H154 individuals were identical and differed strikingly from those observed for individuals bearing other PRNP genotypes (Fig. 1): the PrPres bands displayed apparently lower molecular weights. Immunoblots probed with the 12B2 antibody indicated that in H/R154 goats, PK digestion resulted in an N-terminal cleavage of PrPres (amino acids 93 to 97) that differed from the case in the other genotype groups.
FIG 1.
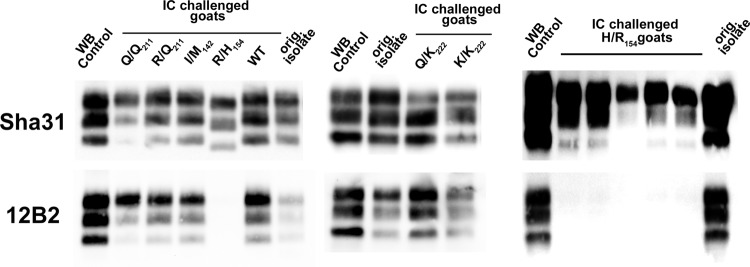
PrPres Western blot patterns for samples from the brains of goats intracerebrally challenged with a classical scrapie isolate. Ten percent tissue homogenates were prepared from brains of goats that had been intracerebrally challenged with a classical scrapie goat isolate and then developed the disease (Table 3). Abnormal PK-resistant PrP (PrPres) was detected following Western blotting using the Sha31 (YEDRYYRE epitope) and 12B2 (WGQGG epitope) antibodies. In each gel, a classical scrapie sheep isolate (WB control) and the original isolate (orig. isolate) were used as controls.
Among the i.c. challenged heterozygous Q/K222 goats, three animals died of intercurrent disease, at 568, 898, and 1,062 dpi. No PrPSc accumulation was observed in any of the investigated tissues from these goats. However, the two remaining animals developed clinical TSEs, after 1,980 and 2,134 dpi, and in those two individuals, PrPSc deposits were observed (by IHC and WB) in the central nervous system but not in lymphoid organs.
One of the five i.c. challenged homozygous K222 animals developed a clinical TSE and was euthanized after 2,101 dpi. Abnormal PrP deposition was detected (by IHC and WB) in the central nervous system but not in the lymphoid tissues. The four remaining K/K222 animals were still apparently healthy at the time of writing (>2,400 dpi).
Using the Sha31 antibody, Q/K222- and K/K222-positive individuals displayed similar PrPres glycoform ratios. The PrPres glycoprofiles of these individuals displayed dominant monoglycosylated bands which clearly differed from the patterns observed for the goats with other genotypes (Fig. 2).
FIG 2.
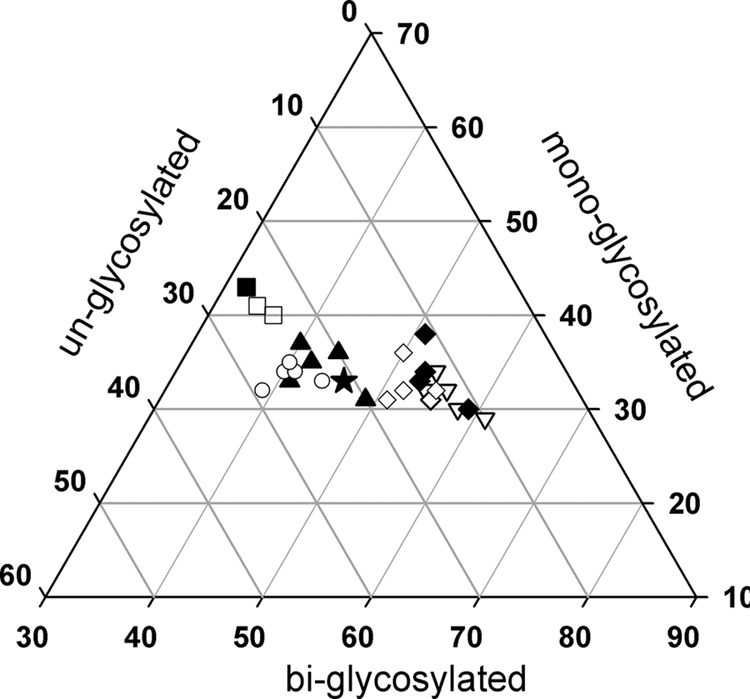
PrPres glycoprofiles of the brains of goats intracerebrally challenged with a classical scrapie isolate. Ten percent tissue homogenates were prepared using posterior brain stems from goats that had been intracerebrally challenged with a classical scrapie goat isolate and then developed the disease (Table 3). Abnormal PK-resistant PrP (PrPres) was detected following Western blotting using the Sha31 antibody (YEDRYYRE epitope). The signal volumes and relative percentages associated with monoglycosylated, biglycosylated, and unglycosylated bands were established using Quantity One software (Bio-Rad). Filled star, original isolate; ▲, wild type; ○, I/M142; □, Q/K222; ■, K/K222; ◊, R/Q211; ⧫, Q/Q211; ▽, H/R154.
More generally, the presence of apparently different PrPres WB signatures for i.c. challenged goats that harbored different genotypes suggests that different TSE agents propagated in those animals. However, it is our opinion that bioassays (which are currently ongoing) remain necessary before making conclusions on that point.
DISCUSSION
Case-control studies of classical scrapie-affected herds (13–18) and limited data from experimental challenges (intracerebral routes) (19, 27) support the view that PrP K222 allele goats might be strongly resistant to classical scrapie infection. Rare cases of the disease (n = 3) were reported for heterozygous K222 goats belonging to one single flock that displayed a high disease prevalence (27.4%), and no case has been reported so far for homozygous K222 animals (14, 21).
Our study indicated that a classical scrapie isolate failed to propagate in heterozygous K222 goats following oral challenge. However, it also demonstrated that the same classical scrapie isolate could propagate in heterozygous and a proportion of homozygous K222 animals following i.c. challenge, but with an incubation period that exceeded those observed in WT animals by 4 to 5 times.
These results for K222 goats are very evocative of those obtained with A136R154R171 allele sheep carriers that were naturally or experimentally exposed to TSE agents. After oral experimental challenge of homozygous and heterozygous ARR sheep, no or poorly efficient propagation of classical scrapie and BSE agents was observed (28–30). In heterozygous ARR sheep that were i.c. challenged with a classical scrapie agent, the disease occurred, but with significantly longer incubation periods than those in homozygous ARQ (wild-type PRNP genotype) sheep (30). Clinical TSEs occurred in a proportion of homozygous ARR sheep that were i.c. challenged with the cattle BSE agent, and the occurrence of rare natural classical scrapie cases was reported for animals harboring this genotype (31, 32).
All these results support the view that like the ARR allele in sheep, the K222 allele is associated with high-level but not absolute resistance to scrapie.
For I/M142 allele carriers, the i.c. and oral challenge results indicate that this allele is not associated with substantial resistance to the classical scrapie isolate we used. These observations are consistent with data collected from naturally infected goat herds (14, 21) and with the observations previously reported by Goldmann et al. for goats challenged with cattle BSE isolates, CH1641, and ME7 passaged in sheep (15).
No transmission or PrPSc deposition could be observed in orally challenged heterozygous H154 and Q211 animals. However, a 100% attack rate was observed in animals bearing those genotypes following i.c. challenge, albeit with longer incubation periods than those in WT animals.
These results were similar to those observed in heterozygous K222 animals. However, unlike homozygous K222 goats, the homozygous Q211 animals developed the disease with a 100% attack rate following i.c. challenge, with shorter incubation periods than those in heterozygous Q211 animals. This indicates that the Q211 allele cannot be considered to provide the same level of resistance against scrapie as the K222 allele.
The lack of homozygous H154 goats in the intracerebral inoculation experiment clearly limits our capacity to draw final conclusions concerning the level of resistance/susceptibility to classical scrapie associated with this PRNP allele. Nevertheless, the risk of atypical scrapie occurrence has been shown to be significantly higher in both H154 allele carrier sheep and goats (the same PrPC sequence is present in goats and sheep) than in WT animals (7, 33). This higher level of susceptibility to atypical scrapie represents a major argument against the selection of the H154 PRNP allele in goat populations.
Beyond this, the main limitation of this experiment is the fact that only one classical scrapie goat isolate was used to test the relative susceptibilities of the different genotypes. The diversity of TSE agents in small ruminants has been documented for several decades (26, 34, 35). In sheep, the susceptibility to TSE infection was shown to be influenced by both the nature of the TSE strain and the PRNP polymorphisms (36). Considering the time and resources necessary to carry out bioassays in large animals, testing of several classical scrapie agents in parallel in this model was simply not feasible. In this context, the inoculation of a variety of TSE agents into transgenic mice that express the WT and K222 PRNP goat alleles will play a pivotal role in confirming the apparently low susceptibility/high resistance associated with the K222 allele, and such work is reported in the accompanying article by Aguilar-Calvo et al. (37).
Finally, it should be noticed that the experimental approach we used allowed only estimations of the impact of individual PRNP polymorphisms on susceptibility to disease. For obvious material reasons, it was not possible to investigate the effects of PRNP haplotype combinations (such as individuals bearing both the Q211 and K222 alleles).
The development of PRNP genotype selection programs is now being considered by the European Union authorities as a potential tool to control and eradicate scrapie in commercial goat populations. In sheep, the diffusion of the ARR allele in the general population and, in particular, its introduction into classical scrapie-affected herds have proven its efficacy for the long-term control of the disease (10–12). The data that we report concur with the view that the K222 allele in goats provides a level of resistance against scrapie infection similar to that seen with the ARR allele in sheep.
ACKNOWLEDGMENTS
We thank Frédéric Bouvier and his team (INRA Domaine de la Sapinière) for producing goats with appropriate PRNP genotypes.
This work was funded by GIS PRION grant 31B06134, INRA grant AIP P00297, Poitou-Charentes Region grants 04/RPC-A-103 and 05/RPC-A-13, and the European Union (grants FOOD-CT-2006-36353 and 219235 FP7 ERA-NET EMIDA).
We declare no competing financial interests.
Footnotes
Published ahead of print 27 November 2013
REFERENCES
- 1.Bolton DC, McKinley MP, Prusiner SB. 1982. Identification of a protein that purifies with the scrapie prion. Science 218:1309–1311. 10.1126/science.6815801 [DOI] [PubMed] [Google Scholar]
- 2.Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, Tongue S, Gravenor MB. 2004. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 85:2735–2740. 10.1099/vir.0.79876-0 [DOI] [PubMed] [Google Scholar]
- 3.Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, Khang JV, Poivey JP, Lantier F, Laplanche JL. 1999. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch. Virol. 144:431–445. 10.1007/s007050050516 [DOI] [PubMed] [Google Scholar]
- 4.Hunter N, Moore L, Hosie BD, Dingwall WS, Greig A. 1997. Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland. Vet. Rec. 140:59–63. 10.1136/vr.140.3.59 [DOI] [PubMed] [Google Scholar]
- 5.Benestad SL, Arsac JN, Goldmann W, Noremark M. 2008. Atypical/Nor98 scrapie: properties of the agent, genetics, and epidemiology. Vet. Res. 39:19. 10.1051/vetres:2007056 [DOI] [PubMed] [Google Scholar]
- 6.Fediaevsky A, Maurella C, Noremark M, Ingravalle F, Thorgeirsdottir S, Orge L, Poizat R, Hautaniemi M, Liam B, Calavas D, Ru G, Hopp P. 2010. The prevalence of atypical scrapie in sheep from positive flocks is not higher than in the general sheep population in 11 European countries. BMC Vet. Res. 6:9. 10.1186/1746-6148-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno CR, Moazami-Goudarzi K, Laurent P, Cazeau G, Andreoletti O, Chadi S, Elsen JM, Calavas D. 2007. Which PrP haplotypes in a French sheep population are the most susceptible to atypical scrapie? Arch. Virol. 152:1229–1232. 10.1007/s00705-007-0956-7 [DOI] [PubMed] [Google Scholar]
- 8.Moum T, Olsaker I, Hopp P, Moldal T, Valheim M, Moum T, Benestad SL. 2005. Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. J. Gen. Virol. 86:231–235. 10.1099/vir.0.80437-0 [DOI] [PubMed] [Google Scholar]
- 9.Nodelijk G, van Roermund HJ, van Keulen LJ, Engel B, Vellema P, Hagenaars TJ. 2011. Breeding with resistant rams leads to rapid control of classical scrapie in affected sheep flocks. Vet. Res. 42:5. 10.1186/1297-9716-42-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson M, Moore RC, Bishop SC. 2008. Progress and limits of PrP gene selection policy. Vet. Res. 39:25. 10.1051/vetres:2007064 [DOI] [PubMed] [Google Scholar]
- 11.Fediaevsky A, Tongue SC, Noremark M, Calavas D, Ru G, Hopp P. 2008. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet. Res. 4:19. 10.1186/1746-6148-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagenaars TJ, Melchior MB, Bossers A, Davidse A, Engel B, van Zijderveld FG. 2010. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Vet. Res. 6:25. 10.1186/1746-6148-6-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J. Gen. Virol. 87:1029–1033. 10.1099/vir.0.81440-0 [DOI] [PubMed] [Google Scholar]
- 14.Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami-Goudarzi K, Rupp R, Babilliot JM, Lacroux C, Lugan S, Schelcher F, Chartier C, Corbiere F, Andreoletti O, Perrin-Chauvineau C. 2009. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222 and 240 in French Alpine and Saanen breeds and their association with classical scrapie. J. Gen. Virol. 90:769–776. 10.1099/vir.0.006114-0 [DOI] [PubMed] [Google Scholar]
- 15.Goldmann W, Martin T, Foster J, Hughes S, Smith G, Hughes K, Dawson M, Hunter N. 1996. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J. Gen. Virol. 77:2885–2891. 10.1099/0022-1317-77-11-2885 [DOI] [PubMed] [Google Scholar]
- 16.Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. 2007. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet. J. 173:459–462. 10.1016/j.tvjl.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 17.Papasavva-Stylianou P, Windl O, Saunders G, Mavrikiou P, Toumazos P, Kakoyiannis C. 2011. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie. Vet. J. 187:245–250. 10.1016/j.tvjl.2009.10.015 [DOI] [PubMed] [Google Scholar]
- 18.Vaccari G, Di Bari MA, Morelli L, Nonno R, Chiappini B, Antonucci G, Marcon S, Esposito E, Fazzi P, Palazzini N, Troiano P, Petrella A, Di Guardo G, Agrimi U. 2006. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J. Gen. Virol. 87:1395–1402. 10.1099/vir.0.81485-0 [DOI] [PubMed] [Google Scholar]
- 19.Acutis PL, Martucci F, D'Angelo A, Peletto S, Colussi S, Maurella C, Porcario C, Iulini B, Mazza M, Dell'atti L, Zuccon F, Corona C, Martinelli N, Casalone C, Caramelli M, Lombardi G. 2012. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Vet. Res. 43:8. 10.1186/1297-9716-43-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabouret G, Lacroux C, Lugan S, Corbiere F, Weisbecker JL, Costes P, Schelcher F, Andreoletti O. 2010. Relevance of oral experimental challenge with classical scrapie in sheep. J. Gen. Virol. 91:2139–2144. 10.1099/vir.0.021311-0 [DOI] [PubMed] [Google Scholar]
- 21.Corbiere F, Perrin-Chauvineau C, Lacroux C, Costes P, Thomas M, Bremaud I, Martin S, Lugan S, Chartier C, Schelcher F, Barillet F, Andreoletti O. 2013. PrP-associated resistance to scrapie in five highly infected goat herds. J. Gen. Virol. 94:241–245. 10.1099/vir.0.047225-0 [DOI] [PubMed] [Google Scholar]
- 22.Arsac JN, Andreoletti O, Bilheude JM, Lacroux C, Benestad SL, Baron T. 2007. Similar biochemical signatures and prion protein genotypes in atypical scrapie and Nor98 cases, France and Norway. Emerg. Infect. Dis. 13:58–65. 10.3201/eid1301.060393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacroux C, Corbiere F, Tabouret G, Lugan S, Costes P, Mathey J, Delmas JM, Weisbecker JL, Foucras G, Cassard H, Elsen JM, Schelcher F, Andreoletti O. 2007. Dynamics and genetics of PrPSc placental accumulation in sheep. J. Gen. Virol. 88:1056–1061. 10.1099/vir.0.82218-0 [DOI] [PubMed] [Google Scholar]
- 24.Andreoletti O, Orge L, Benestad SL, Beringue V, Litaise C, Simon S, Le Dur A, Laude H, Simmons H, Lugan S, Corbiere F, Costes P, Morel N, Schelcher F, Lacroux C. 2011. Atypical/Nor98 scrapie infectivity in sheep peripheral tissues. PLoS Pathog. 7:e1001285. 10.1371/journal.ppat.1001285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feraudet C, Morel N, Simon S, Volland H, Frobert Y, Creminon C, Vilette D, Lehmann S, Grassi J. 2005. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 280:11247–11258. 10.1074/jbc.M407006200 [DOI] [PubMed] [Google Scholar]
- 26.Andreoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen L, Schelcher F, Elsen JM, Lantier F. 2000. Early accumulation of PrP(Sc) in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J. Gen. Virol. 81:3115–3126 http://vir.sgmjournals.org/content/81/12/3115.long [DOI] [PubMed] [Google Scholar]
- 27.White SN, Reynolds JO, Waldron DF, Schneider DA, O'Rourke KI. 2012. Extended scrapie incubation time in goats singly heterozygous for PRNP S146 or K222. Gene 501:49–51. 10.1016/j.gene.2012.03.068 [DOI] [PubMed] [Google Scholar]
- 28.Andreoletti O, Morel N, Lacroux C, Rouillon V, Barc C, Tabouret G, Sarradin P, Berthon P, Bernardet P, Mathey J, Lugan S, Costes P, Corbiere F, Espinosa JC, Torres JM, Grassi J, Schelcher F, Lantier F. 2006. Bovine spongiform encephalopathy agent in spleen from an ARR/ARR orally exposed sheep. J. Gen. Virol. 87:1043–1046. 10.1099/vir.0.81318-0 [DOI] [PubMed] [Google Scholar]
- 29.Foster JD, Parnham D, Chong A, Goldmann W, Hunter N. 2001. Clinical signs, histopathology and genetics of experimental transmission of BSE and natural scrapie to sheep and goats. Vet. Rec. 148:165–171. 10.1136/vr.148.6.165 [DOI] [PubMed] [Google Scholar]
- 30.Goldmann W, Hunter N, Smith G, Foster J, Hope J. 1994. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 75:989–995. 10.1099/0022-1317-75-5-989 [DOI] [PubMed] [Google Scholar]
- 31.Groschup MH, Lacroux C, Buschmann A, Luhken G, Mathey J, Eiden M, Lugan S, Hoffmann C, Espinosa JC, Baron T, Torres JM, Erhardt G, Andreoletti O. 2007. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerg. Infect. Dis. 13:1201–1207. 10.3201/eid1308.070077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houston F, Goldmann W, Chong A, Jeffrey M, Gonzalez L, Foster J, Parnham D, Hunter N. 2003. Prion diseases: BSE in sheep bred for resistance to infection. Nature 423:498. 10.1038/423498a [DOI] [PubMed] [Google Scholar]
- 33.Colussi S, Vaccari G, Maurella C, Bona C, Lorenzetti R, Troiano P, Casalinuovo F, Di Sarno A, Maniaci MG, Zuccon F, Nonno R, Casalone C, Mazza M, Ru G, Caramelli M, Agrimi U, Acutis PL. 2008. Histidine at codon 154 of the prion protein gene is a risk factor for Nor98 scrapie in goats. J. Gen. Virol. 89:3173–3176. 10.1099/vir.0.2008/004150-0 [DOI] [PubMed] [Google Scholar]
- 34.Bruce ME, Boyle A, Cousens S, McConnell I, Foster J, Goldmann W, Fraser H. 2002. Strain characterization of natural sheep scrapie and comparison with BSE. J. Gen. Virol. 83:695–704 http://vir.sgmjournals.org/content/83/3/695.long [DOI] [PubMed] [Google Scholar]
- 35.Pattison IH, Millson GC. 1962. Distribution of the scrapie agent in the tissues of experimentally inoculated goats. J. Comp. Pathol. 72:233–244 [DOI] [PubMed] [Google Scholar]
- 36.Beringue V, Vilotte JL, Laude H. 2008. Prion agent diversity and species barrier. Vet. Res. 39:47. 10.1051/vetres:2008024 [DOI] [PubMed] [Google Scholar]
- 37.Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O, Torres JM. 2014. Role of the goat K222-PrPC polymorphic variant in prion infection resistance. J. Virol. 88:2670–2676. 10.1128/JVI.02074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


