Abstract
Vesicular stomatitis virus (VSV) is an oncolytic virus that induces cancer cell death through activation of the apoptotic pathway. Intrinsic resistance to oncolysis is found in some cell lines and many primary tumors as a consequence of residual innate immunity to VSV. In resistant-tumor models, VSV oncolytic potential can be reversibly stimulated by combination with epigenetic modulators, such as the histone deacetylase inhibitor vorinostat. Based on this reversible effect of vorinostat, we reasoned that critical host genes involved in oncolysis may likewise be reversibly regulated by vorinostat. A transcriptome analysis in prostate cancer PC3 cells identified a subset of NF-κB target genes reversibly regulated by vorinostat, as well as a group of interferon (IFN)-stimulated genes (ISGs). Consistent with the induction of NF-κB target genes, vorinostat-mediated enhancement of VSV oncolysis increased hyperacetylation of NF-κB RELA/p65. Additional bioinformatics analysis revealed that NF-κB signaling also increased the expression of several autophagy-related genes. Kinetically, autophagy preceded apoptosis, and apoptosis was observed only when cells were treated with both VSV and vorinostat. VSV replication and cell killing were suppressed when NF-κB signaling was inhibited using pharmacological or genetic approaches. Inhibition of autophagy by 3-methyladenine (3-MA) enhanced expression of ISGs, and either 3-MA treatment or genetic ablation of the autophagic marker Atg5 decreased VSV replication and oncolysis. Together, these data demonstrate that vorinostat stimulates NF-κB activity in a reversible manner via modulation of RELA/p65 signaling, leading to induction of autophagy, suppression of the IFN-mediated response, and subsequent enhancement of VSV replication and apoptosis.
INTRODUCTION
Oncolytic viruses (OVs) represent a promising biotherapeutic approach to cancer treatment, with demonstrated antitumor efficacy and safety in phase I to III clinical trials (1–3). Vesicular stomatitis virus (VSV) is an enveloped, negative-sense RNA virus of the family Rhabdoviridae that is a potent OV candidate (4). VSV induces cell death primarily through activation of the apoptotic pathway, and both intrinsic and extrinsic apoptotic mechanisms have been described as contributing to VSV-induced apoptosis (5–7). Upon VSV infection, innate sensing of viral RNA by the RIG-I like receptors leads to induction of the innate immune response and production of interferons (IFNs). Newly synthesized IFN proteins act in an autocrine and paracrine fashion to upregulate the expression of hundreds of IFN-stimulated genes (ISGs) that contribute to the development of the host antiviral state, which in normal cells and tissues restricts virus multiplication (reviewed in reference 8).
During the evolution of malignancies, genetic abnormalities accumulate and, while providing the cancer cells with growth and survival advantages, compromise the normal antiviral program of transformed cells. Defects in the IFN antiviral signaling network within transformed cells have been implicated in tumor-specific oncolysis, a strategy termed virotherapy. IFN-related defects allow VSV and other oncolytic viruses to replicate to high titers uninterrupted by the host antiviral response, resulting in high virus production and virus-induced lysis. The tumor mass essentially becomes a selective cellular niche for virus replication, leading to virus-mediated cell death. The same viruses are unable to replicate efficiently in normal tissues and thus have a superior therapeutic index in tumor cells (9; reviewed in reference 3). The therapeutic index of OVs can be further improved by genetic engineering or by expression of a transgene (10, 11). For example, when IFN-β is expressed by VSV, increased selectivity for replication in tumor cells is observed (12); this recombinant VSV is currently being tested in phase I clinical trials as a single agent in patients who are refractory to standard therapeutics (13). A small-plaque variant VSV contains a Δ51 deletion in the viral matrix (M) protein and was shown previously to enhance the safety profile of VSV (14, 15), because the attenuated mutant is a potent inducer of the IFN response in healthy cells that does not block nuclear export of host cell antiviral mRNAs (14, 16, 17).
Several factors limit the efficacy of viral vectors, including intrinsic tumor resistance to oncolysis and limitations in the development of adaptive immune responses against tumor antigens (14, 18, 19). For example, while VSV-based therapy was effective in vivo against an androgen-dependent LNCaP prostate cancer xenograft model, androgen-independent PC3 cells were relatively resistant to oncolysis both in vitro and in vivo (20). We previously characterized a synergistic strategy in prostate cancer that involves the use of histone deacetylase inhibitors (HDIs), such as suberoylanilide hydroxamic acid (SAHA) (vorinostat) or MS-275, together with oncolytic VSV, in the treatment of androgen-independent prostate cancer (19). HDIs manipulate the innate immune response by influencing epigenetic modifications of chromatin and altering gene expression (21, 22). Because of their effect on immune suppression, we and others have reasoned that pretreatment of tumors with HDIs would enhance the replication and spread of OV within malignancies (19, 23). In tumor cell lines, small-animal tumor models, and ex vivo primary tumor tissues, HDIs markedly enhanced the spread and replication of VSV. This increased oncolytic activity was correlated with a time-dependent decrease in the expression of IRF3, IRF7, IFN-β, and MX1 and increased caspase 3, 8, and 9 cleavage in vorinostat-plus-VSV- and MS-275-plus-VSV-treated PC3 cells (19). Interestingly, the effects of HDIs on viral spread were reversible, and removal of HDIs led to a decrease in viral replication within malignant cells both in vitro and in vivo.
Vorinostat has been shown to modulate NF-κB-related signaling, inhibit IFN signaling, and induce cancer cell differentiation by inhibiting deacetylation of cellular proteins and histones (19, 24, 25). Treatment of tumor cells with HDIs frequently induces apoptosis through caspase activation (26–28), although significant levels of apoptosis in some cell lines, such as PC3, are observed at relatively high concentrations and with longer exposures to vorinostat (28, 29). More recently, vorinostat was also shown to induce autophagy in tumor cells (30, 31). Autophagy is a catabolic process by which cytosolic material is targeted for lysosomal degradation by means of double-membrane cytosolic vesicles, termed autophagosomes (reviewed in reference 32). In addition to its role in cellular homeostasis, autophagy also plays a critical role in the intrinsic, innate, and adaptive responses to pathogens, with evidence indicating that multiple viruses can either subvert or hijack the host autophagic machinery to support their own replication (33–35). A number of recent studies highlight the significant role of autophagy in mediating the antitumor effects of oncolytic viruses, although the mechanism is largely unknown (reviewed in reference 36).
The purpose of the present study was to elucidate the molecular mechanism(s) involved in the vorinostat-mediated development of VSV permissiveness in prostate cancer cells. Our results demonstrate that vorinostat not only blocks IFN responses, but also stimulates expression of a subset of NF-κB target genes that enhance VSV replication in part via stimulation of NF-κB-mediated autophagy.
MATERIALS AND METHODS
Cell lines and reagents.
PC3, DU145, and HCT116 cells were purchased from ATCC (Bethesda, MD) and were grown in RPMI medium (PC3), Eagle's minimal essential medium (EMEM) (DU145), or McCoy's medium (HCT116) supplemented with 10% serum (Life Technologies, Carlsbad, CA). Atg5 wild-type (WT) (Atg5+/+) and Atg5 knockout (KO) (Atg5−/−) mouse embryonic fibroblasts (MEFs) were obtained from Nathalie Grandvaux, CRCHUM-Centre Hospitalier de l'Université de Montréal, Montréal, Canada, with the permission of Noboru Mizushima, who generated the original Atg5 WT and KO MEFs. PC3 cells stably transfected with short hairpin RNA (shRNA) targeting luciferase control (Ctrl-Luc) or RELA/p65 (shRel) (37) were a kind gift from Thorsten Schinke, University Medical Center Hamburg Eppendorf, Hamburg, Germany. Vorinostat, MS-275, and BAY11-7082 were purchased from Cayman Chemicals (Ann Arbor, MI). 3-Methyladenine (3-MA) and all other chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
Virus production, quantification, and infection.
VSV stock was grown in Vero cells (ATCC, Bethesda, MD), concentrated from cell-free supernatants by centrifugation, and titrated in duplicate by standard plaque assay. PC3, DU145, or HCT116 cells were infected at various multiplicities of infection (MOIs) and incubated with complete medium at 37°C for the indicated times.
Protein extraction and Western blot analysis.
Cells were washed twice with ice-cold phosphate-buffered saline and lysed in buffer (50 mM Tris-HCl, pH 8, 1% sodium deoxycholate, 1% NP-40, 5 mM EDTA, 5 mM EGTA, and 150 mM NaCl), and the insoluble fraction was removed by centrifugation at 22,000 × g for 20 min. The soluble fractions were separated by SDS-PAGE on 10% gels. Proteins were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane, blocked with 10% bovine serum albumin, and probed with various primary antibodies. All immunoblots were visualized by enhanced chemiluminescence. The protein concentration was determined with Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA).
Immunoprecipitation.
Twenty micrograms of anti-RELA/p65 (Santa Cruz Biotechnology, Santa Cruz, CA) was cross-linked to 300 μl of Protein AG-Plus agarose beads using 0.2 mol/liter triethanolamine, pH 8.0. Cells were lysed with 1% {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (CHAPS) lysis buffer (10 mmol/liter HEPES, 150 mM NaCl, 1% CHAPS, pH 7.4) containing protease inhibitors, and total protein was incubated with cross-linked antibody in 1% CHAPS lysis buffer at 4°C overnight. Immunoprecipitates were collected by centrifugation and washed three times with 1% CHAPS lysis buffer, the beads were boiled in loading buffer, and bound protein was analyzed by immunoblotting. Samples with antibody alone, lysate alone, or an irrelevant isotype-matched immunoglobulin G antibody were used as negative controls. Proteins were separated by electrophoresis and transferred to nitrocellulose membranes. Antibodies for total acetylated lysine (Cell Signaling Technologies, Danvers, MA) or RELA/p65 (Santa Cruz Biotechnology, Santa Cruz, CA) were used to determine protein levels.
Chromatin isolation.
Approximately 3 × 106 cells were harvested and centrifuged, and the pellet was resuspended in buffer A (100 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 10 mM NaF, 1 mM Na2VO3, 1 mM dithiothreitol [DTT], and protease inhibitor cocktail). Triton X-100 was added to a final concentration of 0.1%, and the cells were incubated on ice for 5 min. Upon centrifugation, the supernatant was separated from the pellet, which contained the nuclei. The supernatant was further cleared by high-speed centrifugation at 20,000 × g. The resulting supernatant, which represents the cytosolic fraction, was collected, and the pellet was discarded. The pellet fraction was resuspended in 100 μl of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitor cocktail) and lysed for 30 min on ice. The pellet fraction was then centrifuged at 1,700 × g. The resulting supernatant containing the soluble nuclear proteins was separated from the pellet containing the chromatin. The chromatin fraction was washed once with buffer B and centrifuged at 10,000 × g. The pellet was then resuspended in 1% NP-40 protein extraction buffer, sonicated, and centrifuged at 20,000 × g. The final supernatant contained the chromatin-bound proteins.
Nuclear extraction.
Cells (1 × 107 to 2 × 107) were washed and harvested with cold phosphate-buffered saline (PBS) with 1 mM EDTA. The cell pellet was incubated in ice-cold buffer A (10 mM Tris, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 μM DTT) for 15 min, and 25 μl of 10% NP-40 was added. The cell suspension was vortexed, and after a quick spin at 13,300 × g, the supernatant (cytosolic fraction) was separated from the pellet (nuclear fraction). The nuclear fraction was further solubilized by resuspending the pellet in ice-cold buffer C (20 mM Tris, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, pH 7.9) and shaking vigorously at 4°C for 1 h; insoluble debris was removed by centrifugation (13,300 × g) at 4°C.
Mass spectrometry analysis.
PC3 cells were transfected with an empty vector or S-tagged WT RELA/p65 and treated with vorinostat (5 μM) for 48 h. S-tagged protein was then precipitated using the S-tag purification technique as described previously (38), resolved on SDS-PAGE, fixed, and stained with Coomassie brilliant blue. The corresponding protein bands were excised, destained, and washed with a series of three washing buffers (50 mM ammonium bicarbonate, 50% acetonitrile, and 80% acetonitrile). The bound proteins were reduced with 40 mM dithiothreitol, the gels were rinsed with 50 mM ammonium bicarbonate buffer, and the reduced proteins were alkylated with 1 ml of 50 mM iodoacetamide. The gel-bound proteins were digested with trypsin (20 ng/μl; Promega, Madison, WI) in 5 mM ammonium bicarbonate buffer at 37°C with constant mixing for 12 h. The eluent containing the tryptic peptides was dried and stored at 4°C prior to mass spectrometric analysis. In order to identify proteins, the dried samples were dissolved with 20 μl of 0.1% formic acid-water, and 2 μl of each sample was analyzed by liquid chromatography-electrospray ionization tandem mass spectrometry LC-ESI–MS-MS using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with an Easy NanoLC-1000 system using data-dependent acquisition with dynamic exclusion (DE = 1) settings. The data-dependent acquisition settings used were a top 12 higher-energy collisional dissociation (HCD) for the Q-Exactive MS. The resolving power for Q-Exactive was set at 70,000 for the full MS scan and 17,500 for the MS-MS scan at m/z 200. LC-ESI–MS-MS analysis was conducted using a C18 column (75 μm by 150 mm). The mobile phases for the reverse-phase chromatography were 0.1% HCOOH in water (A) and 0.1% HCOOH in acetonitrile (B). A four-step linear gradient was used for the LC separation (2% to 30% B in the first 47 min, followed by 80% B in the next 1 min and holding at 80% B for 12 min). The Sequest algorithm was used to identify peptides from the resulting MS-MS spectra by searching against the combined human protein database (a total of 22,673 proteins) extracted from Swissprot (version 57) with taxonomy “Homo sapiens” using Proteome Discoverer (version 1.3; Thermo Fisher Scientific, San Jose, CA). The search parameters for parent and fragment ion tolerances were set at 20 ppm and 30 millimass units (mmu) for the Q-Exactive MS. Other parameters used were a fixed modification of carbamidomethylation-modified Cys and variable modifications of acetylation (K) and oxidation (Met). Trypsin was set as the protease, with a maximum of 2 missed cleavages. Raw files were searched against the RELA protein (3 different isoforms, along with 500 other random proteins and reversed proteins as decoys) using Byonic (39) with a peptide tolerance of 15 ppm, an MS-MS tolerance of 20 ppm for HCD data, the carbamidomethylated cysteine as a fixed modification, and oxidation of methionine and acetylation (K) as variable modifications. Byonic scoring gives an indication of whether modifications are localized with confidence.
Autophagy analysis by flow cytometry.
Autophagy was assessed by Cyto-ID staining by flow cytometry (LSRFortessa; BD Biosciences, San Jose, CA). Cyto-ID was used according to the manufacturer's instructions (Enzo Life Sciences, Farmingdale, NY).
Confocal microscopy.
PC3 cells were seeded onto 22-mm-diameter coverslips in 6-well plates at 1 × 105 cells/well. The cells were fixed in 4% paraformaldehyde and permeabilized with methanol. The coverslips were blocked with 3% BSA-PBS and incubated with primary antibody in 3% BSA-PBS overnight at 4°C. Upon removal of the primary antibody and washing of cells with PBS-0.1% Tween 20 and PBS alone, the cells were incubated in secondary antibody (Alexa Fluor 546; Life Technologies, Carlsbad, CA) for 1 h at room temperature, followed by 3% BSA-PBS and PBS washes. The coverslips were mounted on slides using Immu-Mount (Thermo Scientific, Waltham, MA) and left to dry overnight at room temperature in the dark. The intensity of cell fluorescence was calculated for the corrected total cell fluorescence using the program ImageJ (National Institutes of Health, Bethesda, MD) (40).
Real-time reverse transcription (RT)-PCR analysis.
Total RNA was isolated using an RNeasy kit according to the manufacturer's instructions (Qiagen, Valencia, CA). RNA was reverse transcribed with oligo(dT) primers and SuperScript II reverse transcriptase (Life Technologies, Carlsbad, CA). PCR was performed using Taq polymerase (GE Healthcare, Piscataway Township, NJ), and amplification was carried out for 25 to 30 cycles. All data are presented as relative quantifications, with efficiency correction based on the relative expression of target genes versus the β-actin gene as the reference gene. cDNA was amplified using SyBR Green I PCR master mix (Applied Biosystems, Foster City, CA), and the data were collected using the AB 7500 Real-Time PCR System and analyzed by the comparative threshold cycle (CT) method using the SDS v1.3.1 relative quantification software. For the Fluidigm BioMark assay, intron-spanning PCR primers and probes were designed using the Roche Universal Probe Library Assay Design Center. The standard BioMark protocol was used to preamplify cDNA samples for 14 cycles using TaqMan PreAmp Master Mix according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). Quantitative PCRs (qPCRs) were performed using 48.48 BioMark Dynamic Arrays (Fluidigm, South San Francisco, CA), enabling quantitative measurement of up to 48 different mRNAs in 48 samples under identical reaction conditions. Runs were 40 cycles (15 s at 95°C, 5 s at 70°C, and 60 s at 60°C). Raw CT values were calculated with the Real-Time PCR Analysis Software (Fluidigm), and software-designated failed reactions were discarded from analysis.
Intracellular staining.
PC3 cells were exposed to VSV with and without vorinostat for 48 h and fixed in Fix I buffer (BD Biosciences, San Jose, CA). The cells were then washed with PBS containing 4% fetal bovine serum (FBS) and resuspended in Perm II buffer (BD Biosciences, San Jose, CA). After washing, the cells were stained with 1 μg/100 μl of primary antibody for 30 min at room temperature, washed twice, and incubated with Alexa Fluor 647-conjugated anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) for 25 min at room temperature. After two more washes, samples were analyzed by flow cytometry using Diva software (BD Biosciences, San Jose, CA).
Microarray analyses.
Total RNA was isolated from PC3 cells (n = 4 per treatment group) using RNeasy Micro kits (Qiagen, Valencia, CA). Universal human reference RNA was used as a reference. Samples were amplified using Illumina TotalPrep RNA amplification kits (Life Technologies, Carlsbad, CA). The microarray analysis was conducted using 750 ng of biotinylated cRNA hybridized to human reference 8 version 3 BeadChips (Illumina, San Diego, CA) at 58°C for 20 h. The arrays were scanned using Illumina iSCAN and quantified using Genome Studio (Illumina, San Diego, CA). The analyses of the Genome Studio output data were conducted using the R and Bioconductor software packages. A significance analysis of microarray (SAM) test was then performed on each experimental condition versus the control. SAM identifies statistically significant genes by carrying out gene-specific t tests and computes a statistic, dj, for each gene j, which measures the strength of the relationship between gene expression and a response variable. This analysis uses nonparametric statistics, since the data may not follow a normal distribution. The response variable describes and groups the data based on experimental conditions. In this method, repeated permutations of the data are used to determine if the expression of any gene is significantly related to the response. The use of permutation-based analysis accounts for correlations in genes and avoids parametric assumptions about the distribution of individual genes. This generated a list of differentially expressed genes in which we input a threshold of +1.3/−1.3-fold change and a P value of less than or equal to 0.01 to be considered significant. A manual search using Boston University's NF-κB target gene database (http://www.nf-kb.org) was then conducted to create a list of differentially regulated NF-κB target genes. Quantile normalization was applied, followed by a log2 transformation. The LIMMA package (www.bioconductor.org) was used to fit a linear model to each probe and to perform (moderated) t tests or F tests on the groups being compared. To control the expected proportions of false positives, the false discovery rate for each unadjusted P value was calculated using the Benjamini and Hochberg procedure, which relies on on the P values being uniformly distributed under the null hypothesis. Multidimensional scaling was used as a dimensionality reduction method in R to generate plots for the evaluation of similarities or dissimilarities between data sets.
Statistical analysis.
Graphics and statistical analyses were executed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). Differences among the treatment groups were analyzed by paired t tests. P values of <0.05 were considered statistically significant. Average values were expressed as means and standard deviations (SD). Fluidigm BioMark assay data were analyzed with Fluidigm Real-Time PCR Analysis software (Fluidigm, South San Francisco, CA), and Z scores were generated using Microsoft Excel.
RESULTS
Gene expression profiles in PC3 cells treated with VSV and vorinostat.
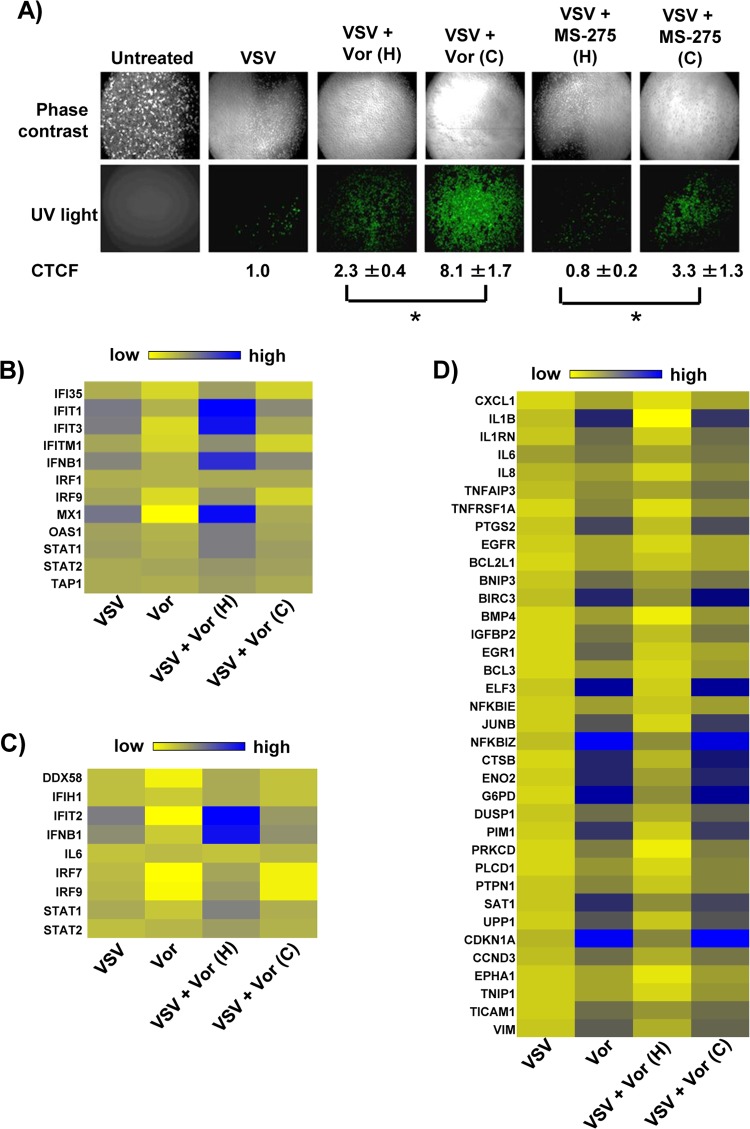
To determine genes and pathways responsible for HDI potentiation of VSV oncolysis, we performed total transcriptome analysis of PC3 cells exposed to the combination of VSV and vorinostat. The effects of vorinostat or MS-275 on both gene transcription and viral replication were reversible; upon removal of vorinostat or MS-275, VSV replication decreased in PC3 cells, as previously demonstrated in vivo in different murine tumor models (19), and is illustrated by a 3- to 4-fold-lower number of green fluorescent protein (GFP)-positive cells (P < 0.05) in wells from which vorinostat or MS-275 was removed prior to VSV-GFP infection (Fig. 1A). Based on previous experiments, we reasoned that critical gene sets involved in VSV replication would likewise be reversibly regulated by vorinostat. To identify such genes, PC3 cells were treated with vorinostat continuously (48 h), or alternatively, vorinostat treatment was halted at the time of VSV infection (24 h). In agreement with previous results, we observed increases in the expression of IFN and several IFN-stimulated genes in the VSV, VSV-plus-vorinostat (continuous), and VSV-plus-vorinostat (halted) groups (Fig. 1B and C). However, the increase in such genes was most prominent in the “halted” group, in which the removal of vorinostat activated antiviral responses and inhibited viral replication (Fig. 1A). Additional bioinformatics analysis of gene expression profiles between treatment groups revealed a subset of reversibly regulated genes, many of which were well-known NF-κB targets (Fig. 1D). Because expression levels of NF-κB target genes were similar in vorinostat- and vorinostat-plus-VSV-treated cells, NF-κB activation appeared to be a direct consequence of vorinostat treatment.
FIG 1.
Vorinostat treatment potentiates VSV replication, induces expression of NF-κB-regulated genes, and suppresses interferon response. (A) PC3 cells were pretreated with MS-275 (2 μM) or vorinostat (Vor) (5 μM) for 24 h and then infected with VSV-GFP at an MOI of 0.01. Following infection, the culture medium was supplemented with either complete medium containing MS-275 or vorinostat (C [continuous treatment]) or complete medium alone (H [halted treatment]). VSV replication was assessed by fluorescence microscopy for GFP expression 24 h after VSV infection, and the fluorescence signal was calculated for corrected total cell fluorescence (CTCF) using ImageJ. *, P ≤ 0.05. (B and C) Heat maps showing differentially regulated genes belonging to the IFN signaling pathway (B) and activation of IRF by cytosolic pattern recognition receptors (C). (D) Heat map showing differentially regulated genes that are targets of the NF-κB transcription factor. The genes in panels B and C were selected based on Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) gene assignment.
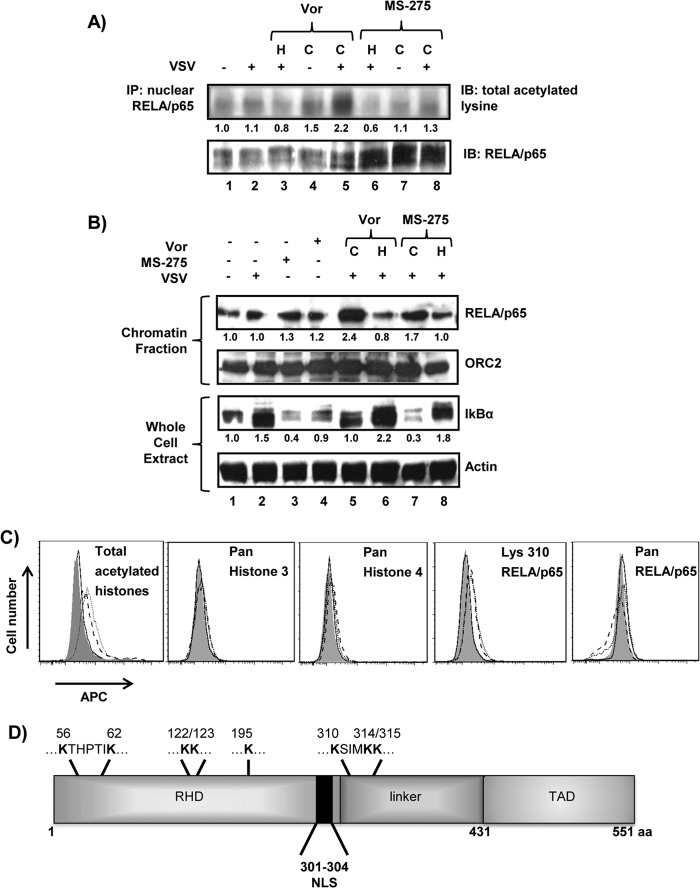
Vorinostat treatment increases chromatin binding and acetylation of RELA/p65.
Vorinostat treatment increases global protein acetylation (reviewed in reference 41) and specifically targets acetylation of NF-κB subunit RELA/p65, leading to increased nuclear localization and DNA binding (42). To evaluate the specific effect of vorinostat or MS-275 on RELA/p65 activity, acetylation and DNA binding of RELA/p65 were evaluated (Fig. 2). Immunoblot analysis of RELA/p65 revealed a modest but significant 1.3- to 2.2-fold increase in total acetylated lysine residues with continuous vorinostat or MS-275 treatment (P < 0.01) (Fig. 2A, lanes 5 and 8). Chromatin fractionation also revealed a 1.7- to 2.4-fold increase in DNA binding of RELA/p65 following continuous treatment (Fig. 2B, lanes 5 and 7). Total IκBα protein levels as an indirect measure of NF-κB activity also displayed a higher turnover rate with continuous vorinostat (2.2-fold higher) or MS-275 (6-fold higher) treatment (P < 0.01). The effect of vorinostat on increased protein acetylation was confirmed by ∼2-fold increases in the mean fluorescence intensity (MFI) for total acetylated histones and ∼1.3- to 1.4-fold increases in the MFI for acetylated K310 of RELA/p65 in vorinostat- and vorinostat-plus-VSV-treated cells (Fig. 2C). No increase in fluorescence intensities was observed in experiments using panspecific antibodies targeting histones 3 and 4 or the RELA/p65 protein. To identify the RELA/p65 lysines acetylated as a consequence of vorinostat treatment, S-tagged WT RELA/p65 expressed in PC3 cells was analyzed by ESI–MS-MS. Eight lysine residues—K56, K62, K122, K123, K195, K310, K314, and K315—demonstrated increased acetylation with vorinostat, including K310 acetylation, which is linked to increased nuclear retention and transcriptional activity of RELA/p65 (43) (Fig. 2D). These data indicate that vorinostat treatment led to prolonged acetylation of 8 of 18 lysine residues in RELA/p65, resulting in increased nuclear localization and DNA binding.
FIG 2.
Vorinostat or MS-275 treatment leads to hyperacetylation and increased DNA binding of nuclear RELA/p65. (A) PC3 cells were exposed to the treatments indicated above the blots, and nuclear extracts were immunoprecipitated for RELA/p65 and assessed for acetylated lysine residues or total RELA/p65 protein. IP, immunoprecipitation; IB, immunoblotting. (B) PC3 cells were subjected to the indicated experimental conditions for 24 h, and the chromatin-bound fraction was immunoblotted for RELA/p65. (Top) ORC2 was used as a loading control for chromatin-bound proteins. (Bottom) Proteins from whole-cell lysates were immunoblotted for IκBα. (A and B) The numbers below the Western blots indicate protein/loading control signal ratios relative to the vehicle-treated cells. (C) PC3 cells were exposed to DMSO (dimethyl sulfoxide) (shaded histogram), VSV (MOI = 0.1) (solid line), vorinostat (5 μM) (dashed line), or VSV plus vorinostat (dotted line) for 24 h, and intracellular staining was performed for total acetylated histones or total histone 3 and histone 4 proteins (left), while antibody targeting K310 of RELA/p65 or pan-RELA/p65 antibody was used for detection of acetylated lysine or total protein, respectively (right). (D) Schematic representation of RELA/p65 with acetylated lysines indicated at the top, as determined by mass spectrometry analysis. C, continuous treatment; H, halted treatment; APC, allophycocyanin; RHD, Rel homology domain; TAD, transactivation domain; NLS, nuclear localization signal; aa, amino acids.
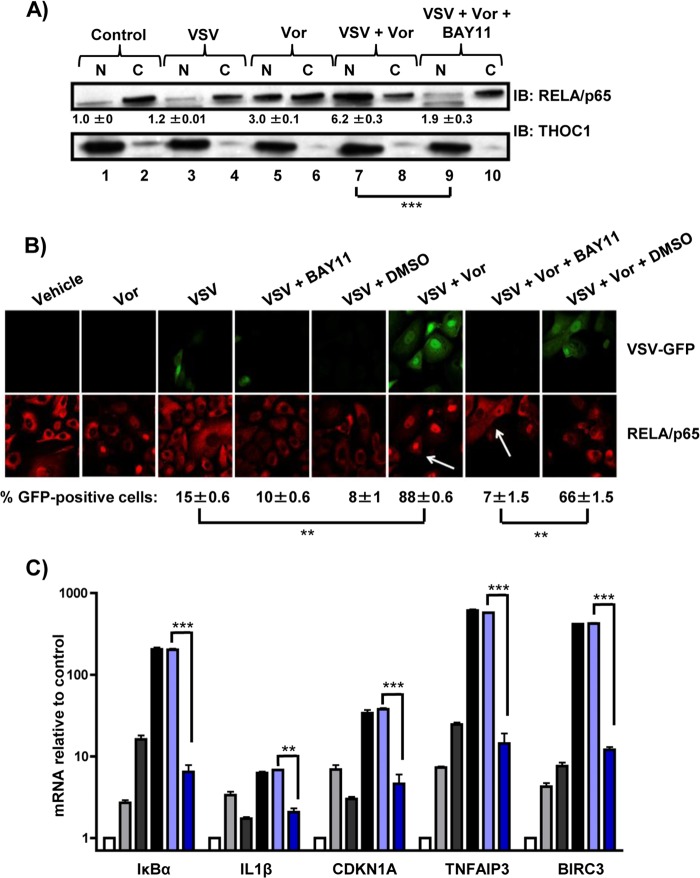
NF-κB inhibition decreases VSV replication and oncolysis.
To further characterize the effects of NF-κB signaling on VSV replication and RELA/p65 nuclear retention, a specific inhibitor of IκB kinase (IKK), BAY11-7082 (BAY11), was used to block NF-κB activity (44). Compared to control or VSV-treated cells (Fig. 3A, lanes 1 and 3), a 3- to 6-fold increase (P < 0.005) in nuclear retention of RELA/p65 was observed in cells treated with vorinostat or vorinostat plus VSV (Fig. 3A, lanes 5 and 7). However, increased nuclear retention of RELA/p65 was reversed in cells treated with vorinostat, VSV, and BAY11, demonstrating that BAY11 efficiently inhibited RELA/p65 nuclear translocation/retention stimulated by vorinostat-plus-VSV treatment (Fig. 3A, lane 9). By immunofluorescence, the combination of vorinostat plus VSV led to a 6-fold increase in the number of GFP-positive cells (P < 0.01) and in nuclear localization of RELA/p65, as indicated by higher nuclear signal for REL/p65 than for the control (Fig. 3B, arrows); cotreatment with BAY11 dramatically decreased the number of GFP-positive cells, as well as the nuclear signal for RELA/p65 (Fig. 3B right), indicating decreases in viral replication and nuclear retention of RELA/p65, respectively. The inhibitory effect of BAY11 on NF-κB target gene expression was also examined by measuring mRNA levels of several NF-κB target genes (Fig. 3C). While increased expression of IκBα, IL-1β, CDKN1A, TNFAIP3, or BIRC3 mRNA was observed in cells treated with vorinostat plus VSV, cotreatment with BAY11 led to an ∼50% decrease in the expression of these genes (P < 0.005). Altogether, these data indicate that BAY11 can efficiently decrease VSV replication, RELA/p65 nuclear retention, and NF-κB target gene expression in PC3 cells.
FIG 3.
The IKK inhibitor BAY11 blocks vorinostat-mediated nuclear translocation of p65 and decreases NF-κB target gene expression. PC3 cells treated with VSV (MOI = 0.01) with or without vorinostat (5 μM) for 24 h were also pretreated with 10 μM BAY11 for 1 h where indicated. (A) Nuclear (N) and cytosolic (C) extracts were separated by gel electrophoresis and immunoblotted for RELA/p65. THOC1 was used as a loading control for the nuclear fraction. The numbers below the Western blots indicate nuclear RELA/p65/THOC1 signal ratios relative to the vehicle-treated cells ± standard errors. (B) Confocal microscopy analysis of PC3 cells that were infected with VSV-GFP (green), treated as indicated, and immunostained for RELA/p65 (red). (C) Relative mRNA levels of several NF-κB target genes in PC3 cells exposed to the indicated treatments and assessed by qPCR. White bars, vehicle (DMSO) treatment; light-gray bars, vorinostat treatment; dark-gray bars, VSV treatment; black bars, VSV-plus-vorinostat treatment; light-blue bars, VSV-plus-vorinostat-plus DMSO treatment; dark-blue bars, vorinostat-plus-VSV-plus BAY11 treatment. **, P ≤ 0.01; ***, P ≤ 0.005. The error bars indicate SD.
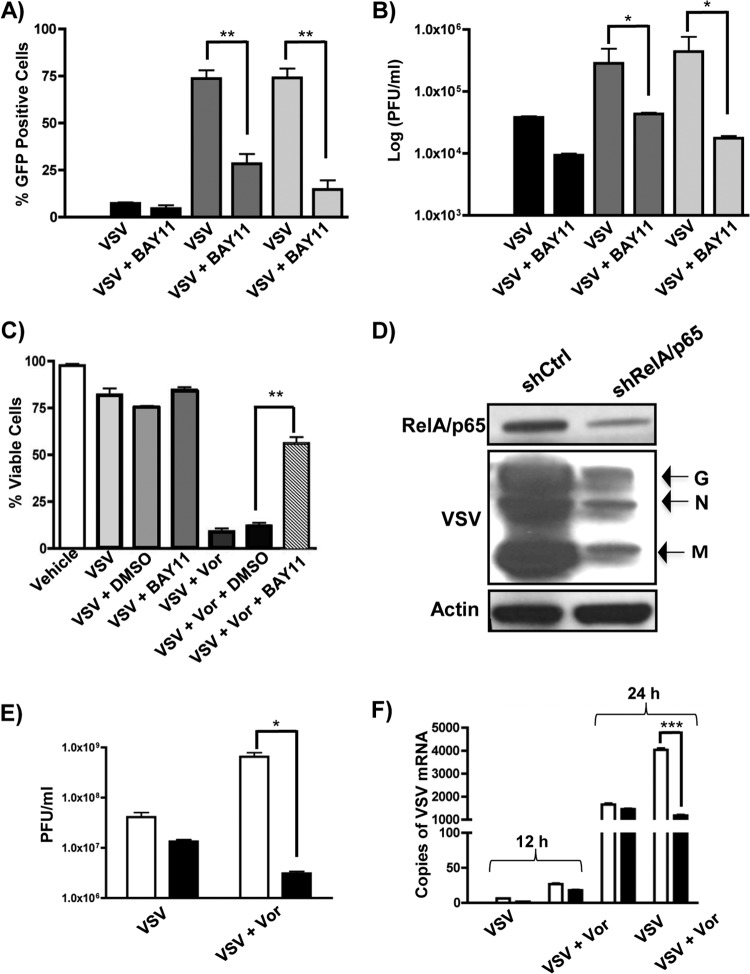
To determine the effect of NF-κB inhibition on VSV replication and oncolysis, PC3 cells were infected with VSV-GFP in the presence of vorinostat or MS-275 and BAY11 (Fig. 4). An ∼50 to 70% decrease in the number of GFP-positive cells (Fig. 4A) and 1- to 2-log-unit decrease in virion release (Fig. 4B) were observed in PC3 cells that were additionally treated with BAY11. Cell viability, as measured by the percentage of GFP-, annexin V, and 7-aminoactinomycin D (7-AAD) triple-negative cells, was also increased (∼80%) with additional BAY11 treatment compared to cells exposed to VSV plus vorinostat only (Fig. 4C).
FIG 4.
BAY11 or knockdown of RELA/p65 reverses vorinostat or MS-275-mediated enhancement of VSV oncolysis. (A and B) PC3 cells were pretreated with DMSO (black bars), MS-275 (2 μM) (dark-gray bars), or vorinostat (5 μM) (light-gray bars) with or without BAY11 (10 μM) and infected with VSV-GFP (MOI = 0.01), as indicated below the graphs, for 24 h. Viral replication was quantified by flow cytometry (A) or plaque assay (B). (C) Cell viability of PC3 cells was measured by flow cytometry as a percentage of GFP and annexin V double-negative cells. (D and E) Quantification of VSV replication by immunoblotting (D) or plaque assay (E) in Ctrl-Luc (white bars) and shRel (black bars) cells exposed to VSV with or without vorinostat. (F) Quantification of VSV genomic mRNA by qPCR in Luc-Ctrl (white bars) and shRel (black bars) cells exposed to VSV with or without vorinostat at 12 and 24 h postinfection. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. The error bars indicate SD.
We also examined the effects of decreased expression of RELA/p65 in PC3 cells on VSV replication using a genetic approach (Fig. 4D and E). PC3 cells stably transfected with LeGO-1xT vector expressing shRNA that targets expression of RELA/p65 (shRel) or a control shRNA targeting expression of luciferase (Ctrl-Luc) (37) were infected with VSV, and viral replication was measured by immunoblotting (Fig. 4D), plaque assay (Fig. 4E), and qPCR of viral RNA (Fig. 4F). Knockdown of RELA/p65 efficiently inhibited VSV replication, as shown by an ∼1-log-unit decrease in VSV replication in the knockdown cells (black bars) compared to control cells (Fig. 4E and F, white bars). Pretreatment with vorinostat did not rescue viral replication, indicating that the vorinostat-mediated effect required functional NF-κB signaling. Altogether, these data demonstrate that pharmacologic or genetic inhibition of NF-κB signaling leads to a decrease in VSV replication and oncolysis, as well as an increase in cell survival.
Vorinostat potentiates VSV oncolysis by increasing autophagy.
To investigate further the mechanistic basis of NF-κB-mediated potentiation of VSV oncolysis, we searched for potential downstream effectors of NF-κB signaling. From the global expression data set, we identified increased expression (up to 2.7-fold) of genes that belong to the autophagy pathway in PC3 cells after treatment with vorinostat alone or with vorinostat plus VSV (Fig. 5A). Several of the autophagy-related genes (indicated by stars in Fig. 5A) were found to possess one or more RELA/p65 binding motifs, based on a search of their promoter regions (45). In agreement with this observation, the levels of mRNA for ATG101, ATG7, and GABARAPL1 increased 6- to 30-fold in PC3 cells exposed to vorinostat or vorinostat plus VSV, whereas an ∼4-fold decrease (P < 0.01) was observed following cotreatment with BAY11 (Fig. 5B). Furthermore, expression levels of ATG101, ATG7, and GABARAPL1 were 2- to 5-fold lower in vorinostat- or vorinostat-plus-VSV-treated cells stably knocked down for RELA/p65 (Fig. 5C, black bars) than in control cells (white bars).
FIG 5.
Vorinostat treatment upregulates the autophagy pathway. (A) Fold changes of autophagy-related genes assessed by global genome expression profiling in four experimental groups relative to nontreated (control) PC3 cells. The autophagy-related genes indicated by the stars were found to possess one or more RELA/p65 binding motifs. (B and C) qPCR assessment of mRNA levels for ATG101, ATG7, and GABARAPL1 genes in PC3 cells exposed to VSV (MOI = 0.01) with or without vorinostat (5 μM) in the presence of BAY11 (10 μM), as indicated below the graphs (B) or in Ctrl-Luc and shRel cells exposed to VSV with or without vorinostat for 24 h (C). White bars, Ctrl-Luc cells; black bars, shRel cells. (D and E) Ctrl-Luc or shRel stably transfected PC3 cells were treated with VSV with or without vorinostat for 24 h (D) or 48 h (E); the levels of autophagic vesicles or apoptosis were measured by flow cytometry using the Cyto-ID Autophagy Detection Kit (black bars) or annexin V staining (white bars). H, halted; C, continuous. **, P ≤ 0.01. The error bars indicate SD.
To determine the effect of RELA/p65 knockdown on autophagy, shRel or Ctrl-Luc PC3 cells were examined for induction of autophagy by Cyto-ID staining of autophagic vesicles (46) and for apoptosis by annexin V staining at 24 or 48 h after exposure to VSV with or without vorinostat. At 24 h, no significant increase in apoptosis (annexin V staining) was observed in any of the treatment groups (Fig. 5D, white bars). However, induction of autophagy was observed for vorinostat-treated cells with or without VSV infection (Fig. 5D, black bars). For these treatment groups, between 75 and 83% of the Ctrl-Luc cells and only ∼15% of the shRel cells were Cyto-ID positive. At 48 h postinfection (Fig. 5E), levels of autophagic and apoptotic cells in the vorinostat-only and VSV-only treatment groups were similar to those in their respective treatment groups at 24 h postinfection. However, the combination treatment of vorinostat plus VSV altered the amount of autophagic and apoptotic cells 48 h postinfection. The number of annexin V-positive Ctrl-Luc cells dramatically increased to ∼66%, while the number of annexin V-positive shRel cells increased by ∼15%. Only a modest increase in the number of shRel autophagic cells was observed (∼35%), indicating that functional RelA expression was required to observe autophagy or apoptosis. Taken together, these data indicate that induction of autophagy in vorinostat-plus-VSV-treated cells (at 24 h) precedes induction of apoptosis (at 48 h), and apoptosis was observed only in cells exposed to VSV plus vorinostat. This also reveals that apoptosis is a direct consequence of VSV oncolysis and not vorinostat-induced autophagy. In agreement with the relative resistance of PC3 cells to vorinostat-induced apoptosis (29), at 48 h, only ∼15% of vorinostat-treated Ctrl-Luc cells were annexin V positive.
To further confirm the role of autophagy in the vorinostat-mediated increase in VSV cytotoxicity, autophagy-deficient mouse embryonic fibroblasts (Atg5−/− MEFs) were infected with VSV in the presence of increasing concentrations of vorinostat (Fig. 6A). While addition of vorinostat led to increased VSV replication in WT MEFs, no increase in VSV replication was observed upon addition of vorinostat to Atg5−/− MEFs (Fig. 6A). Furthermore, in PC3 cells, addition of the autophagy inhibitor 3-MA led to a decrease in the number of GFP-positive cells, even in the presence of vorinostat (Fig. 6B). Because autophagy can positively or negatively modulate the innate immune response to viral infection (47), we evaluated the expression of several innate antiviral response genes using a Fluidigm BioMark assay (Fig. 6C). Treatment with 3-MA in the presence of vorinostat increased the mRNA levels of IFN-stimulated genes—CXCL10, IFIT1, IFIT2, IFITM1, MX1, and MX2 genes—in VSV-infected cells, indicating that the inhibition of autophagy increased IFN antiviral responses. The increase in mRNA was specific for IFN-regulated genes, as no increase in the levels of proinflammatory interleukin 1A (IL-1A), IL-6, or IL-10 genes was observed. The reversal of the immunosuppressive effect of vorinostat by 3-MA was further confirmed by immunoblotting for phosphorylated STAT1, IFIT1, MX1, and ISG15 in Ctrl-Luc and shRel cells. These proteins were upregulated in shRel cells in response to VSV plus vorinostat, and a similar trend was observed in Ctrl-Luc cells that were treated with VSV plus vorinostat plus 3-MA, whereas Ctrl-Luc cells treated with VSV plus vorinostat maintained the ability to downregulate these proteins. Similarly, expression of VSV proteins (data not shown) and markers of apoptosis (cleaved PARP and cleaved caspase 3) was upregulated in Luc-Ctrl cells, but not in shRel or Ctrl-Luc cells treated with 3-MA (Fig. 6D). Taken together, these data demonstrate that NF-κB-dependent autophagy plays a major role in vorinostat-mediated increase in VSV oncolysis by decreasing IFN-mediated antiviral responses and facilitating apoptosis.
FIG 6.
Inhibition of autophagy decreases VSV replication and increases antiviral signaling. (A) Atg5+/+ (black bars) or Atg5−/− (white bars) MEFs were exposed to the indicated concentrations of vorinostat and infected with VSV-GFP (MOI = 0.0001), and viral replication was assessed 24 h later by flow cytometry. (B) PC3 cells were treated with 0 mM (black bars), 5 mM (dark-gray bars), 7.5 mM (light-gray bars), or 10 mM (white bars) 3-MA for 1 h prior to VSV-GFP (MOI = 0.01) infection, and viral replication was assessed by flow cytometry 24 h later. (C) Gene expression analysis using the Dynamic Arrays platform for several genes (indicated on the left) in PC3 cells exposed to VSV (MOI = 0.01) and vorinostat (5 μM) with or without 5, 7.5, or 10 mM 3-MA. (D) PC3 cells were exposed to the treatments indicated at the top, and antiviral signaling and apoptotic markers were assessed by immunoblotting. **, P ≤ 0.01; ***, P ≤ 0.005. The error bars indicate SD.
To further confirm the generality of these observations, the role of NF-κB signaling in VSV oncolysis was also examined using two additional cell lines, prostate cancer DU145 and colon cancer HCT116 cells (Fig. 7). Decreases in cell survival, increased numbers of infected cells, and increased virion release were all observed in DU145 (white bars) and HCT116 (black bars) cells upon VSV-plus-vorinostat treatment compared to VSV infection alone (Fig. 7A to C). Addition of BAY11 increased cell survival and decreased viral replication by approximately 2 log units. To assess whether vorinostat treatment induces autophagy in DU145 and HCT116 cells, we examined two markers of autophagy, LC3 lipidation and degradation of SQSTM1 (48) (Fig. 7D). In both cell lines, vorinostat treatment alone or in combination with VSV induced LC3 lipidation and degradation of SQST1, indicating upregulation of autophagy markers in vorinostat-treated cells.
FIG 7.
Vorinostat promotes VSV replication and induces autophagy in nonpermissive cells. DU145 (black bars) and HCT116 (white bars) cells were exposed to VSV-GFP with or without 5 μM SAHA, in combination with 10 μM BAY11 where indicated, for 24 h. (A) Cell viability was measured by flow cytometry as a percentage of GFP and annexin V double-negative cells. (B and C) VSV replication was assessed by measuring the number of GFP-positive cells (B) and plaque assay (C). (D) DU145 and HCT116 cells were exposed to the treatments indicated above the gels, and autophagy markers were assessed by immunoblotting. The error bars indicate SD.
DISCUSSION
In previous studies, we and others (19, 49–51) characterized a combination strategy using HDIs to potentiate oncolysis in cancer cells that are not highly permissive to OV replication. However, the antitumor mechanism of the therapeutic combination remained elusive, and with the present study, we sought to investigate the molecular basis of VSV oncolysis in combination with the histone deacetylase inhibitor vorinostat. We demonstrate that vorinostat in combination with VSV (i) induced reversible expression of a subset of NF-κB-regulated genes, as well as IFN-stimulated genes; (ii) induced hyperacetylation and increased nuclear retention of RELA/p65; (iii) increased expression of autophagy-related genes in an NF-κB-dependent manner; and (iv) triggered autophagy, which preceded and facilitated VSV oncolysis in PC3 cells.
Global genome expression analysis of differentially regulated genes in PC3 cells exposed to vorinostat with or without VSV revealed increased expression of numerous NF-κB-regulated genes. Subsequent biochemical analysis revealed increased acetylation and nuclear retention of NF-κB subunit RELA/p65 in the presence of vorinostat, and mass spectrometry analysis of RELA/p65 identified acetylation of eight lysine residues in RELA/p65. The functional roles of several acetylated lysines of RELA/p65 are known: acetylation of K310 is required for full transcriptional activity (43), acetylation of K314 and K315 regulates the specificity of gene expression (52), and acetylation of K122 and K123 reduces DNA binding capacity (53). We also detected for the first time increased acetylation of K56, K62, and K195, although the functional roles of these sites are currently unknown. Based on our data, these lysines may play a role in nuclear shuttling and sequestration, specificity of target gene expression, and/or regulation of DNA binding affinity.
Further bioinformatics analysis of differentially regulated genes revealed increased expression of several autophagy-related genes in PC3 cells after treatment with vorinostat or vorinostat plus VSV. This observation led us to further examine the association between NF-κB and autophagy. Increased expression of three autophagy-related genes—ATG101, ATG7, and GABARAPL1—was observed in cells where NF-κB signaling was stimulated with vorinostat; conversely, inhibition of NF-κB by BAY11 or shRNA-mediated knockdown of RELA/p65 led to decreased expression of ATG101, ATG7, and GABARAPL1. The role of autophagy in viral infection is context dependent and may involve elimination of viral particles or suppression of apoptosis and/or innate immune responses, providing a suitable environment for viral replication. Stimulation of pattern recognition receptors induces autophagy by unknown mechanisms, and autophagy may also be a mechanism by which viruses and other pathogens escape innate immune responses (47). A number of viruses, such as coronavirus, dengue virus, and coxsackievirus, can utilize the autophagic machinery as a scaffold to promote viral replication (reviewed in reference 36). Autophagy can also contribute to oncolysis; for example, a combination of oncolytic adenovirus and autophagy inducers greatly improved OV antitumor efficacy in both glioblastoma cell lines and U87MG-derived glioma xenograft models (54). We also previously described a role for autophagy as an antitumor mechanism in chronic lymphocytic leukemia (CLL) cells, where treatment of CLL cells with the combination of Bcl-2 inhibitors and VSV led to induction of metabolic stress and subsequent disruption of anti-autophagic protein-protein interactions (55).
Activation of the canonical NF-κB pathway is also closely associated with regulation of autophagy. Transforming growth factor beta (TGF-β)-activated kinase 1 (TAK1) activates IKK, which in turn phosphorylates IκBα, the inhibitory subunit of the NF-κB complex, leading to activation of NF-κB-mediated signaling. If TAK1 or any other component of the IKK complex is inhibited, induction of autophagy by upstream stimuli is impaired (56). In agreement with this, we observed increased expression of three autophagy-related genes—ATG101, ATG7, and GABARAPL1 genes—in cells where NF-κB signaling was stimulated with vorinostat; conversely, inhibition of NF-κB by BAY11 or shRNA-mediated knockdown of RELA/p65 led to decreased expression of ATG101, ATG7, and GABARAPL1 genes. Although the main function of NF-κB is to regulate the expression of specific genes in the cells of the immune system, some viruses utilize anti-apoptotic properties of NF-κB to divert its function from regulation of immune responses to modulation of cell growth and apoptosis, thus providing a survival advantage to the virus (reviewed in reference 57).
In this study, we uncovered a function of NF-κB that facilitates VSV replication: the NF-κB-regulated suppression of innate immune responses mediated by the autophagic pathway. Pharmacologic inhibition of autophagy by 3-MA led to the upregulation of both type I IFN and cytokine expression and a subsequent decrease in VSV replication even in the presence of vorinostat. These data are in agreement with a recent study in which knockdown of several autophagy genes in immortalized human hepatocytes inhibited the growth of hepatitis C virus and increased IFN-mediated responses (58). Nevertheless, exactly how autophagy decreases innate responses following VSV-plus-vorinostat infection remains unclear; Jounai et al. offered a potential mechanism in which autophagy proteins constitutively associate with the RIG-I and MAVS proteins and thus restrict virus-induced type I IFN response (59).
In summary, we provide new evidence that vorinostat potentiates VSV oncolysis, not only by direct suppression of innate immune responses, but also by indirectly stimulating induction of the autophagic pathway leading to VSV-mediated apoptosis in cells with functional NF-κB signaling. With a number of OVs entering clinical trials and promising data emerging from phase II and III clinical trials (3, 60), understanding the cellular processes that modulate tumor responses to virus-based therapies becomes an essential component of the successful clinical implementation of OVs.
ACKNOWLEDGMENTS
We thank David Olagnier for critical readings of the manuscript and Anthony Cooper and Cheryl Cameron for help with histone acetylation experiments.
This work was supported by grants from the Canadian Institutes of Health Research (MOP 42562) and the Terry Fox Foundation, with the support of the Canadian Cancer Society, and by Department of Defense grant LC110658 to J.H. L.S. is supported by a Fondation de Recherche Sante de Quebec Fellowship award and Prostate Cancer Canada.
We declare no conflict of interest.
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Rowan K. 2010. Oncolytic viruses move forward in clinical trials. J. Natl. Cancer Inst. 102:590–595. 10.1093/jnci/djq165 [DOI] [PubMed] [Google Scholar]
- 2.Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, Nieva J, Hwang TH, Moon A, Patt R, Pelusio A, Le Boeuf F, Burns J, Evgin L, De Silva N, Cvancic S, Robertson T, Je JE, Lee YS, Parato K, Diallo JS, Fenster A, Daneshmand M, Bell JC, Kirn DH. 2011. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature 477:99–102. 10.1038/nature10358 [DOI] [PubMed] [Google Scholar]
- 3.Russell SJ, Peng KW, Bell JC. 2012. Oncolytic virotherapy. Nat. Biotechnol. 30:658–670. 10.1038/nbt.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hastie E, Grdzelishvili VZ. 2012. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J. Gen. Virol. 93:2529–2545. 10.1099/vir.0.046672-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary ZD, Willingham MC, Lyles DS. 2011. Oncolytic vesicular stomatitis virus induces apoptosis in U87 glioblastoma cells by a type II death receptor mechanism and induces cell death and tumor clearance in vivo. J. Virol. 85:5708–5717. 10.1128/JVI.02393-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce AF, Lyles DS. 2009. Vesicular stomatitis virus induces apoptosis primarily through Bak rather than Bax by inactivating Mcl-1 and Bcl-XL. J. Virol. 83:9102–9112. 10.1128/JVI.00436-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaddy DF, Lyles DS. 2005. Vesicular stomatitis viruses expressing wild-type or mutant M proteins activate apoptosis through distinct pathways. J. Virol. 79:4170–4179. 10.1128/JVI.79.7.4170-4179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goubau D, Deddouche S, Reis ESC. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullen JT, Tanabe KK. 2002. Viral oncolysis. Oncologist 7:106–119. 10.1634/theoncologist.7-2-106 [DOI] [PubMed] [Google Scholar]
- 10.Kelly E, Russell SJ. 2007. History of oncolytic viruses: genesis to genetic engineering. Mol. Ther. 15:651–659. 10.1038/sj.mt.6300108 [DOI] [PubMed] [Google Scholar]
- 11.Sinkovics JG, Horvath JC. 2008. Natural and genetically engineered viral agents for oncolysis and gene therapy of human cancers. Arch. Immunol. Ther. Exp. (Warsz) 56(Suppl 1):3s–59s. 10.1007/s00005-008-0047-9 [DOI] [PubMed] [Google Scholar]
- 12.Kurisetty VV, Heiber J, Myers R, Pereira GS, Goodwin JW, Federspiel MJ, Russell SJ, Peng KW, Barber G, Merchan JR. 2013. Preclinical safety and activity of recombinant VSV-IFN-beta in an immunocompetent model of squamous cell carcinoma of the head and neck. Head Neck. 10.1002/hed.23502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naik S, Nace R, Barber GN, Russell SJ. 2012. Potent systemic therapy of multiple myeloma utilizing oncolytic vesicular stomatitis virus coding for interferon-beta. Cancer Gene Ther. 19:443–450. 10.1038/cgt.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojdl DF, Lichty BD, ten Oever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. 10.1016/S1535-6108(03)00241-1 [DOI] [PubMed] [Google Scholar]
- 15.Stojdl DF, Lichty B, Knowles S, Marius R, Atkins H, Sonenberg N, Bell JC. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6:821–825. 10.1038/77558 [DOI] [PubMed] [Google Scholar]
- 16.Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93–102. 10.1016/j.molcel.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 17.Barber GN. 2005. VSV-tumor selective replication and protein translation. Oncogene 24:7710–7719. 10.1038/sj.onc.1209042 [DOI] [PubMed] [Google Scholar]
- 18.Russell SJ, Peng KW. 2007. Viruses as anticancer drugs. Trends Pharmacol. Sci. 28:326–333. 10.1016/j.tips.2007.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TL, Abdelbary H, Arguello M, Breitbach C, Leveille S, Diallo JS, Yasmeen A, Bismar TA, Kirn D, Falls T, Snoulten VE, Vanderhyden BC, Werier J, Atkins H, Vähä-Koskela MJ, Stojdl DF, Bell JC, Hiscott J. 2008. Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc. Natl. Acad. Sci. U. S. A. 105:14981–14986. 10.1073/pnas.0803988105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed M, Cramer SD, Lyles DS. 2004. Sensitivity of prostate tumors to wild type and M protein mutant vesicular stomatitis viruses. Virology 330:34–49. 10.1016/j.virol.2004.08.039 [DOI] [PubMed] [Google Scholar]
- 21.Nguyen TL, Wilson MG, Hiscott J. 2010. Oncolytic viruses and histone deacetylase inhibitors—a multi-pronged strategy to target tumor cells. Cytokine Growth Factor Rev. 21:153–159. 10.1016/j.cytogfr.2010.03.002 [DOI] [PubMed] [Google Scholar]
- 22.Glozak MA, Seto E. 2007. Histone deacetylases and cancer. Oncogene 26:5420–5432. 10.1038/sj.onc.1210610 [DOI] [PubMed] [Google Scholar]
- 23.Katsura T, Iwai S, Ota Y, Shimizu H, Ikuta K, Yura Y. 2009. The effects of trichostatin A on the oncolytic ability of herpes simplex virus for oral squamous cell carcinoma cells. Cancer Gene Ther. 16:237–245. 10.1038/cgt.2008.81 [DOI] [PubMed] [Google Scholar]
- 24.Pei XY, Dai Y, Grant S. 2004. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clin. Cancer Res. 10:3839–3852. 10.1158/1078-0432.CCR-03-0561 [DOI] [PubMed] [Google Scholar]
- 25.Suliman BA, Xu D, Williams BR. 2012. HDACi: molecular mechanisms and therapeutic implications in the innate immune system. Immunol. Cell Biol. 90:23–32. 10.1038/icb.2011.92 [DOI] [PubMed] [Google Scholar]
- 26.Laurenzana A, Balliu M, Cellai C, Romanelli MN, Paoletti F. 2013. Effectiveness of the histone deacetylase inhibitor (S)-2 against LNCaP and PC3 human prostate cancer cells. PLoS One 8:e58267. 10.1371/journal.pone.0058267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang JJ, Kim YS, Kim T, Kim MJ, Jeong IG, Lee JH, Choi J, Jang S, Ro S, Kim CS. 2012. A novel histone deacetylase inhibitor, CG200745, potentiates anticancer effect of docetaxel in prostate cancer via decreasing Mcl-1 and Bcl-XL. Invest. New Drugs 30:1434–1442. 10.1007/s10637-011-9718-1 [DOI] [PubMed] [Google Scholar]
- 28.Kortenhorst MS, Zahurak M, Shabbeer S, Kachhap S, Galloway N, Parmigiani G, Verheul HM, Carducci MA. 2008. A multiple-loop, double-cube microarray design applied to prostate cancer cell lines with variable sensitivity to histone deacetylase inhibitors. Clin. Cancer Res. 14:6886–6894. 10.1158/1078-0432.CCR-08-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. 2006. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc. Natl. Acad. Sci. U. S. A. 103:15540–15545. 10.1073/pnas.0607518103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. 2012. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc. Natl. Acad. Sci. U. S. A. 109:6561–6565. 10.1073/pnas.1204429109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YJ, Won AJ, Lee J, Jung JH, Yoon S, Lee BM, Kim HS. 2012. Molecular mechanism of SAHA on regulation of autophagic cell death in tamoxifen-resistant MCF-7 breast cancer cells. Int. J. Med. Sci. 9:881–893. 10.7150/ijms.5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi AM, Ryter SW, Levine B. 2013. Autophagy in human health and disease. N. Engl. J. Med. 368:651–662. 10.1056/NEJMra1205406 [DOI] [PubMed] [Google Scholar]
- 33.Kirkegaard K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335:323–333. 10.1007/978-3-642-00302-8_16 [DOI] [PubMed] [Google Scholar]
- 34.Kudchodkar SB, Levine B. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359–378. 10.1002/rmv.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307. 10.4161/cc.9.7.11109 [DOI] [PubMed] [Google Scholar]
- 36.Meng S, Xu J, Wu Y, Ding C. 2013. Targeting autophagy to enhance oncolytic virus-based cancer therapy. Exp. Opin. Biol. Ther. 13:863–873. 10.1517/14712598.2013.774365 [DOI] [PubMed] [Google Scholar]
- 37.Schulze J, Weber K, Baranowsky A, Streichert T, Lange T, Spiro AS, Albers J, Seitz S, Zustin J, Amling M, Fehse B, Schinke T. 2012. p65-Dependent production of interleukin-1beta by osteolytic prostate cancer cells causes an induction of chemokine expression in osteoblasts. Cancer Lett. 317:106–113. 10.1016/j.canlet.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 38.Belgnaoui SM, Paz S, Samuel S, Goulet ML, Sun Q, Kikkert M, Iwai K, Dikic I, Hiscott J, Lin R. 2012. Linear ubiquitination of NEMO negatively regulates the interferon antiviral response through disruption of the MAVS-TRAF3 complex. Cell Host Microbe 12:211–222. 10.1016/j.chom.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 39.Bern M, Cai Y, Goldberg D. 2007. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal. Chem. 79:1393–1400. 10.1021/ac0617013 [DOI] [PubMed] [Google Scholar]
- 40.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. U. S. A. 107:12564–12569. 10.1073/pnas.0914191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.New M, Olzscha H, La Thangue NB. 2012. HDAC inhibitor-based therapies: can we interpret the code? Mol. Oncol. 6:637–656. 10.1016/j.molonc.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greene WC, Chen LF. 2004. Regulation of NF-kappaB action by reversible acetylation. Novartis Found. Symp. 259:208–225. 10.1002/0470862637.ch15 [DOI] [PubMed] [Google Scholar]
- 43.Chen LF, Mu Y, Greene WC. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539–6548. 10.1093/emboj/cdf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Rhee MH, Kim E, Cho JY. 2012. BAY 11-7082 is a broad-spectrum inhibitor with anti-inflammatory activity against multiple targets. Mediators Inflamm. 2012:416036. 10.1155/2012/416036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D. 2010. GeneCards Version 3: the human gene integrator. Database 2010:baq020. 10.1093/database/baq020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan LL, Shen D, Wilkinson AR, Patton W, Lai N, Chan E, Kuksin D, Lin B, Qiu J. 2012. A novel image-based cytometry method for autophagy detection in living cells. Autophagy 8:1371–1382. 10.4161/auto.21028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richetta C, Faure M. 2013. Autophagy in antiviral innate immunity. Cell Microbiol. 15:368–376. 10.1111/cmi.12043 [DOI] [PubMed] [Google Scholar]
- 48.Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL, Finkbeiner S, Fueyo-Margareto J, Gewirtz D, Jäättelä M, Kroemer G, Levine B, Melia TJ, Mizushima N, Rubinsztein DC, Simonsen A, Thorburn A, Thumm M, Tooze SA. 2011. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy 7:1273–1294. 10.4161/auto.7.11.17661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Breckenridge CA, Yu J, Price R, Wei M, Wang Y, Nowicki MO, Ha YP, Bergin S, Hwang C, Fernandez SA, Kaur B, Caligiuri MA, Chiocca EA. 2012. The histone deacetylase inhibitor valproic acid lessens NK cell action against oncolytic virus-infected glioblastoma cells by inhibition of STAT5/T-BET signaling and generation of gamma interferon. J. Virol. 86:4566–4577. 10.1128/JVI.05545-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacTavish H, Diallo JS, Huang B, Stanford M, Le Boeuf F, De Silva N, Cox J, Simmons JG, Guimond T, Falls T, McCart JA, Atkins H, Breitbach C, Kirn D, Thorne S, Bell JC. 2010. Enhancement of vaccinia virus based oncolysis with histone deacetylase inhibitors. PLoS One 5:e14462. 10.1371/journal.pone.0014462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe T, Hioki M, Fujiwara T, Nishizaki M, Kagawa S, Taki M, Kishimoto H, Endo Y, Urata Y, Tanaka N, Fujiwara T. 2006. Histone deacetylase inhibitor FR901228 enhances the antitumor effect of telomerase-specific replication-selective adenoviral agent OBP-301 in human lung cancer cells. Exp. Cell Res. 312:256–265. 10.1016/j.yexcr.2005.10.026 [DOI] [PubMed] [Google Scholar]
- 52.Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, Lane WS, Hottiger MO. 2008. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 36:1665–1680. 10.1093/nar/gkn003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758–2766. 10.1074/jbc.M209572200 [DOI] [PubMed] [Google Scholar]
- 54.Alonso MM, Jiang H, Yokoyama T, Xu J, Bekele NB, Lang FF, Kondo S, Gomez-Manzano C, Fueyo J. 2008. Delta-24-RGD in combination with RAD001 induces enhanced anti-glioma effect via autophagic cell death. Mol. Ther. 16:487–493. 10.1038/sj.mt.6300400 [DOI] [PubMed] [Google Scholar]
- 55.Samuel S, Beljanski V, Van Grevenynghe J, Richards S, Ben Yebdri F, He Z, Nichols C, Belgnaoui SM, Steel C, Goulet ML, Shamy A, Brown D, Abesada G, Haddad EK, Hiscott J. 2013. BCL-2 inhibitors sensitize therapy-resistant chronic lymphocytic leukemia cells to VSV oncolysis. Mol. Ther. 21:1413–1423. 10.1038/mt.2013.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niso-Santano M, Criollo A, Malik SA, Michaud M, Morselli E, Marino G, Lachkar S, Galluzzi L, Maiuri MC, Kroemer G. 2012. Direct molecular interactions between Beclin 1 and the canonical NFkappaB activation pathway. Autophagy 8:268–270. 10.4161/auto.8.2.18845 [DOI] [PubMed] [Google Scholar]
- 57.Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S. 2006. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene 25:6844–6867. 10.1038/sj.onc.1209941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. 2011. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53:406–414. 10.1002/hep.24073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 104:14050–14055. 10.1073/pnas.0704014104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel MR, Kratzke RA. 2013. Oncolytic virus therapy for cancer: the first wave of translational clinical trials. Transl. Res. 161:355–364. 10.1016/j.trsl.2012.12.010 [DOI] [PubMed] [Google Scholar]