Abstract
Knowledge on the morphogenesis of pestiviruses is limited due to low virus production in infected cells. In order to localize virion morphogenesis and replication sites of pestiviruses and to examine intracellular virion transport, a cell culture model was established to facilitate ultrastructural studies. Based on results of virus growth kinetic analysis and quantification of viral RNA, pestivirus strain Giraffe-1 turned out to be a suitable candidate for studies on virion generation and export from culture cells. Using conventional transmission electron microscopy and single-tilt electron tomography, we found virions located predominately in the lumen of the endoplasmic reticulum (ER) in infected cells and were able to depict the budding process of virions at ER membranes. Colocalization of the viral core protein and the envelope glycoprotein E2 with the ER marker protein disulfide isomerase (PDI) was demonstrated by immunogold labeling of cryosections. Moreover, pestivirions could be shown in transport vesicles and the Golgi complex and during exocytosis. Interestingly, viral capsid protein and double-stranded RNA (dsRNA) were detected in multivesicular bodies (MVBs), which implies that the endosomal compartment plays a role in pestiviral replication. Significant cellular membrane alterations such as those described for members of the Flavivirus and Hepacivirus genera were not found. Based on the gained morphological data, we present a consistent model of pestivirus morphogenesis.
INTRODUCTION
Pestiviruses are important pathogens of livestock and belong to the family Flaviviridae, together with the genera Hepacivirus and Flavivirus. The genus Pestivirus includes bovine viral diarrhea viruses 1 and 2 (BVDV-1 and -2), border disease virus (BDV), classical swine fever virus (CSFV), the tentative virus species “Giraffe,” and several unassigned viruses (1).
Pestiviruses are enveloped viruses with a single-stranded RNA genome of positive polarity and with a sequence of approximately 12.3 kb. The viral RNA contains one large open reading frame (ORF) flanked by nontranslated regions (NTRs) (2, 3). The envelope glycoproteins are termed Erns, E1, and E2. The enveloped virions have a size of 40 to 50 nm in electron micrographs and have been proposed to display icosahedral symmetry (4–6). For members of the genus Flavivirus, an icosahedral symmetry of the envelope has been demonstrated by image reconstructions from cryo-electron micrographs (7). The pestiviral capsid has a diameter of about 25 nm (6) and is composed of viral RNA and a single protein (C [core]), which are assumed to form a histone-like protein-RNA aggregate (8).
Pestiviruses are found as two biotypes. Infection of cultured cells with cytopathogenic (cp) virus leads to a cytopathic effect (CPE), while replication of noncytopathogenic (ncp) virus has no visible effects (4). Cell death after infection with cp strains is caused by apoptosis (9), which may be mediated by endoplasmic reticulum (ER) stress signals and correlates with increased intracellular viral RNA accumulation (10, 11). Replication of the viral genome starts 4 to 6 h after infection of the cell and is thought to occur in association with viral membranes (12, 13).
The morphogenesis of pestivirions is poorly understood. Assembly of virions is assumed to be initiated at ER-derived membranes by budding of viral capsids at sites of accumulation of viral envelope proteins (14). Enzymes located within the ER lumen have been shown to influence pestivirus envelope glycoprotein folding, and hence the synthesis of virions (15, 16). Golgi complex-mediated modification of oligosaccharide chains linked to the E2 protein was disclosed by resistance of virion-associated E2 toward treatment with endoglycosidase H (17). Other studies showed that treatment of pestivirus-infected cells with brefeldin A inhibited release of virions from culture cells, whereas infectious virus particles could still be obtained from cell lysates (18). Taken together, these studies suggest that virions are transported by the membrane system of the host cell secretory pathway to the cell surface, where they are released, probably by exocytosis (19, 20). Until now, these assumptions have not been confirmed at the ultrastructural level. Studies on morphogenesis of pestiviruses have been hampered by the low rate of virion synthesis. So far, there are only two reports of pestivirus particles in infected cells shown by transmission electron microscopy (TEM), with particles shown (i) inside ER structures (6) and (ii) in small cytoplasmic vesicles (21). Assembly of virions or virus release from cells has not been observed so far. For CSFV and BVDV, detection of extracellular pestivirions as well as immunogold labeling of the envelope protein Erns was achieved (5, 22).
In contrast to the limited number of studies on pestiviruses, morphogenesis of members of the Flavivirus genus has been investigated thoroughly at the ultrastructural level. Viral replication induces intracellular membrane alterations which colocalize with viral proteins and double-stranded RNA (dsRNA). Virions are found in close association with these complexes (23–27). Recently, new insight into the detailed structure of the viral replication complexes was gained by three-dimensional analysis of conventional ultrathin sections by use of electron tomography (28).
In this study, we addressed the issue of the intracellular location of pestivirus replication and assembly and the pathway of subsequent maturation at the ultrastructural level. Different infection models were tested, and based on results of virus growth kinetic analysis and viral RNA quantification by real-time PCR, the ultrastructural studies focused on the early stages of the pestiviral replication cycle. We found that the pestivirus strain Giraffe-1 is a particularly suitable candidate for studies on virion morphogenesis in cell culture. The combination of conventional EM techniques, single-tilt electron tomography, and immuno-EM on thawed cryosections leads to an experimentally supported model of distinct steps in pestiviral morphogenesis.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK; ATCC CCL-22) cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were grown on 10-cm tissue culture plates and passaged twice a week. They were tested regularly for the absence of pestiviruses by indirect immunofluorescence assay.
Pestivirus strain Giraffe-1 was a gift from D. J. Paton, Animal Health and Veterinary Laboratory Agency (AHVLA), Weybridge, United Kingdom. BVDV-1 strain NADL was obtained from M. Collett (29), and BVDV-2 strain 890 was provided by J. F. Ridpath, Virology Cattle Research, National Animal Disease Center, USDA, Ames, IA.
Antibodies.
The monoclonal antibody (MAb) GRS-C3, directed against the pestiviral capsid protein (30; unpublished data), was kindly provided by T. Rümenapf, Vienna, Austria. Generation of MAbs SCR25 and SCR60, directed against the pestiviral envelope protein E2, has been described previously (31). MAb J2, recognizing dsRNA, was purchased from English & Scientific Consulting, Szirak, Hungary. Rabbit polyclonal antibody (pAb) against protein disulfide isomerase (PDI) was obtained from Stressgene, San Diego, CA. Rabbit pAb against mouse IgG(H+L) was purchased from Rockland, Gilbertsville, PA. Protein A gold (PAG; 5, 10, and 15 nm) was purchased from CMC, University Medical Center, Utrecht, The Netherlands.
Growth kinetics.
MDBK cells were infected in a synchronized way. Briefly, medium containing the respective pestivirus strains was adsorbed to cells in suspension at 4°C for 1 h at a multiplicity of infection (MOI) of 10, followed by a temperature shift to 37°C. After incubation for 1 h, the inoculum was removed and cells were washed three times with phosphate-buffered saline without Ca2+ and Mg2+ (PBS def.). Subsequently, cells were seeded on 3.5-cm culture plates, and samples were harvested at 2-h intervals from 4 h to 24 h postinfection (p.i.). Samples were divided into culture supernatants, which were stored at −70°C for virus titration, and cell monolayers, which were used directly for RNA preparation. The virus titers in supernatants were determined by endpoint titration and expressed as 50% tissue culture infective doses (TCID50)/ml.
Quantitative real-time RT-PCR (qPCR).
Cell monolayers were harvested and total cellular RNA was collected using QIAshredder homogenization columns and an RNeasy RNA preparation kit (both from Qiagen, Hilden, Germany) as recommended by the supplier. For pestiviral genome amplification, a fragment (147 bp) from the 5′NTR (31) was used, and for the 18S RNA housekeeping gene, a fragment of 74 bp was chosen. After the reverse transcription (RT) step, using the pestivirus-specific reverse primer pv03R (5′-TCCATGTGCCATGTACAGCAG-3′), the 18S RNA-specific reverse primer 18s02R (5′-GGGTCGGGAGTGGGTAATTT-3′), and Superscript II reverse transcriptase (Invitrogen, Karlsruhe, Germany), quantitative real-time PCR was performed on an ABI Prism 7000 sequence detection system in “relative quantification” mode, using TaqMan Universal PCR master mix (both from Applied Biosystems, Branchburg, NJ). For PCR amplification, the pestivirus-specific nucleic acid primer pair pv03R plus pv02 (5′-GTGGACGAGGGCATGCC-3′) was used. For PCR amplification of the 18S RNA housekeeping gene, the primer pair 18s02R plus 18s01 (5′-CGGAGAGGGAGCCTGAGAA-3′) was employed. The TaqMan probes used for pestivirus detection were pvTaq01, for BVDV-1 NADL (5′-6-carboxyfluorescein [FAM]-ACAGTCTGATAGGATGCTGCAGAGGCCC-6-carboxytetramethylrhodamine [TAMRA]-3′), and pvTaq02, for BVDV-2 890 and Giraffe-1 (VIC-CAGGTCTCTGCTACACCCTATCAGGCTGTG-TAMRA) (31); the 18S RNA gene was detected by use of the 18sTaq01 probe (VIC-ACCACATCCAAGGAAGGCAGCAGG-TAMRA). Cycling conditions were 2 min at 50°C and 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. All samples were run as triplicates. Calculation of the relative amount of viral RNA was performed automatically by the software of the ABI Prism 7000 sequence detection system, based on the ΔΔCT method (32). The sample obtained at 0 h p.i. was chosen as a calibrator. The value for this sample was set as 1, and the values for all other samples were expressed as x-fold amounts of RNA compared to the calibrator.
Validation experiments showed amplification efficiencies of 2.03 (Epvtaq01), 1.97 (Epvtaq02), and 2.01 (E18SRNA) (data not shown), which allowed combined use of both PCRs in the “relative quantification” mode (32).
Electron microscopy.
MDBK cells were infected with pestivirus Giraffe-1 as described above. After the inoculation time of 1 h, fresh medium was added and cells were seeded in 3.5-cm culture plates and fixed at individually chosen time points. For flat embedding of a double monolayer (sandwich method) and vertical cell sectioning, cells were fixed on the plate with 2.5% (wt/vol) glutardialdehyde (GA) in 0.1 M cacodylate buffer for 5 min at room temperature and with fresh GA for 20 min at 4°C.
After washing, postfixation and prestaining were done with 2% (wt/vol) osmium tetroxide–Aqua Dest water for 1 h at room temperature. Monolayers were dehydrated with graded ethanol concentrations (30, 50, 70, 90, 96, and 100% [vol/vol]) and then infiltrated with epoxy resin, and polymerization was performed at 60°C for 48 h followed by 40°C for 24 h. After polymerization, the culture dish was removed mechanically from the resin. The final blocks for production of vertical cell sections were prepared by sawing two small blocks out of the resin plate, which were glued together with epoxy resin and subjected to another polymerization step. Thin sections were cut on an ultramicrotome (Ultracut; Reichert), collected on rhodium-copper grids, and stained with 2% (wt/vol) uranyl acetate in Aqua Dest water, eventually followed by staining with Reynolds lead citrate (33). Sections were examined with a Zeiss 910 transmission electron microscope at 80 kV.
Electron tomography.
Grids with thin sections prepared as described above were mounted in a tomography holder (FEI, Eindhoven, The Netherlands) and then analyzed using a Tecnai Spirit TEM (FEI) with a BioTWIN lens configuration and an LaB6 filament operating at an acceleration voltage of 120 kV. Series of micrographs (tilt series) were recorded, aligned, reconstructed, and visualized as described elsewhere (34).
Immunoelectron microscopy (IEM).
To perform labeling experiments of viral proteins at the ultrastructural level, cells were embedded and processed according to a modified Tokuyasu technique (35, 36). Monolayers of MDBK cells were infected with pestivirus Giraffe-1 on 10-cm tissue culture plates at an MOI of 10. After incubation for 1 h, fresh medium was added. Cells were fixed at 13 h p.i. with 2% (wt/vol) paraformaldehyde (PFA), 0.2% (wt/vol) GA in 0.1 M phosphate buffer for 2 h at room temperature. After washing steps with PBS def. and 0.1% (wt/vol) glycine in PBS def., cells were carefully scraped from the plate into 1% (wt/vol) gelatin in PBS def. Cells were pelleted by centrifugation (3,000 rpm, 4 min). The cell pellet was resuspended in preheated 10% (wt/vol) gelatin in PBS def. and, after incubation at 37°C for 10 min, pelleted again by centrifugation (7,000 rpm, 5 min). Subsequently, the pellet was put on ice, and after solidifying, the gelatin was cut with a razor blade into small blocks of approximately 1 mm in length, which were incubated overnight in 2.3 M sucrose in 0.1 M phosphate buffer. The next day, blocks were placed on small aluminum pins, quickly frozen, and stored in liquid nitrogen for further use. Ultrathin cryosections were cut on a cryoultramicrotome (UC/FC6; Leica, Wetzlar, Germany) at −110/−120°C and collected with a drop of 1.15 M sucrose in 0.05 M phosphate buffer and 2% (wt/vol) methylcellulose (25 cP) in Aqua Dest water on rhodium-copper grids. Sections were stored at 4°C for further use. After removal of the gelatin from the sections by incubation of the grids on prewarmed PBS def. at 40°C for 20 min, immunogold labeling was performed on small droplets of reagents by using the following steps: (i) blocking of unspecific binding sites with 0.1% (wt/vol) glycine in PBS def. (3 times for 2 min each) and 1% (wt/vol) bovine serum albumin (BSA) in PBS def. (3 min), (ii) incubation with primary antibodies at appropriate dilutions in 1% (wt/vol) BSA in PBS def. (30 min), (iii) washing steps with 0.1% (wt/vol) BSA in PBS def. (5 times for 2 min each), (iv) incubation with a bridging rabbit antibody (in the case of a mouse primary antibody) at an appropriate dilution in 1% (wt/vol) BSA in PBS def. (30 min), (v) washing steps with 0.1% (wt/vol) BSA in PBS def. (5 times for 2 min each), (vi) PAG labeling (5- or 10-nm gold) of rabbit antibody for 20 min at an appropriate dilution in 1% (wt/vol) BSA in PBS def., (vii) washing steps with PBS def. (5 times for 2 min each), and (viii) a fixation step with 1% (wt/vol) GA in PBS def. In case of double labeling, the protocol was repeated for the PDI antibody, using 15-nm PAG. Finally, grids were washed with Aqua Dest water (10 times for 1 min each), stained with a 10:1 mixture of 2% (wt/vol) methylcellulose in Aqua Dest water and 4% (wt/vol) uranyl acetate in Aqua Dest water for 5 min on ice, and subsequently dried in a small metal loop. For each labeling experiment, uninfected cells were included as controls, as well as samples in which the first antibody was replaced by buffer solution. Sections were examined with a Zeiss 910 transmission electron microscope at 80 kV.
RESULTS
Viral RNA levels and infectivity titers of different pestiviruses in MDBK cells indicate that early time points after infection are best suited for ultrastructural studies.
The main objective of the initial infection experiments was to determine an optimal time frame for ultrastructural studies of pestivirus assembly. Pestivirus strains which are known to induce relatively high virus titers in cell culture were selected. A combination of qPCR to monitor intracellular viral RNA synthesis and infectivity assays of cell culture supernatants was used to determine initial time points of virus egress from cells. Both parameters were determined within 24 h after infection for pestivirus strains BVDV-1 NADL, BVDV-2 890, and Giraffe-1 in two independent experiments under conditions of synchronized infection.
The three candidate viruses were compared with regard to RNA levels and infectivity titers (Fig. 1). Eight hours after infection, a distinct increase in the viral RNA load of infected cells was observed. At 14 h p.i., the RNA level reached a plateau 30-fold (NADL) or 100-fold (Giraffe-1) above the level directly after infection (calibrator value). BVDV-2 890 showed the highest increase in viral RNA at 8 h p.i., with a plateau about 50 times higher than the calibrator value (Fig. 1).
FIG 1.
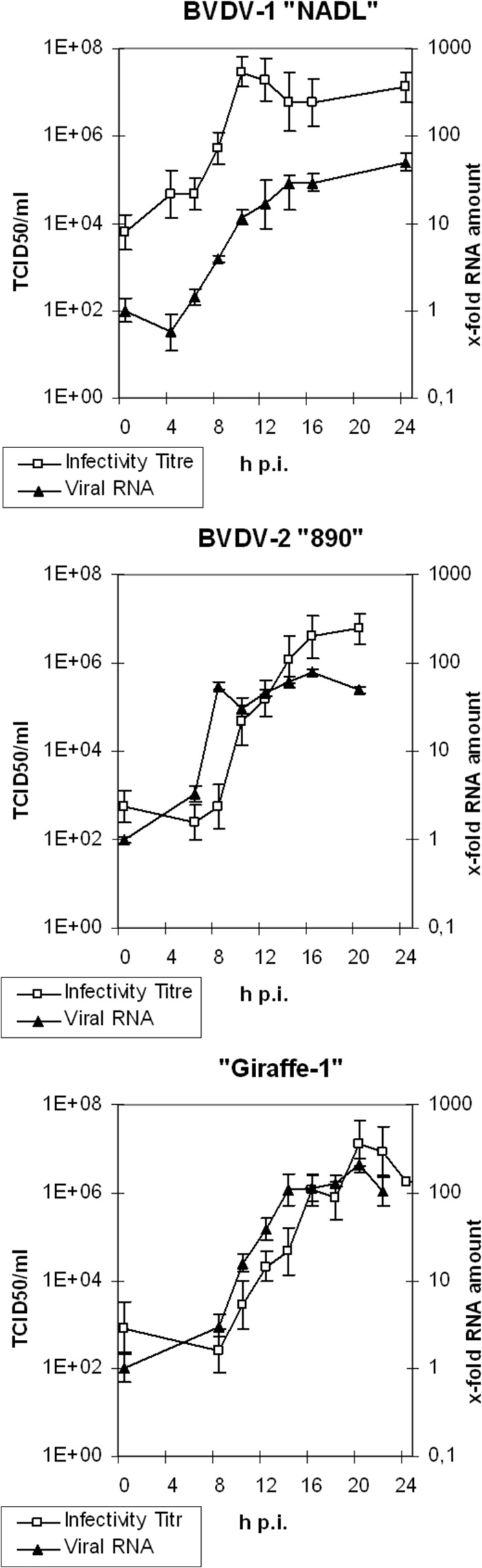
Determination of viral RNA levels and infectivity titers during the first 24 h after infection of MDBK cells with three different pestivirus strains at an MOI of 10. Left y axis, virus titers in supernatants, determined by endpoint titration and expressed as TCID50/ml; right y axis, relative amounts of viral nucleic acid in cell lysates, determined by qPCR using the ΔΔCT method. The calibrator was the sample from 0 h p.i.
Initial release of infectious virus in the cell culture supernatant occurred between 6 and 12 h p.i. CPE was first observed at 16 h p.i. in cells infected with either NADL or Giraffe-1.
Based on the results of the assays of growth kinetics, time points for ultrastructural examination were chosen within the phase of exponentially rising RNA levels and virus titers: 10 h p.i. for BVDV-1 NADL, 10 h, 14 h, and 16 h p.i. for BVDV-2 890, and 12 h, 13 h, and 15 h p.i. for pestivirus Giraffe-1.
Ultrastructural studies of the infection model with Giraffe-1 and MDBK cells disclose pestivirus morphogenesis.
In a comparative ultrahistological study, cells infected with BVDV-1 NADL, BVDV-2 890, and pestivirus Giraffe-1 were examined for events of viral morphogenesis at the time points indicated above. While only a few virions were detected in cells infected with BVDV-1 NADL and BVDV-2 890 (data not shown), MDBK cells infected with Giraffe-1 harbored numerous virus particles, which made it the best candidate for investigating aspects of pestivirus morphogenesis. The morphology of virions, with a diameter of 40 to 50 nm, a darkly stained homogenous inner capsid of 30 nm, and a single visible membrane layer surrounding the inner structures (Fig. 2), matched earlier descriptions (6, 22). In conventional electron micrographs, the outer layer of the envelopes of extracellular particles often appeared slightly deformed or dilated (Fig. 2B, arrows). However, in such cases, electron tomography revealed a complete detachment of the double lipid envelope membrane from the capsid (Fig. 2C, arrows; see Movie S1 in the supplemental material). No virions were observed in noninfected MDBK cells (data not shown). The concentrations of intracellular and extracellular virus particles increased from 12 to 15 h p.i. and were accompanied by a growing number of cells displaying cytopathic effects. Early signs of cell death were dark staining (condensation) and vacuolization of cytoplasm (data not shown). At the chosen time points, classical morphological signs of apoptosis, such as chromatin condensation and “blebbing” of the plasma membrane, were not observed. Accordingly, we found the infection model of Giraffe-1 and MDBK cells to be well suited for studies of pestivirus morphogenesis, despite the cytopathic effects on the host cell.
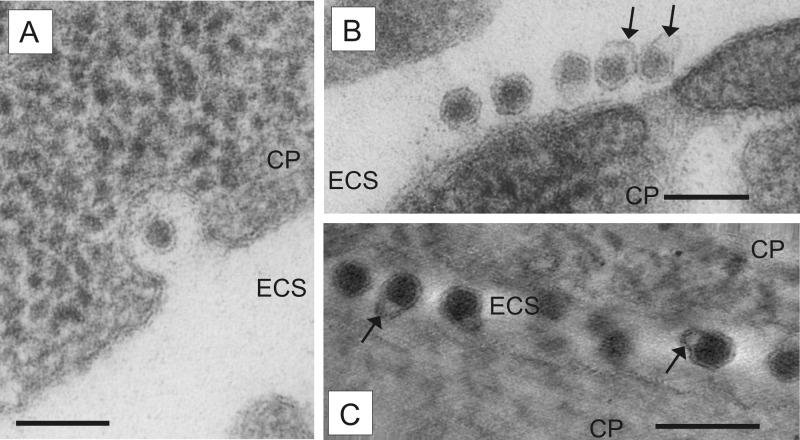
FIG 2.
Morphology of extracellular virions. MDBK cells infected with pestivirus Giraffe-1 were analyzed by EM. CP, cytoplasm; ECS, extracellular space. Bars = 100 nm. (A) Virion in a vesicle opened to the extracellular space at 13 h p.i. (B) Several extracellular virions attached to the plasma membrane, with a loosely arranged outer layer of envelope (arrows) at 15 h p.i. (C) Several extracellular virions between two cells, with a completely detached double lipid membrane (arrows) at 15 h p.i. The orthoslice picture was obtained by electron tomography.
The ER is the initial place of pestivirus assembly.
Budding of pestivirions was observed by a protrusion of the membrane into the ER lumen. This finding was best depicted by electron tomography and defines the ER as the place for pestivirus assembly (Fig. 3A and B; see Movie S2 in the supplemental material). Accordingly, the ER lumen was the first cellular compartment where new virions could be detected (starting at 12 h p.i.) (Fig. 3C). Interestingly, this compartment still harbored the largest amount of intracellular virus particles at later stages of infection (13 h and 15 h p.i.). However, only a few virions were detected in one ER luminal section. Apart from dilatation of the ER lumen, no distinct alterations of intracellular membranes were observed (data not shown).
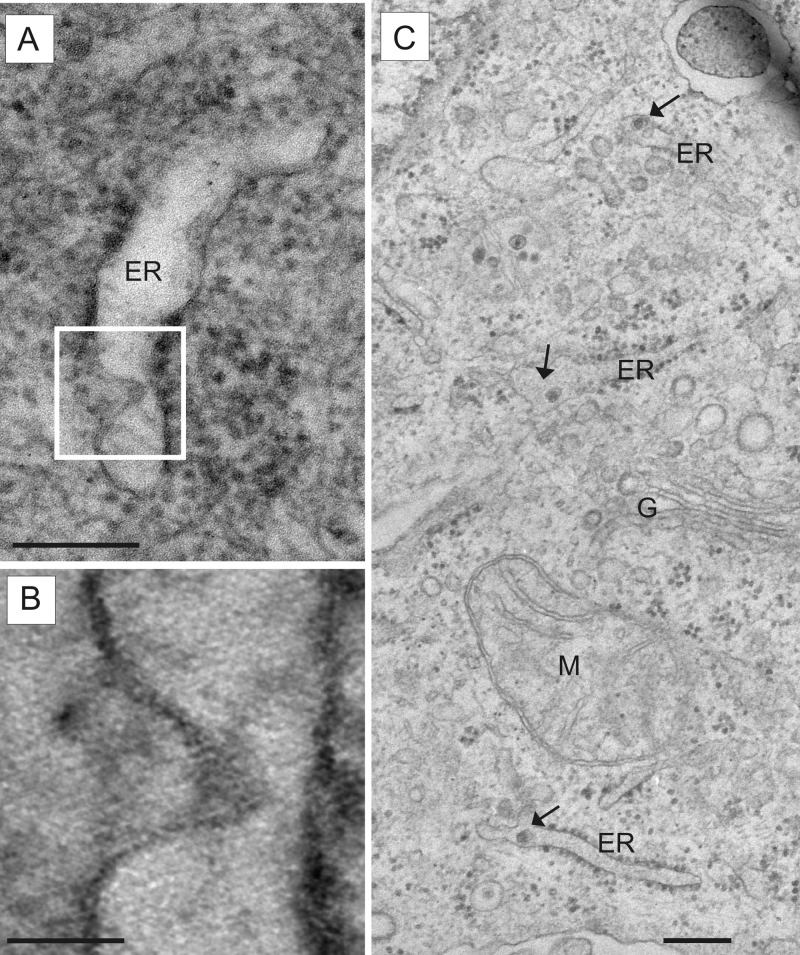
FIG 3.
Detection of budding and pestivirus particles in the ER compartment. MDBK cells infected with pestivirus Giraffe-1 were analyzed by EM. ER, endoplasmic reticulum; G, Golgi complex; M, mitochondrion. (A) Virion budding into the ER lumen at 15 h p.i. The white frame indicates the section depicted in panel B. Bar = 200 nm. (B) Detailed view of double lipid membrane protrusion of arising virion. The orthoslice picture was obtained by electron tomography. Bar = 50 nm. (C) Virions in separate ER lumina (arrows) at 12 h p.i. Bar = 200 nm.
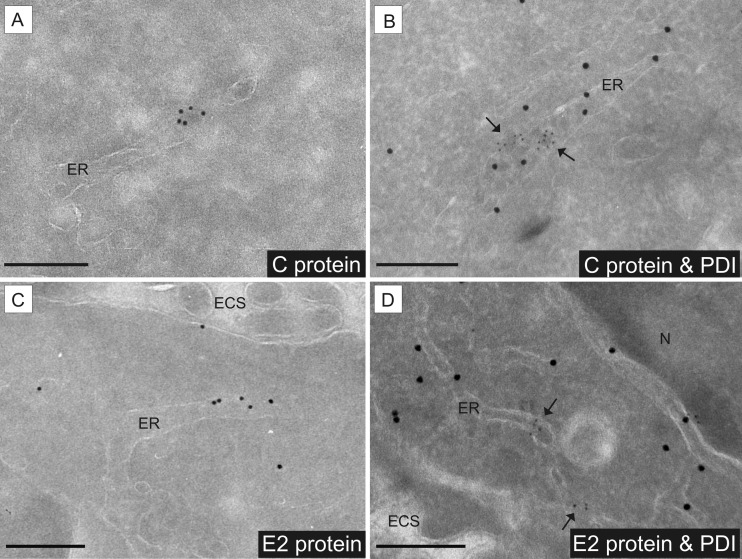
Using immunogold labeling of ultrathin cryosections, the intracellular localization of the pestiviral capsid protein (C) and the structural glycoprotein E2 was investigated. In the case of the capsid protein, a punctate clustering of gold particles closely associated with the ER as well as within the ER lumen was found (Fig. 4A). The envelope protein E2 was found mainly associated with ER membranes and in the ER lumen (Fig. 4C). For both C and E2, the ER localization was confirmed by double immunogold labeling and colocalization with PDI as an ER marker (Fig. 4B and D). Taken together, the results of conventional electron microscopy, electron tomography, and immunoelectron microscopy define the ER as the place of pestivirus particle assembly.
FIG 4.
Detection of pestiviral structural proteins in the ER compartment by IEM. Giraffe-1 and MDBK cells at 13 h p.i. were analyzed by IEM. ECS, extracellular space; ER, endoplasmic reticulum; N, nucleus. Bars = 200 nm. (A) Viral capsid protein C was detected inside the ER lumen by using MAb GRS-C3, an anti-mouse bridging antibody, and protein A gold (10 nm). (B) Intraluminal presence of capsid protein C and of viral particles, shown by colocalization with the ER marker PDI. Detection of C was performed by using MAb GRS-C3, an anti-mouse bridging antibody, and protein A gold (5 nm). Detection of PDI was done with rabbit anti-PDI and protein A gold (15 nm). Arrows point at C protein labeling. (C) Viral envelope protein E2 was detected in association with ER membranes by using MAb SCR60, an anti-mouse bridging antibody, and protein A gold (10 nm). (D) Presence of E2 at ER membranes was shown by colocalization with the ER marker PDI. Detection of E2 was performed by using MAb SCR60, an anti-mouse bridging antibody, and protein A gold (5 nm). Detection of PDI was done with rabbit anti-PDI and protein A gold (15 nm). Arrows point at E2 protein labeling.
Pestivirions pass the Golgi complex and leave the cell in small exocytotic vesicles.
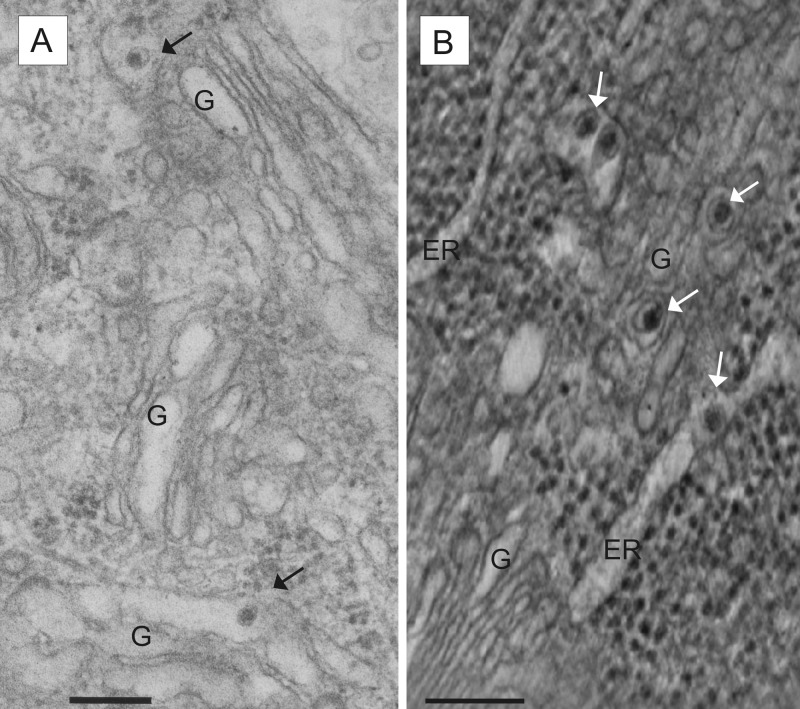
At 13 h and 15 h p.i., virions were found not only within ER lumina but also inside the lumina of Golgi stacks and in vesicles closely associated with the Golgi compartment (Fig. 5). In addition, virions were found in small vesicles close to the cell membrane or open to the extracellular space (Fig. 2A). The latter represent exocytotic vesicles, which differ in their morphology from clathrin-coated pits by the lack of a structured clathrin border. This differentiation is important, because clathrin-coated pits are assumed to mediate endocytosis of pestivirus particles (37, 38). At all chosen time points, the majority of virions were located inside the ER or within small vesicles morphologically not assigned to a specific cellular compartment. At 15 h p.i., virions trapped between cells started to accumulate in the extracellular space (Fig. 2B and C). Extracellular virions were detected at the basal, lateral, and apical plasma membranes of the cells.
FIG 5.
Detection of virions in the cellular secretory pathway. MDBK cells infected with pestivirus Giraffe-1 were analyzed at 13 h p.i. by EM (A) and electron tomography (B). ER, endoplasmic reticulum; G, Golgi complex. Bars = 200 nm. (A) Virions in the lumina of Golgi complexes and associated vesicles. (B) Several virions (white arrows) associated with the Golgi compartment and ER. The orthoslice picture was obtained by electron tomography.
In summary, pestivirions can be detected in the Golgi compartment as well as in exocytotic vesicles; these findings substantiate the hypothesis that virus egress occurs via the cellular secretory pathway.
Pestivirus proteins and dsRNA are present in the endosomal compartment in infected cells.
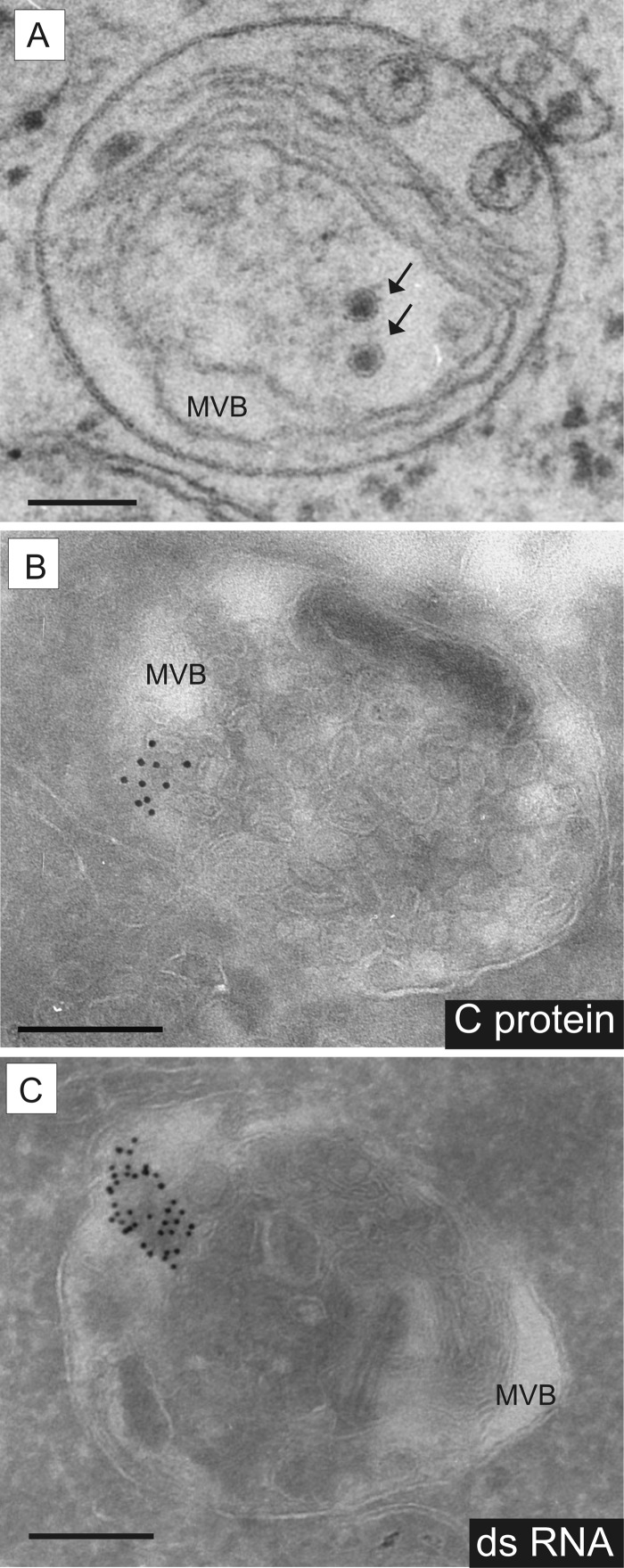
In the course of our studies, viral C protein was regularly detected by IEM inside the endosomal compartment, namely, in the lumina of multivesicular bodies (MVBs) (Fig. 6B). Moreover, particles inside MVBs and matching the morphology criteria for pestivirions were detected. However, unequivocal identification of these particles as pestivirions is hampered by the fact that MVBs of noninfected cells also harbor particles containing proteins for lysosomal degradation, termed intraluminal vesicles (ILVs), that display a morphology very similar to that of pestivirions. Some particles observed inside MVBs of infected cells displayed a hexagonal shape of the envelope. These differed from ILVs and suggested the presence of pestivirions (Fig. 6A). The E2 protein could not be detected inside MVBs.
FIG 6.
Presence of pestivirus proteins and dsRNA in MVBs of the endosomal compartment. MDBK cells infected with pestivirus Giraffe-1 were analyzed at 13 h p.i. by EM (A) and IEM (B and C). MVB, multivesicular body. Bars = 200 nm. (A) Virus-like particles (VLPs; black arrows) found in MVBs of MDBK cells infected with Giraffe-1. VLPs display a hexagonal outline of the envelope and can therefore be identified as virions. (B) Viral core protein C was detected inside the MVB lumen by using MAb GRS-C3, an anti-mouse bridging antibody, and protein A gold (10 nm). (C) dsRNA was detected inside the MVB lumen by using MAb J2, an anti-mouse bridging antibody, and protein A gold (10 nm).
Furthermore, cells were screened by IEM for the presence of dsRNA, which represents an intermediate product of viral RNA replication. Infected cells displayed an intensive signal of clustered gold particles inside the lumina and outer membranes of MVBs (Fig. 6C). Labeling in smaller vesicles and free in the cytoplasm was rarely observed. There was no dsRNA labeling in noninfected cells.
DISCUSSION
Pestiviruses are important pathogens of farm animals. In the last 20 years, significant progress has been made with regard to understanding several aspects of pestivirus biology. However, formation of virions within infected cells was still only poorly understood, as investigations on morphogenesis of pestiviruses at the ultrastructural level were impeded by the low rate of virion synthesis in infected cells.
In this study, we overcame the problem of low intracellular virion numbers by careful selection of an infection model for ultrastructural examination of virion morphogenesis. Our model included synchronized infection of MDBK cells with pestiviruses at a high MOI as well as selection of time frames for ultrastructural examination based on results of qPCR and virus infectivity titers. Accordingly, the time frames selected for EM studies were early after infection and correlated with the beginning of the exponential phase of virion synthesis. For pestiviral RNA synthesis, similar data were reported by others before (13). Former ultrastructural studies on BVDV and BDV in cell culture, which led to the first pictures of pestivirions in the ER lumen (6), and studies presenting extracellular virions of CSFV and BVDV in cell culture (5, 22) were performed with an MOI of 1 and fixation of cells as late as 46 h p.i. We concluded that in these studies, a rather low MOI together with fixation at late time points after infection impeded observation of relevant steps of pestiviral assembly, such as intracellular transport and release.
Based on our results, we developed a consistent model of pestivirus morphogenesis and egress. Pestivirions assemble directly at membranes of the ER by budding of the preformed capsid into the ER lumen at places where the envelope glycoproteins have accumulated. Once they are in the ER lumen, virions are presumably taken up at so-called ER exit sites into local transport vesicles (COP II-coated vesicles) and are transferred to the ERGIC (ER-Golgi intermediate compartment). Accumulation of virions in the ER during the course of infection supports the thesis that transfer from the ER has a low efficiency, presumably due to a lack of specific transport signals. Small transport vesicles containing single virions were assigned to the ERGIC by their close association with the ER or Golgi compartment. Direct proof based on double immunogold staining could not be provided because suitable antibodies against marker proteins for the ERGIC, such as p23, ERGIC53, or KDEL receptor, were not available for IEM on bovine cells. There is, however, already biochemical evidence for pestivirions passing through the Golgi compartment (17, 18, 39). We were able to show the Golgi passage of pestivirions by demonstrating virions inside Golgi membrane stacks. In the next step, virions are transported from the Golgi compartment to the cell surface, most likely by use of the constitutive cellular pathway, resulting in single-particle exocytosis.
It has been reported by others that pestivirions are released from large vacuoles in large numbers and/or aggregates (21, 40). However, the described vacuoles may represent MVBs, and the observed particles may represent ILVs instead of virions, especially since the authors observed particles with identical morphology in vacuoles of uninfected cells.
Based on our findings, transport from the ER to the cell surface either is inefficient or occurs very fast, and due to small virion numbers, it is difficult to depict. The same applies to the moment of exocytosis, since the vesicle membrane is fused to the plasma membrane and the pit on the surface is straightened quickly.
Extracellular virions pictured by conventional electron microscopy have been described by others (6, 22). In agreement with our studies, these are often found deformed, with a loosely arranged single envelope membrane. By electron tomography, we now show that there is in fact a complete detachment of the double lipid envelope membrane from the homogenously darkly stained inner core in these deformed virions. Whether this finding indicates a loose connection between capsid and envelope or represents an artifact caused by treatment of sample material during preparation is not clear.
Interestingly, replication of pestiviral RNA did not correlate with distinct cellular membrane rearrangements. Only slight dilatation of ER lumina was noticed. Nonstructural proteins involved in pestiviral replication were not detected in this study by use of available antibodies, which were probably not well suited for IEM (data not shown). This impeded further studies on the place and shape of the pestiviral replication complex.
For flaviviruses, it is known that replication takes place in close association with virus-induced intracellular membrane alterations that originate around the viral replication complex (24, 26, 27, 41, 42). Recently, it was shown by electron tomography that virus-induced membrane alterations are part of an ER-derived network. Small vesicular pores inside the ER lumen have been identified and were postulated to be the key to the flavivirus RNA replication complex (28). For the genus Hepacivirus, formation of a so-called membranous web harboring the viral replication complex has been described in the same context (43–45). The absence of significant membrane rearrangements in our studies apparently discloses a major difference between members of the genus Pestivirus and other Flaviviridae.
Interestingly, we were able to detect capsid protein inside MVBs. MVBs constitute a connective link between early and late endosomes; accordingly, they play a major role in intracellular protein and membrane sorting (46). Moreover, particles with pestivirus morphology were observed in this compartment. Another striking result was the localization of dsRNA, a marker for viral replication complexes (27, 47), which was mainly restricted to MVBs. These findings led to the assumption that viral RNA, virus proteins, and virions are transferred by the cell to this compartment for disposal and lysosomal degradation and are detected as a side effect of the infection. The vacuoles containing dsRNA may represent autophagosomes or amphisomes (the latter derived from fusion of endosomes and autophagosomes). Formation of autophagosomes is strictly regulated and complex; among others, viruses and bacteria are considered trigger factors (48, 49). On the other hand, the removal of dsRNA from the cytoplasm may be advantageous for the virus, since this facilitates an escape from induction of the cellular interferon response. Furthermore, the detection of C protein and virions might represent the moment during virus entry before the virus envelope fuses with the endosomal membrane to release RNA into the cytoplasm for replication (37). Finally, it is possible that the virus utilizes this compartment for replication and virion formation. Such a role of the endosomal compartment in viral morphogenesis has already been postulated for human immunodeficiency virus, in the family Retroviridae (50), and for Marburg virus, a member of the family Filoviridae (51, 52). It will be of interest to conduct further IEM studies employing marker proteins of the endosomal compartment to characterize the vacuoles and to elucidate their role in pestivirus replication.
In this study, we addressed the issues of pestivirus replication, assembly, and subsequent transport sites at the ultrastructural level. Low virus loads after pestivirus infections were overcome by studying pestivirus growth kinetics and pestiviral RNA quantities to set optimal conditions for subsequent experiments. The pestivirus Giraffe-1 turned out to be a particularly suitable candidate for establishing a consistent model of pestivirus morphogenesis. By a combination of conventional EM techniques, single-tilt electron tomography, and immuno-EM on cryosections, we were able to demonstrate that pestiviral assembly occurs directly at ER membranes. Virus egress from the cell could be shown by single virions inside the Golgi compartment and in exocytotic vesicles. IEM opened a new dimension for studies on the subcellular distribution of pestiviral proteins, which substantiated our model of pestiviral morphogenesis and disclosed a link to the endosomal compartment. It will be interesting for future studies to integrate the latter into the current model of pestivirus replication and morphogenesis.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the Stiftung der Eheleute Dr. Med. Vet. Hans-Joachim und Gertrud Engemann of the Justus-Liebig-Universität Giessen. Electron tomography studies were supported by the short-term mission program of EPIZONE (EU Network of Excellence).
We thank Wiebke Möbius (MPI für Experimentelle Medizin, Göttingen, Germany) for her kind advice and support with IEM techniques. We thank M. Hardt (EM Unit of the ZBB, Justus-Liebig-Universität Giessen) for providing the technical facilities for cryoultramicrotomy.
Footnotes
Published ahead of print 18 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03237-13.
REFERENCES
- 1.Simmonds P, Becher P, Collett MS, Gould EA, Heinz FX, Meyers G, Monath A, Pletnev A, Rice CM, Stiasny K, Thiel H-J, Weiner A, Bukh J. 2012. Family Flaviviridae, p 1003–1020 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA [Google Scholar]
- 2.Brock KV, Deng R, Riblet SM. 1992. Nucleotide sequencing of 5′ and 3′ termini of bovine viral diarrhea virus by RNA ligation and PCR. J. Virol. Methods 38:39–46. 10.1016/0166-0934(92)90167-C [DOI] [PubMed] [Google Scholar]
- 3.Collett MS, Anderson DK, Retzel E. 1988. Comparisons of the pestivirus bovine viral diarrhoea virus with members of the Flaviviridae. J. Gen. Virol. 69:2637–2643. 10.1099/0022-1317-69-10-2637 [DOI] [PubMed] [Google Scholar]
- 4.Meyers G, Thiel H-J. 1996. Molecular characterisation of pestiviruses. Adv. Virus Res. 47:53–118 [DOI] [PubMed] [Google Scholar]
- 5.Weiland E, Ahl R, Stark R, Weiland F, Thiel H-J. 1992. A second envelope glycoprotein mediates neutralization of a pestivirus, hog cholera virus. J. Virol. 66:3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray EW, Nettleton PF. 1987. The ultrastructure of cell cultures infected with border disease and bovine virus diarrhoea viruses. J. Gen. Virol. 68:2339–2346. 10.1099/0022-1317-68-9-2339 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604–2613. 10.1093/emboj/cdg270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riedel C, Lamp B, Heimann M, Rumenapf T. 2010. Characterization of essential domains and plasticity of the classical swine fever virus core protein. J. Virol. 84:11523–11531. 10.1128/JVI.00699-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoff HS, Donis RO. 1997. Induction of apoptosis and cleavage of poly(ADP-ribose) polymerase by cytopathic bovine viral diarrhea virus infection. Virus Res. 49:101–113. 10.1016/S0168-1702(97)01460-3 [DOI] [PubMed] [Google Scholar]
- 10.Jordan R, Wang L, Graczyk TM, Block TM, Romano PR. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588–9599. 10.1128/JVI.76.19.9588-9599.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassilev VB, Donis RO. 2000. Bovine viral diarrhea virus induced apoptosis correlates with increased intracellular viral RNA accumulation. Virus Res. 69:95–107. 10.1016/S0168-1702(00)00176-3 [DOI] [PubMed] [Google Scholar]
- 12.Gong Y, Shannon AD, Westaway EG, Gowans EJ. 1998. The replicative intermediate molecule of bovine viral diarrhoea virus contains multiple nascent strands. Arch. Virol. 143:399–404. 10.1007/s007050050296 [DOI] [PubMed] [Google Scholar]
- 13.Gong Y, Trowbridge R, Macnaughton TB, Westaway EG, Shannon AD, Gowans EJ. 1996. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J. Gen. Virol. 77:2729–2736. 10.1099/0022-1317-77-11-2729 [DOI] [PubMed] [Google Scholar]
- 14.Grummer B, Beer M, Liebler-Tenorio E, Greiser-Wilke I. 2001. Localization of viral proteins in cells infected with bovine viral diarrhoea virus. J. Gen. Virol. 82:2597–2605 [DOI] [PubMed] [Google Scholar]
- 15.Branza-Nichita N, Durantel D, Carrouée-Durantel S, Dwek RA, Zitzmann N. 2001. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J. Virol. 75:3527–3536. 10.1128/JVI.75.8.3527-3536.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durantel D, Carrouée-Durantel S, Branza-Nichita N, Dwek RA, Zitzmann N. 2004. Effects of interferon, ribavirin, and iminosugar derivates on cells persistently infected with noncytopathic bovine viral diarrhea virus. Antimicrob. Agents Chemother. 48:497–504. 10.1128/AAC.48.2.497-504.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan R, Nikolaeva OV, Wang L, Conyers B, Mehta A, Dwek RA, Block TM. 2002. Inhibition of host ER glucosidase activity prevents Golgi processing of virion-associated bovine viral diarrhea virus E2 glycoproteins and reduces infectivity of secreted virions. Virology 295:10–19. 10.1006/viro.2002.1370 [DOI] [PubMed] [Google Scholar]
- 18.Macovei A, Zitzmann N, Lazar C, Dwek RA, Branza-Nichita N. 2006. Brefeldin A inhibits pestivirus release from infected cells, without affecting its assembly and infectivity. Biochem. Biophys. Res. Commun. 346:1083–1090. 10.1016/j.bbrc.2006.06.023 [DOI] [PubMed] [Google Scholar]
- 19.Murray CL, Jones CT, Rice CM. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 6:699–708. 10.1038/nrmicro1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rümenapf T, Thiel H-J. 2008. Molecular biology of pestiviruses, p 39–96 In Mettenleiter TC, Sobrino F. (ed), Animal viruses: molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 21.Bielefeldt Ohmann H, Ronsholt L, Bloch B. 1987. Demonstration of bovine viral diarrhoea virus in peripheral blood mononuclear cells of persistently infected, clinically normal cattle. J. Gen. Virol. 68:1971–1982. 10.1099/0022-1317-68-7-1971 [DOI] [PubMed] [Google Scholar]
- 22.Weiland F, Weiland E, Unger G, Saalmüller A, Thiel H-J. 1999. Localization of pestiviral envelope proteins Erns and E2 at the cell surface and on isolated particles. J. Gen. Virol. 80:1157–1165 [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie JM. 2005. Wrapping things up about virus RNA replication. Traffic 6:967–977. 10.1111/j.1600-0854.2005.00339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie JM, Westaway EG. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787–10799. 10.1128/JVI.75.22.10787-10799.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882. 10.1074/jbc.M609919200 [DOI] [PubMed] [Google Scholar]
- 26.Westaway EG, Khromykh AA, Kenney MT, Mackenzie JM, Jones MK. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31–41. 10.1006/viro.1997.8629 [DOI] [PubMed] [Google Scholar]
- 27.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collett MS, Larson R, Belzer SK, Retzel E. 1988. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology 165:200–208. 10.1016/0042-6822(88)90673-3 [DOI] [PubMed] [Google Scholar]
- 30.Roman-Sosa G. 2007. Charakterisierung des Core-Proteins von Pestiviren. Dissertation. Justus-Liebig-Universität, Giessen, Germany [Google Scholar]
- 31.Cedillo-Rosales S. 2004. Charakterisierung ruminanter Pestiviren mittels Polymerasekettenreaktion und monoklonaler Antikörper. Justus-Liebig-Universität, Giessen, Germany [Google Scholar]
- 32.Applied Biosystems. 2001. Relative quantitation of gene expression. ABI Prism 7700 sequence detection system user bulletin 2. Applied Biosystems, Branchburg, NJ [Google Scholar]
- 33.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Doren EA, De Temmerman PJ, Francisco MA, Mast J. 2011. Determination of the volume-specific surface area by using transmission electron tomography for characterization and definition of nanomaterials. J. Nanobiotechnol. 9:17. 10.1186/1477-3155-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liou W, Geuze HJ, Slot JW. 1996. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106:41–58. 10.1007/BF02473201 [DOI] [PubMed] [Google Scholar]
- 36.Tokuyasu KT. 1973. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57:551–565. 10.1083/jcb.57.2.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krey T, Thiel H-J, Rümenapf T. 2005. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 79:4191–4200. 10.1128/JVI.79.7.4191-4200.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecot S, Belouzard S, Dubuisson J, Rouille Y. 2005. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 79:10826–10829. 10.1128/JVI.79.16.10826-10829.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unger G. 1993. Proteinbiochemische und strukturelle Analysen des Virus der klassischen Schweinepest. Eberhard-Karls-Universität, Tübingen, Germany [Google Scholar]
- 40.Bielefeldt Ohmann H, Bloch B. 1982. Electron microscopic studies of bovine viral diarrhea virus in tissues of diseased calves and in cell cultures. Arch. Virol. 71:57–74. 10.1007/BF01315175 [DOI] [PubMed] [Google Scholar]
- 41.Mackenzie JM, Jones MK, Westaway EG. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240. 10.1006/viro.1996.0307 [DOI] [PubMed] [Google Scholar]
- 43.Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984. 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8:e1003056. 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409. 10.1128/JVI.78.14.7400-7409.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonifacino JS, Glick BS. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153–166. 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- 47.Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240. 10.1006/viro.1996.0307 [DOI] [PubMed] [Google Scholar]
- 48.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2008. Molecular biology of the cell, 5th ed. Garland Science, New York, NY [Google Scholar]
- 49.Kirkegaard K, Taylor MP, Jackson WT. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301–314. 10.1038/nrmicro865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welsch S, Muller B, Krausslich HG. 2007. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 581:2089–2097. 10.1016/j.febslet.2007.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolesnikova L, Berghöfer B, Bamberg S, Becker S. 2004. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J. Virol. 2004:12277–12287. 10.1128/JVI.78.22.12277-12287.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mittler E, Kolesnikova L, Strecker T, Garten W, Becker S. 2007. Role of the transmembrane domain of Marburg virus surface protein GP in assembly of the viral envelope. J. Virol. 81:3942–3948. 10.1128/JVI.02263-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.