Abstract
We measured plasma levels of the oxidative DNA damage marker 8-hydroxy-2′-deoxyguanosine (8-OHdG) and leucocyte mRNA expression levels of the genes encoding the 8-OHdG repair enzyme human 8-oxoguanine DNA glycosylase 1 (hOGG1), the anti-oxidant enzymes copper/zinc superoxide dismutase (Cu/ZnSOD), manganese superoxide dismutase (MnSOD), catalase, glutathione peroxidase-1 (GPx-1), GPx-4, glutathione reductase (GR) and glutathione synthetase (GS), the mitochondrial biogenesis-related proteins mtDNA-encoded ND 1 polypeptide (ND1), ND6, ATPase 6, mitochondrial transcription factor A (Tfam), nuclear respiratory factor 1(NRF-1), pyruvate dehydrogenase E1 component alpha subunit (PDHA1), pyruvate dehydrogenase kinase isoenzyme 1 (PDK-1) and hypoxia inducible factor-1α (HIF-1α) and the glycolytic enzymes hexokinase-II (HK-II), glucose 6-phosphate isomerase (GPI), phosphofructokinase (PFK), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and lactate dehydrogenase A (LDHa). We analysed their relevance to oxidative damage in 85 systemic lupus erythematosus (SLE) patients, four complicated SLE patients undergoing rituximab treatment and 45 healthy individuals. SLE patients had higher plasma 8-OHdG levels (P < 0·01) but lower leucocyte expression of the genes encoding hOGG1(P < 0·01), anti-oxidant enzymes (P < 0·05), mitochondrial biogenesis-related proteins (P < 0·05) and glycolytic enzymes (P < 0·05) than healthy individuals. The increase in plasma 8-OHdG was correlated positively with the elevation of leucocyte expression of the genes encoding hOGG1 (P < 0·05), anti-oxidant enzymes (P < 0·05), several mitochondrial biogenesis-related proteins (P < 0·05) and glycolytic enzymes (P < 0·05) in lupus patients. The patients, whose leucocyte mtDNA harboured D310 heteroplasmy, exhibited a positive correlation between the mtDNA copy number and expression of ND1, ND6 and ATPase 6 (P < 0·05) and a negative correlation between mtDNA copy number and systemic lupus erythematosus disease activity index (SLEDAI) (P < 0·05), as well as plasma 8-OHdG (P < 0·05). In particular, four complicated SLE patients with increased expression of the genes encoding the anti-oxidant enzymes, GAPDH, Tfam and PDHA1, experienced better therapeutic outcomes after rituximab therapy. In conclusion, higher oxidative damage with suboptimal increases in DNA repair, anti-oxidant capacity, mitochondrial biogenesis and glucose metabolism may be implicated in SLE deterioration, and this impairment might be improved by targeted biological therapy.
Keywords: 8-hydroxy-2′-deoxyguanosine (8-OHdG), anti-oxidants, glycolysis, mitochondrial biogenesis, systemic lupus erythematosus (SLE)
Introduction
Systemic lupus erythematosus (SLE) is characterized by the deregulation of both innate and adaptive immunity and the production of pathogenic autoantibodies, leading to systemic inflammation and organ destruction [1]. Reactive oxygen species (ROS) and ROS-induced oxidative damage have been suggested to play important roles in systemic lupus erythematosus (SLE) pathogenesis. Several studies have demonstrated that SLE patients not only exhibit higher oxidative stress than non-affected individuals, but also have lower anti-oxidant capacity [2–5]. Although most ROS are from the surrounding environment, the mitochondria, due to electron leakage from respiratory chain reactions with O2, are the main source of endogenous ROS and immediate culprits in SLE pathogenesis [6]. Without adequate scavengers, excessive ROS may cause mitochondrial impairment that further worsens the electron leak and forms a ROS cascade. Previously, we reported that a progressive decrease in leucocyte mtDNA copy number, suggesting diminished mitochondrial biogenesis [6] and an increase in heteroplasmic D310 distribution in leucocyte mitochondrial deoxyribonucleic acid (mtDNA), suggesting oxidative DNA damage [7], correlate with the disease severity of SLE [8]. Compared with healthy controls, reduced leucocyte mRNA expression for mtDNA-encoded mtDNA-encoded ND 1 polypeptide (ND1) and mtDNA-encoded ATPase 6 polypeptide (ATPase 6) polypeptides occurred in SLE patients [8]. These findings suggested a decline in mitochondrial function in SLE leucocytes. Whether or not this decline is related to the oxidative damage in SLE patients warrants further study.
ROS can attack all cellular biomolecules, including DNA, ribonucleic acid (RNA), lipids and proteins. The formation of 8-hydroxy-2′-deoxyguanosine (8-OHdG), oxidative damage to guanine (G), is the most common form of oxidative DNA damage. Without repair, adenine (A) can pair incorrectly with 8-OHdG in place of cytosine (C) and cause a G : C to T : A transverse mutation. In human cells, 8-OHdG is repaired primarily by human 8-oxoguanine DNA glycosylase 1 (hOGG1) through a base excision repair mechanism [9]. Furthermore, several enzymatic and non-enzymatic anti-oxidants can remove ROS from the cells [10]. Superoxide anions are converted to hydrogen peroxide (H2O2) by copper/zinc superoxide dismutase (Cu/ZnSOD) in the cytosol and manganese superoxide dismutase (MnSOD) in the mitochondria. H2O2 is further reduced to H2O by catalase or glutathione (GSH)-supported glutathione peroxidase (GPx)-1 in the cytosol and by GPx-4 in the mitochondria. GSH, which is synthesized by GS, is the major non-enzymatic anti-oxidant and a redox modulator in human cells. Through GPx, GSH can be oxidized to glutathione disulphide (GSSG) to couple with the reduction of H2O2 to H2O. GSSG is then reduced to GSH by GR to maintain a sufficient GSH level. Therefore, it is of great interest to unravel the role of oxidative stress in SLE by analysing the level of 8-OHdG and the expression levels of the genes encoding hOGG1 and anti-oxidant enzymes.
Mitochondria are the main organelles responsible for ATP production through the integration of glycolysis in the cytosol and the Krebs cycle, electron transport and oxidative-phosphorylation in the mitochondria [6]. Although most respiratory enzyme complexes (approximately 90 polypeptides) are encoded by nDNA, 13 polypeptides are encoded by mtDNA. Thus, the mtDNA copy number and expression of the mtDNA-encoded polypeptides may be crucial for energy supply of cells. Generally, the replication and transcription of mtDNA are controlled by mitochondrial transcription factor A (Tfam) and regulated further by nuclear respiratory factor (NRF)-1/NRF-2 [6]. In glycolysis, the enzymes hexokinase-II (HK-II), glucose 6-phosphate isomerase (GPI), and phosphofructokinase (PFK) that regulates the rate-limiting step in glycolysis, and along with glyceraldehyde 3-phosphate dehydrogenase (GAPDH), these enzymes are involved in the conversion of glucose to pyruvate, which is further metabolized to acetyl-CoA by PDH to enter the Krebs cycle [11]. However, in some extreme conditions, e.g. inadequate oxygen supply or impaired mitochondrial function, pyruvate is transiently reduced to lactate by lactate dehydrogenase (LDH) under the assistance of stabilized hypoxia inducible factor-1α (HIF-1α) in the cytosol [12]. HIF-1α can either increase the expression of glycolytic genes or directly activate the PDK-1 gene, which in turn inhibits PDH [13,14]. Whether these physiological processes are altered in SLE remains unclear.
The mainstays of SLE treatment include non-steroidal anti-inflammatory drugs (NSAIDs), anti-malarial drugs, glucocorticoids and occasionally immunosuppressants or cytotoxic agents. For patients with severe clinical manifestations refractory to conventional treatments, the administration of a chimeric monoclonal antibody to CD20, such as rituximab, promotes a good response [15]. This response is based probably on the depletion of B cells mediated by apoptosis [16]. However, the biological effects on the redox status in the leucocytes of SLE patients after the administration of rituximab remain speculative.
In the present study, we measured various parameters in clinical samples from 89 SLE patients, including four patients receiving rituximab treatment. These parameters included the plasma concentration of 8-OHdG, the leucocyte mRNA expression levels of genes encoding hOGG1, anti-oxidant enzymes and various essential regulators or enzymes involved in mitochondrial biogenesis or glucose metabolism. We aimed to understand the roles of oxidative DNA damage, DNA repair enzymes, anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in the pathogenesis of SLE.
Materials and methods
Patient recruitment and sample preparation
According to the 1997 American College of Rheumatology updated criteria [17] and the 2012 Systemic Lupus International Collaborating Clinics (SLICC) criteria for the classification of SLE [18], samples from 89 SLE patients, including groups I and II, were collected. Initially, 85 SLE patients (73 females) with a mean age of 44·6 years (group I) from the out-patient clinic of the Division of Allergy, Immunology and Rheumatology, Taipei Veterans General Hospital, and 45 age-matched healthy controls (38 females) with a mean age of 42·6 years were recruited. Another four SLE patients (group II, fulfilling the SLICC criteria) with severe clinical symptoms who were treated with intravenous rituximab (Mabthera™) were also recruited, including one with lupus nephritis and interstitial lung disease, one with acute lupus nephritis and rapidly progressive glomerulonephritis, one with lupus nephritis, pleuritis and pericarditis and one with severe pulmonary arterial hypertension. The therapeutic regimen consisted of intravenous rituximab (500 mg) on days 1 and 14. Blood samples were collected before the rituximab treatments on days 1 and 14. The demographic data, immunological profiles and characteristics of leucocyte mtDNA, including copy number and D310 pattern, of the 45 healthy controls and 89 SLE patients are summarized in the Supporting information, Table S1 [8]. Approval from the Institutional Review Board of Taipei Veterans General Hospital was obtained to conduct this study. Approximately 10 ml of venous blood was drawn into a tube (Vacuette®, Greiner Bio-one Monroe, NC, USA) containing ethylenediamine tetraacetic acid (EDTA). After centrifugation at 3000 g for 10 min at 4°C, plasma and leucocyte-enriched buffy coats were collected separately. Using erythrocyte lysis buffer (Bioman Scientific, Taiwan), leucocyte DNA was extracted through standard phenol–chloroform procedures and kept at −20°C until use [19]. In this study, the leucocytes from 48 SLE patients, including 44 from group I and four from group II, and 26 healthy controls were preserved for RNA extraction using the TRI™ reagent (Sigma-Aldrich Chemical Co., St Louis, MO, USA) [20]. By removing residual DNA with DNase, 5 μg of RNA was further purified. Using oligo-dT primers in a 50 μl reaction buffer, 2 μg of purified DNA-free RNA was reverse-transcribed to cDNA using a Ready-To-Go reverse transcription–polymerase chain reaction (RT–PCR) kit (GE Healthcare, Amersham, UK) and kept at −20°C until use.
Determination of 8-OHdG levels in plasma by enzyme-linked immunosorbent assay ( ELISA)
Approximately 200 μl of plasma, filtered through an ultra filter (Centricon®, Ultracel YM-10 membrane; Millipore, Amicon, Billerica, MA, USA) with a cut-off molecular weight of 10 kDa, was subjected to centrifugation at 10 000 g at 4°C for 2 h to eliminate the interfering substances. Then, 50 μl of filtered plasma was used for determining the 8-OHdG concentration by an ELISA (high sensitive 8-OHdG check ELISA; Japan Institute for the Control of Aging, Nikken Seil Co., Ltd, Haruoka, Fukuroi, Shizuoka, Japan) according to the manufacturer's instructions [21]. Each reaction was performed in duplicate and the mean value was used for data presentation.
Determination of leucocyte mtDNA copy number and leucocyte mRNA expression levels of target genes
Quantitative PCR (qPCR) using SYBR Green I (Roche Applied Science, Mannheim, Germany) was carried out to determine the threshold cycle (Ct) and calculate the leucocyte mtDNA copy number, as well as to detect the transcript levels of specific target genes [8,22]. The mtDNA copy number is defined as the number of mtDNA copies divided by the number of 18S-rRNA DNA copies after adjusting the mtDNA copy number of 143B cells to 1·000. Specific gene mRNA expression was defined as the number of target gene cDNA copies divided by the number of 18S-rRNA cDNA copies after adjusting the target gene mRNA expression of 143B cells to 1·000. Briefly, 1 μl of sample DNA (10 ng/μl) or cDNA (×16 dilution) was amplified in a 10 μl reaction containing 0·25 μl (20 μM) of each primer (Table S2), 1·2 μl of 3 mM MgCl2, 1 μl of LightCycler SYBR Green I and 6·3 μl of PCR-grade H2O through a hot start at 95°C for 10 min and 35–45 cycles (depending on the target gene) of 95°C for 20 s, 62°C for 20 s and 72°C for 20 s. Simultaneously, 1 μl of DNA (1 ng/μl) or cDNA (16× dilution) of 143B cells and PCR-grade H2O were included as the positive and negative controls, respectively. Each reaction was performed in duplicate, and the mean value was used for data presentation. Using the data from the reactions with the 143B osteosarcoma cells, the sequences of the primers, replication efficiency, dilution range, squared regression coefficient (R2) of the standard curve, melting temperature and size of PCR product of each gene are listed in the Supporting information, Table S2. The target genes were classified into four categories: 8-OHdG repair enzymes, anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes.
Statistical analysis
The results are presented as the mean ± standard deviation (s.d.) after adjusting the mean value of healthy controls or day 1 to 1·000. The continuous variables between two groups or paired groups were compared using Student's t-test, the Mann–Whitney U-test, paired t-test or Wilcoxon's signed-rank test, as appropriate. The categorical variables between two groups were compared using the χ2 test or Fisher's exact test as appropriate. Relationships between two continuous variables were evaluated using Pearson's correlations. Differences were considered significant at P-values less than 0·05.
Results
Higher 8-OHdG levels in plasma and lower hOGG1 and anti-oxidant enzyme mRNA expression in leucocytes from group I SLE patients
Compared with the healthy controls (Table 1), group I SLE patients not only had higher levels of plasma 8-OHdG (P < 0·001) but also lower expression of hOGG1 (P < 0·001) and the genes encoding anti-oxidant enzymes, including Cu/ZnSOD (P = 0·002), MnSOD (P = 0·051), catalase (P = 0·003), GPx-4 (P = 0·001), GR (P < 0·001) and GS (P < 0·001) in leucocytes. However, no obvious difference was found in GPx-1 (P = 0·268), the cellular form of GPx.
Table 1.
Plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG), relative leucocyte mRNA expression of human 8-oxoguanine DNA glycosylase 1 (hOGG1), anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in healthy controls and group I systemic lupus erythematosus (SLE) patients.
| Healthy controls | Group I SLE patients | P-value | |
|---|---|---|---|
| Case number | n = 45 | n = 85 | |
| Leucocyte mtDNA copy number | 0·193 ± 0·065 | 0·214 ± 0·113 | 0·169 |
| Degree of plasma oxidative DNA damage | |||
| Plasma 8-OHdG (ng/ml, mean ± s.d.) | 0·157 ± 0·038 | 0·225 ± 0·082 | <0·001 |
| Case number | n = 26 | n = 44 | |
| Relative leucocyte mRNA expression† | |||
| DNA repair enzyme | |||
| hOGG1 (mean ± s.d.) | 1·000 ± 0·512 | 0·337 ± 0·180 | <0·001 |
| Anti-oxidant enzymes | |||
| Cu/ZnSOD (mean ± s.d.) | 1·000 ± 0·541 | 0·622 ± 0·425 | 0·002 |
| MnSOD (mean ± s.d.) | 1·000 ± 0·632 | 0·720 ± 0·570 | 0·051 |
| Catalase (mean ± s.d.) | 1·000 ± 0·647 | 0·557 ± 0·417 | 0·003 |
| GPx-1 (mean ± s.d.) | 1·000 ± 0·491 | 0·850 ± 0·567 | 0·268 |
| GPx-4 (mean ± s.d.) | 1·000 ± 0·569 | 0·553 ± 0·334 | 0·001 |
| GR (mean ± s.d.) | 1·000 ± 0·412 | 0·493 ± 0·295 | <0·001 |
| GS (mean ± s.d.) | 1·000 ± 0·536 | 0·524 ± 0·346 | <0·001 |
| Mitochondrial biogenesis-related proteins | |||
| ND1 (mean ± s.d.) | 1·000 ± 0·542 | 0·396 ± 0·283 | <0·001 |
| ATPase 6 (mean ± s.d.) | 1·000 ± 0·594 | 0·401 ± 0·280 | <0·001 |
| ND6 (mean ± s.d.) | 1·000 ± 0·643 | 0·410 ± 0·355 | <0·001 |
| Tfam (mean ± s.d.) | 1·000 ± 0·509 | 0·693 ± 0·376 | 0·005 |
| NRF-1 (mean ± s.d.) | 1·000 ± 0·381 | 0·460 ± 0·231 | <0·001 |
| PDHA1 (mean ± s.d.) | 1·000 ± 0·478 | 0·522 ± 0·316 | <0·001 |
| PDK-1 (mean ± s.d.) | 1·000 ± 0·645 | 0·725 ± 0·500 | 0·050 |
| HIF-1α (mean ± s.d.) | 1·000 ± 0·459 | 0·958 ± 0·509 | 0·732 |
| Glycolytic enzymes | |||
| HK-II (mean ± s.d.) | 1·000 ± 0·717 | 0·295 ± 0·201 | <0·001 |
| GPI (mean ± s.d.) | 1·000 ± 0·455 | 0·569 ± 0·363 | <0·001 |
| PFK (mean ± s.d.) | 1·000 ± 0·859 | 0·505 ± 0·333 | 0·010 |
| GAPDH (mean ± s.d.) | 1·000 ± 0·907 | 0·593 ± 0·727 | 0·033 |
| LDHa (mean ± s.d.) | 1·000 ± 0·477 | 1·059 ± 0·660 | 0·710 |
Adjusting the mean leucocyte mRNA expression level of specific target gene in healthy controls as 1·000. Cu/ZnSOD = copper/zinc superoxide dismutase; 8-OHdG = 8-hydroxy-2′-deoxyguanosine; MnSOD = manganese superoxide dismutase; GPx = glutathione peroxidase; GR = glutathione reductase; GS = glutathione synthetase; ND1 = mtDNA-encoded ND1 polypeptide; ATPase 6 = mtDNA-encoded ATPase 6 polypeptide; Tfam = mitochondrial transcription factor A; NRF-1 = ; nuclear respiratory factor 1; PDHA1 = pyruvate dehydrogenase E1 component alpha subunit; PDK-1 = pyruvate dehydrogenase kinase isoenzyme 1; HIF-1α = hypoxia inducible factor-1α; HK-II = hexokinase-II; GPI = glucose 6-phosphate isomerase; PFK = phosphofructokinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; LDHa = lactate dehydrogenase A; s.d. = standard deviation.
Lower mRNA expression of genes encoding mitochondrial biogenesis-related proteins and glycolytic enzymes in leucocytes from group I SLE patients
We found that the transcripts of mitochondrial biogenesis-related proteins, including the mtDNA-encoded polypeptides ND1 (P < 0·001), ATPase 6 (P < 0·001) and ND6 (P < 0·001), Tfam (P = 0·005), NRF-1 (P < 0·001), pyruvate dehydrogenase E1 component alpha subunit (PDHA1) (P < 0·001) and PDK1 (P = 0·050) and the glycolytic enzymes HK-II (P < 0·001), GPI (P < 0·001), PFK (P = 0·010) and GAPDH (P = 0·033) were all decreased in leucocytes of group I SLE patients compared with healthy controls (Table 1). However, no obvious differences in HIF-1α (P = 0·732) or LDHa (P = 0·710), the anaerobic glycolysis and lactate fermentation markers, respectively, were noted.
Positive correlations between plasma 8-OHdG levels and leucocyte mRNA expression of genes encoding hOGG1, anti-oxidant enzymes, mitochondrial biogenesis-related protein and glycolytic enzymes in group I SLE patients
In Table 2, positive correlations between the level of plasma 8-OHdG and leucocyte mRNA expression of various genes from group I SLE patients are shown. These genes include hOGG1 [P = 0·014, Pearson's correlation coefficient (Pcc) = 0·367, Fig. 1a] and genes encoding anti-oxidant enzymes, such as Cu/ZnSOD (P = 0·032, Pcc = 0·324, Fig. 1b), MnSOD (P = 0·003, Pcc = 0·442, Fig. 1c), catalase (P = 0·015, Pcc = 0·365, Fig. 1d), GPx-1 (P = 0·041, Pcc = 0·309, Fig. 1e), GR (P = 0·003, Pcc = 0·441, Fig. 1f) and GS (P = 0·005, Pcc = 0·413, Fig. 1g), mitochondrial biogenesis-related proteins, including NRF-1 (P = 0·026, Pcc = 0·335), PDHA1 (P = 0·018, Pcc = 0·354), PDK1 (P = 0·005, Pcc = 0·412) and HIF-1α (P = 0·037, Pcc = 0·315) and glycolytic enzymes, including HK-II (P = 0·008, Pcc = 0·398), GPI (P = 0·011, Pcc = 0·378), PFK (P = 0·001, Pcc = 0·478), GAPDH (P = 0·001, Pcc = 0·477) and LDHa (P < 0·001, Pcc = 0·564). However, such positive correlations were not observed for GPx4 (P = 0·573, Pcc = −0·087, Fig. 1h), the mitochondrial form of GPx, Tfam (P = 0·095, Pcc = 0·225), the regulator for mtDNA replication and transcription or ND1 (P = 0·801, Pcc = −0·039), ATPase6 (P = 0·775, Pcc = −0·044) or ND6 (P = 0·538, Pcc = −0·095), which are mtDNA-encoded polypeptides.
Table 2.
Associations between the plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG) and leucocyte mRNA expression of human 8-oxoguanine DNA glycosylase 1 (hOGG1), anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes among 44 of group I systemic lupus erythematosus (SLE) patients.
| Leucocyte mRNA expression level | Pearson's correlation coefficient | P-value |
|---|---|---|
| DNA repair enzyme | Correlation to plasma 8-OHdG | |
| hOGG1 | 0·367 | 0·014 |
| Anti-oxidant enzymes | Correlation to plasma 8-OHdG | |
| MnSOD | 0·442 | 0·003 |
| Cu/ZnSOD | 0·324 | 0·032 |
| Catalase | 0·365 | 0·015 |
| GPx-1 | 0·309 | 0·041 |
| GPx-4 | −0·087 | 0·573 |
| GR | 0·441 | 0·003 |
| GS | 0·413 | 0·005 |
| Mitochondrial biogenesis-related proteins | Correlation to plasma 8-OHdG | |
| ND1 | −0·039 | 0·801 |
| ATPase 6 | −0·044 | 0·775 |
| ND6 | −0·095 | 0·538 |
| Tfam | 0·255 | 0·095 |
| NRF-1 | 0·335 | 0·026 |
| PDHA1 | 0·354 | 0·018 |
| PDK1 | 0·412 | 0·005 |
| HIF-1α | 0·315 | 0·037 |
| Glycolytic enzymes | Correlation to plasma 8-OHdG | |
| HK-II | 0·398 | 0·008 |
| GPI | 0·378 | 0·011 |
| PFK | 0·478 | 0·001 |
| GAPDH | 0·477 | 0·001 |
| LDHa | 0·564 | <0·001 |
Cu/ZnSOD = copper/zinc superoxide dismutase; MnSOD = manganese superoxide dismutase; GPx = glutathione peroxidase; GR = glutathione reductase; GS = glutathione synthetase; ND1 = mtDNA-encoded ND1 polypeptide; ATPase 6 = mtDNA-encoded ATPase 6 polypeptide; Tfam = mitochondrial transcription factor A; NRF-1 = ; nuclear respiratory factor 1; PDHA1 = pyruvate dehydrogenase E1 component alpha subunit; PDK-1 = pyruvate dehydrogenase kinase isoenzyme 1; HIF-1α = hypoxia inducible factor-1α; HK-II = hexokinase-II; GPI = glucose 6-phosphate isomerase; PFK = phosphofructokinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; LDHa = lactate dehydrogenase A.
Figure 1.
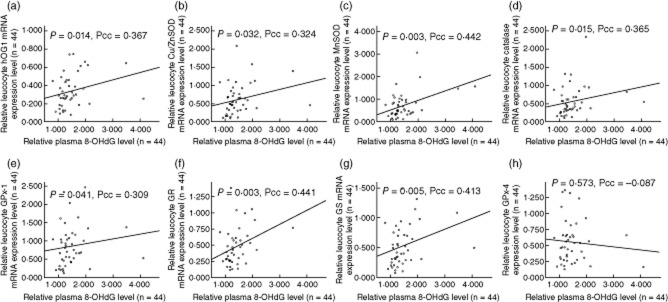
Illustrations demonstrated positive correlations between the relative plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG) level (adjust the mean of health controls as to 1·000, x-axis) and relative leucocyte mRNA expression level of various anti-oxidant genes (adjust the mean of healthy controls to 1·000, y-axis) in group I systemic lupus erythematosus (SLE) patients, including: (a) human 8-oxoguanine DNA glycosylase 1 (hOGG1) [P = 0·014, Pearson's correlation coefficient (Pcc) = 0·367]; (b) copper/zinc superoxide dismutase (Cu/ZnSOD) (P = 0·032, Pcc = 0·324); (c) manganese superoxide dismutase (MnSOD) (P = 0·003, Pcc = 0·442); (d) catalase (P = 0·015, Pcc = 0·365); (e) glutathione peroxidase-1 (GPx-1) (P = 0·041, Pcc = 0·309); (f) glutathione reductase (GR) (P = 0·003, Pcc = 0·441); and (g) glutathione synthetase (GS) (P = 0·005, Pcc = 0·413). However, such a positive correlation was not observed in GPx-4. (h) GPx-4 (P = 0·573, Pcc = −0·087).
Positive correlations between the mRNA expression of HIF-1α and downstream genes responsible for glycolysis in leucocytes of group I SLE patients
In group I SLE leucocytes, positive correlations between the mRNA expression of HIF-1α and downstream genes involved in glycolysis including HK-II (P < 0·001, Pcc = 0·573), GPI (P < 0·001, Pcc = 0·744), PFK (P < 0·001, Pcc = 0·716), GAPDH (P = 0·024, Pcc = 0·339) and LDHa (P < 0·001, Pcc = 0·819) were observed (Table 3).
Table 3.
Associations between the leucocyte mRNA expression level of hypoxia inducible factor-1α (HIF-1α) and glycolytic enzymes in 44 group I systemic lupus erythematosus (SLE) patients.
| Leucocyte mRNA expression level | Pearson's correlation coefficient | P-value |
|---|---|---|
| Glycolytic enzymes | Correlation to HIF-1α | |
| HK-II | 0·573 | <0·001 |
| GPI | 0·744 | <0·001 |
| PFK | 0·716 | <0·001 |
| GAPDH | 0·339 | 0·024 |
| LDHa | 0·819 | <0·001 |
HK-II = hexokinase-II; GPI = glucose 6-phosphate isomerase; PFK = phosphofructokinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; LDHa = lactate dehydrogenase A.
Positive correlation of mtDNA-encoded genes and negative correlation of plasma 8-OHdG levels and SLE disease activity index (SLEDAI) with the mtDNA copy number in SLE patients harbouring D310 heteroplasmy
As shown in the Supporting information, Table S1 [8], 59 group I SLE patients harboured heteroplasmic D310 distribution in leucocyte mtDNA. This particular heteroplasmy denotes oxidative mtDNA damage [7]. Further examination of these patients revealed that their mtDNA copy number correlated positively with the expression of the mtDNA-encoded ND1 (P = 0·035, Pcc = 0·429), ATPase 6 (P = 0·018, Pcc = 0·452) and ND6 (P = 0·031, Pcc = 0·413) polypeptides in leucocytes. However, mtDNA copy number was correlated negatively with the SLEDAI (P = 0·040, Pcc = −0·268) and plasma 8-OHdG levels (P = 0·020, Pcc = −0·302) (Table 4).
Table 4.
Associations between leucocyte mtDNA copy number and mRNA expression level of mtDNA-encoded polypeptides, systemic lupus erythematosus disease activity index (SLEDAI) and plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG) in 59 group I systemic lupus erythematosus (SLE) patients harbouring heteroplasmic D310 distribution of leucocyte mtDNA.
| Association with leucocyte mtDNA copy number | Pearson's correlation coefficient | P-value |
|---|---|---|
| mtDNA-encoded ND1 polypeptide (n = 27) | 0·429 | 0·035 |
| mtDNA-encoded ATPase 6 polypeptide (n = 27) | 0·452 | 0·018 |
| mtDNA-encoded ND6 polypeptides (n = 27) | 0·413 | 0·032 |
| SLEDAI (n = 59) | −0·268 | 0·040 |
| Plasma 8-OHdG (n = 59) | −0·302 | 0·020 |
ND1 = mtDNA-encoded ND1 polypeptide; ATPase 6 = mtDNA-encoded ATPase 6 polypeptide.
Higher plasma 8-OHdG levels and lower leucocyte mtDNA copy number in group II SLE patients with severe disease status
The plasma 8-OHdG levels of the group II SLE patients at days 1 (0·641 ± 0·207 ng/ml) and 14 (0·830 ± 0·281 ng/ml) were both much higher than healthy controls (0·157 ± 0·038, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test) or the group I SLE patients (0·225 ± 0·082, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test) (Supporting information, Table S1 and Table 5, footnote2). Furthermore, the mtDNA copy numbers of group II SLE patients at days 1 (0·077 ± 0·021) and 14 (0·080 ± 0·009) were much lower than those of healthy controls (0·193 ± 0·065, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test) or the group I SLE patients (0·214 ± 0·113, P = 0·002 for day 1 and P = 0·002 for day 14, Mann–Whitney U-test) (Supporting information, Table S1 and Table 5, footnote1) [8].
Table 5.
Distributions and alterations of plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG), relative leucocyte mRNA expression of human 8-oxoguanine DNA glycosylase 1 (hOGG1), anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in group II systemic lupus erythematosus (SLE) patients who underwent rituximab treatment.
| Case number (n = 4)/variables | Distributions |
Alterations |
P-value‡ |
||
|---|---|---|---|---|---|
| Day 1† | Day 14† | Increase (ratio† > 1·000) | Decrease (ratio† ≤ 1·000) | P-value‡ | |
| Leucocyte mtDNA copy number1 | 0·077 ± 0·021 | 0·080 ± 0·009 | 2 | 2 | 0·715 |
| Degree of plasma oxidative DNA damage2 | |||||
| Plasma 8-OHdG (ng/ml, mean ± s.d.) | 0·641 ± 0·207 | 0·830 ± 0·281 | 3 | 1 | 0·465 |
| Relative leucocyte mRNA expression† | |||||
| DNA repair enzyme | |||||
| hOGG1 (mean ± s.d.) | 1·000 ± 0·000 | 0·638 ± 0·483 | 1 | 3 | 0·144 |
| Anti-oxidant enzymes | |||||
| Cu/ZnSOD (mean ± s.d.) | 1·000 ± 0·000 | 1·099 ± 0·886 | 1 | 3 | 0·465 |
| MnSOD (mean ± s.d.) | 1·000 ± 0·000 | 0·659 ± 0·276 | 0 | 4 | 0·068 |
| Catalase (mean ± s.d.) | 1·000 ± 0·000 | 0·900 ± 0·433 | 1 | 3 | 0·273 |
| GPx-1 (mean ± s.d.) | 1·000 ± 0·000 | 1·284 ± 0·832 | 2 | 2 | 1·000 |
| GPx-4 (mean ± s.d.) | 1·000 ± 0·000 | 1·255 ± 0·866 | 2 | 2 | 0·715 |
| GR (mean ± s.d.) | 1·000 ± 0·000 | 1·441 ± 0·433 | 3 | 1 | 0·144 |
| GS (mean ± s.d.) | 1·000 ± 0·000 | 1·545 ± 1·040 | 3 | 1 | 0·465 |
| Mitochondrial biogenesis-related proteins | |||||
| ND1 (mean ± s.d.) | 1·000 ± 0·000 | 0·257 ± 0·150 | 0 | 4 | 0·068 |
| ATPase 6 (mean ± s.d.) | 1·000 ± 0·000 | 0·225 ± 0·116 | 0 | 4 | 0·068 |
| ND6 (mean ± s.d.) | 1·000 ± 0·000 | 0·174 ± 0·135 | 0 | 4 | 0·068 |
| Tfam (mean ± s.d.) | 1·000 ± 0·000 | 0·544 ± 0·172 | 0 | 4 | 0·068 |
| NRF-1 (mean ± s.d.) | 1·000 ± 0·000 | 1·477 ± 0·434 | 4 | 0 | 0·068 |
| PDHA1 (mean ± s.d.) | 1·000 ± 0·000 | 0·271 ± 0·100 | 0 | 4 | 0·068 |
| PDK-1 (mean ± s.d.) | 1·000 ± 0·000 | 1·057 ± 0·469 | 2 | 2 | 1·000 |
| HIF-1α (mean ± s.d.) | 1·000 ± 0·000 | 1·088 ± 0·454 | 2 | 2 | 1·000 |
| Glycolytic enzymes | |||||
| HK-II (mean ± s.d.) | 1·000 ± 0·000 | 0·496 ± 0·531 | 1 | 3 | 0·144 |
| GPI (mean ± s.d.) | 1·000 ± 0·000 | 0·784 ± 0·242 | 1 | 3 | 0·144 |
| PFK (mean ± s.d.) | 1·000 ± 0·000 | 0·435 ± 0·278 | 0 | 4 | 0·068 |
| GAPDH (mean ± s.d.) | 1·000 ± 0·000 | 1·223 ± 0·587 | 3 | 1 | 0·465 |
| LDHa (mean ± s.d.) | 1·000 ± 0·000 | 0·795 ± 0·295 | 1 | 3 | 0·144 |
Adjusting the analysed variables on day 1 as 1·000.
Wilcoxon's signed ranks test; ratio = day 14/day 1.
The mtDNA copy numbers of the four group II SLE patients with severe clinical presentations on days 1 and 14 were much lower than that of health controls (n = 45, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test) and group I SLE patients (n = 85, P = 0·002 for day 1 and P = 0·002 for day 14, Mann–Whitney U-test), respectively.
The plasma 8-OHdG of the four group II SLE patients with severe clinical presentations on days 1 and 14 were much higher than that of health controls (n = 45, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test) and group I SLE patients (n = 85, P = 0·001 for day 1 and P = 0·001 for day 14, Mann–Whitney U-test), respectively. Cu/ZnSOD = copper/zinc superoxide dismutase; 8-OHdG = 8-hydroxy-2′-deoxyguanosine; MnSOD = manganese superoxide dismutase; GPx = glutathione peroxidase; GR = glutathione reductase; GS = glutathione synthetase; ND1 = mtDNA-encoded ND1 polypeptide; ATPase 6 = mtDNA-encoded ATPase 6 polypeptide; Tfam = mitochondrial transcription factor A; NRF-1 = ; nuclear respiratory factor 1; PDHA1 = pyruvate dehydrogenase E1 component alpha subunit; PDK-1 = pyruvate dehydrogenase kinase isoenzyme 1; HIF-1α = hypoxia inducible factor-1α; HK-II = hexokinase-II; GPI = glucose 6-phosphate isomerase; PFK = phosphofructokinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; LDHa = lactate dehydrogenase A; s.d. = standard deviation.
Decreases in relative leucocyte mRNA expression of genes encoding mitochondrial biogenesis-related proteins, mitochondrial-specific anti-oxidant-related proteins and rate-limiting glycolytic enzymes in group II SLE patients after rituximab therapy
As shown in Table 5, the leucocyte mRNA expression of specific mitochondrial biogenesis-related proteins, including mtDNA-encoded ND1 (P = 0·068), ND6 (P = 0·068) and ATPase 6 (P = 0·068), PDHA1 (P = 0·068), which converts pyruvate to acetyl-CoA, and Tfam (P = 0·068), which directly regulates the replication and transcription of mtDNA, were decreased simultaneously after rituximab administration. Additionally, decreases in the transcripts of MnSOD, the mitochondria-specific SOD, and PFK, which catalyzes the rate-limiting step in glycolysis, were observed.
Higher leucocyte mRNA levels of genes encoding several anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes were correlated with a better clinical response after rituximab therapy in group II SLE patients with severe disease
Among the four patients in group II, two showed good clinical responses after rituximab administration. As shown in Table 6, these two patients presented with higher leucocyte mRNA levels of the genes encoding anti-oxidant enzymes, including MnSOD, Cu/ZnSOD, catalase, GPx-1, GPx-4 and GR, higher ratios of Tfam and PDHA1, which regulate mitochondrial biogenesis, and higher ratios of some glycolytic enzymes, including GPI, GAPDH (a housekeeping gene) and LDHa (a lactate fermentation marker).
Table 6.
Alterations of plasma 8-hydroxy-2′-deoxyguanosine (8-OHdG), relative leucocyte mtDNA copy number, relative leucocyte mRNA expression of human 8-oxoguanine DNA glycosylase 1 (hOGG1), anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes and their differences based on the clinical treatment response in group II systemic lupus erythematosus (SLE) patients.
| Variables | Alteration of the analysed variables (day 14 versus day 1) |
P-value† | |||||
|---|---|---|---|---|---|---|---|
| Good response (n = 2) |
Poor response (n = 2) |
||||||
| Ratio‡ | > Median | ≤ Median | Ratio‡ | > Median | ≤ Median | ||
| Leucocyte mtDNA copy number‡ | 1·189 ± 0·279 | 1 | 1 | 0·984 ± 0·144 | 1 | 1 | 0·667 |
| Degree of plasma oxidative DNA damage | 1·776 ± 0·813 | 1 | 1 | 1·032 ± 0·407 | 1 | 1 | 0·667 |
| Relative leucocyte mRNA expression‡ | |||||||
| DNA repair enzyme | |||||||
| hOGG1 (mean ± s.d.) | 0·833 ± 0·726 | 1 | 1 | 0·443 ± 0·147 | 1 | 1 | 1·000 |
| Anti-oxidant enzymes | |||||||
| Cu/ZnSOD (mean ± s.d.) | 1·607 ± 1·123 | 2 | 0 | 0·590 ± 0·242 | 0 | 2 | 0·333 |
| MnSOD (mean ± s.d.) | 0·848 ± 0·133 | 2 | 0 | 0·470 ± 0·260 | 0 | 2 | 0·333 |
| Catalase (mean ± s.d.) | 1·206 ± 0·427 | 2 | 0 | 0·593 ± 0·065 | 0 | 2 | 0·333 |
| GPx-1 (mean ± s.d.) | 1·878 ± 0·804 | 2 | 0 | 0·689 ± 0·130 | 0 | 2 | 0·333 |
| GPx-4 (mean ± s.d.) | 1·793 ± 1·016 | 2 | 0 | 0·718 ± 0·242 | 0 | 2 | 0·333 |
| GR (mean ± s.d.) | 1·721 ± 0·023 | 2 | 0 | 1·162 ± 0·498 | 0 | 2 | 0·333 |
| GS (mean ± s.d.) | 2·099 ± 1·400 | 1 | 1 | 0·991 ± 0·237 | 1 | 1 | 0·667 |
| Mitochondrial biogenesis-related proteins | |||||||
| ND1 (mean ± s.d.) | 0·271 ± 0·177 | 1 | 1 | 0·244 ± 0·188 | 1 | 1 | 0·667 |
| ATPase 6 (mean ± s.d.) | 0·223 ± 0·101 | 1 | 1 | 0·227 ± 0·174 | 1 | 1 | 1·000 |
| ND6 (mean ± s.d.) | 0·171 ± 0·192 | 1 | 1 | 0·178 ± 0·135 | 1 | 1 | 1·000 |
| Tfam (mean ± s.d.) | 0·691 ± 0·051 | 2 | 0 | 0·398 ± 0·011 | 0 | 2 | 0·333 |
| NRF-1 (mean ± s.d.) | 1·692 ± 0·599 | 1 | 1 | 1·263 ± 0·146 | 1 | 1 | 0·667 |
| PDHA1 (mean ± s.d.) | 0·328 ± 0·130 | 2 | 0 | 0·213 ± 0·006 | 0 | 2 | 0·333 |
| PDK-1 (mean ± s.d.) | 1·422 ± 0·298 | 2 | 0 | 0·693 ± 0·202 | 0 | 2 | 0·333 |
| HIF-1α (mean ± s.d.) | 1·151 ± 0·392 | 1 | 1 | 1·025 ± 0·669 | 1 | 1 | 1·000 |
| Glycolytic enzymes | |||||||
| HK-II (mean ± s.d.) | 0·758 ± 0·753 | 1 | 1 | 0·233 ± 0·052 | 1 | 1 | 0·667 |
| GPI (mean ± s.d.) | 0·971 ± 0·177 | 2 | 0 | 0·596 ± 0·065 | 0 | 2 | 0·333 |
| PFK (mean ± s.d.) | 0·553 ± 0·403 | 1 | 1 | 0·316 ± 0·113 | 1 | 1 | 0·667 |
| GAPDH (mean ± s.d.) | 1·604 ± 0·562 | 2 | 0 | 0·841 ± 0·370 | 0 | 2 | 0·333 |
| LDHa (mean ± s.d.) | 1·003 ± 0·090 | 2 | 0 | 0·585 ± 0·278 | 0 | 2 | 0·333 |
Mann–Whitney U-test.
Adjusting the analysed variables on day 1 before the administration of rituximab as 1·000. Cu/ZnSOD = copper/zinc superoxide dismutase; MnSOD = manganese superoxide dismutase; GPx = glutathione peroxidase; GR = glutathione reductase; GS = glutathione synthetase; ND1 = mtDNA-encoded ND1 polypeptide; ATPase 6 = mtDNA-encoded ATPase 6 polypeptide; Tfam = mitochondrial transcription factor A; NRF-1 = ; nuclear respiratory factor 1; PDHA1 = pyruvate dehydrogenase E1 component alpha subunit; PDK-1 = pyruvate dehydrogenase kinase isoenzyme 1; HIF-1α = hypoxia inducible factor-1α; HK-II = hexokinase-II; GPI = glucose 6-phosphate isomerase; PFK = phosphofructokinase; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; LDHa = lactate dehydrogenase A; s.d. = standard deviation.
Discussion
Figure 2 shows that increased oxidative damage in SLE patients is reflected by decreased mRNA expression of hOGG1 and the genes encoding anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in leucocytes. Increased oxidative damage and decreased anti-oxidant capacity in SLE patients have been reported by several research groups [2–5]. Similarly, we demonstrated an elevated level of plasma 8-OHdG combined with decreased leucocyte mRNA expression of hOGG1 and the genes encoding anti-oxidant enzymes in SLE patients compared with healthy controls (Table 1). In addition, a positive correlation was observed between the level of plasma 8-OHdG and leucocyte mRNA expression of hOGG1 and the genes encoding anti-oxidant enzymes in SLE patients (Table 2, Fig. 1a–g). Thus, SLE patients may suffer a higher level of oxidative stress and exhibit a compensatory mechanism that increases the expression of the DNA repair enzyme hOGG1 and genes encoding anti-oxidant enzymes to counter offending stimuli. However, these compensations are suboptimal. Interestingly, the expression level of the GPx isoenzyme, as shown in Tables 1 and 2, may vary depending on the subcellular location. For example, GPx-1, which is primarily a cytoplasmic enzyme, exhibited activity that was correlated with the plasma level of 8-OHdG, as did that of the healthy controls; however, GPx-4, which is mainly a mitochondrial enzyme, exhibited a lower level of expression than that in the healthy controls but did not change in response to the increase in plasma 8-OHdG. GPx4 participates mainly in the oxidation of GSH to reduce H2O2 into H2O, thus guaranteeing ATP production in the mitochondria [23]. Improving GPx-4 expression in mitochondria may be a good therapeutic strategy for SLE patients. Generally, the efficiency of GPx is supported by GSH, which is a tripeptide of cysteine, glutamate and glycine, and the plasma concentration of cysteine is the rate-limiting factor for its synthesis. A recent study showed that the administration of cysteine significantly reduced oxidative stress and improved the symptoms of lupus disease [24]. Thus, an ample supply of cysteine may increase the mitochondrial anti-oxidant capacity in SLE patients by increasing GSH and facilitating its interaction with GPx-4.
Figure 2.
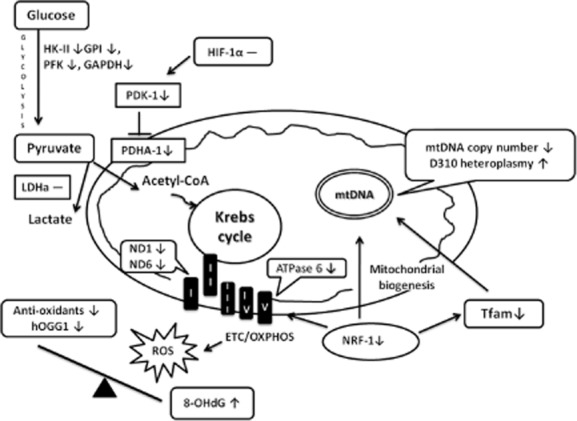
Summary of the relationship between oxidative damage, expression of anti-oxidant enzymes, mitochondrial biogenesis-related proteins and glycolytic enzymes in patients with systemic lupus erythematosus (SLE). ↑ = significant increase; ↓ = significant decrease; – = no significant difference; I∼V = mitochondrial respiratory enzyme complexes I∼V; ETC = electron transport chain; OXPHOS = oxidative phosphorylation; anti-oxidant enzymes listed in this figure include manganese superoxide dismutase (MnSOD), copper/zinc superoxide dismutase (Cu/ZnSOD), catalase, glutathione peroxidase-4 (GPx-4), glutathione reductase (GR) and glutathione synthetase (GS).
Much research has aimed to unravel mitochondrial dysfunction in SLE patients because impaired mitochondrial function is assumed to generate endogenous ROS and thus enhance oxidative stress [25,26]. Although we did not directly measure the activity of the enzymes that participate in mitochondrial respiration, we showed that SLE leucocytes exhibited decreased levels of transcripts of genes encoding mitochondrial biogenesis-related proteins that are implicated in electron transport (ND1 and ND6), oxidative phosphorylation (ATPase 6), the replication and transcription of mtDNA (Tfam and NRF-1) and the transformation of pyruvate to acetyl-CoA (PDHA1) for the Krebs cycle (Table 1). Conversely, we did not observe a compensatory increase in leucocyte mRNA expression of the gene encoding Tfam, which regulates mtDNA replication and transcription, or the mtDNA-encoded ND1, ATPase 6, and ND6, in parallel with the increase in the plasma 8-OHdG level in SLE patients (Table 2). These results suggest that leucocyte mitochondria function is impaired in SLE patients.
However, the causal relationship between oxidative stress and impaired leucocyte mitochondrial function in SLE has remained obscure. Compared with nDNA, mtDNA is far more susceptible to oxidative damage, especially in the D310 region. Oxidative mtDNA damage is suspected when the nucleotide sequences in the D310 region exhibit a heteroplasmic distribution [7]. As we reported previously, when SLE leucocyte mtDNA harbours a heteroplasmic D310 distribution, the mtDNA copy number decreases noticeably along with the severity of SLEDAI [8]. In the present investigation, we demonstrated that a decrease in the leucocyte mtDNA copy number in SLE patients was related closely to an increase in plasma 8-OHdG and decreases in mtDNA-encoded polypeptides (Table 4). Similarly, the four group II SLE patients who had more severe clinical symptoms and higher SLEDAI exhibited much higher plasma 8-OHdG levels and lower leucocyte mtDNA copy numbers (Supporting information, Table S1 and Table 5, footnotes1,2). These results suggest that ROS may cause mitochondrial dysfunction via oxidative mtDNA damage, resulting in an mtDNA copy number decrease, and that this dysfunction may further amplify oxidative stress in SLE.
Because mitochondria are the main powerhouses of human cells, it will be intriguing to determine whether accelerated glucose metabolism or increased lactate fermentation exists to counterbalance the effect of defective mitochondria in SLE leucocytes. Altered glucose metabolism, anaerobic glycolysis and lactate fermentation, which sustain energy production, have been widely recognized to occur in human tissues under hypoxic conditions or in neoplasm through LDHa and HIF-1α [14]. Nevertheless, our data showed that the leucocyte mRNA expression of genes encoding glycolytic enzymes in group I SLE patients was lower than that of the healthy controls (Table 2). The mRNA expression levels of HIF-1α and LDHa in leucocytes from group I SLE patients and the healthy controls were similar. Furthermore, leucocyte mRNA expression levels of genes encoding glycolytic enzymes correlated positively with the levels of plasma 8-OHdG and HIF-1α (Tables 2 and 3). It is reasonable to speculate that oxidative damage causes mitochondrial dysfunction, leading to a hypoxic environment that enhances the expression of HIF-1α to regulate glycolytic enzymes. However, such regulation is not sufficient to cause a metabolic switch, as observed in human cancers [14]. In line with a previous report [27], this assertion could be supported by our finding that excluded the possibility of a glucose metabolic switch to compensate for the decreased mitochondrial function in SLE patients. Overall, both mitochondrial biogenesis and lactate fermentation for energy production are universally decreased in leucocytes from SLE patients. All these findings prompted us to conclude that the overall energy production through glucose metabolism is decreased in SLE patients.
In addition to glucose, glutamine is an energy source that may participate in energy production to compensate for mitochondrial dysfunction in SLE patients. Several novel roles for L-glutamine in keeping cells alive have been identified. These include a supply of α-ketoglutarate to the Krebs cycle, an essential amino donor, and a molecule involved in de-novo GSH biosynthesis to overcome oxidative stress [28]. Although we did not examine the role of glutamine and its relationship to glucose metabolism in this study, increased glutamine and altered glucose metabolism were observed in the brain, an organ with high energy demand, of lupus-prone mice [29]. With the enzymatic deamination by glutaminase in mitochondria, glutamine can be converted to α-ketoglutarate to maintain mitochondrial function in T lymphocytes [30]. The exact relationship of glutamine to the present findings of altered mitochondrial function in SLE patients deserves further in-depth study.
Because of the potentially serious toxicity of conventional immunosuppressive agents, such as glucocorticoid, azathioprine, mycophenolate mofetil and cyclophosphamide, rituximab has been regarded as an alternative therapeutic agent in advanced SLE patients [15]. Although the mechanism by which rituximab depletes CD20-positive B cells remains speculative, antibody-dependent cell-mediated cytotoxicity, complement-dependent cytotoxicity and direct signalling-related apoptosis are plausible pathways [16]. We are interested in the alterations in enzymes in leucocytes that are important in maintaining the redox status, mitochondrial biogenesis and glycolysis in SLE patients undergoing rituximab therapy. Table 5 shows that four SLE patients with severe disease (group II) exhibited a decrease in the mRNA expression of the genes encoding MnSOD, which is the mitochondria-specific SOD, ND1, ND6, ATPase6, Tfam and PDHA1, which are mitochondrial biogenesis-related proteins, and PFK, which catalyzes the rate limiting step in glycolysis. These results suggest that energy deprivation leads to cell death through a mitochondrial-dependent pathway in lupus disease. Thus, signalling-related apoptosis or activation-induced cell death may operate through the mitochondria to cause autoimmune damage in SLE patients. As reported previously, mitochondria-mediated apoptosis by rituximab is important in the pathogenesis of non-Hodgkin lymphoma [31,32]. Among the four patients in group II, two showed good response to rituximab treatment. A further analysis of these two patients showed that they had higher levels of the genes encoding MnSOD, Cu/ZnSOD, catalase, GPx-1, Gpx-4 and GR, which are necessary for cellular anti-oxidant capacity, Tfam and PDHA1, which are necessary for mitochondrial biogenesis, and GAPDH, which is necessary for housekeeping functions of glycolysis. These findings imply that an increase in the anti-oxidant capacity and improvements in mitochondrial biogenesis and glycolysis are crucial for energy production and the recovery of leucocyte function in patients undergoing rituximab treatment. Although there was no obvious reduction in the plasma 8-OHdG level, the increased anti-oxidant capacity might have been sufficient to respond to the oxidative challenge. Because our preliminary results focused on only four patients, future studies are warranted.
In conclusion, an increase in oxidative DNA damage, decreases in the DNA repair and anti-oxidant enzyme capacity and mitochondrial and glucose metabolism dysfunction may play important roles in SLE pathogenesis. Rituximab, which improves anti-oxidant capacity, glycolysis and mitochondrial function for increased energy production, may be an effective remedy for SLE patients with severe disease.
Acknowledgments
This study was supported by grants (NSC-97-2320-B-010-013-MY3, NSC100-2320-B-010-024-MY3 and NSC102-2314-B-195-020) from the National Science Council, Taiwan, and (DOH101-HO-1006) from Taipei Hospital, Department of Health, Executive Yuan, Taiwan.
Disclosures
The authors have no financial conflicts of interest.
Author contributions
All authors were involved in drafting the paper or critically reviewing and revising it for important intellectual content, and all authors approved the final version to be published. H.-T. L. had full access to all the data obtained from Taipei Veterans General Hospital during the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. For study conception and design: Hui-Ting Lee, Chen-Sung Lin, Chyou-Shen Lee, Chang-Youh Tsai, and Yau-Huei Wei; for data acquisition: H.-T. L. and C.-Y. T.; for analysis and interpretation of data: H.-T. L. and C.-S. L.; and for manuscript preparation: H.-T. L., C.-S. L., C.-S. L., C.-Y. T. and Y.-H. W.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web-site:
Table S1. Demographic data, immunological profile, degree of oxidative damage and leucocyte mtDNA characteristics of the 45 health controls and 89 systemic lupus erythematosus (SLE) patients.
Table S2. Summary of the analysed target genes and their primer information.
References
- 1.Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity. 2010;43:1–6. doi: 10.3109/08916930903374741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansour RB, Lassoued S, Gargouri B, et al. Increased levels of autoantibodies against catalase and superoxide dismutase associated with oxidative stress in patients with rheumatoid arthritis and systemic lupus erythematosus. Scand J Rheumatol. 2008;37:103–108. doi: 10.1080/03009740701772465. [DOI] [PubMed] [Google Scholar]
- 3.Morgan PE, Sturgess AD, Davies MJ. Evidence for chronically elevated serum protein oxidation in systemic lupus erythematosus patients. Free Radic Res. 2009;43:117–127. doi: 10.1080/10715760802623896. [DOI] [PubMed] [Google Scholar]
- 4.Shah D, Wanchu A, Bhatnagar A. Interaction between oxidative stress and chemokines: possible pathogenic role in systemic lupus erythematosus and rheumatoid arthritis. Immunobiology. 2011;216:1010–1017. doi: 10.1016/j.imbio.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Pierangeli SS, Papalardo E, Ansari GA, Khan MF. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthritis Rheum. 2010;62:2064–2072. doi: 10.1002/art.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci USA. 2003;100:1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HT, Lin CS, Chen WS, Liao HT, Tsai CY, Wei YH. Leukocyte mitochondrial DNA alteration in systemic lupus erythematosus and its relevance to the susceptibility to lupus nephritis. Int J Mol Sci. 2012;13:8853–8868. doi: 10.3390/ijms13078853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 10.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 14.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 15.Galarza C, Valencia D, Tobon GJ, et al. Should rituximab be considered as the first-choice treatment for severe autoimmune rheumatic diseases? Clin Rev Allergy Immunol. 2008;34:124–128. doi: 10.1007/s12016-007-8028-z. [DOI] [PubMed] [Google Scholar]
- 16.Weiner GJ. Rituximab: mechanism of action. Semin Hematol. 2010;47:115–123. doi: 10.1053/j.seminhematol.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 18.Petri M, Orbai AM, Alarcón GS, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64:2677–2686. doi: 10.1002/art.34473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CS, Kuo CL, Cheng WL, Huang CS, Lee CF, Wei YH. Alteration of the copy number of mitochondrial DNA in leukocytes of patients with hyperlipidemia. Ann NY Acad Sci. 2005;1042:70–75. doi: 10.1196/annals.1338.008. [DOI] [PubMed] [Google Scholar]
- 20.Chomczynski P, Mackey K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide-and proteoglycan-rich sources. Biotechniques. 1995;19:942–945. [PubMed] [Google Scholar]
- 21.Sato T, Takeda H, Otake S, et al. Increased plasma levels of 8-hydroxydeoxyguanosine are associated with development of colorectal tumors. J Clin Biochem Nutr. 2010;47:59–63. doi: 10.3164/jcbn.10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin CS, Lee HT, Lee SY, et al. High mitochondrial DNA copy number and bioenergetic function are associated with tumor invasion of esophageal squamous cell carcinoma cell lines. Int J Mol Sci. 2012;13:11228–11246. doi: 10.3390/ijms130911228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang H, Van Remmen H, Frohlich V, Lechleiter J, Richardson A, Ran Q. Gpx4 protects mitochondrial ATP generation against oxidative damage. Biochem Biophys Res Commun. 2007;356:893–898. doi: 10.1016/j.bbrc.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 24.Lai ZW, Hanczko R, Bonilla E, et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012;64:2937–2946. doi: 10.1002/art.34502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez D, Perl A. Metabolic control of T cell activation and death in SLE. Autoimmun Rev. 2009;8:184–189. doi: 10.1016/j.autrev.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HM, Sugino H, Aoki C, Nishimoto N. Underexpression of mitochondrial-DNA encoded ATP synthesis-related genes and DNA repair genes in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:R63. doi: 10.1186/ar3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu T, Xie C, Han J, et al. Metabolic disturbances associated with systemic lupus erythematosus. PLOS One. 2012;7:e37210. doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexander JJ, Zwingmann C, Quigg R. MRL/lpr mice have alterations in brain metabolism as shown with [1H-13C] NMR spectroscopy. Neurochem Int. 2005;47:143–151. doi: 10.1016/j.neuint.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Maciver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eeva J, Nuutinen U, Ropponen A, et al. The involvement of mitochondria and the caspase-9 activation pathway in rituximab-induced apoptosis in FL cells. Apoptosis. 2009;14:687–698. doi: 10.1007/s10495-009-0337-7. [DOI] [PubMed] [Google Scholar]
- 32.Alas S, Ng CP, Bonavida B. Rituximab modifies the cisplatin-mitochondrial signaling pathway, resulting in apoptosis in cisplatin-resistant non-Hodgkin's lymphoma. Clin Cancer Res. 2002;8:836–845. [PubMed] [Google Scholar]
- 33.Stolz C, Hess G, Hahnel PS, et al. Targeting Bcl-2 family proteins modulates the sensitivity of B-cell lymphoma to rituximab-induced apoptosis. Blood. 2008;112:3312–3321. doi: 10.1182/blood-2007-11-124487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Demographic data, immunological profile, degree of oxidative damage and leucocyte mtDNA characteristics of the 45 health controls and 89 systemic lupus erythematosus (SLE) patients.
Table S2. Summary of the analysed target genes and their primer information.


