Abstract
GM (γ marker) allotypes, genetic variants of immunoglobulin γ chains, have been reported to be associated strongly with susceptibility to lung cancer, but the mechanism(s) underlying this association is not known. One mechanism could involve their contribution to humoral immunity to lung tumour-associated antigens. In this study, we aimed to determine whether particular GM and KM (κ marker) allotypes were associated with antibody responsiveness to XAGE-1b, a highly immunogenic lung tumour-associated cancer-testis antigen. Sera from 89 patients with non-small cell lung cancer (NSCLC) were allotyped for eight GM and two KM determinants and characterized for antibodies to a synthetic XAGE-1b protein. The distribution of various GM phenotypes was significantly different between XAGE-1b antibody-positive and-negative patients (P = 0·023), as well as in the subgroup of XAGE-1b antigen-positive advanced NSCLC (P = 0·007). None of the patients with the GM 1,17 21 phenotype was positive for the XAGE-1b antibody. In patients with antigen-positive advanced disease, the prevalence of GM 1,2,17 21 was significantly higher in the antibody-positive group than in those who lacked the XAGE-1b antibody (P = 0·026). This phenotype also interacted with a particular KM phenotype: subjects with GM 1,2,17 21 and KM 3,3 phenotypes were almost four times (odds ratio = 3·8) as likely to be positive for the XAGE-1b antibody as the subjects who lacked these phenotypes. This is the first report presenting evidence for the involvement of immunoglobulin allotypes in immunity to a cancer-testis antigen, which has important implications for XAGE-1b-based immunotherapeutic interventions in lung adenocarcinoma.
Keywords: cancer-testis antigen, GM/KM allotypes, humoral immunity, non-small cell lung cancer, XAGE-1b (GAGED2a)
Introduction
Genetic variants of immunoglobulin G (IgG) heavy chains are called GM allotypes. They are encoded by three very closely linked genes – immunoglobulin heavy chain G1 (IGHG1), IGHG2 and IGHG3 – on chromosome 14q32. They are expressed on the constant regions of γ1, γ2 and γ3 chains. There are striking qualitative and quantitative differences in the distribution of GM allotypes among different racial groups. In addition, there is almost complete linkage disequilibrium between particular GM determinants within a race, and every major racial group is characterized by a distinct array of GM haplotypes [1],[2]. Using hypothesis-driven candidate gene approaches, several studies have identified particular GM genes/genotypes as risk factors for many malignant diseases [2]–[7]. In lung cancer, a highly significant association was found between the GM 1,2 13,15,16,21 phenotype and susceptibility to this malignancy in a Japanese population [8]. The mechanism(s) underlying this association is not known.
One mechanism underlying the reported GM gene–lung cancer association could involve the contribution of GM determinants to humoral immunity to lung tumour-associated antigens, as GM genes are known to influence immunity to several self and non-self antigens, including tumour-associated antigens mucin 1 and human epidermal growth factor receptor 2 [9]–[14]. In this investigation, we aimed to determine whether GM allotypes are associated with antibody responsiveness to XAGE-1b, a highly immunogenic lung tumour-associated antigen that belongs to the cancer-testis antigen gene families [15]–[17]. A recent comprehensive analysis of human gene expression has identified the Ig κ constant (IGKC) gene as a strong prognostic marker in human solid tumours, including lung cancer [18]. Identification of tumour-infiltrating plasma cells as the source of IGKC expression in this study strongly suggests a role for humoral immunity in lung cancer and provides a compelling rationale for investigating the role of KM alleles, genetic variants of IGKC, in humoral immunity to lung tumour-associated antigens.
There is increasing evidence that genes do not act in isolation, and that epistasis – modification of the action of a gene by one or more other genes – plays a significant role in human diseases. Genes expressed on the Ig heavy and light chains are probably some of the most likely candidates for gene–gene interactions in the human genome. Therefore, the aim of the present investigation was to determine whether GM and KM allotypes – individually or in particular epistatic combinations – contribute to antibody responsiveness to XAGE-1b in patients with non-small cell lung cancer (NSCLC).
Materials and methods
Blood samples
The study population is described in detail elsewhere [17]. The Institutional Review Boards of the respective institutions approved the study protocol. Blood samples from 89 Japanese patients with NSCLC were included in this investigation. Of these, 80 patients were diagnosed histologically examining available tumour specimens and nine were diagnosed cytologically using tumour cells in pleural effusion, sputum or bronchoalveolar fluid (BALF) because tumour tissue was not available.
Anti-XAGE-1b antibody determinations
These antibodies were measured by a previously described enzyme-linked immunosorbent assay (ELISA) [16],[17]. Briefly, synthetic XAGE-1b (GAGED2a) protein (1 μg/ml) in coating buffer was adsorbed onto a 96-well ELISA plate (Nunc, Roskilde, Denmark) and incubated overnight at 4°C. Plates were washed with phosphate-buffered saline (PBS) and blocked with 5% fetal calf serum (FCS)/PBS (200 μl/well) for 1 h at 37°C. After washing, 100 μl of serially diluted serum was added to each well and incubated for 2 h at 4°C; horseradish peroxidase (HRP)-conjugated goat anti-human IgG (MBL) was then added to the wells, and the plates were incubated for 1 h at 37°C. After washing and development, absorbance [optical density (OD)] was read at 490 nm. Sera with OD values exceeding 1·0 at a dilution of 1:300 were considered positive for the XAGE-1b antibody, while those with OD values less than 0·2 were considered negative for this antibody. Patients who showed OD values between 0·2 and 1·0 were excluded. Of the 89 NSCLC patients, 29 were positive for the XAGE-1b antibody and 60 were negative.
Immunohistochemistry
Tumour specimens from 80 patients were also examined by immunohistochemistry. Surgically resected tissues were fixed with buffered formalin and embedded in paraffin. Five-micrometre sections were deparaffinized with xylene and ethanol. Antigen retrieval and inactivation of endogenous peroxidase have been described previously [16]. After incubation with 0·1% Tween 20/5% FCS/PBS for 1 h, the USO 9–13 monoclonal antibody (mAb) was placed at a concentration of 2 μg/ml and incubated for 1 h at room temperature. Immunofluorescence staining was performed as described above. For intracellular localization, rhodamine-conjugated wheat germ agglutinin (WGA) (Vector Laboratories, Burlingame, CA, USA) and 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) were used. The stained cells were visualized under a digital high-definition microscopic system (model BZ-9000 for the magnification of ×40; Keyence, Osaka, Japan).
Of 80 patients, 46 were XAGE-1b antigen-positive and 34 were antigen-negative. Detailed clinical information was not available for three antigen-positive patients. Of the remaining 43 antigen-positive patients, 26 were antibody-positive and 17 were antibody-negative.
GM and KM allotyping
Serum samples were typed for G1M (1/a, 2/x, 3/f, 17/z), G2M (23/n), G3M (5/b1, 13/b3, 21/g) and KM 1 and 3 allotypes by a standard haemagglutination-inhibition method [19]. In brief, a mixture containing human blood group O rhesus-positive (ORh+) erythrocytes coated with anti-Rh antibodies of known GM/KM allotypes, the test sera and monospecific anti-allotype antibodies were incubated in a microtitre plate. Test sera containing IgG of the particular allotype inhibited haemagglutination by the anti-allotype antibody, whereas negative sera did not. The notation for GM allotypes follows the international system for human gene nomenclature, in which haplotypes and phenotypes are written by grouping together the markers that belong to each IgG subclass, by the numerical order of the marker and of the subclass; markers belonging to different subclasses are separated by a space, while allotypes within a subclass are separated by commas.
Three alleles – KM 1, KM 1,2 and KM 3 – segregate at the KM locus on chromosome 2p12. More than 98% of people positive for KM 1 are also positive for KM 2. The KM 1 allele, without KM 2, is extremely rare. Here, and in most other investigations, positivity for KM 1 includes both KM 1 and KM 1,2 alleles.
Statistical analysis
The significance of the association between GM and KM phenotypes and the prevalence of antibodies to XAGE-1b in NSCLC patients was analysed using Fisher's exact test and Pearson's χ2 test. Subjects with very unusual GM phenotypes and those whose frequency was <4% were combined as ‘other’, in order not to have a test with too many degrees of freedom. Associations between the prevalence of antibodies and GM phenotypes and patient survival were assessed using a Cox regression model. Statistical significance was defined as P < 0·05. All reported P-values are two-sided.
Results
Table 1 presents the distribution of GM and KM phenotypes in XAGE-1b antibody-positive and-negative patients with lung adenocarcinoma. The majority of the subjects possessed typical Japanese GM phenotypes, which can be explained by postulating the segregation of four haplotypes present in this population: GM 1,17 21, GM 1,2,17 21, GM 1,17 13 and GM 1,3 23 5,13. The frequency of KM phenotypes observed was also typical of this population.
Table 1.
Distribution of GM* and KM phenotypes in the XAGE-1b antibody-positive and-negative patients with lung adenocarcinoma (n = 89).
| Phenotype | XAGE-1b antibody |
||||
|---|---|---|---|---|---|
| Positive (n = 29) | (%) | Negative (n = 60) | (%) | P-value | |
| GM 1,17 21 | 0 | 0 | 10 | 16·7 | 0·027 |
| GM 1,2,17 21 | 15 | 51·7 | 19 | 31·7 | 0·06 |
| GM 1,17 13,21 | 0 | 0 | 4 | 6·7 | 0·30 |
| GM 1,2,17 13,21 | 1 | 3·4 | 8 | 13·3 | 0·26 |
| GM 1,2,3,17 23 5,13,21 | 2 | 6·9 | 5 | 8·3 | 1·0 |
| Other GM | 11 | 37·9 | 14 | 23·3 | 0·21 |
| KM 1 | 3 | 10·3 | 6 | 10·0 | 0·61 |
| KM 1,3 | 8 | 27·6 | 26 | 43·3 | 0·15 |
| KM 3 | 11 | 62·1 | 28 | 46·7 | 0·17 |
Fisher's exact test (6 × 2), P = 0·023.
A global Fisher's exact test, considering all GM phenotypes, shows that there is a significant difference in the distribution of various phenotypes between the XAGE-1b antibody-positive and-negative groups of patients (P = 0·023). Further dissection of this association elucidates that the discrepancy in the distribution of GM 1,17 21 and GM 1,2,17 21 phenotypes contributed most to the total variation. None of the subjects with the GM 1,17 21 phenotype was positive for the XAGE-1b antibody (P = 0·027). The frequency of the GM 1,2,17 21 phenotype in the antibody-positive group was higher than in the antibody-negative group, but it did not reach statistical significance (52 versus 32%; Pearson's χ2 = 3·3; P = 0·06). However, in subjects who were also homozygous for the KM 3 allele, this GM phenotype contributed significantly to the antibody responsiveness: subjects with GM 1,2,17 21 and KM 3,3 phenotypes were almost four times [odds ratio (OR) = 3·8] as likely to be positive for the XAGE-1b antibody as the subjects who lacked both these phenotypes (Table 2). No other significant interactions were found. Also, none of the KM phenotypes alone was associated with anti-XAGE-1b antibody responsiveness.
Table 2.
Distribution of combined GM 1,2,17 21 and KM 3,3 phenotypes in antibody-positive and-negative patients in relation to existence of XAGE-1b antibody (n = 89).
| Phenotype | XAGE-1b antibody |
|||
|---|---|---|---|---|
| Positive n = 29 (%) | Negative n = 60 (%) | OR (95% CI) | P-value | |
| GM 1,2,17 21(+)/KM 3,3 (+) | 11 (37·9) | 9 (15·0) | 3·8 (1·1–13·1) | 0·04 |
| GM 1,2,17 21(+)/KM 3,3 (−) | 4 (13·8) | 10 (16·7) | 1·3 (0·3–5·3) | 1·0 |
| GM 1,2,17 21(−)/KM 3,3 (+) | 7 (24·1) | 19 (31·7) | 1·2 (0·3–3·9) | 1·0 |
| GM 1,2,17 21(−)/KM 3,3 (−) | 7 (24·1) | 22 (36·7) | 1·0 | |
CI: confidence interval; OR: odds ratio.
Subsequent analyses were restricted to patients with XAGE-1b antigen-positive advanced (IIIB/IV) lung cancer. The clinical and demographic characteristics of these patients are presented in Table 3. The prevalence of anti-XAGE-1b antibodies was higher in patients with less advanced disease (P = 0·030). Other characteristics, except age, were not significantly different in the two groups of patients. A global Fisher's exact test, considering all GM phenotypes, shows that there is a significant difference in the distribution of various phenotypes between the XAGE-1b antibody-positive and-negative groups of patients with XAGE-1b antigen-positive advanced lung cancer (P = 0·007). There were only three patients with the GM 1,17 21 phenotype in this group, and all were negative for the XAGE-1b (P = 0·055, Table 4). The prevalence of GM 1,2,17 21 was significantly higher in the antibody-positive group than in those who lacked the XAGE-1b antibody (54 versus 18%; P = 0·026). The only allotype different between the responder and non-responder phenotypes is the γ1 determinant GM 2, prompting us to analyse the interindividual variation in antibody responsiveness in relation to the GM 2 status of the subjects. No significant associations were found in the whole group (P = 0·34) as well as in the XAGE-1b antigen-positive group (P = 0·18). Thus, it appears that the influence of GM 2 on antibody responsiveness is manifested only when it is in a complex with γ1 determinants GM 1 and 17 and the γ3 determinant GM 21. Although a significant interactive effect of GM 1,2,17 21 with KM 3 homozygosity was observed (OR = 10; P = 0·04), this association should be viewed with caution, as the number of subjects in some categories was very small, resulting in a wide confidence interval (data not shown).
Table 3.
Characteristics of the patients with XAGE-1b antigen-positive advanced lung cancer (n = 43).
| Characteristic | XAGE-1b antibody |
||
|---|---|---|---|
| Positive (n = 26) | Negative (n = 17) | P-value | |
| Sex, no. (%) | |||
| Male/female | 13/13 (50·0) | 13/4 (76·5) | 0·11 |
| Age, years | |||
| Mean | 76·5 ± 7·6 | 69·8 ± 10·1 | 0·018 |
| Smoking status, no. (%) | |||
| Never smoked | 10 (38·5) | 5 (29·4) | 0·75 |
| ECOG performance status score, no. (%) | |||
| 0–1 | 21 (80·8) | 11 (64·7) | 0·30 |
| Clinical stage, no. (%) | |||
| IIIB/IV | 10/16 (38·5) | 1/16 (5·9) | 0·030 |
| Brain metastasis, no. (%) | |||
| positive/negative | 9/17 (34·6) | 5/12 (29·4) | 0·75 |
| EGFR mutation, no. (%) | |||
| Positive/negative | 13/13 (50·0) | 4/13 (23·5) | 0·12 |
ECOG: Eastern Cooperative Oncology Group; EGFR: epidermal growth factor receptor.
Table 4.
Distribution of GM* and KM phenotypes in the XAGE-1b antibody-positive and-negative patients with XAGE-1b antigen-positive advanced lung adenocarcinoma (n = 43).
| Phenotype | XAGE-1b antibody |
||||
|---|---|---|---|---|---|
| Positive (n = 26) | (%) | Negative (n = 17) | (%) | P-value | |
| GM 1,17 21 | 0 | 0 | 3 | 17·6 | 0·055 |
| GM 1,2,17 21 | 14 | 53·8 | 3 | 17·6 | 0·026 |
| GM 1,17 13,21 | 0 | 0 | 1 | 5·9 | 0·40 |
| GM 1,2,17 13,21 | 1 | 3·8 | 4 | 23·5 | 0·07 |
| GM 1,2,3,17 23 5,13,21 | 1 | 3·8 | 1 | 5·9 | 1·0 |
| Other GM | 10 | 38·5 | 5 | 29·4 | 0·75 |
| KM 1 | 3 | 11·5 | 2 | 11·7 | 1·0 |
| KM 1,3 | 8 | 30·8 | 7 | 41·2 | 0·53 |
| KM 3 | 15 | 57·7 | 8 | 47·1 | 0·55 |
Fisher's exact test (6 × 2), P = 0·007.
Of the 43 patients with antigen-positive tumours, 17 were negative for the XAGE-1b antibody; however, only one of these belonged to the clinical stage IIIB, the rest being clinical stage IV. Therefore, survival curves were plotted with the stage IV patients as well as with the combined group of patients with clinical stages IIIB and IV. As shown in Fig. 1, the anti-XAGE-1b antibody positivity was associated significantly with enhanced overall survival in both groups of patients, the antibody-positive subjects surviving more than twice as long as those who lacked this antibody (stage IIIB/IV: 33 versus 14 months, P = 0·007; stage IV: 33 versus 13 months, P = 0·039). Although not statistically significant (due possibly to the small sample size), stage IIIB/IV subjects with the GM 1,2,17 21 phenotype, which was associated with a higher prevalence of anti-XAGE-1b antibodies, survived longer than those expressing the GM 1,17 21 phenotype, which was associated with the lack of antibodies to XAGE-1b (31 versus 15 months, P = 0·29, Fig. 2).
Figure 1.
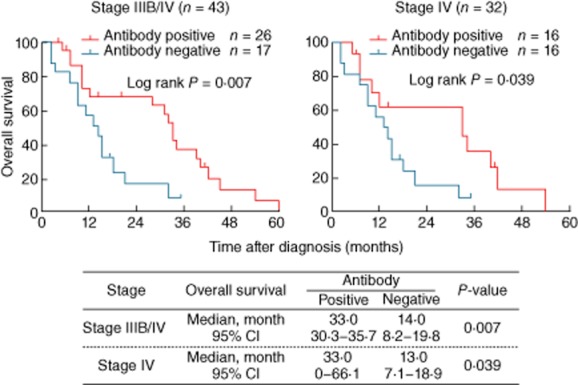
Kaplan–Meier survival plots of XAGE-1b antigen-positive advanced lung adenocarcinoma patients as a function of XAGE-1b antibody.
Figure 2.
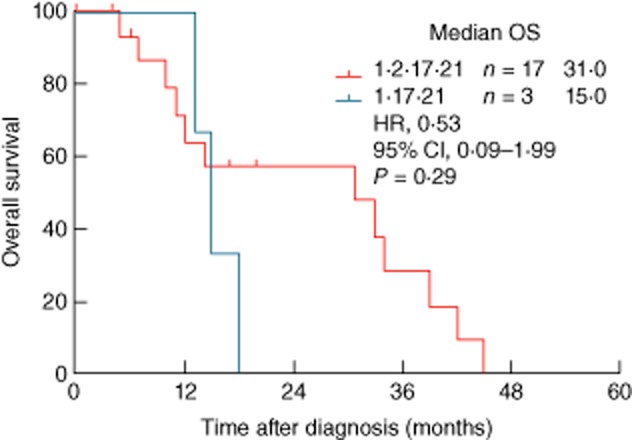
Kaplan–Meier survival plots of XAGE-1b antigen-positive stage IIIB/IV lung adenocarcinoma patients as a function of GM 1,2,17 21 and GM 1,17 21 phenotypes.
Discussion
The results presented here show that the Ig GM 1,2,17 21 phenotype is associated with the presence of naturally occurring antibodies to the cancer-testis antigen XAGE-1b, while the GM 1,17 21 phenotype is associated with the lack of such antibodies. One mechanism underlying this association could involve GM allotypes being part of the recognition structures for the immunogenic epitopes of the XAGE-1b protein. Perhaps membrane-bound IgG (mIgG) molecules with GM 1,2,17 21 allotypes are more efficient in the uptake, processing and subsequent presentation of XAGE-1b epitopes to the collaborating T cells, resulting in strong humoral immunity, whereas the mIgG molecules with the GM 1,17 21 phenotype form a lower affinity receptor for the critical epitopes of this protein. Additionally – and contrary to the prevalent belief in immunology – these constant-region determinants could directly influence anti-XAGE-1b antibody specificity by causing conformational changes in the antigen-binding site in the Ig variable region. There is convincing evidence that the Ig constant region can influence antibody affinity and specificity [20]. Thus, constant regions expressing different GM allotypes, even when combined with identical variable region sequences, can generate new antibody molecules with new functions. They could also influence the expression of idiotypes involved in XAGE-1b immunity. The contribution of both variable and constant regions in the formation of idiotypic determinants has been clearly documented for the T15 system in mice, and such isotype-restricted idiotypes have been postulated to be involved in the regulation of class-specific antibody responses [21].
We also found that subjects with GM 1,2,17 21 and KM 3,3 phenotypes were significantly more likely to generate anti-XAGE-1b antibodies than subjects who lacked both these phenotypes. The simultaneous involvement of both GM and KM alleles on antibody responsiveness would suggest that the association of γ and κ chains in IgG antibodies directed against XAGE-1b might not be random. Only γ and κ chains carrying specific GM and KM allotypes might form a paratope with the necessary quaternary structure for an effective recognition of the XAGE-1b epitopes. Non-random pairing of heavy and light chains has been reported in experimental animals [22],[23].
As mentioned previously, the XAGE-1b antigen is highly immunogenic and, therefore, an excellent vaccine candidate for active immunotherapy. In XAGE-1b antibody-positive patients, specific T cell responses were also frequently observed [17]. If the results presented here are confirmed in an independent study, they could aid in identifying subjects (GM 1,2,17 21) who are more likely to benefit from XAGE-1b-based vaccines. For those with the non-responder (GM 1,17 21) phenotype, XAGE-1b could be fused with appropriate adjuvants, such as heat shock proteins or flagellin, to overcome the allotypic restriction in immune responsiveness. It is relevant to note that antibody responses to certain heat shock proteins as well as to flagellin are also influenced by GM genotypes [24],[25], making it conceivable to formulate a fusion XAGE-1b–heat shock protein/flagellin vaccine that could potentially generate high antibody responses in the majority of the population. Identification of the natural responders/non-responders to XAGE-1b would also be helpful in the proper evaluation of any future vaccine efficacy trials.
Associations observed in this report can also be explained by postulating as-yet unidentified immune response genes for XAGE-1b whose alleles might be in linkage disequilibrium with those of GM and KM loci.
Although the results reported here are statistically significant, they could also be the result of chance fluctuations, as the P-values for the associations were not adjusted for multiple testing. Such adjustment is controversial [26], and in the present investigation would be overly punitive, as the multiple tests performed are not independent due to significant linkage disequilibrium in the GM gene complex. This is the first study of its kind, and needs to be replicated and extended by independent investigations.
It is relevant to point out that the highly significant GM phenotype–lung cancer association that was reported more than three decades ago [8] has not been confirmed or refuted by modern genome-wide association studies (GWAS) of this malignancy [27]. One contributing factor for this omission might be the absence of GM gene probes in most genotyping platforms. GWAS are assumed to be able to detect/tag all single nucleotide polymorphisms (SNPs) in the genome whose frequency is at least 5%. This, however, is not true. Most GM alleles are common within a racial group (some with allele frequency >70%), but the IGHG gene segments harbouring them are highly homologous and apparently not amenable to the high-throughput genotyping technology used in GWAS. Because these genes were not typed in the HapMap or the 1000 Genomes projects, they cannot be imputed or tagged (through linkage disequilibrium) by any SNPs that are included in the genotyping platforms. Therefore, a candidate gene approach would be necessary to confirm/refute the findings reported here.
It is hoped that these results, coupled with those identifying the IGKC gene as a strong prognostic marker in human solid tumours [18], would inspire large-scale studies to determine conclusively the contribution of Ig GM and KM alleles in humoral immunity to XAGE-1b. It would also be of interest to investigate the role of these determinants in immunity to NY-ESO-1, a prototype cancer-testis antigen. Results from such investigations would be extremely valuable in devising novel immunotherapeutic interventions in patients with lung adenocarcinoma.
Acknowledgments
The study was supported by Project for Development of Innovative Research on Cancer Therapeutics of Ministry of Education, Culture Sports Science and Technology of Japan.
Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Grubb R. Advances in human immunoglobulin allotypes. Exp Clin Immunogenet. 1995;12:191–197. doi: 10.1159/000424871. [DOI] [PubMed] [Google Scholar]
- 2.Pandey JP, Li Z. The forgotten tale of immunoglobulin allotypes in cancer risk and treatment. Exp Hematol Oncol. 2013;2:6. doi: 10.1186/2162-3619-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey JP, Kistner-Griffin E, Iwasaki M, et al. Genetic markers of immunoglobulin G and susceptibility to breast cancer. Hum Immunol. 2012;73:1155–1158. doi: 10.1016/j.humimm.2012.07.340. [DOI] [PubMed] [Google Scholar]
- 4.Morell A, Scherz R, Käser H, Skvaril F. Evidence for an association between uncommon Gm phenotypes and neuroblastoma. Lancet. 1977;1:23–24. doi: 10.1016/s0140-6736(77)91657-9. [DOI] [PubMed] [Google Scholar]
- 5.Ilić V, Milosević-Jovcić N, Marković D, et al. A biased Gm haplotype and Gm paraprotein allotype in multiple myeloma suggests a role for the Gm system in myeloma development. Int J Immunogenet. 2007;34:119–125. doi: 10.1111/j.1744-313X.2007.00673.x. [DOI] [PubMed] [Google Scholar]
- 6.Pandey JP, Johnson AH, Fudenberg HH, et al. HLA antigens and immunoglobulin allotypes in patients with malignant melanoma. Hum Immunol. 1981;2:185–190. doi: 10.1016/0198-8859(81)90010-0. [DOI] [PubMed] [Google Scholar]
- 7.Pandey JP, Ebbesen P, Bülow S, et al. IgG heavy-chain (Gm) allotypes in familial polyposis coli. Am J Hum Genet. 1986;39:133–136. [PMC free article] [PubMed] [Google Scholar]
- 8.Nakao Y, Matsumoto H, Miyazaki T, et al. Immunoglobulin G heavy-chain allotypes as possible genetic markers for human cancer. J Natl Cancer Inst. 1981;67:47–50. [PubMed] [Google Scholar]
- 9.Pandey JP, Namboodiri AM, Kistner-Griffin E. IgG and FcγR genotypes and humoral immunity to mucin 1 in prostate cancer. Hum Immunol. 2013;74:1030–1033. doi: 10.1016/j.humimm.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Pandey JP, Nietert PJ, Mensdorff-Pouilly S, et al. Immunoglobulin allotypes influence antibody responses to mucin 1 in patients with gastric cancer. Cancer Res. 2008;68:4442–4446. doi: 10.1158/0008-5472.CAN-07-5607. [DOI] [PubMed] [Google Scholar]
- 11.Pandey JP, Nietert PJ, Klaamas K, et al. A genetic variant of immunoglobulin γ2 is strongly associated with natural immunity to mucin 1 in patients with breast cancer. Cancer Immunol Immunother. 2009;58:2025–2029. doi: 10.1007/s00262-009-0709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey JP, Namboodiri AM, Kurtenkov O, et al. Genetic regulation of antibody responses to human epidermal growth factor receptor 2 (HER-2) in breast cancer. Hum Immunol. 2010;71:1124–1127. doi: 10.1016/j.humimm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey JP, Namboodiri AM, Kistner-Griffin E, et al. Racially restricted contribution of immunoglobulin Fcγ and Fcγ receptor genotypes to humoral immunity to human epidermal growth factor receptor 2 in breast cancer. Clin Exp Immunol. 2013;171:273–277. doi: 10.1111/cei.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey JP, Shannon BT, Tsang KY, et al. Heterozygosity at Gm loci associated with humoral immunity to osteosarcoma. J Exp Med. 1982;155:1228–1232. doi: 10.1084/jem.155.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali Eldib AM, Ono T, Shimono M, et al. Immunoscreening of a cDNA library from a lung cancer cell line using autologous patient serum: identification of XAGE-1b as a dominant antigen and its immunogenicity in lung adenocarcinoma. Int J Cancer. 2004;108:558–563. doi: 10.1002/ijc.11587. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa K, Noguchi Y, Uenaka A, et al. XAGE-1 expression in non-small cell lung cancer and antibody response in patients. Clin Cancer Res. 2005;11:5496–5503. doi: 10.1158/1078-0432.CCR-05-0216. [DOI] [PubMed] [Google Scholar]
- 17.Ohue Y, Eikawa S, Okazaki N, et al. Spontaneous antibody, and CD4 and CD8 T-cell responses against XAGE-1b (GAGED2a) in non-small cell lung cancer patients. Int J Cancer. 2012;131:E649–658. doi: 10.1002/ijc.27359. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18:2695–2704. doi: 10.1158/1078-0432.CCR-11-2210. [DOI] [PubMed] [Google Scholar]
- 19.Schanfield MS, Loghem van E. Human immunoglobulin allotypes. In: Weir DM, editor. Handbook of experimental immunology. Boston, MA: Blackwell; 1986. pp. 94.1–18. [Google Scholar]
- 20.Casadevall A, Pirofski L-A. A new synthesis for antibody-mediated immunity. Nat Immunol. 2012;13:21–28. doi: 10.1038/ni.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morahan G, Berek C, Miller JFAP. An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature. 1983;301:720–722. doi: 10.1038/301720a0. [DOI] [PubMed] [Google Scholar]
- 22.Czerwinski M, Siemaszko D, Siegel DL, et al. Only selected light chains combine with a given heavy chain to confer specificity for a model glycopeptide antigen. J Immunol. 1998;160:4406–4417. [PubMed] [Google Scholar]
- 23.Primi D, Drapier AM, Cazenave PA. Highly preferential VH–VL pairing in normal B cells results in antigen-independent selection of the available repertoire. J Immunol. 1987;138:1607–1612. [PubMed] [Google Scholar]
- 24.Pandey JP, Prohászka Z, Veres A, et al. Epistatic effects of genes encoding immunoglobulin GM allotypes and interleukin-6 on the production of autoantibodies to 60-and 65-kDa heat-shock proteins. Genes Immun. 2004;5:68–71. doi: 10.1038/sj.gene.6364033. [DOI] [PubMed] [Google Scholar]
- 25.Pandey JP. Comment on ‘Flagellin as an adjuvant: cellular mechanisms and potential’. J Immunol. 2011;186:1299. doi: 10.4049/jimmunol.1090134. [DOI] [PubMed] [Google Scholar]
- 26.Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiraishi K, Kunitoh H, Daigo Y, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–903. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]


