Abstract
Perillyl derivatives are increasingly important due to their flavouring and antimicrobial properties as well as their potential as anticancer agents. These terpenoid species, which are present in limited amounts in plants, may be obtained via bioconversion of selected monoterpene hydrocarbons. In this study, seventeen yeast strains were screened for their ability to oxidize the exocyclic methyl group in the p-menthene moiety of limonene into perillic acid. Of the yeast tested, the highest efficiency was observed for Yarrowia lipolytica ATCC 18942. The conversion of R (+)-limonene by Y. lipolytica was evaluated by varying the pH (3 to 8) and the temperature (25 to 30 °C) in a reaction medium containing 0.5% v/v limonene and 10 g/L of stationary phase cells (dry weight). The best results, corresponding to 564 mg/L of perillic acid, were obtained in buffered medium at pH 7.1 that was incubated at 25 °C for 48 h. The stepwise addition of limonene increased the perillic acid concentration by over 50%, reaching 855 mg/L, whereas the addition of glucose or surfactant to the reaction medium did not improve the bioconversion process. The use of Y. lipolytica showed promise for ease of further downstream processing, as perillic acid was the sole oxidised product of the bioconversion reaction. Moreover, bioprocesses using safe and easy to cultivate yeast cells have been favoured in industry.
Keywords: Yarrowia lipolytica, bioconversion, limonene, perillic acid
Introduction
The introduction of new and innovative medicines based on small-molecule candidates remains a focus of the pharmaceutical industry (Buckland et al., 2000). Within this context, biotransformation processes offer a plethora of possibilities for obtaining regioselective and stereoselective molecules of industrial interest (Krings and Berger, 1998; Liese et al., 2006; Pollard and Woodley, 2007). The biotechnological generation of these compounds via bioconversion with isolated enzymes, microorganisms or plant cells has been increasingly used as a tool to explore new and cost-effective methods of biocatalysis using selected metabolic pathways from cells. Moreover, “green processes”, in which gentle conditions are employed, are mostly based on biotransformation reactions (Matsuyama et al., 2002; Persidis, 1997).
The microbial transformation of terpenes has been widely studied for the production of an array of molecules. Within this context, the oxidative pathways leading to regioselective products are dependent on the metabolic characteristics of the selected cell systems and the external reaction conditions (Van der Werf et al., 1997). Examples include transformation by bacteria, fungi and yeasts of compounds such as linalool (Madyastha et al., 1977), cineole and α-pinene (Trudgill, 1990) and limonene (Trytek and Fiedurek, 2005) to their respective epoxides, alcohols and diols, aldehydes, ketones and acids (Duetz et al., 2003; Tan and Day, 1998).
Because of the abundant availability of isomeric R-(+)-limonene from industrial citrus residues, the microbial bioconversion of this compound has been extensively explored to establish cost-effective methods for the production of aromatics (e.g., α-terpineol) (Bicas et al., 2009; Cadwallader et al., 1989; Tan and Day, 1998) and potentially pharmacologically active derivatives, such as perillyl alcohol and perillic acid (Cadwallader et al., 1989; Mirata et al., 2009; Speelmans et al., 1998), which have been studied as antimicrobial and anticancer drugs (Da Fonseca et al., 2011; Yeruva et al., 2007).
The selective oxidation of the R-(+)-limonene exocyclic methyl group produces various perillyl derivatives, a transformation that has been accomplished using bacteria (e.g., the Pseudomonas species Bacillus stearothermophylus and Mycobacterium sp.) (Duetz et al., 2003; Mirata et al., 2009; Van Beilen et al., 2005), fungi (e.g., Aspergillus species) (Duetz et al., 2003) and yeasts (e.g., Yarrowia lipolytica and Arxula adeninivorans) (Van Rensburg et al., 1997). The use of safe and easy to cultivate yeasts, such as the two mentioned above (Fickers et al., 2005; Gellissen et al., 2005), for the production of potentially non-toxic anticancer agents has driven increasing interest in this specific bioconversion route.
The present study screened yeasts for their ability to convert (R)-(+)-limonene to perillic acid. A strain of Yarrowia lipolytica (ATCC 18942) was shown to successfully produce perillic acid as the sole oxidised terpene product from limonene. Several parameters for the bioconversion were investigated to improve the perillic acid yield and to better understand the oxidative enzymatic system involved in the reaction.
Materials and Methods
Materials and equipment
Commercial standards of R-(+)-limonene 96% (art. 62120), S-(−)-limonene 95% (art. 62130) and (+)-perillaldehyde 98% (art. 77301) were purchased from Fluka (Buchs, Switzerland). (−)-Perillyl alcohol 96% (art. 218391) was obtained from Aldrich (Steinheim, Germany). (S)-( −)-Perillic acid 95% (art. 218359) and anisaldehyde were obtained from Aldrich (St. Louis, MO, USA). Thin layer chromatography (TLC) was performed on silica gel 60 F254 plates obtained from Merck (Darmstadt, Germany). Solvents for TLC elution were obtained from Vetec (Rio de Janeiro, Brazil). Yeast nitrogen base without amino acids and ammonium sulphate (YNB) was obtained from Difco 0335-15 (Detroit, MI, USA). Bioprocess flasks were stirred on a rotary shaker (Scientific/Innova 4340, New Brunswick Scientific. Edison, NJ, USA). The pH was measured using a Beckman Φ560 pH meter. The optical density was obtained with a Shimadzu-UV 1601 spectrophotometer (Kyoto, Japan) operating at 600 nm.
Microorganisms
The yeast strains Arxula adeninivorans ATCC (American Type Culture Collection) 76597, Candida famata ATCC 62894, C. glabrata ATCC 2001, C. inconspicua ATCC 16783, C. krusei ATCC 34135, C. lusitaniae ATCC 34449, C. stellata ATCC 10673, C. tropicalis ATCC 13803, C. guilliermondi ATCC 6260, C. parapsilosis ATCC 22019, Kodamaea ohmeri ATCC 46053, Komagataella pastoris ATCC 28485, Pichia angusta ATCC 28485, P. anomala ATCC 16763, Saccharomyces cerevisiae ATCC 9080 and Yarrowia lipolytica ATCC 18942 (strain A) were used in this study and were obtained from the Brazilian National Institute for Quality Control in Health. Y. lipolytica strain B was isolated from a polluted estuary of Guanabara Bay by Dr. Allen Hagler and deposited in the yeast collection of the Microbiology Institute of the Federal University of Rio de Janeiro under no. 50678.
Screening procedure
The microorganisms were cultivated in 250 mL Erlenmeyer flasks containing 50 mL of YMB medium (10 g/L glucose, 3.0 g/L yeast extract, 3.0 g/L malt extract and 5.0 g/L peptone) on a rotary shaker (250 rpm) at 25 °C for 48 h. For the standard biotransformation assay, the grown cells were centrifuged and re-suspended at a final optical density at 600 nm (OD600) of 20 in 25 mL of 50 mM phosphate buffer (pH 7.0) containing 100 mM glucose and 0.5% v/v R-(+)-limonene. The reaction mixture was incubated at 25 °C with shaking at 250 rpm. Aliquots were withdrawn at 24 and 48 h, and cells were removed by centrifugation at 3000 g. The supernatant was assayed for R-(+)-limonene conversion without further processing.
Effect of carbon and nitrogen sources on cell growth and product profile
The selected yeast strains C. parapsilosis and Y. lipolytica ATCC 18942 were grown in growth media containing either glucose, glycerol or ethanol (20 g/L) as the carbon source, either ammonium sulphate or urea (2 g/L) as the nitrogen source and YNB (2 g/L). Cells were incubated at 28 °C with shaking at 250 rpm. Samples were withdrawn at 4, 8, 24 and 48 h and assayed for the conversion of R-(+)-limonene using the standard method described above. In control experiments, cells were grown in YMB medium.
Limonene toxicity to Y. lipolytica cell growth
The selected strain Y. lipolytica ATCC 18942 was grown in the YMB medium supplemented with 0.5 or 0.1% limonene. Cells were also grown in YMB medium in which glucose was substituted by 0.5% limonene. Cell growth was monitored by OD600.
Preliminary assessments of the Y. lipolytica bioconversion pathway
S-(−)-Limonene (0.5% v/v), perillaldehyde (0.1% v/v) and perillyl alcohol (0.1% v/v) were also tested as substrates for oxidation by Y. lipolytica ATCC 18942 with the standard conditions. Bioconversion of R-(+)-limonene, perillaldehyde, perillyl alcohol and perillic acid was also assayed in the absence of oxygen by incubating the reaction mixture in a screw cap flask under a nitrogen atmosphere. Trapping less-oxidised perillyl derivatives produced in the limonene bioconversion reaction was accomplished by adding inhibitors of alcohol oxidase and alcohol dehydrogenase (22): Cu2+ (5 mM CuSO4•5H2O), Zn2+ (2 mM ZnSO4•7H2O), Ni2+ (2 mM NiSO4•6H2O), Ag1+ (5 μM AgNO3), Hg2+ (5 μM HgCl2), Cu2+ (5 mM CuSO4•5H2O), Zn2+ (2 mM ZnSO4•7H2O), Ni2+ (2 mM NiSO4•6H2O), Ag1+ (5 μM AgNO3), Hg2+ (5 μM HgCl2), ketoconazole (5, 10 and 15 μM), cysteine (5 mM), EDTA (5 nM), DMSO (5 mM), oxidised glutathione (4 mM), or reduced glutathione (5 mM). Samples were taken out at 24 and 48 h and analysed for the production of perillic compounds.
Bioconversion assays using Y. lipolytica ATCC 18942
Cell mass for biotransformation reactions using Y. lipolytica ATCC 18942 was obtained as described in the screening procedure. The bioconversion assays were performed in the following conditions unless otherwise stated: a cell mass concentration of 10 g/L, a pH of 7.0 (50 mM phosphate buffer), 0.5% (v/v) R-(+)-limonene and a temperature of 25 °C. The reaction mixtures (25 mL in a 250 mL Erlenmeyer flask) were incubated with shaking at 250 rpm for 48 h. Reaction conditions for the limonene bioconversion to perillyl derivatives assay were as follows: (i) temperatures of 20, 25, 28 and 30 °C; (ii) pH from 5.0 to 8.0 in phosphate buffer (50 mM) and from 3.0 to 4.0 in phosphate-citrate buffer (50 mM); (iii) presence or absence of glucose (100 mM) and phosphate buffer (50 mM) at pH 7.0; (iv) addition of tween 20 or tween 80 (0.1%); (v) direct addition of limonene (0.5%) to the 48 h YMB grown cell culture whose pH was adjusted to 7.0 (vi) and the stepwise addition of limonene (0.5% distributed in 6 dosages). Testing of conditions (i) and (ii) was performed in the presence of 100 mM glucose.
Biomass quantification
The cell mass concentration was determined at OD600 with reference to a calibration curve. One OD600 unit corresponded to a dry cell concentration of 0.444 g/L of Y. lipolytica.
Identification and quantification of perillic derivatives
The production of perillic derivatives was monitored by TLC. For this purpose, commercial standards of perillic acid, perillaldehyde, perillyl alcohol (1 μL from a 10 mM solution in ethanol) and the culture supernatant (5 μL) were directly spotted on silica plates (5 × 6.5 cm), run with ethyl acetate:n-hexane (1:1) and stained with acidic p-anisaldehyde in ethanol. The retention values for perillic acid, perillaldehyde and perillyl alcohol were 0.36, 0.63 and 0.48, respectively.
Perillic acid was quantified by gas chromatography with flame ionisation detection (GC-FID). An aliquot (1 mL) of the culture supernatant was transferred to a centrifuge tube and 50 μL of 0.6 M HCl was added to induce precipitation. After centrifugation at 10,000 rpm for 10 min, the precipitate was dissolved in 2 mL of ethyl acetate. Analyses were performed on an HP gas chromatograph model 6890N Network with an HP-Innowax capillary column (30 m × 0.25 mm × 0.25 mm film thickness), a flow rate of 2.0 mL/min and a 1/15-split ratio mode (split/splitless injector). The temperature program was as follows: inicial temperature of 50 °C held for 2 min, final temperature of 250 °C held for 8 min and a heating rate of 10 °C/min. Helium was used as the carrier gas. The injector temperature was set to 270 °C. Detection was performed with a flame ionization detector (FID) at 270 °C. A calibration curve was built using a concentration range of commercial perillic acid of 160–1000 μg/mL with the determination coefficient R2 = 0.9997. The method of sample preparation was evaluated using an artificial preparation of commercial perillic acid. The result was a 98.1% of recovery from the ethyl acetate extraction.
The formation of perillic acid was confirmed by gas chromatography-mass spectrometry (GC-MS). Analyses were performed at the same conditions used in the FID analysis, except for the use of an HP-5 MS column. Perillic acid was eluted from the column at 13.03 min and showed a mass fragmentation pattern identical to the commercial standard [m/z 166 (M+), 121 (M+ - CO2H), 105, 93, 79, 68].
Results and Discussion
Yeast strain selection
Acccording to TLC analysis, of the seventeen yeast strains evaluated, A. adeninivorans, C. parapsilosis and both Y. lipolytica strains were able to convert (R)-(+)-limonene to perillic acid. Conversion efficiencies varied, with A. adeninivorans effecting poor conversion, in agreement with the literature (Van Rensburg et al., 1997). A slightly better result was obtained with C. parapsilosis. Far better results were observed for Y. lipolytica ATCC 18942, as a single very strong spot was observed upon reaction of the TLC plate with p-anisaldehyde. This result was confirmed by GC-MS analysis, demonstrating the potential of this yeast for the selective oxidation of limonene to perillic acid.
Effect of culture medium on cell grow and the product profile
Growth media containing different sources of carbon and nitrogen were assayed for the cultivation of the selected yeasts C. parapsilosis and Y. lipolytica ATCC 18942. Both strains presented the best biomass yield when cultivated in the glucose- and protein-containing YMB medium. Y. lipolytica was also able to grow well in media containing glucose-ammonia, glycerol-ammonia or glycerol-urea as carbon and nitrogen sources. Cells grown in YMB were more efficient at limonene conversion, although promising results were also shown by urea-grown cells. TLC data demonstrated that Y. lipolytica was far better than C. parapsilosis at limonene bioconversion. Amongst all possible oxidised perillyl derivatives, perillic acid was the only one produced, regardless of the reaction conditions. Therefore, Y. lipolytica ATCC 18942 was chosen for the continuation of the study.
Limonene toxicity to cell growth
Y. lipolytica cell growth was fully inhibited when glucose in the YMB medium was substituted by 0.5% limonene or when 0.1% or 0.5% limonene was added to the complete YMB medium immediately after cell inoculation, indicating the toxicity of limonene to Y. lipolytica cells. However, the presence of 0.5% (R)-limonene in the biotransformation reaction mixtures did not impair the ability of the resting yeast cells to perform the bioconversion reaction.
Preliminary assessments of the bioconversion pathway
According to Mirata et al. (2009), limonene bioconversion to perillic acid performed by the bacterium Pseudomonas putida involves three oxidation steps catalysed by a monooxygenase, an alcohol dehydrogenase and an aldehyde dehydrogenase, leading in series to perillyl alcohol, perillaldehyde and perillic acid (Figure 1). In Mycobacterium sp. strain HXN-1500, the production of perillyl alcohol via limonene oxidation was accomplished by a multi-component electron transfer chain containing cytochrome P450 (P450), a ferredoxin and a ferredoxin reductase (Van Beilen et al., 2005). This Mycobacterium strain was selected in a screening program that showed a positive correlation between alkane degrading bacteria and the ability to hydroxylate limonene. Furthermore, according to the authors, the physiological function of the P450 responsible for limonene oxidation was the hydroxylation of alkanes.
Figure 1.
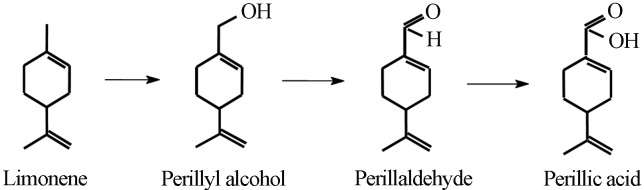
Stepwise oxidation of limonene to perillic acid.
The oxidative steps required for limonene bioconversion by yeasts have not yet been identified, with the exception, according to the literature, that the process is initiated by P450 monooxygenases (Duetz et al., 2003). Notably, Y. lipolytica is an alkane-assimilating yeast. The primary alkane oxidation involves a terminal hydroxylation catalysed by a P450-dependent alkane monooxygenase system. This is followed by the action of alcohol-oxidising enzymes (alcohol oxidases or alcohol dehydrogenases) and aldehyde dehydrogenases, resulting in the corresponding acid (Fickers et al., 2005). Therefore, it would be reasonable to assume that the P450 system would be responsible for the oxidation of limonene by this yeast.
To better understand the limonene biotransformation process carried out by Y. lipolytica, intermediates of the oxidative reaction were assayed for their conversion. Y. lipolytica was able to recognize perillyl alcohol as a substrate, which was then oxidized to perillaldehyde and perillic acid. It was also able to recognize perillaldehyde, which was transformed into perillic acid, as shown by TLC analysis. In the absence of O2, only perillaldehyde reacted and was reduced to perillyl alcohol. These results collectively suggest that limonene oxidation to perillic acid is a stepwise and multi-enzymatic oxidative pathway. Moreover, as the alcohol and aldehyde intermediates were not detected in all of the limonene bioconversion conditions tested, the first oxidation step, i.e., the oxidation of limonene to perillyl alcohol, would be the pacemaker reaction.
Inhibitors of the enzymes alcohol oxidase and alcohol dehydrogenase (White and White, 1997) were added to the limonene bioconversion medium in an attempt to trap the alcohol intermediate. The addition of zinc sulphate, ketoconazole, EDTA, cysteine or DMSO did not have any effect on the oxidation of limonene to perillic acid. Although dimercury dichloride, nickel(II) sulphate and oxidised or reduced glutathione decreased the formation of perillic acid, this was still the sole product of the reaction, and as such, the selective inhibition of the limonene oxidative steps was not observed. Copper(II) sulphate and silver nitrate completely inhibited the conversion, possibly by interfering with the relevant oxidative system.
The oxidation to perillic acid took place with either (R)- or (S)-limonene, although the later afforded a lower yield, as was qualitatively indicated by TLC analysis. The enantiomeric properties of the products were not determined.
Bioconversion assays
According to GC-FID analysis, the bioconversion of limonene at 25, 28 and 30 °C resulted in the formation of 564, 442 and 338 mg/L of perillic acid, respectively. The decreasing yield pattern observed with the temperature increase might be related either to a higher evaporation rate of the volatile limonene from the reaction medium or to the effect of the temperature on the enzymatic system involved in the reaction.
The effect of the initial pH of the culture on limonene bioconversion was evaluated by assaying the reaction at pH values in the range of 3.0 to 8.0. TLC analysis showed that bioconversion at pH 3.0 and 4.0 (phosphate-citrate buffer 50 mM) resulted in poor perillic acid formation in comparison to results at higher pH values (data not shown). Conversions within the pH range 5.7 to 7.4 (phosphate buffer 50 mM) showed that 7.1 was the best pH for bioconversion, as determined using GC-FID analysis, resulting in the formation of 562 mg/L of perillic acid (Table 1).
Table 1.
Influence of pH on limonene bioconversion to perillic acid by Y. lipolytica. Measured by GC-FID. Assays were performed at 25 °C, 200 rpm and the reaction mixtures contained 0.5% v/v initial R-(+)-limonene, 50 mM phosphate buffer at the desired pH, and 10 g/L cell mass.
| Initial pH | Perillic acid (mg/L) | |
|---|---|---|
|
|
||
| 24 h | 48 h | |
| 5.7 | 316.7 | 406.1 |
| 6.4 | 387.6 | 525.6 |
| 7.1 | 419.0 | 562.0 |
| 7.4 | 391.2 | 478.4 |
Table 2 shows the relative conversion of limonene to perillic acid in the presence or absence of glucose (100 mM) and phosphate buffer (50 mM). Yeast cells did not consume the sugar, the concentration of which remained constant throughout the 48 hours reaction time, demonstrating that the use of glucose in the reaction medium to reduce co-factors was not necessary for bioconversion. However, the production of the oxidised limonene derivative was reduced by 20% after 24 h and 30% after 48 h in non-buffered media, demonstrating that phosphate is required for efficient bioconversion. In all four experiments, slight increases in the final pH were observed (Table 2), indicating that the decrease in the reaction yields in the non-buffered medium was not related to pH variation.
Table 2.
Influence of glucose and/or buffer addition on limonene bioconversion to perillic acid by Y. lipolytica. Measured by GC-FID.
| Experiment | Relative conversion rate (%) | pH | ||
|---|---|---|---|---|
|
|
|
|||
| 24 h | 48 h | Initial | 48 h | |
| A(control) | 82.2 | 100.0 | 7.1 | 7.3 |
| B | 78.3 | 104.1 | 7.0 | 7.4 |
| C | 63.3 | 75.1 | 6.4 | 7.5 |
| D | 65.2 | 68.3 | 6.9 | 7.5 |
A: addition of 100 mM glucose and 50 mM pH 7.0 phosphate buffer; B: addition of 50 mM pH 7.0 phosphate buffer, no glucose; C: addition of 100 mM glucose no buffer; D: no glucose, no buffer. Assays were performed at 25 °C, 200 rpm and the reaction mixtures contained 0.5% v/v initial R-(+)-limonene and 10 g/L cell mass.
Addition of 0.1% tween 20 or tween 80 led to a 22% and 8.5% decrease in the production of perillic acid, respectively. The direct addition of 0.5% (v/v) limonene to the stationary-phase yeast culture (48 h) without previously centrifuging the cells decreased product formation by 25%. The stepwise addition of limonene to the bioconversion medium (6 doses distributed over 48 h) resulted in a 50% increase in perillic acid concentration, reaching 855 mg/L, compared to the usual one-step addition at the beginning of the experiment (data not shown). In summary, the reaction conditions that promoted the highest limonene conversion (16.7%) after a 48 h reaction time were as follows: a temperature of 25 °C, a pH of 7.1 (50 mM phosphate buffer) and gradual addition of limonene (0.5% distributed in 6 dosages over 48 h).
As described by Mirata et al. (2009), growing cells of P. putida DSM 12264 (using 5 g/L glycerol as a co-substrate) produced up to 3.0 g/L of perillic acid from 20.4 g/L limonene in a 120 h batch culture. This corresponded to a 12% limonene conversion. Upon process optimization in fed-batch culture, 10.6 g/L of perillic acid was achieved. A method for in situ product recovery resulted in a cumulative perillic acid concentration of 31 g/L after 7 days.
There are few articles that address the oxidation of limonene mediated by yeast. Although the process of converting limonene to perillic acid using Y. lipolytica has been reported, the reaction has not yet been characterized (Van Rensburg et al., 1997). Although the identification of the relevant metabolic pathway that performs the reaction in yeast cells and its location in the cell structure is still an open issue, the results presented here are a step forward in understanding the parameters that affect limonene bio-oxidation to perillic acid and the involvement of P450 as the enzymatic oxidative system. It should also be emphasised that the use of Y. lipolytica allows the formation of a single, stable, excreted product. These initial studies on the oxidation of limonene by this yeast will be expanded via experimental bioprocess optimization and the improvement of cell catalytic abilities to achieve higher conversion yields and productivity. The development of general processes using this yeast, which is known for its highly efficient metabolism of hydrophobic substrates and whose genome has already been sequenced, would help to establish a pharmaceutically safe microorganism that can synthesize pharmacologically important molecules.
Acknowledgments
The present study was supported by the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
References
- Bicas JL, Dionísio AP, Pastore GM. Bio-oxidation of terpenes: An approach for the flavor industry. Chem Rev. 2009;109:4518–4531. doi: 10.1021/cr800190y. [DOI] [PubMed] [Google Scholar]
- Buckland BC, Robinson DK, Chartrain M. Biocatalysis for pharmaceuticals – status and prospects for a key technology. Metab Eng. 2000;2:42–48. doi: 10.1006/mben.1999.0138. [DOI] [PubMed] [Google Scholar]
- Cadwallader KR, Braddock RJ, Parish ME, Higgins DP. Bioconversion of d-limonene by Pseudomonas gladioli. J Food Sci. 1989;54:1241–1245. [Google Scholar]
- Da Fonseca CO, Simão M, Lins IR, Caetano RO, Futuro D, Quirico-Santos T. Efficacy of monoterpene perillyl alcohol upon survival rate of patients with recurrent glioblastoma. J Cancer Res Clin Oncol. 2011;137:287–293. doi: 10.1007/s00432-010-0873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz WA, Bouwmeester H, Van Beilen JB, Witholt B. Biotransformation of limonene by bacteria, fungi, yeasts, and plants. Appl Microbiol Biotechnol. 2003;61:269–277. doi: 10.1007/s00253-003-1221-y. [DOI] [PubMed] [Google Scholar]
- Fickers P, Benetti PH, Wache Y, Marty A, Mauersberger S, Smit MS, Nicaud JM. Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005;5:527–543. doi: 10.1016/j.femsyr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Gellissen G, Kunze G, Gaillardin C, Cregg JM, Berardi E, Veenhuis M, Van der Klei I. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica – A comparison. FEMS Yeast Res. 2005;5:1079–1096. doi: 10.1016/j.femsyr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Krings U, Berger RG. Biotechnological production of flavours and fragrance. Appl Microbiol Biotechnol. 1998;49:1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- Liese A, Seelbach K, Wandrey C. Industrial Biotransformations. Wiley-VCH; Weinheim: 2006. [Google Scholar]
- Madyastha KM, Bhattacharyya PK, Vaidyanathan CS. Metabolism of a monoterpene alcohol, linalool, by a soil pseudomonad. Can J Microbiol. 1977;23:231–238. doi: 10.1139/m77-035. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Yamamoto H, Kobayashi Y. Practical application of recombinant whole-cell biocatalysts for the manufacturing of pharmaceutical intermediates such as chiral alcohols. Org Proc Res & Develop. 2002;6:558–561. [Google Scholar]
- Mirata MA, Heerd D, Schrader J. Integrated bioprocess for the oxidation of limonene to perillic acid with Pseudomonas putida DSM 12264. Process Biochem. 2009;44:764–771. [Google Scholar]
- Persidis A. Chiral-based therapeutics. Nat Biotechnol. 1997;15:594–595. doi: 10.1038/nbt0697-594. [DOI] [PubMed] [Google Scholar]
- Pollard DJ, Woodley JM. Biocatalysis for pharmaceutical intermediates, the future is now. Trends Biotechnol. 2007;25:66–73. doi: 10.1016/j.tibtech.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Speelmans G, Bijlsma A, Eggink G. Limonene bioconversion to high concentrations of a single and stable product, perillic acid, by a solvent-resistant Pseudomonas putida strain. Appl Microbiol Biotechnol. 1998;50:538–544. [Google Scholar]
- Tan Q, Day DF. Bioconversion of limonene to alphaterpineol by immobilized Penicillium digitatum. Appl Microbiol Biotechnol. 1998;49:96–101. [Google Scholar]
- Trudgill PW. Microbial metabolism of monoterpenes - Recent developments. Biodegradation. 1990;1:93–105. doi: 10.1007/BF00058829. [DOI] [PubMed] [Google Scholar]
- Trytek M, Fiedurek J. A novel psychrotrophic fungus, Mortierella minutissima, for D-limonene biotransformation. Biotechnol Lett. 2005;27:149–153. doi: 10.1007/s10529-004-7347-x. [DOI] [PubMed] [Google Scholar]
- Van Beilen JB, Holtackers R, Lüscher D, Bauer U, Witholt B, Duetz WA. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl Environ Microbiol. 2005;71:1737–1744. doi: 10.1128/AEM.71.4.1737-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Werf MJ, De Bont JAM, Leak DJ. Opportunities in microbial biotransformation of monoterpenes. Adv Biochem Eng Biotechnol. 1997;55:147–177. [Google Scholar]
- Van Rensburg E, Molekeli N, Van der Walt JP, Botes PJ, Van Dyk MS. Biotransformation of (+) limonene and (−) piperitone by yeasts and yeast-like fungi. Biotechnol Lett. 1997;19:779–782. [Google Scholar]
- White JS, White DC. Source Book of Enzymes. CRC Press; Boca Raton: 1997. [Google Scholar]
- Yeruva L, Pierre KJ, Elegbede A, Wang RC, Carper SW. Perillyl alcohol and perillic acid induced cell cycle and apoptosis in non small cell lung cancer. Cancer Letters. 2007;257:216–226. doi: 10.1016/j.canlet.2007.07.020. [DOI] [PubMed] [Google Scholar]


