Abstract
Campylobacter jejuni isolates of different origins (bovine, broiler meat, human) were screened by polymerase chain reaction for the presence of 4 genes cdtB, cst-II, ggt, and virB11, previously linked to virulence such as adherence, invasion, colonization, molecular mimicry, and cytotoxin production. In addition, the isolates were screened for the presence of the global gene regulator csrA linked to oxidative stress responses, biofilms formation, and cell adhesion. All the C. jejuni isolates were positive for cdtB gene. The csrA gene was detected in 100% and 92% of C. jejuni isolates from human and animal origin and the virB11 gene was detected in 7.3% and 3.6% isolates from chicken and human respectively. All isolates from bovine were negative for the virB11 gene. The isolates showed a wide variation for the presence of the remaining genes. Of the C. jejuni recovered from human 83.6%, and 32.7% were positive for cst-II, and ggt respectively. Out of the isolates from chicken 40% and 5.5% isolates revealed the presence of cst-II, and ggt, respectively. Finally of the C. jejuni isolates from bovine, 97.7% and 22.7% were positive for cst-II, and ggt respectively. We conclude that the genes of this study circulate among humans and animals. These results led us to hypothesize that the isolates associated with enteritis (cdtB positives) are not selected by environmental or host-specific factors. On the other hand, the high frequencies of csrA gene in C. jejuni show that this gene is important for the survival of C. jejuni in animals and humans.
Keywords: Campylobacter jejuni, virulence, Guillain Barré syndrome, broiler, bovine
Introduction
Campylobacter spp. is the leading cause of bacterial foodborne enteric disease in Europe (European Union, European Food Safety Authority and European Centre for Disease Prevention and Control, 2012) while in the USA, Campylobacter spp. has been ranked among the most important bacterial foodborne pathogens (Scallan et al., 2011). Clinical syndromes vary from mild to severe and from enterocolitis to extraintestinal diseases such as the Guillain-Barré Syndrome (GBS) (Islam et al., 2009). Fifty to 80% of human infection may be attributed to the chicken (European Union, European Food Safety Authority, 2010), but also cattle-related cases (via undercooked beef or unpasteurized milk and dairy products) have been reported (Sheppard et al., 2009). A study in Chile showed that 38–68% of broiler meat after chilling is contaminated with C. jejuni (Figueroa et al., 2009). Studies from our laboratory, recently detected genetically indistinguishable isolates between broiler meat and human campylobacteriosis in Chile by pulsed-field gel electrophoresis (PFGE) (González-Hein et al., 2013). In 2010, Chileans on average consumed 33.3kg of poultry meat being the main source of animal protein (Chile, Office of Agricultural Policies, Trades and Information, 2012). All this emphasizes the importance of chickens as a potential reservoir and source of C. jejuni infection in Chile.
The natural heterogeneity of C. jejuni has made studying the pathogenicity of this pathogen particularly challenging (Croinín and Backert, 2012). However, in recent years, significant progress has been made to increase our understanding of the role of several key factors associated with bacterial virulence mechanisms such as the cytolethal distending toxin (CDT) (Ge et al., 2008) as well as the molecular mimicry process in GBS (Koga et al., 2006; Louwen et al., 2008).
Cell invasion of epithelial cells and CDT production are important bacterial virulence mechanisms that induce enterocolitis. Cell invasion could result in cellular injury, leading to reduced absorptive capacity of the intestine, whereas CDT production is important for interleukin-8 (IL-8) release by intestinal cells in vitro which plays an important role in the host mucosal inflammatory response caused by C. jejuni (Hickey et al., 2000; Deun et al., 2007). CDT is composed of three subunits: the catalytic subunit CdtB, which is encoded by the cdtB gene, and has DNase I-like activity, whereas CdtA, and CdtC are binding proteins for delivering CdtB into target cells. Translocation of CdtB to the nucleus induces genotoxic effects on host DNA, triggering DNA repair cascades that lead to cell cycle arrest and eventual cell death. In addition it has also been suggested that CDT may play a role in adhesion and invasion (Konkel et al., 2001; Jain et al., 2008).
C. jejuni is also the major cause of the GBS, a post infectious autoimmune -mediated neuropathy (Koga et al., 2006). The development of this autoimmune neuropathy after C. jejuni infection is thought to be primarily related to sialylated lipooligosaccharides (LOS) on the cell surface of C. jejuni that exhibit molecular mimicry with gangliosides on peripheral nerves (Nachamkin et al., 2002). First Van Belkum et al. (2001) showed that a sialyltransferase encoded by the cst-II gene in C. jejuni is associated with risk of developing GBS, and later studies have confirmed this link (Nachamkin et al., 2002; Koga et al., 2006). On the other hand, the cst-II gene has been linked to the invasiveness of C. jejuni for intestinal epithelial cells (Louwen et al., 2008).
The C. jejuni gene ggt encoding the periplasmic gamma-glutamyltranspeptidase (GGT) seems to play a pivotal role in the enteric colonization. GGT has been shown in chicken model to be important in long lasting gut colonization, and in vitro it has been shown that GGT plays a significant role in C. jejuni-mediated apoptosis (Barnes et al., 2007). This genetic determinant has also been proposed as a host associated genetic marker (Gonzalez et al., 2009), hence we were interested in assessing the presence of ggt and in investigating the possible association with C. jejuni strains from broiler origin.
Another virulence gene linked with Campylobacter spp. invasiveness is the invasion-associated marker (virB11) gene. In vitro studies have shown that this genetic marker of C. jejuni strains is associated preferentially with both adherence and invasion (Bacon et al., 2000).
Although many genes related to the pathogenicity of C. jejuni have been reported, the relationships between these genes and the sources of strains are not clear.
C. jejuni is ubiquitous in the aerobic environment and possess regulatory systems to sense and adapt to external stimuli, such as oxidative and aerobic (O2) stress (Gundogdu et al., 2011). Considering the limited contingent of regulatory effectors found in C. jejuni genomes, it has been suspected that the gene encoding the regulatory protein CsrA might play a vital role in the regulation of stress responses and virulence determinants in this pathogen. It was demonstrated that the global posttranscriptional regulator csrA (carbon starvation regulator) favors biofilm formation, adherence of intestinal epithelial cells and survival to oxidative stress, suggesting an important regulatory role for this gene in C. jejuni pathogenesis (Fields and Thompson, 2008).
In this paper, we assess the presence of a set of genes associated with virulence in C. jejuni isolates of different sources (broiler meat, bovine and human) to determine whether host-specific or environmental factors select for or against a set of genes related to virulence in C. jejuni isolates.
Material and Methods
Bacterial isolates
The C. jejuni isolates (n = 154) were obtained from the strain collection at the Microbiology and Probiotics Laboratory of the Food Technology and Nutrition Institute, University of Chile and the Microbiology Laboratory of the Pontificial Catholic University of Chile. All 154 C. jejuni isolates were collected in the Metropolitan Region during 2006 to 2010. Among the isolates, 55 were from stool specimens of diarrheal patients (sporadic cases), the remaining 55 strains were obtained from chicken carcasses and 44 were obtained from bovine rectal swabs. The confirmation of the samples was carried out by standard microbiological methods. The hippurate hydrolysis test was used for determination of the C. jejuni strains. All hippurate-positive isolates were determined as C. jejuni.
PCR of genes associated with virulence
DNA from isolates was extracted by standard molecular biological techniques using the kit: Genomics DNA Purification (Bioingentech, Concepción, Chile). The DNA from all isolates was amplified by PCR as a control for DNA extraction and C. jejuni species confirmation by analysis of the 16SrRNA (cccj gene). Then all isolates were screened by polymerase chain reaction (PCR). Amplification of the cdtB, csrA, cst-II, ggt and virB11, locus were carried out in a master mix volume of 15 μL containing buffer 1X (5X Green GoTaq® Flexi Buffer Promega, Madison, WI, U.S), 0.4 mM each dNTP’s (Promega, Madison, Wisconsin, United States (U.S)), 0.06 U/μL GoTaq® Flexi DNA polymerase (Promega, Madison, Wisconsin, U.S), 2.4 mM Magnesium Chloride Solution(Promega, Madison, Wisconsin, U.S) and 0.7 μM each of forward primer and reverse primer (IDT®, Coralville, Iowa, U.S)). Amplification of the ggt gen varied in the concentrations of MgCl2 (2.0 mM) and primers (0.5 μM). Primers and PCR conditions generated in this study are given in Table 1. Amplified products were visualized in agarose gels (1.5%) stained with ethidium bromide. DNA of C. jejuni 81176 and Staphylococcus aureus ATCC® 25923 were used as positive and negative control respectively. In addition, a reaction control was also included (Mix with IDT® water used as template). The reference strain C. jejuni 81176 was isolated during an outbreak of Campylobacter diarrhea associated with raw milk consumption in Minnesota and it has been widely used in pathogenesis studies (Korlath et al., 1985).
Table 1.
Primers and conditions used in the fragments amplification of the five genes associated with virulence.
| Gen target and PCR conditions | Primers | Sequence (5′–3′) | Amplicon |
|---|---|---|---|
| 16S ribosomal gene de Campylobacter coli and C.jejuni, 94 °C for 3 m/ 4 cycles 94 °C for 30 s, 54 °C x 30 s and 72 °C x 35 s /30 cycles of 94 °C x 30 s, 56 °C x 30 s y 72 °C x 35 s, /5 min x 72 °C/ 65 °C for 5 min | cccjFW cccjRV | GCG TAG GCG GAT TAT CAA GT ATT CCA CTG TGG ACG GTA AC | 896 pb |
| cdtB gene, 94 °C for 3 min/ 4 cycles of 94 °C for 30 s, 54 °C x 30 s y 72 °C x 35 s /30 cycles de 94 °C x 30 s, 56 °C for 30 s and 72 °C x 35 s, /5 min a 72 °C/ 65 °C for 5 min | cdtBFW cdtBRV | CAC GGT TAA AAT CCC CTG CT GCA CTT GGA ATT TGC AAG GC | 495 pb |
| virB11 gene, 94 °C for 3 min/ 3 cycles of 94 °C for 30 s, 55 °C for 30 s y 72 °C for 30 s/ 35 cycles of 94 °C for 35 s, 57 °C for 35 s and 72 °C for 35 s/ 10 min for 72 °C/ 65 °C for 5 min | Vir FW Vir RV | GGT GGA ACA GGA AGT GGA AA AGG TTA TTT CCG CAT TGG GC | 719 pb |
| csrA gene, 94 por 3 min/ 30 cycles of 94 °C x 30 s, 58 °C for 30 s and 72 °C for 30 s / 5 min for 72 °C/ 65 °C for 5 min | csrAFW csrARV | CAC AGT CAG TGA AGG TGC TT ACT CGC ACA ATC GCT ACT TC | 878 pb |
| ggt gene, 94 °C for 3 min/ 30 cycles of 94 °C x 30 s, 58 °C for 30 s and 72 °C for 30 s / 5 min for 72 °C/ 65 °C for 5 min | ggtFW ggt RV | GAG TGC TAT GCT TGA TCG CT TAG GTG GCG ACA TGG AAA TG | 419 pb |
| cstII gene, 94 °C for 3 min/5 cycles of 94 °C by 35 s, 52 °C for 35 s and 72 for 35 s and 72 °C x 35 s/ 5 min for 72 °C /65 °C for 5 min | cstIIFW cstIIRV | CAG CTT TCT ATT GCC CTT GC ACA CAT ATA GAC CCC TGA GG | 570 pb |
Sequencing of PCR products
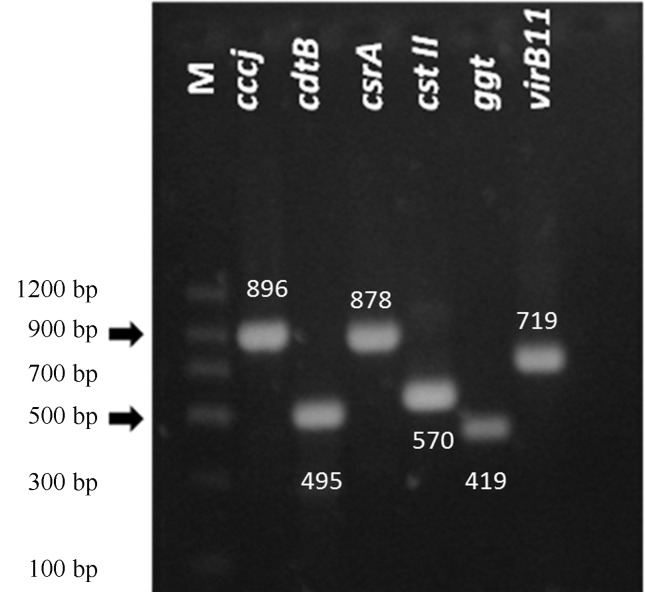
Five PCR products of C. jejuni 81176 were purified using the Kit Wizard® SV gel and PCR clean-up system, (Promega, Madison, Wisconsin, U.S) (Figure 1). Finally, the PCR products were automatically sequenced in both directions at Pontifical Catholic University of Chile. Sequencing was done on an ABI PRISM® 3130 Applied Biosystems. For bioinformatics analysis of the sequences and alignments, Blast and ClustalW2 software were used and matched with the database. The sequences of each gene were shown to be rather conserved (95 to 100% similarity within each gene).
Figure 1.
Purified polymerase chain reaction products from the cccj, cdtB, csrA, cst-II, ggt and virB11 genes. Strain: C. jejuni 81176. M, DNA Molecular Weight Marker.
Statistical analyses
The Chi-square test or Fisher’s exact test when necessary were used to test for similarity in the frequencies of genes within the isolates from different hosts, using alpha level of 0.05.
Results
Distribution of virulence genes
In each of the 154 isolates, the 16SrRNA gene was detected by PCR. Thus, all isolates could be confirmed as C. jejuni. Additionally, this PCR was suitable to control the DNA extraction procedure. The cdtB gene was also present in 100% of C. jejuni isolates tested, regardless of their origin. Similarly the regulator gene csrA was identified as an habitual virulence gene in the C. jejuni isolates. The frequency of detection of csrA, cst-II, and ggt genes varied between human, bovine and chicken isolates. From a total of 154 isolates, 146 (94.8%), 111 (72%) and 32 (20.8%) tested positive for csrA, cst-II, ggt respectively being the three genes more frequently detected in bovine and in diarrheagenic human isolates. Finally, results also indicated that only 3.9% (6 of 154) of the C. jejuni strains were virB11- positive and this gene was not detected in the bovine strains. The identification of these genes by PCR is depicted in Table 2. This figure shows the distribution of these genes associated with virulence according to the source of the isolates.
Table 2.
Presence of virulence-associated genes in Campylobacter jejuni strains isolated from chicken carcasses, bovine and human, Chile.
| Origin | Number of isolates | cdtB | csrA | cst-II | ggt | virB11 |
|---|---|---|---|---|---|---|
| Human | 55 | 55(100%) | 55(100%) | 46(83.6%) | 18(32.7%) | 2(3.6%) |
| Broiler meat | 55 | 55(100%) | 48(87.3%) | 22(40%) | 3(5.5%) | 4(7.3%) |
| Bovine | 44 | 44(100%) | 43(97.7%) | 43(97.7%) | 10(22.7%) | 0(0%) |
| Total | 154 | 154(100%) | 146(94.8%) | 111(72%) | 32(20.8%) | 6 (3.9%) |
Statistical analysis of the distribution of the five genes among the various origins of the isolates: in this analysis, csrA, cst-II and ggt genes had significantly different frequencies (p < 0.05) for isolates from different sources of origin using the Chi-square test or Fisher’s exact test.
In human isolates csrA gene was found with higher frequency than in chicken isolates (p < 0.05). In bovine and human C. jejuni isolates cst-II gene was found with higher frequency than in chicken isolates (p < 0.05). On the other hand, a lower amount of isolates from chicken meat harbored the ggt gene as detected by PCR. Besides, the frequency of bovine and human strains harboring ggt gene was higher than chicken meat isolates (p < 0.05). The distribution between the isolates of different origins harboring genes cdtB, and virB11 was similar in all group tested (p > 0.05).
Discussion
The majority of C. jejuni research has been focused using European isolates or from the U.S. origin. The occurrence of virulence and toxin genes among C. jejuni isolates from different sources has been studied poorly in South America. As far as we know there are some reports in Brazil, where a set of virulence-associated genes were detected in a substantial proportion of children with diarrhoea (Quetz et al., 2012) and less in chicken (Carvalho et al., 2010).
In this study all the strains investigated harbored cdtB gene. It is indeed generally accepted that the cdtB genes are widespread amongst poultry, cattle and human isolates in Denmark, Japan, Poland, and Belgium (Bang et al., 2003; Datta et al., 2003; Rozynek et al., 2005; Deun et al., 2007). However, low percentages of occurrence of cdtB have been reported in humans (28%) and chickens (20%) in India, which could be due to genetic reasons or variation in the isolates from different geographic areas (Rizal et al., 2010).
There are other human infection sources of campylobacteriosis beyond chicken and cattle. However, the equal distribution of cdtB in all human infectious sources of campylobacteriosis, as in chicken, and cattle, and the crucial role of CDT in the intestinal pathology - persistence of infection in the gastrointestinal tract and in the severity of mucosal inflammation (Ge et al., 2008; Jain et al., 2008) - led us to suggest that no selection for or against CDT associated strains of C. jejuni occurs in these animals. Georgiades and Raoult (2011) hypothesized that the only truly identifiable phenomena, witnessing the convergent evolution of the most pathogenic bacteria for humans are the loss of metabolic activities, i.e., the outcome of the loss of regulatory and transcription factors and the presence of protein toxins.
To date, only three studies have included the detection of GBS-related genes for human and animal isolates (Parker et al., 2005; Hardy et al., 2011; Amon et al., 2012). Parker et al. (2005) detected genes responsible for ganglioside mimics in 64% of the human enteric and animal isolates. Recently in Austria, Amon et al. (2012) also frequently detected the cst-II gene in isolates of bovine, poultry and human. Our analysis of the cst-II gene detection indicates the presence of potentially risky C. jejuni strains from bovine feces and contaminated broiler meats. It is not clear why the detection of cst-II from all sources is so high, although these findings seem to suggest that these ganglioside-mimicking LOS structures are advantageous to C. jejuni colonization of various hosts. Louwen et al. (2008) demonstrated that the disruption of cst-II significantly affects the invasiveness of C. jejuni for intestinal epithelial cells. What is clear is that the production of ganglioside-mimicking LOS structures alone is not sufficient to elicit GBS; other bacterial and/or other types of factors as the individual immune system are also required (Amon et al., 2012). The data generated revealed that the gene encoding the regulatory protein CsrA is present in all examined diarrheagenic human strains of C. jejuni, and is highly conserved among animals. A similar situation has been shown by Barnard et al. (2004) who detected the regulator csraA gene in all examined clinical strains of Helicobacter pylori. In this pathogen, a close relative of C. jejuni, CsrA is involved in the regulation of several virulence phenotypes, including motility, oxidative stress resistance, and mouse colonization (Barnard et al., 2004).The high frequency of csrA gene in C. jejuni allows us to hypothesize that this regulator is important for the survival of C. jejuni in the broiler meat, bovine and human. Recently a novel C. jejuni transcriptional regulator, Cj1556 that is involved in oxidative and aerobic stress responses, ability to form biofilms, and survival of C. jejuni was identified (Gundogdu et al., 2011).
The ggt gene has been recognized more frequently in human and chicken isolates (Gonzalez et al., 2009). Our findings diverged from those reported by Gonzalez et al. (2009), who suggested the ggt gene to be chicken associated based on PCR analysis of C. jejuni strains isolated from humans, chickens, and cattle. This may reflect differences between the C. jejuni populations in livestock in Chile and those in Finland; however, our results stress the need to confirm the results obtained using a set of C. jejuni strains from diverse geographical origins. Nevertheless, it is interesting to note that ggt-positive strains predominated in human isolates (33%) as it has been reported by other authors in Europe (Barnes et al., 2007; Gonzalez et al., 2009; Zautner et al., 2011). This is consistent with the hypothesis that C. jejuni isolates with an extended amino acid metabolism are more prevalent in humans (Gonzalez et al., 2009). So the acquisition of a gene encoding a gamma-glutamyltranspeptidase enabled this strain to utilize glutamine and glutathione and enhanced its ability to colonize the intestine (Hofreuter et al., 2008). Although we have found a low frequency of ggt in broiler isolates, it is necessary to stress that it is clear that successful colonization of chicken GI tract is a multifactorial process in which several genes involved in all areas of the colonization process of C. jejuni play a role (Hermans et al., 2011).
The virB11 gene was present equally in the broiler meat strains as in the human strains, suggesting that, at least, some broiler isolates potentially could invade the human intestine. It should be noted, however, that this gene is present in a very small subset of C. jejuni isolates (Bacon et al., 2000) and in this study was not detected in isolates from bovine feces.
The overlap observed in distribution of the C. jejuni genes among human, bovine, and chicken isolates (Table 2), together with the high consumption of meat by the Chilean population, suggests that human campylobacteriosis may be linked with chicken and bovine meats or unpasteurized milk.
The prevalence of C. jejuni virulence genes and their relationship with clinical severity in humans and the expression of virulence factors should be further investigated. It is known that source tracking depends on accurate estimation of the frequency of different genes in each host reservoir. The frequencies in which ggt gene were detected in C. jejuni isolates from cattle and broiler meat in the present study reveal that the potential use of this molecular genetic marker associated to determined hosts (European Union, European Food Safety Authority, 2010; Zautner et al., 2008) is controversial. The use of additional genotypic methods such as PFGE and/or Multilocus sequence typing that provides more discriminatory power is strongly recommended.
Acknowledgments
This study was supported by Grants from the National Commission for Scientific and Technological Research (CONICYT for its acronym in Spanish).
References
- Amon P, Klein D, Springer B, Jelovcan S, Sofka D, Hilbert F. Analysis of Campylobacter jejuni isolates of various sources for loci associated with Guillain-Barré syndrome. Eur J Microbiol Immunol. 2012;2:20–23. doi: 10.1556/EuJMI.2.2012.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon DJ, Alm RA, Burr DH, Hu L, Kopecko DJ, Ewing CP, Trust JT, Guerry P. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang DD, Nielsen EM, Scheutz F, Pedersen K, Handberg K, Madsen K. PCR detection of seven virulence and toxin genes of Campylobacter jejuni and Campylobacter coli isolates from Danish pigs and cattle and cytolethal distending toxin production of the isolates. J Appl Microbiol. 2003;94:1003–1014. doi: 10.1046/j.1365-2672.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- Barnard MF, Loughlin MF, Fainberg HP, Messenger MP, Ussery DW, Williams P, Jenks PJ. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;51:15–32. doi: 10.1046/j.1365-2958.2003.03788.x. [DOI] [PubMed] [Google Scholar]
- Barnes IH, Bagnall MC, Browning DD, Thompson SA, Manning G, Newell DG. Gamma-glutamyl transpeptidase has a role in the persistent colonization of the avian gut by Campylobacter jejuni. Microb Pathog. 2007;43:198–207. doi: 10.1016/j.micpath.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Silva DM, Azevedo SS, Piatti RM, Genovez ME, Scarcelli E. Detection of cytolethal distending toxin genes in strains of Campylobacter jejuni isolated from broiler carcasses. Arq Bras Med Vet Zootec. 2010;62:1054–1061. [Google Scholar]
- Croinín TO, Backert S. Host epithelial cell invasion by Campylobacter jejuni: trigger or zipper mechanism? Front Cell Inf Microbio. 2012 doi: 10.3389/fcimb.2012.00025. http://www.frontiersin.org/Cellular_and_Infection_Microbiology/10.3389/fcimb.2012.00025/abstract. [DOI] [PMC free article] [PubMed]
- Chile, Office of Agricultural Policies, Trades and Information. [Accessed 10 May 2012];Chilean Agriculture Overview. 2012 Available at: http://www.odepa.gob.cl/articulos/MostrarDetalle.action;jsessionid=12E079A48C5A38B6FC8F04F7DC4187EC?idcla=3&idcat=6&idclase=99&idn=5769.
- Datta S, Niwa H, Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J Med Microbiol. 2003;52:345–348. doi: 10.1099/jmm.0.05056-0. [DOI] [PubMed] [Google Scholar]
- Deun KV, Haesebrouck F, Heyndrickx M, Favoreel H, Dewulf J, Ceelen L, Dumez L, Messens W, Leleu S, Immerseel FV, Ducatelleand R, Pasmans F. Virulence properties of Campylobacter jejuni isolates of poultry and human origin. J Med Microbiol. 2007;56:1284–1289. doi: 10.1099/jmm.0.47342-0. [DOI] [PubMed] [Google Scholar]
- European Union, European Food Safety Authority. Scientific opinion on quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J. 2010;8:1437–1526. [Google Scholar]
- European Union, European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2010. EFSA J. 2012;10:2597–3039. [Google Scholar]
- Fields JA, Thompson SA. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J Bact. 2008;190:3411–3416. doi: 10.1128/JB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa G, Troncoso M, López C, Rivas P, Toro M. Occurrence and enumeration of Campylobacter spp. during the processing of Chilean broilers. BMC Microbiology. 2009 doi: 10.1186/1471-2180-9-94. http://www.biomedcentral.com/1471-2180/9/94. [DOI] [PMC free article] [PubMed]
- Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell Microbiol. 2008;10:1599–1607. doi: 10.1111/j.1462-5822.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- Georgiades K, Raoult D. Comparative genomics evidence that only protein toxins are tagging badbugs. Front Cell Inf Microbio. 2011 doi: 10.3389/fcimb.2011.00007. http://www.frontiersin.org/cellular_and_infection_microbiology/10.3389/fcimb.2011.00007/abstract. [DOI] [PMC free article] [PubMed]
- Gonzalez M, Hakkinen M, Rautelin H, Hänninen ML. Bovine Campylobacter jejuni strains differ from human and chicken strains in an analysis of certain molecular genetic markers. Appl Environ. Microbiol. 2009;75:1208–1210. doi: 10.1128/AEM.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hein G, García P, Foerster C, Troncoso M, Figueroa G. Campylobacter jejuni isolated from human cases in Chile showed indistinguishable PFGE profiles with strains isolated from poultry and bovine sources. Cyta J Food. 2013;11:185–189. [Google Scholar]
- Gundogdu O, Mills DC, Elmi A, Martin MJ, Wren BW, Dorrell N. The Campylobacter jejuni transcriptional regulator cj1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. J Bact. 2011;193:4238–4249. doi: 10.1128/JB.05189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CG, Lackey LG, Cannon J, Price LB, Silvergeld EK. Prevalence of potentially neuropathic Campylobacter jejuni strains on commercial broiler chicken products. Int J Food Microbiol. 2011;145:395–399. doi: 10.1016/j.ijfoodmicro.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Hermans D, Van Deun K, Martel A, Immerseel F, Messens W, Heyndrickx M, Haesebrouck F, Pasmans F. Colonization factors of Campylobacter jejuni in the chicken gut. Vet Res. 2011 doi: 10.1186/1297-9716-42-82. http://www.veterinaryresearch.org/content/42/1/82. [DOI] [PMC free article] [PubMed]
- Hickey TE, McVeigh AL, Scott DA, Michielutti RE, Bixby A, Carroll SA, Bourgeois AL, Guerry P. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect Immun. 2000;68:6535–6541. doi: 10.1128/iai.68.12.6535-6541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreuter D, Novik V, Galán JE. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe. 2008;4:425–433. doi: 10.1016/j.chom.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Islam Z, Belkum A, Wagenaar JA, Cody AJ, Boer AG, Tabor H, Jacobs BC, Talukder KA, Endtz HP. Comparative Genotyping of Campylobacter jejuni Strains from Patients with Guillain-Barré Syndrome in Bangladesh. PLoS One. 2009 doi: 10.1371/journal.pone.0007257. http://www.plosone.org/article/info:doi%2F10.1371%2Fjournal.pone.0007257. [DOI] [PMC free article] [PubMed]
- Jain D, Prasad KN, Sinha S, Husain N. Differences in virulence attributes between cytolethal distending toxin positive and negative Campylobacter jejuni strains. J Med Microbiol. 2008;57:267–272. doi: 10.1099/jmm.0.47317-0. [DOI] [PubMed] [Google Scholar]
- Koga M, Gilbert M, Takahashi M, Li J, Koike S, Hirata K, Yuki N. Comprehensive analysis of bacterial risk factors for the development of Guillain-Barre syndrome after Campylobacter jejuni enteritis. J Infect Dis. 2006;193:547–555. doi: 10.1086/499969. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Monteville MR, Rivera-Amill V, Joens LA. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr Issues Intest Microbiol. 2001;2:55–71. [PubMed] [Google Scholar]
- Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- Louwen R, Heikema Astrid, Belkum AV, Ott Alewijn, Gilbert M, Ang W, Endtz HP, Bergman MP, Nieuwenhuis EE. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect Immun. 2008;76:4431–4438. doi: 10.1128/IAI.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachamkin I, Liu J, Li M, Ung H, Moran AP, Prendergast MM, Sheikh K. Campylobacter jejuni from patients with Guillain-Barré syndrome preferentially expresses a GD(1a)- like epitope. Infect Immun. 2002;70:5299–5303. doi: 10.1128/IAI.70.9.5299-5303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Horn ST, Gilbert M, Miller WG, Woodward DL, Mandrell RE. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from variety of sources. J Clin Microbiol. 2005;43:2771–2781. doi: 10.1128/JCM.43.6.2771-2781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetz JS, Lima IF, Havt A, Prata MM, Cavalcante P, Medeiros PH, Cid D, Moraes ML, Rey LC, Soares AM, Mota RM, Weigl BH, Guerrant RL, Lima AA. Campylobacter jejuni infection and virulence-associated genes in children with moderate to severe diarrhea admitted to emergency rooms in northeastern. Brazil J Med Microbiol. 2012;61:507–513. doi: 10.1099/jmm.0.040600-0. [DOI] [PubMed] [Google Scholar]
- Rizal A, Kumar A, Vidyarthi A. Prevalence of pathogenic genes in Campylobacter jejuni isolated from poultry and human. Internet Journal of Food Safety. 2010;12:29–34. [Google Scholar]
- Rozynek E, Dzierzanowska-Fangrat K, Jozwiak P, Popowski J, Korsak D, Dzierzanowska D. Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J Med Microbiol. 2005;54:615–619. doi: 10.1099/jmm.0.45988-0. [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States - Major pathogens. Emerg Infect. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, Gormley FJ, Falush D, Ogden ID, Maiden MC, Forbes KJ. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis. 2009;48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A, Van den Braak N, Godschalk P, Ang W, Jacobs B, Gilbert M, Wakarchuk W, Verbrugh H, Endtz H. A Campylobacter jejuni gene associated with immune-mediated neuropathy. Nat Med. 2001;7:752–753. doi: 10.1038/89831. [DOI] [PubMed] [Google Scholar]
- Zautner AE, Herrmann S, Corso J, Tareen AM, Alter T, Groß U. Epidemiological Association of Different Campylobacter jejuni Groups with Metabolism-Associated Genetic Markers. Appl Environ Microbiol. 2011;77:2359–2365. doi: 10.1128/AEM.02403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]