Abstract
An extracellular alkaline lipase from Pseudomonas aeruginosa mutant has been purified to homogeneity using acetone precipitation followed by anion exchange and gel filtration chromatography and resulted in 27-fold purification with 19.6% final recovery. SDS-PAGE study suggested that the purified lipase has an apparent molecular mass of 67 kDa. The optimum temperature and pH for the purified lipase were 45°C and 8.0, respectively. The enzyme showed considerable stability in pH range of 7.0–11.0 and temperature range 35–55 °C. The metal ions Ca2+, Mg2+ and Na+ tend to increase the enzyme activity, whereas, Fe2+ and Mn2+ ions resulted in discreet decrease in the activity. Divalent cations Ca+2 and Mg+2 seemed to protect the enzyme against thermal denaturation at high temperatures and in presence of Ca+2 (5 mM) the optimum temperature shifted from 45°C to 55°C. The purified lipase displayed significant stability in the presence of several hydrophilic and hydrophobic organic solvents (25%, v/v) up to 168 h. The pure enzyme preparation exhibited significant stability and compatibility with oxidizing agents and commercial detergents as it retained 40–70% of its original activities. The values of Km and Vmax for p-nitrophenyl palmitate (p-NPP) under optimal conditions were determined to be 2.0 mg.mL−1 and 5000 μg.mL−1.min−1, respectively.
Keywords: alkaline lipase, detergent stability, mutant, organic-solvent, p-nitrophenyl palmitate Pseudomonas
Introduction
According to the current opinion of industry, there can be a commercial exploitation of hydrolytic enzymes in the coming years due to their wide range of industrial applications in various fields including organic synthesis, clinical analysis, pharmaceuticals, detergents and food production. Enzymes are progressively swapping the use of harsh chemicals in diverse industrial processes. The reaction specificity of the enzymes leads to minimum by-product formation and thereby application of enzymes offers minimal risk to the environment (Malathu et al., 2008) -.
Lipases (triacylglycerol acylhydrolase, EC 3.1.1.3) comprise a group of hydrolytic enzymes which catalyze reversibly the hydrolysis and synthesis of triacylglycerides in the oil-water interface (Macrae and Hammond, 1985). They have potential applications in oleo-chemical, paper manufacturing, cosmetics, pharmaceuticals and agrochemical industries. However, the biggest market of their use is in the detergent formulation (Jaeger and Reetz, 1998; Rathi et al., 2008). The functional importance of lipases in the detergent industry is related to the removal of fatty residues in laundry, dishwashers as well as for cleaning of clogged drains (Vulfson, 1994). In addition to detergent applications, lipases can be used in versatile chemical reactions like transesterification, enantiomeric separation of alcohols and separate racemic amine mixtures. They have also been used to form aromatic and aliphatic polymers (Shrinivas, 2008).
During industrial fermentation, the production of extracellular enzymes usually yields large volumes of fermentation broth. Therefore, competent downstream processing and purification of these fermentation products are the key factors affecting the capital of the process. Most commercial applications do not demand homogeneous lipase preparations, however, a certain degree of purity facilitates efficient and successful usage in drug targeting, identifying the primary amino acid sequence and, more recently, determining the three-dimensional structure (Taipa et al., 1992; Aires-Barros et al., 1994). Therefore, nowadays industries stare for purification strategies that are economical, rapid, high yielding and amenable to large-scale operations (Kumarevel et al., 2005).
As nature provides an amazing diversity of enzymes, identifying enzyme solutions for a specific problem can be extremely difficult. So, the biochemical characterization of any enzyme, obtained from a particular source becomes crucial to appreciate its maximal catalytic performance, which in turn is indispensable for the best industrial exploitation of that enzyme (Sangeetha et al., 2011).
In the light of above facts, we made an attempt to purify and characterize the extracellular alkaline lipase produced by a mutant strain of Pseudomonas aeruginosa 10,055 with respect to optimal pH and temperature as well as stability, and to evaluate the effect of several metal ions, inhibitors, organic solvents, surfactants and detergents on the enzyme activity for its feasible applications.
Materials and Methods
Microorganism and culture conditions for lipase production
The mutant strain was previously developed in our laboratory by chemical mutagenesis of P. aeruginosa MTCC 10,055 (Bisht et al., 2012) and extracellular alkaline lipase was produced in the fermentation medium optimized by Bisht et al. (2012). The culture broth was clarified by centrifugation (10,000 rpm for 10 min at 4 °C) to recover the supernatant, which was used as enzyme source for further studies.
Analytical methods
Lipase activity was determined spectrophotometrically by following the method of Winkler and Stuckman (1979) with slight modifications. The substrate solution containing 10 mL of isopropanol with 30 mg of p-nitrophenyl palmitate (p-NPP) was mixed with 90 mL of Tris-HCl buffer (50 mM, pH 9.0), containing 0.4% Triton-X-100 and 100 mg of gum arabic. Freshly prepared substrate solution (2.4 mL) was incubated at 37 °C with 25 μL of suitably diluted cell-free supernatant for 15 min. After incubation absorbance was measured at 410 nm by using a spectrophotometer (UV-1601, Shimadzu) against a control with heat inactivated enzyme. One unit of enzyme is defined as the amount of enzyme liberating 1 μg of p-nitrophenol.mL−1.min−1 under assay conditions.
Purification of alkaline lipase
A three step purification method was used to purify the lipase produced by the mutant strain. All the purification steps were conducted at temperatures between 0–4 °C unless and otherwise stated.
Acetone precipitation
The crude enzyme obtained after centrifugation, was precipitated by the addition of different volumes (1:1, 1:2 and 1:3) of enzyme grade chilled acetone (−20 °C) with gentle stirring on ice-bath. The precipitate was stored overnight, at −20 °C and recovered by centrifugation at 10,000 × g for 10 min at 4 °C and the pellet obtained was air dried, so as to remove traces of acetone. The precipitate was then dissolved in suitable volume of Tris-HCl buffer (pH 9.0, 50 mM). Additional chilled acetone was added to the supernatant to bring the saturation to 1:2 and 1:3, the mixture was then left overnight. The corresponding precipitates were recovered, dissolved individually in fresh buffer and assayed for both total protein content and lipase activity.
Ion-exchange chromatography
The active fraction obtained after acetone precipitation, showing maximum specific activity, was further purified by ion exchange chromatography using Q-Sepharose (Sigma-Aldrich, USA) column equilibrated with sodium phosphate buffer (50 mM, pH 7.0). The desired enzyme fraction was allowed to bind with matrix for 2 h at 4 °C. The unbound fraction was collected and analyzed for enzyme activity and for protein content. The bound fractions were eluted with a linear gradient of NaCl (0.1–0.5 M, 10 mL each) in the same buffer.
Gel-filtration chromatography
The partially purified enzyme was applied to gel-filtration chromatography for purification up to homogeneity. The Sephadex-75 column (Sigma Aldrich Pvt. Ltd., USA, 1.5 × 40 cm) was equilibrated with Tris-HCl (pH 8.0, 50 mM) buffer and 1 mL of concentrated sample was applied to the column. The flow rate was adjusted to 5–6 mL.h−1 and fraction of 2 mL each were collected. Lipase activity and estimation of protein content were determined for each individual fraction.
Protein estimation
The protein content of individual fraction obtained after different steps of chromatography was monitored by measuring the extinction at 280 nm. Quantitative estimation of protein content was done by the method of Lowry et al. (1951) using Bovine serum albumin (BSA) as standard and expressed as mg.mL−1.
Electrophoretic analysis for molecular weight determination and homogeneity test
The active fraction, with maximum specific activity, obtained after gel filtration chromatography along with crude, acetone precipitate and anion-exchange chromatography was electrophorezed by Sodium Dodecyl Sulphate-Poly Acrylamide Gel Electrophoresis in a 12.5% polyacrylamide gel according to the method of Laemmli (1970). Approximate molecular weight of the lipase was estimated by SDS-PAGE against the molecular mass markers i.e. lysozyme (14.3 kDa), β-lactoglobulin (20 kDa), Carbonic anhydrase (29 kDa), ovalbumin (43 kDa), bovine serum albumin (66 kDa) and phosphorylase B (97.4 kDa) (Sigma-Aldrich Pvt Ltd., USA) run with the samples.
Characterization of purified enzyme
Effect of pH on enzyme activity and stability
The optimum pH of the lipase was determined by using buffer solutions (50 mM) of different pH (Sodium phosphate, 6.0, 7.0; Tris-HCl, 8.0, 9.0; and Carbonate/bicarbonate, 10.0, 11.0). For the pH stability the enzyme was incubated with buffers (in ratio of 1:1) at 37 °C for 1 h and assayed under standard assay conditions.
Effect of temperature on enzyme activity and stability
The influence of temperature on activity of lipase was studied by incubating the reaction mixture at different temperatures (30–60 °C). The enzyme was incubated at different temperatures 35–55 °C for 1 h to study the stability of the enzyme. The residual lipase activity was measured by conducting the reaction at temperature 37 °C and pH 8.0. The activity of the enzyme was considered as 100% under standard assay conditions.
Effect of metal ions on activity and stability
The effect of various metal ions (2 mM and 5 mM) on enzyme activity was investigated using FeSO4, CaCl2, KCl, NaCl, MgSO4, MnCl2, ZnCl2, CuSO4, HgCl2 and NiCl2. The enzyme was incubated with different metals at 37 °C for 1 h to study metal ion stability and assayed under standard assay conditions.
Effect of organic solvents on enzyme stability
The effect of different organic solvents (methanol, iso-propanol, ethanol, acetone, butanol, toluene, iso-octane, xylene, n-hexane, n-decane and n-dodecane) having log P values ranging from −0.76–6.0, on enzyme stability was investigated by incubating the enzyme with solvents at a concentration of 25% (v/v) at 37 °C for 168 h. The enzyme assay was done after 1 h and thereafter at an interval of 24 h for 7 days. The enzyme activity without incubation with organic solvent was taken as control (100%).
Effect of inhibitors on alkaline lipase activity
The effects of ethylene diamine tetra acetic acid (EDTA), β-mercaptoethanol, Phenyl methyl sulphonyl flouride (PMSF) and urea as inhibitors on alkaline lipase activity were investigated at a concentration of 2 mM and 5 mM in order to characterize enzyme. Purified alkaline lipase was pre-incubated with the above mentioned reagents for 1 h at 37 °C and residual activity (%) was determined under standard assay conditions.
Effect of surfactants, commercial detergents and oxidizing agents on enzyme stability
The lipase sample was incubated with surfactants viz., triton-X-100, Tween-40, Tween-60, Tween-80, SDS (0.1 and 1.0%, v/v), commercial detergents viz., surf, aerial, ghari, henko and fena (0.1 and 1.0%, w/v), and oxidizing agents viz., H2O2, sodium perborate and sodium hypochlorite (0.1 and 1.0%, v/v) for 1 h at 37 °C and then the residual activity (%) was tested under standard assay conditions.
Kinetic study of alkaline lipase
The influence of substrate concentration on the reaction velocity of the purified lipase was studied with p-NPP. The purified lipase was incubated with various concentration of p-NPP. The final concentration ranged from 0.5–5.5 mg.mL−1. In all cases, the enzymatic activity was assayed under standard conditions. The Michaelis constant (Km) and maximum velocity (Vmax) was determined from Lineweaver-Burk plots.
The linear velocity data was plotted as the function of concentration of the substrate by linear transformation of the Michaelis-Menten equation and usual non-linear curve fitting of the Michaelis-Menten equation for the calculation of Km and Vmax of the reaction.
Results and Discussion
Purification of alkaline lipase
The crude enzyme extract was first concentrated by acetone precipitation. Maximum activity was observed in the fraction obtained by the addition of acetone in ratio 1:1 with protein content of 18.05 mg.mL−1. This fraction had 10,314.6 U.mg−1 of specific activity with recovery of 78.6% and with regard to purification it showed 4.2-fold purification.
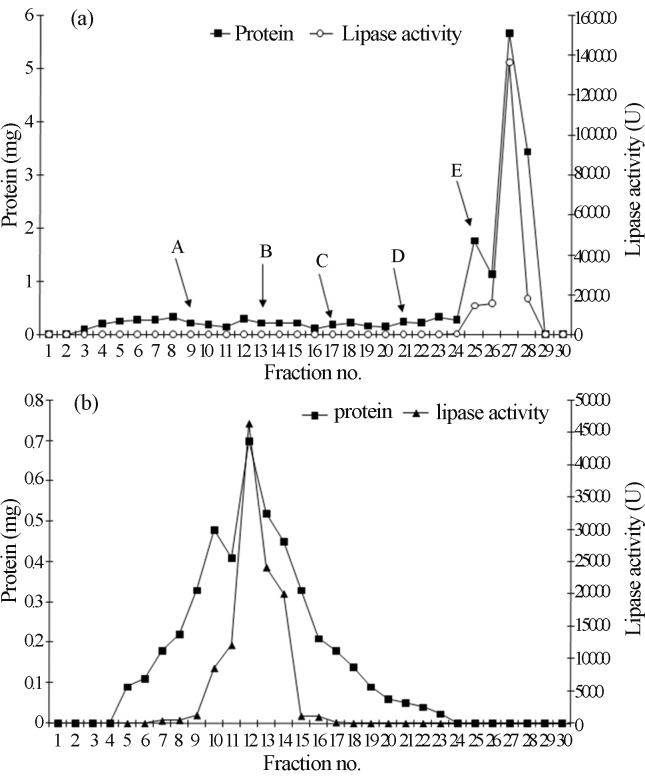
The active fraction of acetone precipitation method was used for further purification by using ion exchange chromatography. Sample (1 mL) was loaded into the Q-Sepharose column pre-equilibrated with sodium phosphate buffer (50 mM, pH 7.0) and allowed to pass through the column. The unbound fraction was collected and analyzed for lipase activity and protein content. There was no lipase activity in the fraction, while 1.9 mg.mL−1 of protein was estimated. The absence of enzyme in unbound fraction suggested that total lipase was bound to matrix. The bound enzyme was eluted by sodium phosphate buffer (50 mM, pH 7.0) having NaCl with increasing concentration at gradient of 0.1 M. Ten mL solution of each concentration of NaCl was used to evade the bound enzyme. The lipase activity was detected in the fraction released by the addition of 0.5 M NaCl Anion-exchange chromatography of lipase on column resulted in one prominent peak at the 27th fraction (Figure 1a).
Figure 1.
(a): Chromatographic purification profile of extracellular alkaline lipase from Pseudomonas aeruginosa MTCC 10,055 on anion-exchange column (Q-Sepharose) equilibrated with sodium phosphate buffer (50 mM, pH 7.0). The lipase was eluted with a gradient of sodium chloride (0.1 M–0.5 M) in sodium phosphate buffer (50 mM, pH 7.0). A-0.1 M, B- 0.2 M, C- 0.3 M, D- 0.4 M, E- 0.5 M. (b): Chromatographic purification profile of extracellular alkaline lipase on Sephadex G-75 gel filtration chromatography. Column was equilibrated with Tris-HCl buffer (50 mM, pH 8.0). The sample was loaded and eluted with the same buffer.
The active fraction was applied on Sephadex G-75 column. Figure 1b shows the fractionation pattern of lipase on Sephadex G-75 column. One distinctive protein peak was appeared that overlapped with the lipase activity. The purification process resulted in 27-fold purification factor and a final recovery of 19.6% of the enzyme with specific activity of 66323.6 U.mg−1 (Table 1). However, Ji et al. (2010) reported 4.3-fold purified lipase with 41.1% recovery by using ammonium sulphate precipitation and ion-exchange chromatography for purification of lipase from P. aeruginosa LX1.
Table 1.
Summary of purification of lipase from mutant of P. aeruginosa.
| Purification Steps | Total activity (U) | Total protein (mg) | Specific activity (U/mg) | Yield (%) | Purification fold |
|---|---|---|---|---|---|
| Crude | 2,36,870 | 96.5 | 2,454.6 | 100 | 1.0 |
| Acetone | 1,86,179.8 | 18.05 | 10,314.6 | 78.6 | 4.2 |
| Q-Sepharose | 1,36151.1 | 5.66 | 24,054.9 | 57.4 | 9.8 |
| Sephadex G-75 | 46,426.5 | 0.70 | 66,323.6 | 19.6 | 27.0 |
Each value presented here is an average of triplicates of three independent trials. The mean standard deviation for each value is < ± 5.0%.
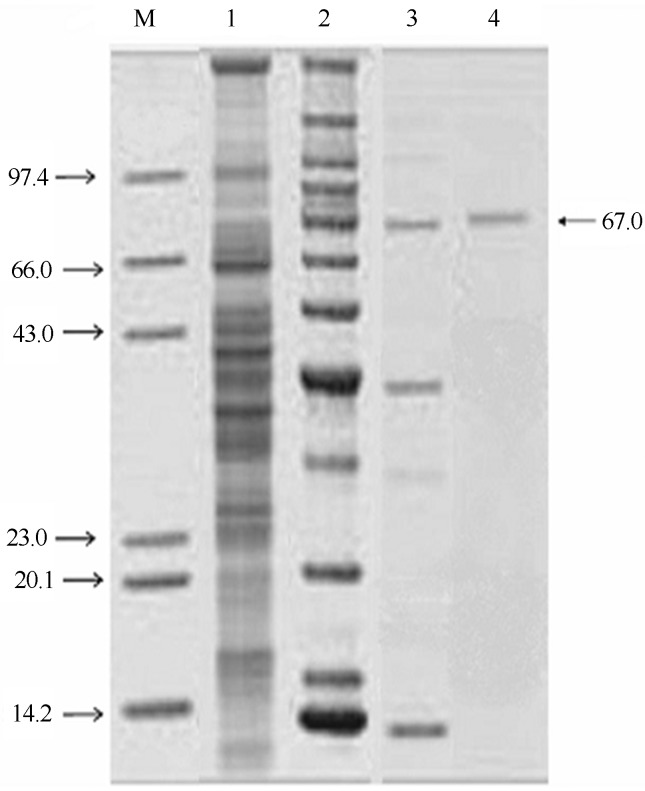
The purity of the enzyme was confirmed by the presence of a single band on SDS-PAGE and its molecular weight was approximately 67 kDa (Figure 2), which was similar to P. aeruginosa lipase (59.4 kDa) (Singh and Banerjee, 2007) but different from P. aeruginosa ATCC 27853 lipase (30 kDa) (Izrael-Zivkovic et al., 2009).
Figure 2.
Photographic representation of the SDS-PAGE of different fractions of alkaline lipase of mutant strain of P. aeruginosa 10,055 obtained during purification steps. Lane 1: Marker proteins; lane 2: Crude supernatant; lane 3: Concentrated enzyme after acetone precipitation; lane 4: Purified enzyme obtained after anion-exchange chromatography; lane 5: Purified enzyme obtained after gel-filtration chromatography. Molecular weights were presented in the form of kDa.
Characterization of purified enzyme
Effect of pH on enzyme activity and stability
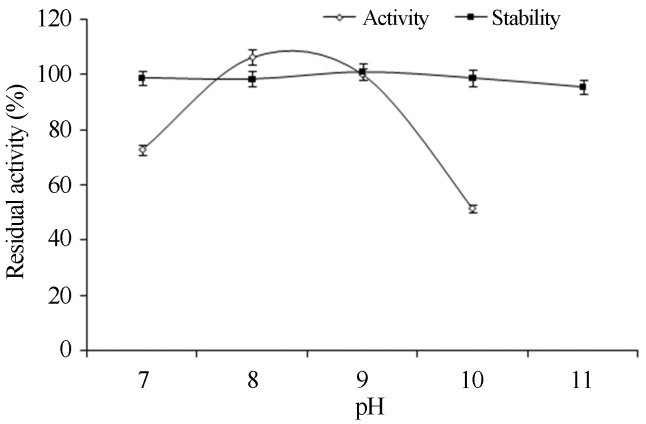
In the present study, the activity profile of the P. aeruginosa 10,055 lipase at different pH showed that the enzyme was active over a wide range of pH 7–10. Maximum activity was obtained at pH 8.0 whereas minimum was obtained at pH 7.0 (Figure 3). However, activity beyond pH 10.0 was not performed because of the difficulties in rate estimation caused by the spontaneous hydrolysis of p-NPP at pH above 10.0. Similarly, Sifour et al. (2010) also reported a higher activity at pH 8.0 by G. stearothermophilius. On the other hand, lipases from P. aeruginosa SRT 9 and Burkholderia sp. had shown maximum lipase activity at pH 6.9 and 8.5 respectively (Park et al., 2007; Borkar et al., 2009).
Figure 3.
Effect of pH on enzyme activity and stability. For enzyme activity the reaction was assayed at respective pH and for stability enzyme was pre-incubated with buffers (50 mM, in ratio 1:1) of different pH (7–11) at 37 °C for 1 h and assayed by standard assay method.
The enzyme showed 100% stability in the pH range 7.0–9.0, however, there was slight decrease in stability at pH 10.0 and 11.0 after 1 h incubation at 37 °C (Figure 3). Similarly, Rahman et al. (2005) reported that the Pseudomonas sp. strain S5 lipase showed great stability at pH 7.0–9.0, however, the activity was reduced drastically at pH 10.0–12.0 after 30 min. The lipase from G. candidum was stable up to pH 8.5 and lost its activity when the pH was raised above 11 (Gopinath et al., 2003).
Effect of temperature on enzyme activity and stability
It is revealed from Figure 4 that enzyme was active in the temperature range of 35–45 °C. The maximum activity of the enzyme was achieved at temperature 45 °C. The enzyme retained 85.5% activity when the temperature increased from 45 to 50 °C, but further increase in temperature, enzyme activity considerably decreased. The enzyme showed 100 and 83.7% stability at 40 and 45 °C, respectively, whereas about 50 and 25% of the activity was retained at 55 °C and 60 °C, respectively.
Figure 4.
Effect of temperature on enzyme activity and stability. For enzyme activity reaction mixture was incubated at different temperatures (30–60 °C) and for stability enzyme was pre-incubated at respective temperatures for 1 h and reaction was conducted as standard assay method.
It can be inferred from the results that lipase from P. aeruginosa MTCC 10,055 showed better activity and stability over the others lipase from P. monteilii TKU009 exhibiting optimum temperature 40 °C and exhibited only 70% of initial activity at temperature 50 °C after 1 h of incubation (Wang et al., 2009a).
Effect of metal ions on activity and stability
Among the metal ions tested, enhancement in the lipase activity was observed in presence of Ca+2 Mg+2, Na+ and K+ with 166.1, 135.9, 127.8 and 124.2% residual activity respectively at 2 mM concentration whereas moderate increase in activity was observed at 5 mM. However, no lipase activity was detected in the presence of Zn+2 and Hg+2 at both the concentration (Table 2). The lipase was significantly stable in the presence of Ca+2, Fe+2, Mg+2, Mn+2, K+ and Na+ at concentration of 2 mM, whereas, considerable decrease in enzyme stability was observed at 5 mM concentration except Na+ and K+. This suggested that this lipase is activated by metal ions which binds to the enzyme and change the conformation of the protein to provide greater stability to the enzyme. But transition metal ions change the conformation of the protein to less stable due to ion toxicity (Rahman et al., 2005).
Table 2.
Effect of metal ions on enzyme activity and stability.
| Metal ions | Concentration (mM) | Residual activity (%) | |
|---|---|---|---|
|
|
|||
| Activity | Stability | ||
| Control | 100.0 | 100.0 | |
| CaCl2 | 2 | 166.5 | 108.2 |
| 5 | 136.4 | 70.3 | |
| NiCl2 | 2 | 124.5 | 82.2 |
| 5 | 41.3 | 68.5 | |
| FeSO4 | 2 | 81.9 | 103.8 |
| 5 | 57.2 | 28.7 | |
| MgCl2 | 2 | 135.9 | 110.6 |
| 5 | 125.1 | 62.3 | |
| CuSO4 | 2 | 89.5 | 84.7 |
| 5 | 0 | 0 | |
| HgCl2 | 2 | 0 | 46.4 |
| 5 | 0 | 0 | |
| MnCl2 | 2 | 78.6 | 113.9 |
| 5 | 62.1 | 51.7 | |
| KCl | 2 | 124.2 | 106.4 |
| 5 | 121.1 | 92.1 | |
| NaCl | 2 | 127.8 | 101.4 |
| 5 | 121.4 | 100.3 | |
| ZnSO4 | 2 | 0 | 4.9 |
| 5 | 0 | 0 | |
Enzyme activity was determined at 45 °C in the presence of metal ions in the reaction mixture directly and for stability enzyme was pre-incubated with different metal ions at 37 °C for 1 h and assayed as standard assay method. The enzyme activity without incubation with metal ions was taken as 100%. Each value presented here is an average of triplicates of three independent trials. Mean standard deviation for all the values is < ± 5.0%.
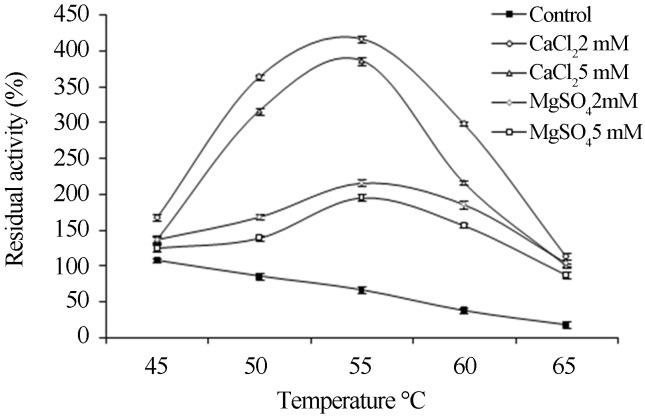
Owing to these results, the effects of calcium and magnesium ions (2 mM and 5 mM) on lipase activity at various temperatures (45–65 °C) were studied. Increase in lipase activity was observed with the increase in temperature and maximum activity was recorded at 55 °C which was 6.2 and 5.8-fold over the control in the presence 2 mM and 5 mM CaCl2, respectively (Figure 5). However, with Mg+2 ions the residual activities were 215.2% (2 mM) and 194.9% (5 mM), respectively at 55 °C. Further increase in reaction temperature resulted in decrease in enzyme activity with both the ions but it was still higher than their respective control. Alvarez and Stella (1989) suggested that the presence of calcium ions facilitated the binding of the enzyme to interface under physico-chemical conditions, resulting in the stimulation of enzyme activity. Our findings are in contrary to Karadzic et al. (2006) where inhibitory effect of calcium ion was reported on the san-ai lipase from P. aeruginosa. However, the present study is in agreement with Sharma et al. (2001), who also reported that Ca+2 ions activated the enzyme, whereas, Fe+3 and Zn+2 strongly inhibited its activity.
Figure 5.
Effect of CaCl2 and MgCl2 on lipase activity at different temperature. Enzyme activity was determined at various temperatures (45–65 °C) in the presence of CaCl2 (2 mM and 5 mM) and MgCl2 (2 mM and 5 mM). The enzyme activity assayed at respective temperatures without metal ions was taken as control.
Effect of organic solvents on enzyme stability
It is well known fact that organic solvents (10–20%) pose adverse effect on lipase activity (Karadzic et al., 2006), however, their effect differ from lipase to lipase (Hun et al., 2003). As shown in Table 3 the enzyme activity was enhanced in all the organic solvents, except iso-propanol, ethanol and xylene after 1 h and retained more than 100% of its activity up to 96 h of incubation. However, the enzyme activity decreased after prolonged incubation, except hexane, butanol, iso-octane, toluene and n-decane. The hydrophilic organic solvents pose more inhibitory effect on lipase activity as compared to hydrophobic solvents. However, interestingly the enzyme was able to hold considerable activity (83–109%) with both the types of organic solvents even after incubation of 168 h.
Table 3.
Stability of purified lipase in presence of various organic solvents
| Organic solvents (25%) | log P | Residual activity (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| 1 h | 24 h | 48 h | 72 h | 96 h | 120 h | 144 h | 168 h | ||
| Methanol | −0.76 | 101.4 | 130.1 | 132.2 | 125.3 | 118.6 | 111.4 | 106.3 | 98.3 |
| Iso-propanol | −0.28 | 90.2 | 101.3 | 99.3 | 92.4 | 91.5 | 90.4 | 85.3 | 83.4 |
| Ethanol | −0.24 | 97 | 107.6 | 106.5 | 101.6 | 98.5 | 95.4 | 90.2 | 87.3 |
| Acetone | −0.23 | 104.1 | 110.2 | 107.6 | 101.1 | 100.3 | 97.3 | 95.4 | 89.9 |
| Butanol | −0.80 | 120.7 | 160.3 | 148.2 | 135.4 | 126.6 | 118.3 | 110 | 103 |
| Toluene | 2.5 | 111.9 | 190.2 | 150.1 | 128.1 | 120.8 | 112.4 | 105.6 | 104.3 |
| Iso-octane | 2.9 | 123.2 | 170.1 | 135.5 | 130.5 | 123.4 | 120.3 | 115.7 | 109.2 |
| Xylene | 3.1 | 90.2 | 115.8 | 120.4 | 115.2 | 109.7 | 102.3 | 98.4 | 92.4 |
| Hexane | 3.6 | 132.4 | 151.7 | 140.3 | 137.4 | 121.4 | 120.1 | 112.4 | 103.3 |
| n-decane | 5.6 | 156.3 | 161.3 | 150.8 | 148.3 | 126.2 | 111.7 | 108.9 | 105.6 |
| n-dodecane | 6.0 | 151.3 | 153.2 | 146.3 | 141.1 | 112.9 | 108.2 | 102.1 | 98.8 |
Enzyme was pre-incubated with different organic solvents at a concentration of 25% (v/v) at 37 °C for different time period and assayed as standard assay method. The enzyme activity without incubation with organic solvent was taken as 100%. Mean standard deviation for all the values is < ± 5.0%.
The activation and stability effects of the organic solvent-tolerant lipase in aqueous-organic mixtures suggested the capability of this enzyme to resist denaturation by organic solvent and to form multiple hydrogen bonds with water for structural flexibility and conformational mobility for optimal catalysis (Klibnov, 2001). Eltaweel et al. (2005) also reported slight enhancement in lipase activity by hydrophobic solvents (Benzene, n-hexane) and reduction by hydrophilic solvents (1-propanol and ethylacetate).
Generally the enzymes showed considerable stability in only one type of solvent, either hydrophobic or hydrophilic. Therefore, stability of our lipase with respect to various organic solvents may be different and interesting to other organic solvent tolerant lipase with regard to its application in organic synthesis.
Effect of inhibitors on alkaline lipase stability
Inhibition studies primarily give an insight into the nature of an enzyme, its cofactor requirements, and the nature of the active center (Sigma and Mooser, 1975). The chelating agent (EDTA) slightly inhibited the lipase activity with 95.8% residual activity at a concentration of 2 mM, indicating that this lipase is not dependent on a metal cofactor. Our results are in agreement with Saeed et al. (2005) and Lin and Ko (2005) who also showed that activity of lipases produced from Bacillus sp. and Pseudomonas sp. were not affected by EDTA. However reducing agent, β-mercaptoethanol (0.1%) enhanced the lipase activity by 19.6% (Table 4). This can be explained as lipases contain very few sulfhydryl groups, which are essential for lipase activity (Gupta et al., 2004).
Table 4.
Effect of different inhibitors on enzyme stability.
| Reagents | Concentration | Residual activity (%) |
|---|---|---|
| Control | 100.0 | |
| β-mercaptoethanol | 0.1 (%) | 119.6 |
| 0.5 (%) | 98.6 | |
| EDTA | 2 mM | 95.8 |
| 5 mM | 86.7 | |
| Urea | 2 mM | 99.3 |
| 5 mM | 90.4 | |
| PMSF | 2 mM | 85.4 |
| 5 mM | 72.5 |
Enzyme was pre-incubated with different oxidizing agents/inhibitors at 37 °C for 1 h and assayed as standard assay method. The enzyme activity without incubation with inhibitors was taken as 100%. Each value presented here is an average of triplicates of three independent trials. Mean standard deviation for all the values is < ± 5.0%.
All known lipases have serine in their active centre; nonetheless, some lipases show resistance to inactivation by serine reactive agents (Abramic et al., 1999). The effect of PMSF at 2 mM concentration gave only a 15% reduction in the lipase activity possibly suggesting the presence of a hydrophobic lid hindering access to the catalytic site (Cote and Shareck, 2008). In accordance with our result, Choo et al. (1998) reported that lipase from Pseudomonas sp. strain B11-1 was not affected by PMSF.
The urea can reduce the enzyme activity and stability by impairing the enzyme structure by direct interaction with the enzyme or by indirect action through changing the properties of surrounding solvents (Enea and Jolicoeur, 1982). However, the enzyme exhibited noteworthy residual activity (99.3%) in the presence of 2 mM urea. In accordance to present study B. licheniformis and B. cepacia lipases were reported to be stable with a significant residual activity with urea (Chakraborty and Paulraj, 2008; Yu et al., 2009).
Effect of surfactants, commercial detergents and oxidizing agents on enzyme stability
As shown in Table 5 the purified enzyme was appreciably stable in the presence of non-ionic surfactants like Tween-40, Tween-60 and Tween-80. However, triton-X-100 slightly inhibited the lipase activity with 84.2% of residual activity at concentration 0.1% (v/v) which further decreased at concentration 1.0%. SDS was found to be a strong inhibitor causing almost complete inhibition of lipase activity at concentration 0.1% (w/v). SDS is known to acts upon the di-sulphide linkages and cause inactivation/denaturation of proteins (Liebeton et al., 2001) which could be the reason of complete inactivation of enzyme. Our results coincide with the results of Park et al. (2007) where complete loss of Burkholderia sp. HY-10 lipase activity was observed in the presence of SDS. Similarly, an extracellular lipase from Yarrowia lipolytica also lost its complete activity in the presence of SDS, however, activity was enhanced in the presence of 0.1% Tween-80 (Yu et al., 2007).
Table 5.
Effect of surfactants, commercial detergents and oxidizing agents on lipase stability.
| Surfactants | Concentration (%) | Residual activity (%) |
|---|---|---|
| Control | 100.0 | |
| Tween-40 | 0.1 | 101.0 |
| 1.0 | 98.6 | |
| Tween-60 | 0.1 | 105.7 |
| 1.0 | 99.1 | |
| Tween-80 | 0.1 | 100.4 |
| 1.0 | 95.5 | |
| Triton-X-100 | 0.1 | 84.2 |
| 1.0 | 78.8 | |
| SDS | 0.1 | 0.0 |
| Ghari | 0.1 | 94.1 |
| 1.0 | 65.2 | |
| Ariel | 0.1 | 84.7 |
| 1.0 | 51.3 | |
| Surf Excel | 0.1 | 81.6 |
| 1.0 | 45.6 | |
| Fena | 0.1 | 91.4 |
| 1.0 | 68.4 | |
| Henko | 0.1 | 89.2 |
| 1.0 | 55.9 | |
| Sodium perborate | 0.1 (%) | 75.8 |
| 0.5 (%) | 56.8 | |
| 1.0 (%) | 45.3 | |
| Sodium hypochlorite | 0.1 (%) | 111.9 |
| 0.5 (%) | 103.2 | |
| 1.0 (%) | 92.4 | |
| H2O2 | 0.1 (%) | 104.5 |
| 0.5 (%) | 96.4 | |
| 1.0 (%) | 85.2 |
Enzyme was pre-incubated with different surfactants, commercial detergents and oxidizing agents at 37 °C for 1 h and assayed as standard assay method. The enzyme activity without incubation with surfactants, commercial detergents and oxidizing agents was taken as 100%. Each value presented here is an average of triplicates of three independent trials. Mean standard deviation for all the values is < ± 5.0%.
The purified alkaline lipase was substantially stable with commercial detergents at lower concentration (0.1%, w/v). However, higher concentration (1.0%, w/v) led to decrease the enzyme activity. Rathi et al. (2001) also reported 57–80% residual activity with lipase from B. cepacia in the presence of commercial detergents. In another study, Bancerz et al. (2007) reported that lipase from Bjerkandera adusta R59 showed sufficient compatibility with commercial detergents.
Among the oxidizing agents tested the lipase activity enhanced in presence of sodium hypochlorite and H2O2 with residual activities 111.9 and 104.5% at concentration 0.1%, whereas, higher concentrations (0.5 and 1.0%) decreased the stability except sodium hypochlorite (103.2% residual activity at concentration 0.5%, w/v). Likewise, Wang et al. (2009b) also reported that lipase from B. cepacia was highly stable in the presence of hydrogen peroxide, sodium hypochlorite and sodium perborate after 1 h at 25 °C.
The stability profile of the lipase in the presence of detergents and oxidizing agents prove its potential application in the detergent formulations as these agents are the active components of house hold detergents (Hajji et al., 2007).
Kinetic study
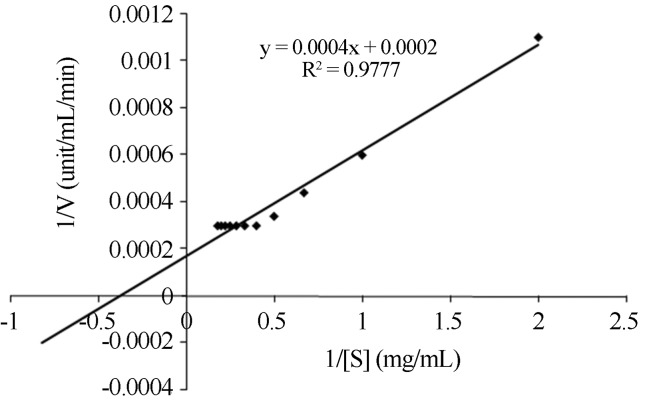
The Lineweaver-Burk plots were linear and indicated that hydrolysis of p-nitrophenyl palmitate by the lipase followed Michaelis-Menten kinetics. Results revealed that increase in substrate concentration from 0.5 to 2.5 mg.mL−1 showed exponential increase in lipase activity; beyond that it was constant which may be attributed to saturation of active sites of lipase enzyme. The Michaelis constant (Km, 2.0 mg.mL−1) and maximum velocity (Vmax, 5000 μg.mL−1.min−1) were determined from Lineweaver-Burk plots, for p-NPP as substrate (Figure 6). The lower apparent Km indicates that the purified lipase has high affinity for p-NPP.
Figure 6.
Lineweaver-burk plot of alkaline lipase of mutant strain of P. aeruginosa.
For a P. cepacia lipase, Pencreac’h and Baratti, (1996) reported Km and Vmax values of 12 mM and 30 mmol.min−1, respectively, when the substrate was p-NPP. For a lipase of R. glutinis, the Km values were 2.7 and 0.7 mM when the substrates were p-nitrophenyl butyrate and p-nitrophenyl laurate, respectively (Hatzinikolaou et al., 1999).
Conclusion
Conclusively, the extracellular alkaline lipase from mutant strain of P. aeruginosa has several properties of prominent industrial importance, in particular, pH and temperature stability. The features of this lipase viz., activity in alkaline pH, high temperature, resistance to many surfactants, and significant stability and compatibility with most of the tested commercial laundry detergents, demonstrating its feasibility for its inclusion in laundry detergent formulation and in food and pharmaceutical industries. It is anticipated that the organic solvent tolerant enzyme secreted by this species will also be applicable as catalysts for reaction in the presence of organic solvents.
Acknowledgments
Financial assistance to the researchers (Deepali Bisht) by University Grants Commission, New Delhi, in the form of Major Research Project is thankfully acknowledged. The assistance provided by the Government of Uttar Pradesh to the department under the scheme of Center of Excellence is duly acknowledged.
References
- Abramic M, Lescic I, Korica T, Vitale L, Saenger W, Pigac J. Purification and properties of extracellular lipase from Streptomyces rimosus. Enz Microb Technol. 1999;25:522–529. [Google Scholar]
- Aires-Barros MR, Taipa MA, Cabral JMS. Isolation and purification of lipases. In: Woolley P, Petersen SB, editors. Lipases: Their Structure, Biochemistry and Application. Cambridge Univ. Press; Cambridge: 1994. pp. 243–270. [Google Scholar]
- Alvarez FJ, Stella VJ. The role of calcium ions and bile salts on the pancreatic lipase-catalyzed hydrolysis of triglyceride emulsions stabilized with lecithin. Pharmacological Res. 1989;6:449–452. doi: 10.1023/a:1015956104500. [DOI] [PubMed] [Google Scholar]
- Bancerz R, Ginalska G. A noble thermostable lipase from Basidiomycete Bjerkandera adusta R59: characterization and esterification studies. J Ind Microbiol Biotechnol. 2007;34:553–560. doi: 10.1007/s10295-007-0232-6. [DOI] [PubMed] [Google Scholar]
- Bisht D, Yadav SK, Darmwal NS. Enhanced production of extracellular alkaline lipase by an improved strain of Pseudomonas aeruginosa MTCC 10,055. Am J Appl Sci. 2012;9:158–167. [Google Scholar]
- Bisht D, Yadav SK, Darmwal NS. Computation of interactive effects and optimization of process parameters for alkaline lipase production by mutant strain of Pseudomonas aeruginosa using response surface methodology. Braz J Microbiol. 2012;44:245–252. doi: 10.1590/S1517-83822013005000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkar PS, Bodade RG, Rao SR, Khobragade CN. Purification and characterization of extracellular lipase from a new strain-Pseudomonas aeruginosa SRT 9. Braz J Microbiol. 2009;40:358–366. doi: 10.1590/S1517-838220090002000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K, Paulraj R. An extracellular alkaline metallolipase from Bacillus lichenformis MTCC 6824: Purification and biochemical characterization. Food Chem. 2008;109:727–736. doi: 10.1016/j.foodchem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Choo DW, Kurihara T, Suzuki T, Soda K, Esaki N. A cold-adapted lipase of Alaskan psychrotroph, Pseudomonas sp. Strain B11-1: Gene cloning and enzyme purification and characterization. Appl Environ Microbiol. 1998;64:486–491. doi: 10.1128/aem.64.2.486-491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote A, Shareck F. Cloning, purification and characterization of two lipases from Streptomyces coelicolor A3(2) Enz Microb Technol. 2008;42:381–388. [Google Scholar]
- Eltaweel MA, Rahman RNZRA, Salleh AB, Basri M. An organic solvent-stable lipase from Bacillus sp. strain 42. Ann Microbiol. 2005;55:187–192. [Google Scholar]
- Enea O, Jolicoeur C. Heat capacities and volumes of several oligopeptides in urea-water mixtures at 25°C. Some implications for protein unfolding. J Phy Chem. 1982;86:3870–3881. [Google Scholar]
- Gopinath SCB, Hilda A, Lakshmi Priya T, Annadurai G, Anbu P. Purification of lipase from Geotrichum candidum: conditions optimized for enzyme production using Box-Behnken design. World J Microbiol Biotechnol. 2003;19:681–689. [Google Scholar]
- Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol. 2004;64:763–781. doi: 10.1007/s00253-004-1568-8. [DOI] [PubMed] [Google Scholar]
- Hajji M, Kanoun S, Nasri M, Gharsallah N. Purification and characterization of an alkaline serine-protease produced by a new isolated Aspergillus clavatus ES1. Process Biochem. 2007;42:791–797. [Google Scholar]
- Hatzinikolaou DG, Kourentzi E, Stamatis H, Christakopoulos P, Kolisis FN, Kekos D, Macris BJ. A novel lipolytic activity of Rhodotorula glutinis cells: production, partial characterization and application in the synthesis of esters. J Biosci Bioeng. 1999;88:53–56. doi: 10.1016/s1389-1723(99)80175-3. [DOI] [PubMed] [Google Scholar]
- Hun CJ, Rahman RNZA, Salleh AB, Basri M. A newly isolated organic solvent tolerant Bacillus sphaericus 205 y producing organic solvent-stable lipase. Biochem Eng J. 2003;15:147–151. [Google Scholar]
- Izrael-Zivkovic LT, Gojgic-Cvijovic GD, Gopcevic KR, Vrvic MM, Karadzic IM. Enzymatic characterization of 30 kDa lipase from Pseudomonas aeruginosa ATCC 27853. J Basic Microbiol. 2009;49:452–462. doi: 10.1002/jobm.200800229. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Reetz MT. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998;16:396–403. doi: 10.1016/s0167-7799(98)01195-0. [DOI] [PubMed] [Google Scholar]
- Ji Q, Xiao S, He B. Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J Mol Catal B Enzymat. 2010;66:264–269. [Google Scholar]
- Karadzic I, Masui A, Zivkovic LI, Fujiwara N. Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metalworking fluid. J Biosci Bioeng. 2006;102:82–89. doi: 10.1263/jbb.102.82. [DOI] [PubMed] [Google Scholar]
- Klibnov AM. Improving enzyme by using them in organic-solvents. Nature. 2001;409:241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- Kumarevel T, Gopinath S, Hilda A, Gautham N, Ponnusamy M. Purification of lipase from Cunninghamella verticillata by stepwise precipitation and optimized conditions for crystallization. World J Microbiol. 2005;21:23. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage. T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liebeton K, Zacharias A, Jaeger KE. Disulfide bond in Pseudomonas aeruginosa lipase stabilizes the structure but is not required for interaction with its foldase. J Bacteriol. 2001;183:597–603. doi: 10.1128/JB.183.2.597-603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ES, Ko HC. Glucose stimulates production of the alkaline-thermostable lipase of the edible Basidiomycete Antrodia cinnamomea. Enz Microb Technol. 2005;37:261–265. [Google Scholar]
- Lowry OH, Rosenbrough MJ, Farr AL, Randell RJ. Protein measurement with folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Macrae AR, Hammond RC. Present and future applications of lipases. Biotech Genet Eng Rev. 1985;3:193–217. [Google Scholar]
- Malathu R, Chowdhury S, Mishra M, Das S, Moharana P, Mitra J, Mukhopadhyay UK, Thakur AR, Chaudhuri SR. Characterization and wash performance analysis of microbial extracellular enzymes from east Calcutta wetland in India. Am J Appl Sci. 2008;5:1650–1661. [Google Scholar]
- Park DS, Oh HW, Heo SY, Jeong WJ, Shin DH, Bas KS, Park HY. Characterization of an extracellular lipase in Burkholderia sp. HY-10 isolated from a longicorn beetle. J Microbiol. 2007;45:409–417. [PubMed] [Google Scholar]
- Pencreac’h G, Baratti JC. Hydrolysis of p-nitrophenyl palmitate in n-heptane by Pseudomonas cepacia lipase: a simple test for the determination of lipase activity in organic media. Enz Microb Technol. 1996;18:417–422. [Google Scholar]
- Rahman RNZRA, Baharum SN, Basri M, Salleh AB. High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Ann Biochem. 2005;341:267–274. doi: 10.1016/j.ab.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Rathi P, Saxena RK, Gupta R. A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochem. 2001;37:187–192. [Google Scholar]
- Saeed HM, Zaghloul TI, Khalil AI, Abdelbaeth MT. Purification and Characterization of Two Extracellular Lipases from Pseudomonas aeruginosa Ps-x. Pol J Microbiol. 2005;54:233–240. [PubMed] [Google Scholar]
- Sangeetha R, Arulpandi I, Geetha A. Bacterial lipases as potential industrial biocatalysts: An overview. Res J Microbiol. 2011;6:1–24. [Google Scholar]
- Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization and applications of lipases. Biotechnol Adv. 2001;19:627–662. doi: 10.1016/s0734-9750(01)00086-6. [DOI] [PubMed] [Google Scholar]
- Shrinivas PK. Industrial enzymes. Reference and Education: Science. 2008 http://ezinearticles.com/?Industrial-Enzymes&id=890345.
- Sifour M, Saeed HM, Zaghloul TI, Berekaa MM, Abdel-Fattah YR. Purification and properties of a lipase from thermophilic Geobacillus stearothermophilus strain-5. Inter J Biol Chem. 2010;4:203–212. doi: 10.1016/j.nbt.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Sigma DS, Mooser G. Chemical studies of enzyme active sites. Annu Rev Biochem. 1975;44:889–931. doi: 10.1146/annurev.bi.44.070175.004325. [DOI] [PubMed] [Google Scholar]
- Singh S, Banerjee UC. Purification and characterization of trans-3(4-methoxyphenyl) glycidic acid methyl ester hydrolyzing lipase from Pseudomonas aeruginosa. Process Biochem. 2007;42:1063–1068. [Google Scholar]
- Taipa MA, Aires-Barros MR, Cabral JMS. Purification of lipases. J Biotechnol. 1992;26:111–142. doi: 10.1016/0168-1656(92)90001-p. [DOI] [PubMed] [Google Scholar]
- Vulfson EN. Industrial applications of lipases. In: Wooley P, Petersen SB, editors. Lipases. Cambridge University Press; Cambridge, Great Britain: 1994. p. 271. [Google Scholar]
- Wang SL, Lin YT, Liang TW, Chio SH, Ming LJ. Purification and characterization of extracellular lipase from Pseudomonas monteilii tku009 by the use of soybeans as the substrate. J Ind Microbiol Biotechnol. 2009a;36:65–73. doi: 10.1007/s10295-008-0473-z. [DOI] [PubMed] [Google Scholar]
- Wang X, Yu X, Xu Y. Homologous expression, purification and characterization of a novel high-alkaline and thermal stable lipase from Burkholderia cepacia ATCC 25416. Enz Microb Technol. 2009b;45:94–102. [Google Scholar]
- Winkler UK, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Xu Y, Yu X. Purification and properties of a highly enantioselective L-menthyl acetate hydrolase from Burkholderia cepacia. J Mol Catal B Enzy. 2009;57:27–33. [Google Scholar]
- Yu M, Qin S, Tan T. Purification and characterization of the extracellular lipase Lip 2 from Yarrowia lipolytica. Process Biochem. 2007;42:384–391. [Google Scholar]