Abstract
Bacteria from the genus Methylobacterium interact symbiotically (endophytically and epiphytically) with different plant species. These interactions can promote plant growth or induce systemic resistance, increasing plant fitness. The plant colonization is guided by molecular communication between bacteria-bacteria and bacteria-plants, where the bacteria recognize specific exuded compounds by other bacteria (e.g. homoserine molecules) and/or by the plant roots (e.g. flavonoids, ethanol and methanol), respectively. In this context, the aim of this study was to evaluate the effect of quorum sensing molecules (N-acyl-homoserine lactones) and plant exudates (including ethanol) in the expression of a series of bacterial genes involved in Methylobacterium-plant interaction. The selected genes are related to bacterial metabolism (mxaF), adaptation to stressful environment (crtI, phoU and sss), to interactions with plant metabolism compounds (acdS) and pathogenicity (patatin and phoU). Under in vitro conditions, our results showed the differential expression of some important genes related to metabolism, stress and pathogenesis, thereby AHL molecules up-regulate all tested genes, except phoU, while plant exudates induce only mxaF gene expression. In the presence of plant exudates there is a lower bacterial density (due the endophytic and epiphytic colonization), which produce less AHL, leading to down regulation of genes when compared to the control. Therefore, bacterial density, more than plant exudate, influences the expression of genes related to plant-bacteria interaction.
Keywords: endophyte, Methylobacterium, quantitative PCR (qPCR), plant-bacteria Interaction, homoserine
Introduction
Endophytes colonize the plant inner tissues, commonly coming from the soil and entering the plants by intercellular spaces or little root fissure. The genus Methylobacterium is constituted by methylotrophic bacteria, which are able to interact symbiotically with different plant species, where they occupy mainly the inner tissues of the host, as endophytes. In agriculture, it has been described that M. nodulans is involved with nitrogen fixation and nodulation of plants (Sy et al., 2003), while other species can promote plant growth or induce systemic resistance (Araújo et al., 2002; Lacava et al., 2004; Madahaiyan et al., 2006). In citrus, it has been shown that endophytic Methylobacterium can interact with Xylella fastidiosa, the causal agent of citrus variegate clorose (CVC) (Araújo et al., 2002; Lacava et al., 2004). Gai et al. (2009) manipulated M. mesophilicum SR1.6/6 to express the green fluorescence protein (GFP) in C. roseus, and observed that the colonization and transmission of M. mesophilicum by Bucephalogonia xanthophis, the X. fastidiosa insect-vector. Their results proved that M. mesophilicum is really transmitted by the insect to host plant living in the same niche of X. fastidiosa inside of C. roseus. Thus, the authors proposed the M. mesophilicum as a candidate to symbiotic control of X. fastidiosa due to this intimate association of SR1.6/6 with the plant, it can be suggested that this bacterium can act in plant, influencing the microbial balance in the plant host and participating on the plant development.
The occurrence of plant colonization by pathogenic or non-pathogenic bacteria is dependent of a communication between plants and microorganisms (Rosenblueth and Martinez-Romero, 2006; Hardoim et al., 2008; Wang et al., 2010), which is believed to be made by plant exudation of 17 compounds, such as flavonoids (Hardoim et al., 2008), ethanol (Williams and Yavitt, 2009) and methanol (Sudtachat et al., 2009; Sy et al., 2005; Jourand et al., 2005) that attracts microorganisms. Bacterial communication during the plant-interaction can be coordinated physiological and adaptive changes in their population (Cha et al., 1998), favoring its adaptation to specific environments (Bauer and Mathesius, 2004; Joint, 2006; Soto et al., 2006; Sanches-Contreras et al., 2007). Some quorum sense (QS) systems use N-acyl-homoserine lactones (AHLs) as signaling molecules, commonly found in Gram-negative bacteria that living in association with plants (Cha et al., 1998; Loh et al., 2002; Camilli and Bassler, 2006; Sanches-Contreras et al., 2007; Barnard et al., 2007; White and Winans, 2007). QS system allows bacteria to function as multicellular organisms, because the extracellular concentration of auto-inducer increases with bacteria population growth, after reaches a determinate number, this molecules diffuse back into the bacteria and regulate the transcription of different genes that may be related with the secretion of virulence factors, biofilm formation, sporulation, exchange of DNA and others (Zhu and Sun, 2008).
Although several studies demonstrate the importance of the association between Methylobacterium-plants (Pirttila et al., 2000; Sy et al., 2001, Lee et al., 2006; Madhaiyan et al., 2009a; 2009b; Dourado et al., 2012a,b) and that members of the Methylobacterium genus produces AHL (Penãlver et al., 2006; Pomini et al., 2009; Poonguzhali et al., 2007), little is known about the role of plant exudates and AHL on the expression of bacterial genes that are involved in bacterium plant-interaction. Studies reported that biofilm production seems to be dependent of the production of AHL (Penãlver et al., 2006), and that biofilm formation on the plant may be the first step towards endophytic colonization by Methylobacterium (Andreote et al., 2006; Rossetto et al., 2011). Despite of the difficulty to understand the complexity of mechanisms involved in plant-microbe interactions, some bacterial genes responsible for metabolism, stress defence and pathogenicity that present an important role on plant bacterial interaction are described. The mxaF gene, responsible for the methylotrofic metabolism, encodes a subunit of the dehydrogenase methanol enzyme (MDH) (Zhang and Lindstrom, 2003) its expression confers adaptive advantages in competitive conditions on the plant surface (Willians and Yavitt, 2009), increasing methylotrophic activity during symbioses (Sy et al., 2005).
The genes phoU and sss are transport genes, where phoU gene is responsible mainly for the phosphate homeostasis regulation, but also controls and interferes on stress, antibiotic production (Li and Zhang, 2007; Gristwood et al., 2009) and virulence gene expression (Cheng et al., 2009) and the sss (sodium solute symporter) gene is responsible for the symport transport of solute with the sodium (Scier, 1998). Genes crtI and acdS genes are associated to the stress response (Sandmann, 2009) and to plant metabolism (Hardoim et al., 2008). Phytoene dehydrogenase gene (crtI) codifies an enzyme that catalyses the denaturation reaction resulting on the lycopene synthesis that protects the cell against oxidative damages, different types of radiations and dissections (Xu et al., 2007) and the acdS gene codifies the enzyme carboxylic acid aminocyclopropane deaminase that degrades ACC (1-carboxilate amineciclopropane), forerunner of plant ethylene (Glick et al., 2007; Hardoim et al., 2008).
The patatin gene is associated to the pathogenic bacteria and is activated during the pathogeneses process, the patatin family genes encode phospholypases enzymes that hydrolyze phospholipids, frequently used by pathogenic bacteria on the effective host colonization hydrolyzing the membrane phospholipids, resulting in a membrane damage and cytotoxicity (Camera et al., 2009), the phoU gene is also involved in the bacteria pathogeneses process (Cheng et al., 2009).
Thus, the aim of the present study was to evaluate the expression of several genes previously related to plant-bacteria and bacteria-bacteria interaction (mxaF, phoU, crtI, acdS, sss and patatin) using as model an endophytic bacterium M. mesophilicum SR1.6/6, quorum sense molecules (AHL (S)-N-dodecanoyl HSL-extracted from this isolate) and rice and eucalyptus plants as well ethanol as rooting exudate carbon source.
Material and Methods
Microorganism and plant material
The selected bacterial strain SR1.6/6, M. mesophilicum, was previously isolated from Citrus sinensis (Araújo et al., 2002). The SR1.6/6 was routinely cultivated at 28 °C in CHOI3 medium (Toyama et al., 1998). The bacterium is stored in dilute liquid CHOI3 supplemented with glycerol at −80 °C. Fresh cultures were started from glycerol stocks for each experiment by plating portions onto CHOI3. The plant hosts used in this study were rice (Oryza sativa) and eucalyptus (Eucalyptus citriodora) seedlings. The axenic seedlings of rice and eucalyptus were obtained from seeds previously disinfected, washing in 70% ethanol for 2 min, sodium hypochlorite solution (2% available Cl−) for 6 min, and again 70% ethanol for 2 min for rice seeds and washed for 1 min in 70% ethanol, 2 min in sodium hypochlorite solution and 1 min in 70% ethanol for eucalyptus seeds, both seeds followed by two rinses in sterile distilled water. The seeds were germinated on the MS medium (Ramakers et al., 2003) in a wet chamber at 28 °C under 16 hours of photoperiod.
AHL (S)-N-dodecanoyl-HSL assay
The AHL (S)-N-dodecanoyl-HSL was purified from the same bacterial strain (SR1.6/6 - M. mesophilicum) and synthesized in Chemistry Institute of Unicamp University (Pomini et al., 2009). To minimize the levels of endogenous AHL in M. mesophilicum SR1.6/6, the bacteria was initially cultivated in 25 mL of CHOI3 medium (Toyama et al., 1998) at 28 °C to O.D.600 of 0.5. Then the cells were pelletized, washed in PBS buffer (10 mM Na2HPO4; 2 mM KH2PO4; 3 mM KCl; 140 mM NaCl; pH 7.4), inoculated into 50 mL of CHOI3 medium and grown at 28 °C to O.D.600 of 0.5. Proceeding as before, the culture was centrifuged; the pellet cells were washed in PBS buffer and transferred to 100 mL of CHOI3 medium supplemented with the amount of AHL naturally produced by this isolated (0.1 g/mL) (Pomini et al., 2009). The treated cultures were incubated at 28 °C to transition to stationary phase (18 hours; O.D.600 −1.7). The experiment was performed in three biological replicates. Triplicate control cultures no added AHL were also grown under the same conditions to RNA extraction.
Root exudates role in the gene expression
After seven and twenty days of rice and eucalyptus respectively disinfected seeds germination, the seedlings were removed from pots and inoculated with M. mesophilicum SR1.6/6 cell by adding 5 mL of bacterial suspension (108 UFC mL−1) on CHOI3 medium (Toyama et al., 1998). Two assays were developed, to observe the biotic factors, all the inoculums were done in liquid MS medium (Murashige and Skoog, 1962), in the first assay SR1.6/6 was inoculated with rice or eucalyptus seedlings and to accesses the effects of carbon source SR1.6/6 in CHOI3, using methanol or ethanol on this medium. All treatments were constituted by three biological repetitions and the incubation period before RNA extraction was 48 hours.
Total RNA isolation and cDNA synthesis
Bacteria cells were collected from all treatment describe above by centrifugation (15 min at 6000 g), and the RNA was isolated from the bacterial pellet according to the Invitrogen protocol (TRIzol, Invitrogen). The RNA was ressuspended in 30 μL of dietyl-pirocarbonate (DEPC) treated water and stored at −80 °C. The integrity and amount of extracted RNA was verified in a denaturant agarose gel 1.2% in FA 1X (MOPS 200 mM, sodium acetate 50 mM and EDTA 10 mM), containing formaldehyde (0.7%) and ethidium bromide (0.3 g.mL−1). The RNA was quantified by O.D.260, measured in spectrophotometer NanoDrop ND-1000 (Thermo Scientific, USA). All material used obtain and treat the RNA was sterilized and/or treated with DEPC to eliminated RNAse. Total RNA (1–10 μg) was reverse-transcribed into cDNA using random hexamer primers (Invitrogen) and 200 U SuperscriptII RNAse H- reverse transcriptase (Invitrogen) according to the procedure supplied with the enzyme. For each RNA sample, a negative RT (no addition of reverse transcriptase) was performed and used as a negative control in subsequent PCRs.
Primers designing and validation
The construction of the primers to amplify the target genes in this study were designed based on the six genomes sequences of Methylobacterium genus (M. extorquens DM4, M. extorquens AM1, M. chloromethanicum CM4, M. populi BJ001, M. nodulans ORS 2060, M. radiotolerans JCM 2831) available at GenBank database (National Center for Biotechnology Information). Sequences from all genes were aligned, and primers were designed, with the software Primer 3 program (v. 0.4.0) (http://frodo.wi.mit.edu/), to anneal in conserved regions of the gene (Table 1). Firstly, the conventional PCR was used to validate the design primers. PCRs were performed in 25 μL reaction containing 1 X enzyme buffer, 3.75 mM of MgCl2, 0.2 mM of each dNTPs, 0.2 μM of each primer and 0.1 U/μL of Taq DNA Polymerase (Invitrogen, Brazil). An initial denaturation was carried out at 94 °C for 5 min, followed by 35 thermal cycles of 30 s at 95 °C, 1 min at 59 °C and 1 min at 72 °C, with a final extension performed at 72 °C for 5 min. All PCR amplificons were checked by electrophoresis on agarose gel (1.5% w/v agarose) and UV visualization of the ethidium bromide stained gels.
Table 1.
Sequencing of designed primers to evaluate the gene expression of the endophytic bacterium M. mesophilicum SR1.6/6 in plants.
| Primer | Target gene | Sequencing (5′-3′) | Fragment length | Reference |
|---|---|---|---|---|
| MxaFqPCRAF | mxaF | CGTCAACGTCATGATGCT(C/G)T | 250 pb | This study |
| MxaFqPCRAR | GATGTCCTTGGCGAG(A/G)TG | This study | ||
| ACC Met1 f | acdS | GACCGGGTCGGCAACATC | 200 pb | This study |
| ACC Met2 r | AGCCCGCCGTACTTGTGC | This study | ||
| PatatinF | Patatin | CTTCAACGCCAACCTGATG | 250 pb | This study |
| PatatinR | CCGATCCGCTCGTAGTTCT | This study | ||
| PhyF | crtI | AATACTTCAAGCCGGTGCTG | 186 pb | This study |
| PhyR | GACATGCCGAGGTACTTGGT | This study | ||
| sssF | sss | ATCGACGCCCTGTACAATTC | 221 pb | This study |
| sssR | ACCGTCGCGTAGTTCGAC | This study | ||
| phoUF | phoU | TTCGACGGGCTGATCTACTC | 189 pb | This study |
| phoUR | GATCAGGTAGAAGGCCACCA | This study | ||
| recAF | recA | CGAACTGCATGGTC(G)ATCTTC | 232 pb | This study |
| recAR | ATGTCGAACTCGACCTGCTT ′ | This study | ||
| zwfrF | zwf | AGCAGCTGGAACATGTGGTT | 231 pb | This study |
| zwfrR | CGACGAGAGCCAGTTCTACC | This study | ||
| rpoDrF | rpoD | ACGACCTCGAGAACAACGTC | 229 pb | This study |
| rpoDrR | ACGACCTCGAGAACAACGTC | This study | ||
| proCF | proC | CCAGCAGGAAGACGTAGGC | 282 pb | This study |
| proCR | ACACGCTCCTCGTCTCGAT | This study | ||
| MMC1 | 16S rRNA | TACGTGGAGAGATTCACGGTC | 390 pb | Lacava et al. (2006) |
| MMC2 | GTACAAGGCCCGGGAACGTAC | Lacava et al. (2006) |
The specificity of each primer pair was tested in 17 bacterial strains of Methylobacterium genus, and also in strains affiliated to Rhizobium, Sinorizobium and Bacillus genera (Table 2). All amplification products of the seven genes (mxaF, acdS, crtI, patatin, phoU, sss and recA) were purified, sequenced and compared to the GenBank data by BLASTn (http://blast.ncbi.nlm.nih.gov/) confirming the identity of the amplified fragment.
Table 2.
Isolates of Methylobacterium spp. and other genera from different plant hosts, identified by partial 16S rRNA gene sequencing, and used to evaluate primers specificity.
| Isolate | Host plant | Genes | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| mxaF | acdS | Patatin | crtI | phoU | sss | recA | ||
| Methylobacterium sp. TC3–6 | Coffee | + | − | + | − | + | + | + |
| Methylobacterium sp. TC3–7 | Coffee | + | − | + | − | + | + | + |
| M. fujisawaense F5 | Sugarcane | + | − | + | − | + | + | + |
| Methylobacterium sp. F7 | Sugarcane | + | − | + | + | + | + | + |
| Methylobacterium sp. F8 | Sugarcane | + | − | − | − | + | + | + |
| Methylobacterium sp. F9 | Sugarcane | + | + | + | − | + | + | + |
| M. mesophilicum SR1.6/6 | Citrus | + | + | + | + | + | + | + |
| Methylobacterium sp. SR3/27 | Citrus | + | − | − | − | + | + | + |
| M. mesophilicum PR1/3 | Citrus | + | + | − | − | + | + | − |
| Methylobacterium sp. PR3/11 | Citrus | + | + | + | + | + | + | − |
| Methylotrophyc bacteria TP7 | Sweet peper | + | + | + | + | + | + | + |
| M. hispanicum TP8 | Sweet peper | + | − | + | − | + | + | + |
| Methylobacterium spp. R3E | Eucalyptus spp. | + | − | − | − | − | + | + |
| Methylobacterium spp. MA2.9 | Laguncularia racemosa | + | + | + | − | + | + | + |
| Methylobacterium spp. MA3.1 | Aviscenia shaueriana | + | + | − | + | + | + | + |
| Methylobacterium spp. MB1.1 | Rhizophora mangle | + | − | + | − | + | − | + |
| Methylobacterium spp. MB1.3 | Rhizophora mangle | + | − | − | − | − | − | − |
| Rhizobium SP | Laboratory collection | − | + | − | − | − | − | − |
| Sinorhizobium SP | Laboratory collection | − | − | − | − | − | − | − |
| Bacillus cereus | Laboratory collection | − | − | − | − | − | − | − |
| Total of 20 | 17 | 8 | 11 | 5 | 15 | 15 | 14 | |
RT qPCR analysis
All the amplification reactions by qPCR used the thermocycler iQTM5 (Bio-Rad) programmed to an initial denaturation of 5 min at 94 °C, followed by 40 cycles of 15 seconds at 94 °C and 1 min at 62 °C. The specificity of qPCR primer sets were evaluated by the melting curve with gradient from 60 to 96 °C ranging 1 °C each 30 s. Each amplification reaction it was used 2 μL of cDNA (100 ng), 10 μM of each primer and the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen). The primer-efficiencies were calculated through LinRegPCR software program (Ramakers et al., 2003).
The selection of endogenous gene, to be used as a normalizer was made by testing the Ct values for five different genes (16S rRNA, recA, zwf, rpoD e proC) (Stevenson and Weimer, 2005). Thereby, the recA presented the lowest deltaCt and the higher amplification efficiency, resulting in its selection as the normalizer for gene expression analysis.
The Pfaffl method (Pfaffl, 2001) was used to determine a relative quantification of the target genes in comparison to the reference gene. The DNA recombination and repair protein - recA gene was used as a reference gene.
Results and Discussion
The designed primers (Table 1) were specific for 17 Methylobacterium isolates exhibiting PCR products with expected size. However, these fragments were not observed from Sinorhizobium sp., Rhizobium sp. and Bacillus cereus, except for the primer that amplifies acdS gene, which amplified the target gene from Rhizobium sp. (Table 2). Therefore, these primers were used to evaluate the effects of bacteria gene expression under the following conditions: 1. AHL (S)-N-dodecanoyl-HSL; 2. Rice root exudates; 3. Eucalyptus root exudates and 4. Ethanol as carbon source.
No differences were observed between the bacterial growth with and without AHL (S)-N-dodecanoyl-HSL as well in the assay with ethanol as carbon source. However, there were less growth in the presence of the plant, due to bacteria colonization. During experimental period all plants exhibited normal growth and healthy aspects, with no lesions or chlorotic spots. In general, bacterial cell growth actively in culture medium with plant roots and after 48 hours some cells were observed attached on root surface. Taking in mind that one of our objective was to evaluate the differential gene expression in response to AHL, root exudates and carbon source, and that the planktonic cells should vary the gene expression response when attached to the root when plant-associated, in the present study only planktonic cells of M. mesophilicum SR1.6/6 under the mentioned conditions were accessed.
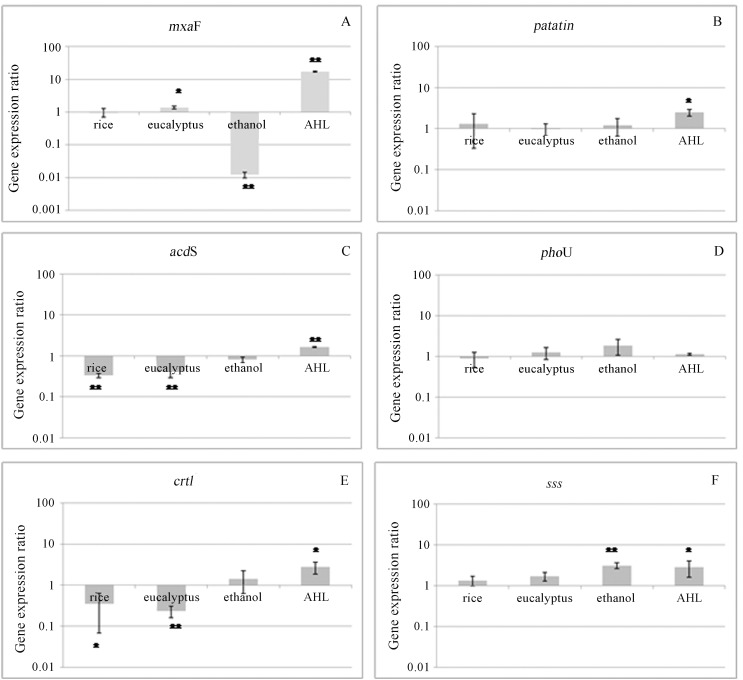
Looking specifically to each analyzed gene, the gene mxaF was over-expressed (more than 10 times) in the presence of AHL, and it was repressed only during the bacterial incubation in medium with ethanol as the carbon source (Figure 1A), the bacteria of eucalyptus treatments presented an increase in gene expression, but no significant differences occurred between the gene expression of M. mesophilicum in control and rice host.
Figure 1.
Relative effects of ethanol, plant exudates and AHL on SR1.6/6 transcription of plant-interaction genes: mxaF (A), patatin(B), acdS (C), phoU (D), crtI (E) and sss (F). The gene expression ration were determined by the method of Pfaffl. The recA as a reference gene. The results are means +/-standard errors of three replicates. Values of asterisks (* or **) differ statistically of control treatment (a = 0.05 and 0.01 respectively) according to t test of Student.
The mxaF gene codifies an enzyme responsible for the transformation of methanol on formaldehyde (McDonald et al., 2005; Zhang and Lindstrom, 2003), and it was over-expressed in the presence of AHL showing that this gene responds to quorum sensing therefore, in the presence of ethanol the expression of this gene was repressed.
On leaves, the production and liberation of methanol is readily metabolized by M. mesophilicum, fact that could confer competitive advantage to the bacterium over other bacteria that do not present such metabolic alternative (Sudtachat et al., 2009; Sy et al., 2005; Jourand et al., 2005), different from rhizosphere, where (considering plant treatments) no significant differences occurred between the gene expression of M. mesophilicum in control and plant hosts, considering the conditions found in this study, where the bacterium was not under competition with other microorganisms, it is possible to generate the hypothesis that the expression of mxaF gene is stimulated mainly by quorum sensing molecules (produced by itself or other bacteria), and in a competing condition, produces mxaF gene as an advantage.
The expression of genes related to plant metabolism and bacterial stress (acdS and crtI genes, respectively) presented the same pattern. It was not affected by ethanol amendment, however it was super expressed in the presence of AHL and repressed in bacterial-rice and Eucalyptus interaction (Figure 1C and E). Considering the genes roles, it can be suggested that on the evaluated conditions, the bacteria did not feel under stressing conditions while in contact with plant roots and its exudates. It is believed that the product of acdS gene is responsible for increasing the plant capacity to support physic and biological stresses. Tittabutr et al. (2008) showed that the introduction of multiples copies of acdS gene increased the enzyme ACC deaminase activity in Rhizobium sp and enhances its symbiotic efficiency on plant host (L. leucocephala). Besides that, it was demonstrated that in some rhizobia strains the acdS gene is regulated by the promoter responsible for the transcription activation of nif genes, which encode for nitrogen fixation (Glick et al., 2007). Therefore, with ACC deaminase enzyme activity there is an increase in plant growth (Cheng et al., 2009).
In this context, Madahaiyan et al. (2006) described that Methyhlobacterium is capable of inducing systemic resistance in rice and sugarcane. However, considering that in the presence of AHL increases acdS expression and in the presence of both plants reduces acdS gene expression, suggesting that this gene is regulated by quorum sense (AHL), and probably the treatment of planktonic bacteria with the plant presented lower bacteria cell density, consequently less AHL, than planktonic bacterial cell without plant, despite of the same initial inoculums, in the presence of the plant, some bacterial colonizes plants (endophytically and epiphytically) (Andreote et al., 2006; Rossetto et al., 2011).
In this way, the results presented suggests that the expression of phytoene dehydrogenase gene (crtI), responsible for the lycopene synthesis that protects the cell against oxidative stress (Xu et al., 2007), in the evaluated conditions were similar to acdS gene. It is associated to the fact that during the bacterium-plant interaction, the microbial defense system is probably not activated due the fact that the plant does not recognize the endophytic bacterium as a pathogen (Figure 1E).
In studied conditions, the expression of genes related to pathogenicity (patatin and phoU genes) are not differently expressed in the presence of plant exudates or ethanol, and it seems to be not regulated by plant interaction. But in the presence of AHL patatin gene is induced, but does not influence the phoU gene expression (Figure 1B and D).
The gene phoU is a transport gene, responsible for stress control (Gristwood et al., 2009; Li and Zhang et al., 2007) and bacteria pathogeneses process (Cheng et al., 2009). While patatin gene is a phospholypases enzymes that hydrolyze phospholipids, activated during the pathogeneses process to hydrolyze the membrane phospholipids (Camera et al., 2009). The expression of patatin and phoU genes, both related to bacterial pathogenicity, do not varies in the presence of plant exudates (Figure 1B and D), corroborating with previous studies that state M. mesophilicum as an endophyte able to colonize plants without cause damages to the host (Barnerji et al., 2008). Also, the absence on the expression variation of these two genes in the different treatments shows that although present, the regulation of such genes in this endophytic bacterium does not function as their homologous found in pathogenic bacteria. However, these pathogenicity genes respond differently to AHL. It induces only patatin expression and does not influence phoU gene.
The sss (sodium solute symporter) gene responsible for the symport transport of solute with the sodium, in studied conditions, seems to be not regulated by plant interaction (Figure 1F). However, the sss gene expression was induced in medium containing ethanol and in the presence of AHL (Figure 1F).
This study allowed a better understanding of the endophytic gene expression during plant recognition and planktonic bacterial cell interaction with rice and eucalyptus, the effect of ethanol (root exudates) and the effect of quorum sense molecule (AHL (S)-N-dodecanoyl-HSL). It was observed that homoserine lactone induces all analyzed genes mxaF, acdS, crtI, sss and patatin, expect for phoU gene (it does not influence its expression). It was observed that the mxaF gene was not induced in rice, only in eucalyptus, possibly due to the differences on plant-bacteria interaction and on using different metabolic routes, also suggesting that the plant can induce the expression of this gene and indirectly increase the bacteria fitness during the plant host interaction.
In addition, the plant-related environment was stated as a free-of-stress niche for bacterial colonization, based on the analysis of the genes crtI, acdS and phoU expression. Hence, such symbiotic interaction was confirmed by the absence of induction for gene related to pathogenesis characteristics (patatin and phoU). Future studies might reveal differences in specific genes, modulated by the plant genotype, what will add information about the differential behavior of bacteria, according to the host plant. Concluding, we remark that more than plant exudates, bacterial density influences the expression of genes related to metabolism, stress and pathogenesis.
Acknowledgments
This work was supported by a grant from the FAPESP - Foundation for Research Assistance of São Paulo State, Brazil (Proc. 2003/14143-3; Proc. 2010/07594-5). We thank CNPq for the Fellowship to M.N.D. and A.C.B. and also FAPESP for the fellowship to M.C.Q. (Proc. 2005/53748-6).
References
- Andreote FD, Lacava PT, Gai CS, Araújo WL, Maccheroni W, Jr, Overbeek LSV, Elsas JDV, Azevedo JL. Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa. NRC Research Press. 2006;52:419–426. doi: 10.1139/w05-142. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Marcon J, Maccheroni W, Jr, Elsas JDV, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plant. Appl Environ. Microbiol. 2002;68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Aurass P, Flieger A. The manifold phospholipases A of Legionella pneumophila -identification, export, regulation, and their link to bacterial virulence. Int J Med Microbiol. 2008;298:169–181. doi: 10.1016/j.ijmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Barnard AML, Bowdwn SD, Burr T, Coulthurst SJ, Monson RE, Salmond GPC. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Phil Trans R Soc B. 2007;362:1165–1183. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer WD, Mathesius U. Plant responses to bacterial quorum sensing signals. Curr Opin Plant Biol. 2004;7:429–433. doi: 10.1016/j.pbi.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Bogacki P, Oldach KH, Williams KJ. Expression profiling and mapping of defense response genes associated with the barley-Pyrenophora teres incompatible interaction. Mol Plant Pathol. 2008;9:645–660. doi: 10.1111/j.1364-3703.2008.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camera S, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, Roby D, Heitz T. The Arabidopsis Patatin-Like Protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol Plant Microbe Interact. 2009;22:469–481. doi: 10.1094/MPMI-22-4-0469. [DOI] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- Cheng C, Tennant SM, Azzopardi KI, Bennett-Wood V, Hartland EL, Robins-Browne RM, Tauschek M. Contribution of the pst-phoU operon to cell adherence by atypical enteropathogenic Escherichia coli and virulence of Citrobacter rodentium. Infect Immun. 2009;77:1936–1944. doi: 10.1128/IAI.01246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Duan J, Hao Y, Mcconkey BJ, Glick BR. Identification of Bacterial Proteins mediating the interactions between Pseudomonas putida UW4 and Brassica napus (Canola) Mol Plant Microbe Interact. 2009;22:686. doi: 10.1094/MPMI-22-6-0686. [DOI] [PubMed] [Google Scholar]
- Dourado MN, Ferreira A, Araújo WL, Azevedo JL, Lacava PT. The diversity of endophytic methylotrophic bacteria in an oil-contaminated and an oil-free mangrove ecosystem and their tolerance to heavy metals. Biotechnol Res Int. 2012a;2012:759865. doi: 10.1155/2012/759865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourado MN, Andreote FD, Dini-Andreote F, Conti R, Araújo JM, Araújo WL. Analysis of 16S rRNA and mxaF genes revealing insights into Methylobacterium niche-specific plant association. Genet Mol Biol. 2012b;35:142–148. doi: 10.1590/s1415-47572012005000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai CS, Lacava PT, Quecine MC, Auriac MC, Lopes JR, Araújo WL, Miller TA, Azevedo JL. Transmission of Methylobacterium mesophilicum by Bucephalogonia xanthophis for paratransgenic control strategy of citrus variegated chlorosis. J Microbiol. 2009;47:448–454. doi: 10.1007/s12275-008-0303-z. [DOI] [PubMed] [Google Scholar]
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, Mcconkey B. Promotion of plant growth by bacterial ACC Deaminase. CRC Crit Rev Plant Sci. 2007;26:227–242. [Google Scholar]
- Gristwood T, Fineran PC, Everson L, Williamson NR, Salmond GP. The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 2009;9:112. doi: 10.1186/1471-2180-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, Overbeek LSV, Elsas JDV. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Holmes DE, Nevin KP, O’neil RA, Ward JE, Adams LA, Woodard TL, Vrionis HA, Lovley DR. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71:6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joint I. Bacterial conversations: talking, listening and eavesdropping. A NERC Discussion Meeting held at the Royal Society on 7 December 2005. J R Soc Interface. 2006;3:459–463. doi: 10.1098/rsif.2006.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourand P, Renier A, Rapior S, Faria SM, Prin Y, Galiana A, Giraud E, Dreyfus B. Role of methylotrophy during symbiosis between Methylobacterium nodulans and Crotalaria podocarpa. Mol Plant Microbe Interact. 2005;18:1061–1068. doi: 10.1094/MPMI-18-1061. [DOI] [PubMed] [Google Scholar]
- Lacava PT, Araújo WL, Marcon J, Maccheroni W, Jr, Azevedo JL. Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa, casual agent of citrus-variegated clorosis. Lett Appl Microbiol. 2004;39:55–59. doi: 10.1111/j.1472-765X.2004.01543.x. [DOI] [PubMed] [Google Scholar]
- Lee HS, Madhaiyan M, Kim CW, Choi SJ, Chung KY, Sa TM. Physiological enhancement of early growth of rice seedlings (Oryza sativa L.) by production of phytohormone of N2-fixing methylotrophic isolates. Biol Fert Soils. 2006;42:402–408. [Google Scholar]
- Li Y, Zhang Y. PhoU is a Persistence Switch Involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob Agents Chemother. 2007;51:2092–2099. doi: 10.1128/AAC.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Carlson RW, York WS, Stacey G. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc Natl Acad Sci USA. 2002;99:144–146. doi: 10.1073/pnas.222336799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madahaiyan M, Reddy BVS, Anandham R, Senthilkumar M, Poonguzhali S, Sundaram SP, Sa TM. Plant growth-promoting Methylobacterium induces defense responses in groundnut (Arachis hypogaea L.) compared with rot pathogens. Curr Microbiol. 2006;53:270–276. doi: 10.1007/s00284-005-0452-9. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Kwon SW, Sa TM. Methylobacterium phyllosphaerae sp. Nov., a pink pigmented, facultative methylotroph from the phyllosphere of rice. Int J Syst Evol Micr. 2009;59:22–27. doi: 10.1099/ijs.0.001693-0. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Poonguzhali S, Senthilkumar M, Sundaram SP, Sa T. Nodulation and plant-growth promotion by methylotrophic bacteria isolated from tropical legumes. Microbiol Res. 2009;164:114–120. doi: 10.1016/j.micres.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Mcdonald IR, Radajewski S, Murrell JC. Stable isotope probing of nucleic acids in methanotrophs and methylotrophs: A review. Org Geochem. 2005;36:779–787. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Penãlver CGN, Morin D, Cantet F, Saurel O, Milon A, Vorholt JA. Methylobacterium extorquens AM1 produces a novel type of acyl homoserine lactone with a double insaturated side chain under methylotrophic growth conditions. FEBS Lett. 2006;580:561–567. doi: 10.1016/j.febslet.2005.12.062. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirttila AM, Laukkanen H, Pospiech H, Myllyla R, Hohtola A. Detection of intracellular bacteria in the buds of scotch pine (Pinus Sylvestris L.) by in situ hybridization. Appl Environ Microbiol. 2000;66:3073–3077. doi: 10.1128/aem.66.7.3073-3077.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomini AM, Cruz PLR, Gai C, Araújo WL, Marsaioli AJ. Long-Chain Acyl-Homoserine Lactones from Methylobacterium mesophilicum: Synthesis and Absolute Configuration. J Nat Prod. 2009;72:2130–2134. doi: 10.1021/np900043j. [DOI] [PubMed] [Google Scholar]
- Poonguzhali S, Madhaiyan M, Sa T. Production of acyl-homoserine lactone quorum-sensing signals is widespread in gram-negative Methylobacterium. J Microbiol Biotech. 2007;17:226–233. [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time PCR data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rosenblueth M, Martinez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- Rossetto PB, Dourado MN, Quecine MC, Andreote FD, Araújo WL, Azevedo JL, Pizzirani-Kleiner AA. Specific plant induced biofilm formation in Methylobacterium species. Braz J Microbiol. 2011;42:878–883. doi: 10.1590/S1517-83822011000300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanches-Contreras M, Bauer WD, Gao M, Robinson JB, Downie A. Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Phil Trans R Soc B. 2007;362:1149–1163. doi: 10.1098/rstb.2007.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann G. Evolution of carotene desaturation: The complication of a simple pathway. Arch Biochem Biophys. 2009;483:169–174. doi: 10.1016/j.abb.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Scier MH. Proposed independent evolution of different channel and carrier families. In: Poole RK, editor. Advanced in Microbial Physiology. Academic press; London: 1998. pp. 81–136. [Google Scholar]
- Soto MJ, Sanjuán J, Olivares J. Rhizobia and plant-pathogenic bacteria: common infection weapons. Microbiology. 2006;152:3167–3174. doi: 10.1099/mic.0.29112-0. [DOI] [PubMed] [Google Scholar]
- Stevenson DM, Weimer PJ. Expression of 17 Genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl Environ Microbiol. 2005;71:4672–4678. doi: 10.1128/AEM.71.8.4672-4678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudtachat N, Ito N, Itakura M, Masuda S, Eda S, Mitsui H, Kawaharada Y, Minamisawa K. Aerobic vanillate degradation and C1 compound metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 2009;75:5012–5017. doi: 10.1128/AEM.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Giraud E, Jourand P, Garcia N, Willems A, De Lajudie P, Prin Y, Neyra M, Gillis M, Bivin-Masson C, Dreyfus B. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol. 2001;183:214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Timmers ATJ, Knief C, Vorholt JA. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microbiol. 2005;71:7245–7252. doi: 10.1128/AEM.71.11.7245-7252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittabutr P, Awaya JD, Li QX, Borthakur D. The cloned 1-aminocyclopropane-1-carboxylate (ACC) deaminase gene from Sinorhizobium sp. strain BL3 in Rhizobium sp. strain TAL1145 promotes nodulation and growth of Leucaena leucocephala. Syst Appl Microbiol. 2008;31:141–150. doi: 10.1016/j.syapm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Toyama H, Anthony C, Lidstrom ME. Construction of insertion and deletion mxa mutants of Methylobacterium extorquens AM1 by electroporation. FEMS Microbiol Lett. 1998;166:1–7. doi: 10.1111/j.1574-6968.1998.tb13175.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu W, Chen X, Tang C, Dong Y, Ma J, Huang X, Wei G, Han Q, Huang L, Kang Z. Differential gene expression in incompatible interaction between wheat and stripe rust fungus revealed by cDNA-AFLP and comparison to compatible interaction. BMC Plant Biol. 2010;10:9. doi: 10.1186/1471-2229-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EW, Winans SC. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Phil Trans R Soc B. 2007;362:1135–1148. doi: 10.1098/rstb.2007.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Yavitt JB. Temperate wetland methanogenesis: The importance of vegetation type and root ethanol production. Soil Sci Soc Am J. 2009;74:317–325. [Google Scholar]
- Xu Z, Tian B, Sun Z, Lin J, Hua Y. Identification and functional analysis of a phytoene desaturase gene from the extremely radioresistant bacterium Deinococcus radiodurans. Microbiology. 2007;153:1642–1652. doi: 10.1099/mic.0.2006/002857-0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Lidstrom ME. Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology. 2003;149:1039–1040. doi: 10.1099/mic.0.26105-0. [DOI] [PubMed] [Google Scholar]
- Zhu H, Sun SJ. Inhibition of bacterial quorum sensing-regulated behaviors by Tremella fuciformis extract. Curr Microbiol. 2008;57:418–422. doi: 10.1007/s00284-008-9215-8. [DOI] [PubMed] [Google Scholar]