Abstract
Many pathologies can be identified by evaluating differences raised in the physical parameters of involved tissues. In a Magnetic Resonance Imaging (MRI) framework, spin-lattice T1 and spin-spin T2 relaxation time parameters play a major role in such an identification. In this manuscript, a theoretical study related to the evaluation of the achievable performances in the estimation of relaxation times in MRI is proposed. After a discussion about the considered acquisition model, an analysis on the ideal imaging acquisition parameters in the case of spin echo sequences, i.e., echo and repetition times, is conducted. In particular, the aim of the manuscript consists in providing an empirical rule for optimal imaging parameter identification with respect to the tissues under investigation. Theoretical results are validated on different datasets in order to show the effectiveness of the presented study and of the proposed methodology.
Keywords: Magnetic Resonance Imaging, complex decomposition, statistical signal processing, Cramer, Rao lower bounds, relaxation time estimation
1. Introduction
Relaxation times define the rate of spin magnetic equilibrium recovery in nuclear magnetic resonance (NMR) [1,2]. For each tissue, several relaxation times can be defined. Besides, the main interest is in the evaluation of two of them: the spin-lattice and the spin-spin relaxation times, commonly referred to as T1 and T2, respectively. Such time constants, together with the hydrogen nuclei abundance, ρ, define the behavior of the signal generated by each resolution element.
It is largely known that the knowledge of relaxation times can provide interesting information about imaged tissues. Concerning the medical diagnostic field, many pathologies have been found to involve a significant variation of the relaxation time constants more than a variation of ρ, such as Alzheimer's disease [3], Parkinson's disease [4] and cancer [5,6]. The evaluation of the tissue relaxation times can be considered an excellent tool for improving clinical diagnosis.
Classic approaches for retrieving relaxation parameter maps of imaged tissue slices propose the estimation of T1 and T2 separately. In particular, the “gold standard” for spin-lattice relaxation time T1 estimation exploits inversion recovery (IR) sequences [7,8]. However, this approach is too slow for in vivo clinical applications. Different evolutions have been proposed in the literature. In particular, the exploitation of spoiled gradient-recalled echo (SPGR) sequences has shown interesting results [9,10]. With respect to spin-spin relaxation time T2 estimation, a widely used imaging sequence is the spin echo (SE) [11,12].
The magnitude of the acquired signal is typically used for relaxation parameter estimation [12–15]. Within this framework, the exponential curve fitting via the least squares (LS) algorithm is the commonly adopted estimator [11,13]. Although being very easy to be implemented and not computationally heavy, it has the disadvantage of producing biased estimations [11,16]. The alternative consists in using a maximum likelihood estimator (MLE) [12]. The MLE is asymptotically unbiased and optimal, but the function to be maximized, which is related to the statistical distribution of the MRI amplitude data, is computationally heavy, as it contains the Bessel function [17].
Recently, new approaches based on the complex decomposition of acquired data have been proposed [10,18]. The exploitation of the complex model leads to a main advantage concerning the estimation: due to the circular Gaussian distribution of the complex noise, the LS-based estimator coincides with the MLE and is asymptotically unbiased and optimal.
While much effort has been directed to improving the estimation procedures, only a little effort has been directed to the choice of the optimal imaging parameter selection (i.e., the optimal choice of the MRI scanner imaging parameters). In particular, in [19], the ideal repetition times have been investigated in the case of saturation recovery spin-lattice measurements at 4.7 T, while in [20], the optimization of T2 measurements in the case of bi-exponential systems is considered. Following the approach proposed by [15], within this paper we investigate the possibility of finding the optimal imaging parameter configuration for relaxation time estimation. As an alternative to [15], we investigate the optimal configuration not only for the T2 time estimation, but for the joint T2 and T1 estimation.
Since the SE sequence-based model allows the simultaneous estimation of both spin-spin and spin-lattice relaxation times, we focus our attention on this imaging sequence. In any case, the theoretical study reported in the following could be easily adapted to different imaging sequences. Considering an SE sequence [2], the two imaging parameters involved in the acquisition procedure are the repetition time, TR, and the echo time, TE. We briefly recall that these two parameters allow the scanner to differently interact with tissues characterized by different T1 and T2 values. By exploiting different TR and TE combinations, it is possible to emphasize the effect of one tissue intrinsic parameter with respect to others, obtaining the well-known ρ-weighted, T1-weighted or T2-weighted images.
Given the previously mentioned motivations, we present a deeper analysis of the complex SE model considered in [18] extended to three parameters (i.e., ρ, T1 and T2). The analysis is conducted exploiting the Cramer–Rao lower bounds (CRLBs) [16]. Since CRLBs provide the best achievable performances in the unbiased estimation of one or more parameters, by minimizing them with respect to the MR scanner imaging parameters, it is possible to find the optimal acquisition configuration for the relaxation time estimation. Practically speaking, we look for the acquisition parameters that allow achieving lower relaxation time estimation errors. The result of the study is the introduction of a general empirical rule for determining the optimal (with respect to CRLBs) MRI scanner parameter configuration. In particular, the identification of these parameters in the case of several tissues has been conducted. The effectiveness of the theoretical results and of the empirical rule is validated and verified on different datasets.
The manuscript is organized as follows: in Section 2, the acquisition model for an MRI spin echo sequence is presented, and in Section 3, the achievable performances of the estimation are analyzed via the CRLBs. In Section 4, the CRLB-based empirical rule for the optimal acquisition parameter configuration is presented. Finally, validation on different datasets is presented in Section 5, and conclusions are drawn.
2. The Model
Let us consider an MRI acquisition system using a spin echo imaging sequence. The amplitude of the recorded complex signal after the image formation process, i.e., after the computation of the 2D Fourier transform, is related to the tissue parameters, ρ, T1 and T2, via [2,21]:
| (1) |
where TE and TR are the echo and repetition time, respectively, which are two imaging parameters that can be set in the MRI scanner, and θ = [ρ T1 T2]T is the vector containing the tissue parameters in which we are interested. Note that Equation (1) is valid in the case of a homogeneous imaged object. In the case of clinical data, the presence of different hydrogen environments within each voxel has to be taken into account. The acquisition model reported in Equation (1), which is a solution to Bloch equations, assuming that TE is short with respect to TR, is related to the noise-free case and does not take into account the dependency on the static magnetic field, B. Considering noise, in the complex domain, the model becomes:
| (2) |
where nR and nI are the real and imaginary parts of the noise samples, which are distributed as independent circularly Gaussian variables [22], and ϕ represents the angle of the complex data [23,24]. Thus, the statistical distributions of the real and imaginary parts of the acquired signal are:
| (3) |
where σ2 is the variance of real and imaginary noise components. Due to the independence of the real and imaginary parts of noise, the joint statistical distribution of yR and yI is the product of the two probability density functions of Equation (3).
Once N acquisitions with different TR and TE combinations have been recorded and collected in the data vector y = [yR, yI], with yR = [yR(1), ⋯ yR(N)] and yI = [yI(1), ⋯ yI(N)], we can derive the likelihood function from the factorization of the Probability Density Functions (PDFs):
| (4) |
Starting from the likelihood function of Equation (4), the CRLBs for θ are derived and analyzed in the following sections.
3. Cramer-Rao Lower Bounds Evaluations
In order to evaluate the performances of the optimal estimator for the model presented in Section 2, the Cramer–Rao lower bounds have to be computed. According to Statistical Estimation Theory [16], given an observation model, the accuracy of any estimator can be evaluated according to its mean and its variance. In particular, in order to be optimal, an estimator needs to have its mean equal to the value to be estimated (i.e., unbiased estimator) and to have the smallest possible variance. CRLBs represent the lower bound of the variance of any unbiased estimator, resulting an interesting and powerful tool for evaluating the achievable performances of a considered model. By computing the CRLBs for different configuration of the parameters involved in the acquisition model, it is possible to find the best parameter configuration, the one that provides the minimum values of CRLBs (i.e., the minimum achievable variances). Considering the vector parameter θ, the minimum variance that any unbiased estimator of parameter θi can reach is provided by the i-th diagonal element of the inverse of matrix I [16]:
| (5) |
with I being the Fisher matrix, which is equal to:
| (6) |
where E [·] is the expected value operator.
A closed form for the second order derivatives of Equation (6) has been derived and reported in the Appendix. The closed form greatly improves the computational accuracy of the CRLB evaluation and decreases the computational burden of the simulations reported in the following.
In order to experimentally compute the matrix of Equation (6), Monte Carlo simulations with 105 iterations have been considered for statistical average computation.
For the following evaluations, we considered a tissue, named A, with parameters θ = [ρ T1 T2]T = [2.5 1600 90]T. Note that within the paper, all relaxation times are expressed in milliseconds, while the proton density is in percentage. The following simulations are reported and analyzed in order to investigate CRLB dependency and behavior with respect to the signal-to-noise ratio (SNR), the number of acquisitions and the scanner acquisition parameters.
3.1. CRLB vs. SNR
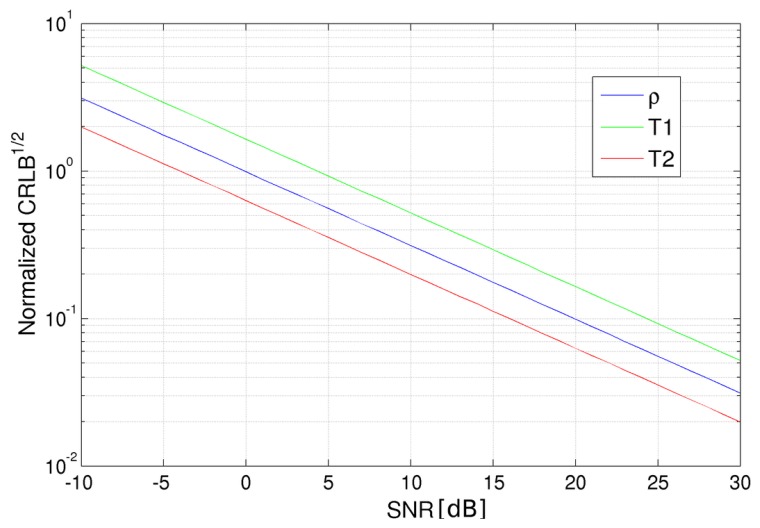
Let us start by computing CRLBs varying the noise standard deviation (i.e., the SNR). Sixteen images have been considered, which refer to the all combinations of four TR and four TE values equally spaced in the intervals [500, 3500] ms and [80, 350] ms, respectively. Note that the lower TE value has been set according to the minimum echo time for the SE sequence accepted by the Philips Achieva 3.0 T, the MR scanner we worked on, while the maximum value of TR has been set in order to limit the global acquisition time. The CRLBs in the case of different SNRs are shown in Figure 1. As expected, the square root of the CRLBs related to all considered parameters decreases with respect to SNR growth, i.e., high SNRs positively affect the estimator performances. In the considered range of SNRs, no saturation appears. Very similar behaviors are obtained varying TR and TE combinations. Globally, it can be stated that SNR linearly affects CRLBs, so in the following the results of each simulation can be easily extended to any SNR configuration.
Figure 1.
Square root of the Cramer–Rao lower bound (CRLB) for proton density (blue), spin-lattice (T1) relaxation time (green) and spin-spin (T2) relaxation time (red) for different signal-to-noise ratio values expressed in decibels (logarithmic scale). CRLB values have been normalized for the parameter values in order to be plotted in the same graph.
3.2. CRLB vs. the Number of Acquisitions
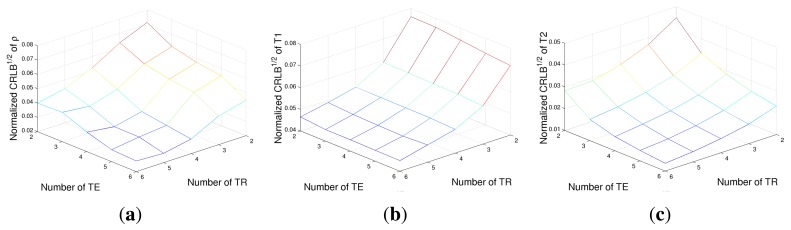
A second case study has been conducted in order to evaluate the advantage of increasing the number of acquisitions. Two vectors of TR and TE, of a length of NR and NE, respectively, have been generated in the [500, 3500] ms (for TR) and [80, 350] ms (for TE) intervals. The square root of CRLBs, i.e., the minimum achievable standard deviations, are reported in Figure 2 for ρ, T1 and T2, respectively, for different NR and NE combinations. The noise variance has been fixed in order to obtain an SNR of 16 dB for the image with the lowest signal intensity (i.e., TR = 500 ms and TE = 80 ms). It can be noted that the number of TR values mainly affects the achievable performances with respect to T1 estimation, while CRLBs of ρ and T2 are dependent on the number of both TE and TR values, with a higher dependency on echo times. The results confirm the strict connections between TR and T1 and also between TE and T2, as expected from the exponential terms of the SE signal model (Equation (1)). However, it is interesting to stress how the CRLB of ρ is very tightly related to TE values rather than to TR ones.
Figure 2.
The square root of the CRLB of proton density ρ (a), T1 relaxation time (b) and T2 relaxation time (c) for different numbers of acquisitions.
3.3. CRLB vs. TR and TE Values
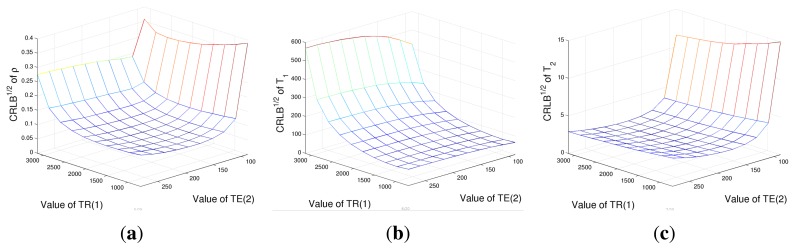
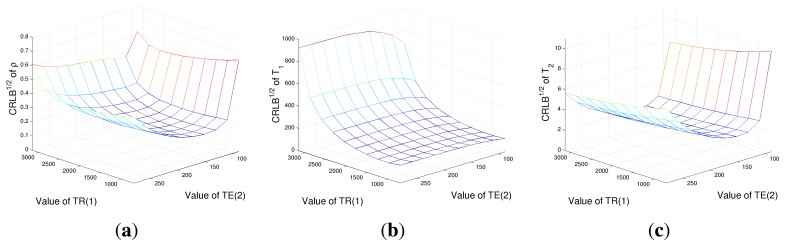
As a further case study, an evaluation of CRLBs with respect to TR and TE values with a fixed number of acquisition has been performed. Four acquisitions have been considered, corresponding to all the combinations of TR = [TR(1), TR(2)] and TE = [TE(1), TE(2)]. CRLBs have been computed while varying TR(1) and TE(2) and considering TR(2)= 3, 500 ms and TE(1) = 80 ms, again in the case of tissue A parameters. Results are reported in Figures 3. Figure 4a shows that the ρ estimation would prefer low TR (1) and high TE(2) values. The behaviors of CRLBs for T1 and T2 differ remarkably from Figure 4a, as it can be noticed that the estimation of T1 is almost unresponsive with respect to TE(2) values, as far as T2 estimation with respect to TR(1). In particular, for the estimation of T1, the ideal TR(1) is as low as possible, while the ideal TE(2) for the estimation of T2 is between 150 and 250 ms. For this experiment, a second dataset has also been considered: the same simulation has been conducted in the case of a second tissue, named B, with parameters θ = [ρ T1 T2] T = [2.8 1800 60]T, in order to know if the results of Figure 3 are always valid or if they are highly dependent on the considered tissue. The results are reported in Figure 4. It can be noticed that the lower regions remain in the same position, although being increased in value, but for CRLBs of ρ and T2, the ideal TE(2) range reduces to [130, 180] ms. This is mainly due to the lower T2 value of tissue B with respect to tissue A. Thus, it can be concluded that the general trend is confirmed, although the position of the global minimum is strictly related to the considered tissue. These two simulations show that the choice of optimal parameters is strictly dependent on the relaxation times of the imaged tissues. In the next section, we investigate the possibility of finding a rule for ideal imaging parameter identification.
Figure 3.
The square root of the CRLB of proton density ρ (a), T1 relaxation time (b) and T2 relaxation time (c) for different combinations of TR and TE values in the case of ρ = 2.5, T1 = 1,600 ms and T2 = 90 ms.
Figure 4.
The square root of the CRLB of proton density ρ (a), T1 relaxation time (b) and T2 relaxation time (c) for different combinations of TR and TE values in the case of ρ = 2.8, T1 = 1,800 ms and T2 = 60 ms.
4. Optimal Parameter Configuration
After the evaluation of ρ, T1 and T2 CRLB behaviors, an analysis dedicated to the computation of optimal TR and TE combinations is presented. In the following, it will be shown that a proper imaging configuration can greatly improve the performances with respect to such a choice. In particular, the aim of this section is to identify the ideal imaging parameters with respect to imaged tissues.
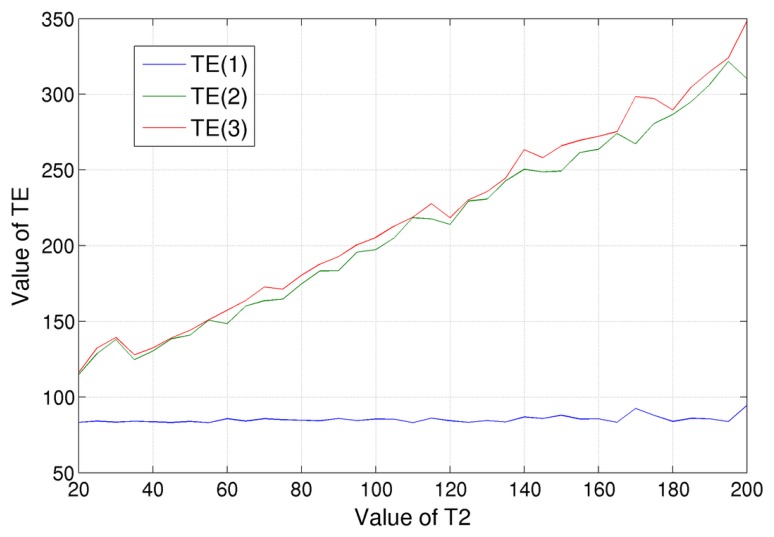
Let us show how the optimal imaging parameters can be determined. Initially, we have focused on the minimization of T2 CRLB, which consist in finding TE values that minimize the element (3, 3) of the inverse of the Fisher matrix, I(θ), of Equation (6) for different spin-spin relaxation times T2. The optimization has been performed by searching the three optimal TE values in the [82, 350] ms range for a fixed value of TR. The evaluation has been done varying the tissue T2 relaxation times in the [20, 200] ms range, obtaining the results shown in Figure 5. Some considerations can be drawn:
there is no TE value combination that is simultaneously ideal for tissues with different spin-spin relaxation times. As a consequence, we can only find the TE combination that is ideal for a specific tissue;
by analyzing Figure 5, it can be noticed that the lowest TE value of the ideal configuration is always equal to the lower bound of the considered variability range, which, in our case, was fixed to 82 ms. As stated before, this value is the minimum echo time for the SE sequence accepted by the Philips Achieva 3.0 T, the MR scanner we worked on;
the two higher TE values, which are the red and the green lines of Figure 5, practically coincide. This can be explained considering that we are interested in the estimation of relaxation times, i.e., of decay rates. In order to achieve such a goal, it is crucial that the measurement of the signal decrease, i.e., the ratio of the signal acquired in two different echo times. Therefore, instead of values TE, it is only important the difference between them. A third echo time, TE(3), equal to TE(2), allows us to compute twice the signal decay, which is the quantity in which we are interested;
the red and the green lines of Figure 5 show a clear trend: their values grow linearly when increasing T2. In particular, we found that the distance with the blue line (i.e., lowest TE, 82 ms) is a little bit bigger than the value of the considered spin-spin relaxation time, T2. For example, in the case of T2 = 100 ms, the ideal echo times were TE = [83, 197, 205] ms; the last two values are approximately 110% of (TE(1) + T2). By considering the other simulated T2 values, we found that this coefficient is 110% ± 10%. Within this range, the CRLB of T2 can be considered constant.
Figure 5.
TE values that minimize the CRLB of T2 for tissues with different spin-spin relaxation times, T2. Three values have been considered: the blue line is for the lowest TE value, the red line for the highest one and green for the intermediate one.
From these simulations, we can derive an empirical rule for the optimal TE selection: the lower one should be fixed to the minimum value accepted by the MR scanner, while the other values should to be set in the range of 100%–120% of the value of (TE(1) + T2), considering the T2 of the tissue in which we are interested.
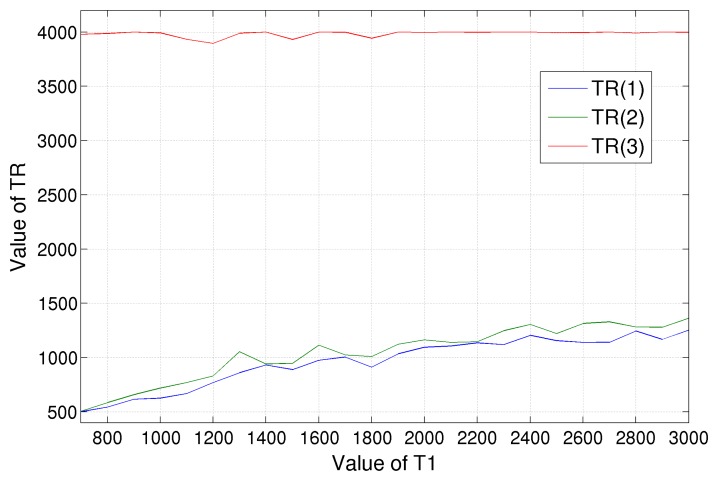
A similar evaluation has been conducted for the minimization of T1 CRLB varying MRI scanner repetition times TR, with a fixed value of TE; the results are shown in Figure 6. The higher TR value is fixed to the right edge of the considered variability range, which we set equal to 4, 000 ms. The intermediate and low TRs have similar values, which, starting from 500 ms in the case of tissue with T1 = 700 ms, grow almost linearly up to 1, 400 ms for tissues with higher T1 (about 3, 000 ms). It is hard to determine an empirical rule in this case; anyway, we can say that a choice of around 1, 000 ms for TR(1) and TR(2) will fit a wide class of tissues, i.e., those with 1, 200 < T1 < 2, 000 ms.
Figure 6.
TR values that minimize the CRLB of T1 for tissues with different spin-lattice relaxation times, T1. Three values have been considered: the red line is for the highest TR value, the blue line for the lowest one and green for the intermediate one.
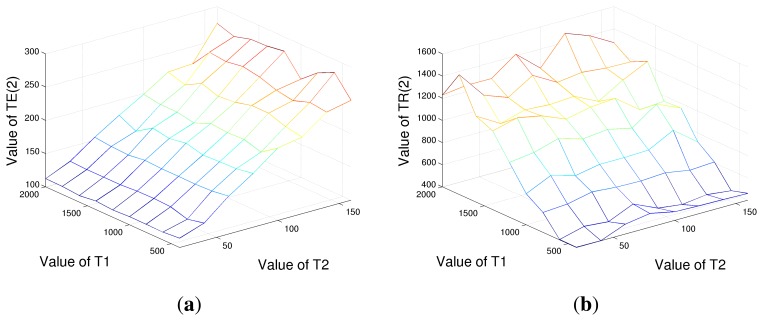
Concluding this section, one more evaluation has been conducted. Instead of optimizing TR and TE values separately, a joint minimization has been done. Nine acquisitions have been considered, related to three repetition and three echo times. Among the three values, the lower and the higher have been fixed to the search range bounds, so only the intermediate TE and TR values were variable. Results are shown in Figure 7, respectively. It is evident from the figure that TE values can be considered independent from T1, as far as TR from T2, proving the correctness of the separate optimization of the echo and repetition times. In particular, from Figure 7a, we can state that tissues with equal T2, but very different T1 values share the same three optimal echo times for T2 estimation, and vice versa. That said, the behaviors of Figures 5 and 6, i.e., the minimization, one parameter at a time, are confirmed.
Figure 7.
Optimal TE(2) (a) and TR(2) (b) values considering nine acquisitions in the case of tissues with different T1 and T2 relaxation times. It can be noticed that the TE(2) value is substantially independent from tissue spin-lattice relaxation time T1, as far as TR(2) from spin-spin relaxation time T2.
In order to easily apply the obtained results, the ideal acquisition parameters for different tissues have been computed exploiting CRLB minimization in the case of a 1.5 T and a 3 T MRI scanner. The results are shown in Tables 1 and 2 for T1 and T2, respectively. According to the results reported in Figure 7, the minimizations have been computed independently for spin-lattice and spin-spin relaxation time estimation. Tissue relaxation times have been simulated according to reference values present in the literature [25], which are reported in Table 3.
Table 1.
Optimal repetition times, TR, for T1 estimation in case of different tissues and numbers of acquisitions at 1.5 T and 3 T.
| Tissue | 1.5T | 3T | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| 2 images | 3 images | 4 images | 2 images | 3 images | 4 images | |
| liver | 490; 4000 | 490; 510; 4,000 | 380; 490; 510; 4,000 | 650; 4,000 | 570; 650; 4,000 | 570; 570; 650; 4,000 |
| skeletal muscle | 720; 4,000 | 720; 720; 4,000 | 680; 720; 720; 4,000 | 1,090; 4,000 | 990; 1,090; 4,000 | 870; 990; 1,090; 4,000 |
| heart | 840; 4,000 | 770; 840; 4,000 | 770; 790; 840; 4,000 | 1,060; 4,000 | 910; 1,060; 4,000 | 770; 910; 1,060; 4,000 |
| kidney | 570; 4,000 | 430; 570; 4,000 | 430; 460; 570; 4,000 | 910; 4,000 | 790; 910; 4,000 | 750; 790; 910; 4,000 |
| cartilage | 760; 4,000 | 690; 760; 4,000 | 630; 690; 760; 4,000 | 880; 4,000 | 770; 880; 4,000 | 770; 780; 880; 4,000 |
| white matter | 690; 4,000 | 690; 710; 4,000 | 640; 690; 710; 4,000 | 850; 4,000 | 780; 850; 4,000 | 730; 780; 850; 4,000 |
| gray matter | 920; 4000 | 840; 920; 4000 | 800; 840; 920; 4,000 | 1,150; 4,000 | 980; 1,150; 4,000 | 910; 980; 1,150; 4,000 |
| optic nerve | 960; 4,000 | 960; 1,060; 4,000 | 960; 1,060; 1,100; 4,000 | 970; 4,000 | 910; 970; 4,000 | 910; 970; 1,030; 4,000 |
| spinal cord | 600; 4,000 | 550; 600; 4,000 | 450; 550; 600; 4,000 | 760; 4,000 | 660; 760; 4,000 | 660; 700; 760; 4,000 |
| blood | 1,120; 4,000 | 840; 1,120; 4,000 | 830; 840; 1,120; 4,000 | 1,120; 4,000 | 1,040; 1,120; 4,000 | 1,030; 1,040; 1,120; 4,000 |
Table 2.
Optimal echo times TE for T2 estimation in case of different tissues and numbers of acquisitions at 1.5 T and 3 T.
| Tissue | 1.5T | 3T | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| 2 images | 3 images | 4 images | 2 images | 3 images | 4 images | |
| liver | 82; 134 | 82; 134; 138 | 82; 134; 138; 146 | 82; 134 | 82; 134; 134 | 82; 134; 134; 142 |
| skeletal muscle | 82; 130 | 82; 130; 138 | 82; 130; 138; 1;400 | 82; 132 | 82; 132; 144 | 82; 132; 144; 146 |
| heart | 82; 124 | 82; 124; 134 | 82; 124; 134; 136 | 82; 132 | 82; 132; 140 | 82; 132; 140; 148 |
| kidney | 82; 158 | 82; 158; 168 | 82; 158; 168; 188 | 82; 144 | 82; 144; 154 | 82; 144; 154; 164 |
| cartilage | 82; 116 | 82; 116; 116 | 82; 116; 116; 122 | 82; 114 | 82; 112; 114 | 82; 112; 114; 120 |
| white matter | 82; 162 | 82; 162; 188 | 82; 162; 188; 208 | 82; 162 | 82; 162; 178 | 82; 162; 178; 214 |
| gray matter | 82; 210 | 82; 210; 244 | 82; 210; 244; 280 | 82; 192 | 82; 192; 218 | 82; 192; 218; 240 |
| optic nerve | 82; 192 | 82; 192; 222 | 82; 192; 222; 250 | 82; 168 | 82; 168; 196 | 82; 168; 196; 240 |
| spinal cord | 82; 160 | 82; 160; 192 | 82; 160; 192; 208 | 82; 174 | 82; 174; 190 | 82; 174; 190; 212 |
| blood | 82; 516 | 82; 516; 558 | 82; 516; 558; 620 | 82; 436 | 82; 436; 562 | 82; 436; 562; 588 |
Table 3.
Mean spin-lattice and spin-spin relaxation times for the considered tissues at 1.5 T and 3 T.
| Tissue | 1.5T | 3T | ||
|---|---|---|---|---|
|
|
|
|||
| T1 | T2 | T1 | T2 | |
| liver | 576 | 46 | 818 | 42 |
| skeletal muscle | 1,008 | 44 | 1,412 | 50 |
| heart | 1,030 | 40 | 1,471 | 47 |
| kidney | 690 | 55 | 1,194 | 56 |
| cartilage | 1,038 | 44 | 1,156 | 43 |
| white matter | 884 | 72 | 1,084 | 69 |
| gray matter | 1,124 | 95 | 1,820 | 99 |
| optic nerve | 815 | 77 | 1,083 | 78 |
| spinal cord | 745 | 74 | 993 | 78 |
| blood | 1,441 | 290 | 1,932 | 275 |
In Table 4, optimal echo times in the case of gray matter T2 estimation for different minimum TE are reported. It can be noticed that the lower optimal echo time is always the minimum and that the empirical rule is confirmed.
Table 4.
Optimal echo times for T2 estimation of gray matter for acquisitions at 1.5 T in the case of different minimum TE values.
| 2 images | 3 images | 4 images | |
|---|---|---|---|
| minimum TE = 82 ms | 82, 210 | 82, 210, 244 | 82, 210, 244, 280 |
| minimum TE = 50 ms | 50, 182 | 50, 182, 212 | 50, 182, 212, 234 |
| minimum TE = 20 ms | 20, 158 | 20, 158, 180 | 20, 158, 180, 210 |
Note that the usefulness of a proper TR and TE selection, besides the lower estimation variance, consists also in reducing the acquisition time. In order to make such an advantage evident, Table 5 reports the achievable performance in the case of 16 images (4 TR and 4 TE values) when moving from equally spaced to optimized acquisition parameters. In particular, the last column of Table 5 shows that 12 acquisitions, with properly chosen parameters, can lead to better results with respect to 16 equally spaced images, while definitely reducing the global acquisition time.
Table 5.
CRLBs for equally and optimally spaced TR and TE values.
| Tissue parameter | CRLB: 16 images Equispaced | CRLB: 16 images Optimized | Improvement (%) | CRLB: 12 images Optimized | Improvement (%) |
|---|---|---|---|---|---|
| ρ | 0.1562 | 0.1291 | 17.34% | 0.1506 | 3.58% |
| T1 | 3144 | 1483 | 52.83% | 1.960 | 37.66% |
| T2 | 2.708 | 1.808 | 33.23% | 2.144 | 20.83% |
5. Numerical Experiments
Within this section, some numerical results are shown in order to validate the advantage of the optimal selection of the imaging parameters according to the previously reported theoretical studies. A tissue with parameters [ρ T1 T2] = [5.5 775 44.5] has been considered. Three noisy datasets (SNR = 30 dB) have been simulated, each one composed of four acquisitions. The parameters of Dataset 1 have been chosen according to the results of Figures 5 and 6 in order to optimize the estimation for the considered tissue. Datasets 2 and 3 have been generated with non-ideal parameters. The dataset characteristics are summarized in Table 6.
Table 6.
Acquisition parameters: three datasets composed of four images.
| Repetition Times (s) | Echo Times (ms) | SNR (dB) | |
|---|---|---|---|
| Dataset 1 | 0.55, 4.0 | 80, 140 | 30 |
| Dataset 2 | 0.75, 4.0 | 80, 170 | 30 |
| Dataset 3 | 0.90, 4.0 | 80, 200 | 30 |
To asses and validate the CRLB studies, the estimation of the relaxation times has been implemented via Monte Carlo simulation. In particular, a maximum likelihood estimator (MLE) has been set up in the complex domain. Considering that the noise is circularly Gaussian distributed, MLE corresponds to a non-linear least squares (NLLS) estimator [18]. It is important to note that the previously reported theoretical studies about the optimal selection of the imaging parameters are valid for any unbiased estimator, since CRLBs are related only to the acquisition model. Among different estimators, NLLS has been chosen thanks to its low computational times and complexity. We recall that the choice of the optimal estimator is not the aim of this paper.
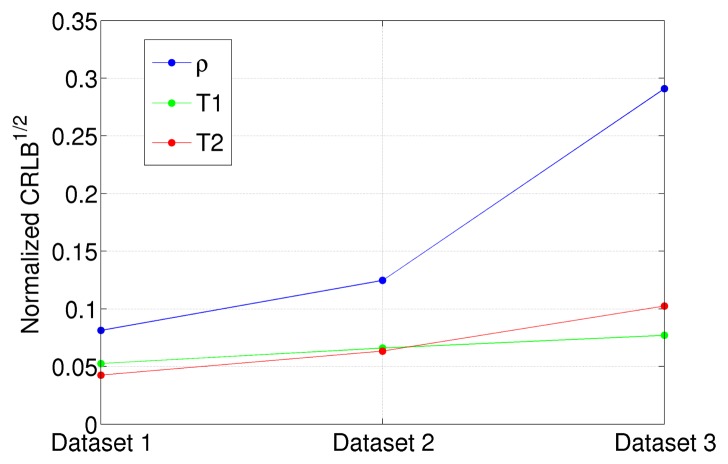
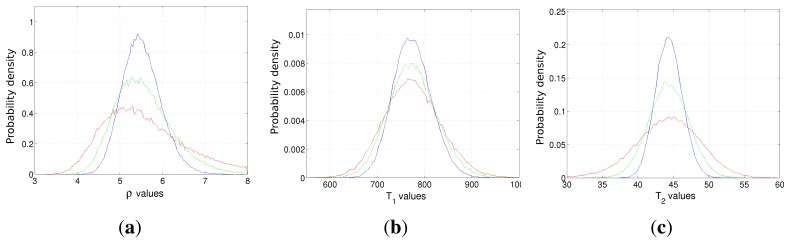
The NLLS estimator for the ρ, T1 and T2 parameters has been implemented on the three datasets of Table 6. A quantitative analysis of the results, in terms of estimation means and variances, has been reported in Table 7. By analyzing it, it is possible to infer that the estimator means are very close, while variances significantly vary from one dataset to the other. In particular, the smallest variances are obtained in the case of Dataset 1. This fully agrees with the theoretical studies reported in Section 4; as a matter of fact, Dataset 1 has been generated by using the previously developed optimal TE and TR parameter selection for the considered relaxation times. It is evident that choosing a non-ideal imaging parameters configuration can lead to very inaccurate results. For example, the T2 estimator variance of Dataset 3 is approximately six times larger than the one of Dataset 1. In order to visualize such results, the normalized standard deviations of ρ, T1 and T2 in the case of Datasets 1, 2 and 3 are reported in Figure 8.
Table 7.
Estimator performances: three datasets composed of four images.
| Parameter | True Value | Dataset 1 | Dataset 2 | Dataset 3 | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Mean | Variance | Mean | Variance | Mean | Variance | ||
| ρ̂ | 5.5 | 5.52 | 0.20 | 5.58 | 0.47 | 5.79 | 2.56 |
| T̂1 | 775 | 776.1 | 1662 | 776.5 | 2615 | 776.5 | 3, 566 |
| T̂2 | 44.5 | 44.54 | 3.58 | 44.49 | 7.95 | 44.32 | 20.82 |
Figure 8.
Square root of the CRLB for proton density (blue), spin-lattice (T1) relaxation time (green) and spin-spin (T2) relaxation time (red) for the dataset with different acquisition parameters. CRLBs values have been normalized for the parameter values in order to be plotted in the same graph.
The higher achievable accuracy in the case of optimal imaging parameters selection can also be inferred from the empirical probability density functions of the estimators, reported in Figure 9. In each image, the blue, the green and the red curves refer to Datasets 1, 2 and 3 of Table 6. As expected, most of the presented estimators follow a Gaussian distribution, with a different width. Blue curves obtained using Dataset 1, characterized by the optimal TR/TE values for the simulated tissue, are always the narrowest (smallest variances). Moving to curves obtained from Datasets 2 and 3, the estimation error becomes larger. Moreover, in the case of the ρ estimator, the results start showing a bias in the case of Dataset 3, i.e., the one with the worst acquisition parameters, and the empirical PDF does not look like a Gaussian function any more.
Figure 9.
The empirical probability density function of the ρ (a), T1 (b) and T2 (c) estimators in the case of Dataset 1 (blue line), Dataset 2 (green line) and Dataset 3 (red line). The true values are ρ = 5.5, T1 = 775 and T2 = 44.5, respectively.
Finally, one further simulation is presented. Signals from two different tissues have been simulated, with parameters [ρ T1 T2] = [5 700 68] (spinal cord) and [ρ T1 T2] = [5 1190 115] (gray matter). Two datasets composed of four acquisitions have been generated, with parameters reported in Table 8. Taking into account the developed procedure, Dataset 1 parameters represent the ideal configuration for the first tissue, while Dataset 2 is the ideal for the second one.
Table 8.
Acquisition parameters in the case of two tissues; the datasets are composed of four images.
| Repetition Times (s) | Echo Times (ms) | SNR (dB) | |
|---|---|---|---|
| Dataset 1 | 0.55 4.0 | 82, 160 | 26 |
| Dataset 2 | 0.80, 4.0 | 82, 250 | 26 |
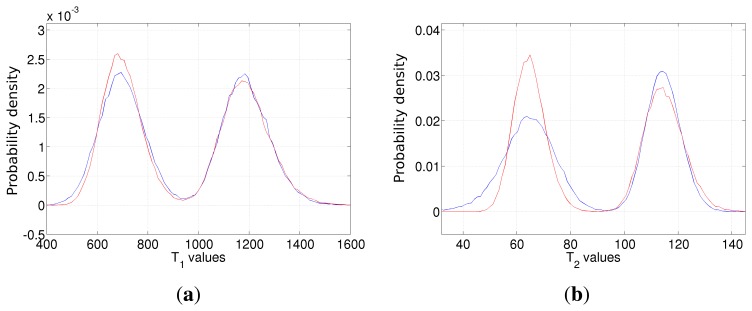
The empirical probability density functions for T1 and T2 estimators have been computed for both datasets and are shown in Figures 10, respectively. Once again, The results validate the theoretical study of Section 4. Estimation based on Dataset 1 (red line) shows lower variance in the case of spinal cord, i.e., the tissue with the lowest relaxation times (the left peaks of Figures 10). Considering gray matter, Dataset 2 (blue line) -based estimation gives better results, although the improvement of the T1 estimator is not pronounced. Once again, the result highlights the need of properly tuning the acquisition parameters.
Figure 10.
The empirical probability density function of the T1 (a) and T2 (b) estimators in the case of Dataset 1 (blue line) and Dataset 2 (red line). Dataset 1 (blue line) imaging parameters are ideal for tissues with lower T1 and T2. On the contrary, Dataset 2 (red line) is ideal for the tissue with higher relaxation times.
6. Conclusions
Within this paper, an analysis on the spin echo signal model in MR imaging has been addressed. In particular, Cramer–Rao lower bounds for relaxation time estimation in the case of a complex Gaussian acquisition model have been evaluated. Several CRLB-based evaluations have been presented in order to investigate the possibility of finding the optimal, in terms of reconstruction accuracy, imaging parameter configuration for the estimation of T1 and T2 maps. According to these theoretical studies, an empirical rule together with the identification of the optimal imaging parameter combination (echo and repetition times) in case of different tissues (different T1 and T2) has been proposed. Moreover, the optimal acquisition parameters for several tissues have been computed for both 1.5 T and 3 T acquisitions. The theoretical results have been numerically validated on different datasets. It should be underlined that such optimal parameters are independent from the implemented estimators, as CRLBs only depend on the signal model. Once the data have been acquired, different estimators proposed in the literature can be applied. It is important to underline that the theoretical studies reported within the paper can be easily adapted to different imaging sequences.
Appendix
From [16], CRLBs may also be expressed in a slightly different form with respect to Equation (6). In particular, it yields:
| (7) |
From Equation (4), the log-likelihood function related to N complex acquisitions is:
where subscript k refers to k-th acquisition, i.e., the MRI scan with parameters TR(k), TE(k). The partial derivatives can be computed as:
where the first order derivatives are:
In order to compute the expected value of Equation (7), Monte Carlo simulations have to be implemented.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Slichter P. Principles of Magnetic Resonance. 3rd ed. Springer; New York, NY, USA: 1996. [Google Scholar]
- 2.Cho Z.H., Jones J., Singh M. Foundations of Medical Imaging. Wiley Interscience; New York, NY, USA: 1993. [Google Scholar]
- 3.Haley A.P., Knight-Scott J., Fuchs K.L., Simnad V.I., Manning C. Shortening of hippocampal spin-spin relaxation time in probable Alzheimer's disease: A 1H magnetic resonance spectroscopy study. Neurosci. Lett. 2004;362:167–170. doi: 10.1016/j.neulet.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 4.Antonini A., Leenders K.L., Meier D., Oertel W.H., Boesiger P., Anliker M. T2 relaxation time in patients with Parkinson's disease. Neurologyn. 1993;43:697–700. doi: 10.1212/wnl.43.4.697. [DOI] [PubMed] [Google Scholar]
- 5.Mariappan S.V.S., Subramanian S., Chandrakumar N., Rajalakshmi K.R., Sukumaran S.S. Proton relaxation times in cancer diagnosis. Magn. Reson. Med. 1988;8:119–128. doi: 10.1002/mrm.1910080202. [DOI] [PubMed] [Google Scholar]
- 6.Roebuck J.R., Haker S.J., Mitsouras D., Rybicki F.J., Tempany C.M., Mulkern R.V. Carr-Purcell-Meiboom-Gill imaging of prostate cancer: Quantitative T2 values for cancer discrimination. Magn. Reson. Imaging. 2009;27:497–502. doi: 10.1016/j.mri.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drain L.E. A direct method of measuring nuclear spin-lattice relaxation times. Proc. Phys. Soc. 1949;62:301. [Google Scholar]
- 8.Barral J.K., Gudmundson E., Stikov N., Etezadi-Amoli M., Stoica P., Nishimura D.G. A robust methodology for in vivo T1 mapping. Magn. Reson. Med. 2010;64:1057–1067. doi: 10.1002/mrm.22497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang L.C., Koay C.G., Basser P.J., Pierpaoli C. Linear least-squares method for unbiased estimation of T1 from SPGR signals. Magn. Reson. Med. 2013;60:496–501. doi: 10.1002/mrm.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trzasko J.D., Mostardi P.M., Riederer S.J., Manduca A. Estimating T1 from multichannel variable flip angle SPGR Sequences. Magn. Reson. Med. 2013;69:1787–1794. doi: 10.1002/mrm.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonny J.M., Zanca M., Boire J.Y., Veyre A. T2 maximum likelihood estimation from multiple spin-echo amplitude images. Magn. Reson. Med. 1996;36:287–293. doi: 10.1002/mrm.1910360216. [DOI] [PubMed] [Google Scholar]
- 12.Sijbers J., den Dekker A.J., Raman E., van Dyck D. Parameter estimation from Magnitude MR images. Int. J. Imaging Syst. Technol. 1999;10:109–114. [Google Scholar]
- 13.Liu J., Nieminen A.O., Koenig J.L. Calculation of T1 T2 and proton spin density in nuclear magnetic resonance imaging. J. Magn. Reson. 1989;85:95–110. [Google Scholar]
- 14.Fennessy F.M., Fedorov A., Gupta S.N., Schmidt E.J., Tempany CM., Mulkern R.V. Practical considerations in T1 mapping of prostate for dynamic contrast enhancement pharmacokinetic analyses. Magn. Reson. Imaging. 2012;30:1224–1233. doi: 10.1016/j.mri.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones A.J., Hodgkinson P., Barker A.L., Hore P.J. Optimal Sampling Strategies for the Measurement of Spin-Spin Relaxation Times. J. Magn. Reson. 1996;113:25–34. [Google Scholar]
- 16.Kay S.M. Fundamentals of Statistical Signal Processing. Prentice Hall; Upper Saddle River, NJ, USA: 1993. [Google Scholar]
- 17.Gudbjartsson H., Patz S. The Rician distribution of noisy MRI data. Magn. Reson. Med. 1995;34:910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baselice F., Ferraioli G., Pascazio V. Relaxation time estimation from complex magnetic resonance images. Sensors. 2010;10:3611–3625. doi: 10.3390/s100403611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spandonis Y., Heese F.P., Hall D.H. High resolution MRI relaxation measurements of water in the articular cartilage of the meniscectomized rat knee at 4.7T. Magn. Reson. Imaging. 2004;22:943–951. doi: 10.1016/j.mri.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Anastasiou A., Hall L.D. Optimisation of T2 and M0 measurements of bi-exponential systems. Magn. Reson. Imaging. 2004;22:67–80. doi: 10.1016/j.mri.2003.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Wright G.A. Magnetic resonance imaging. IEEE Signal Process. Mag. 1997;14:56–66. [Google Scholar]
- 22.Wang Y., Lei T. Statistical Analysis of MR Imaging and Its Applications in Image Modeling. Proceedings of IEEE International Conference Image Processing; Austin, TX, USA. 13–16 November 1994; pp. 866–870. [Google Scholar]
- 23.Baselice F., Ferraioli G., Shabou A. Field map reconstruction in magnetic resonance imaging using Bayesian estimation. Sensors. 2010;10:266–279. doi: 10.3390/s100100266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggers H., Knopp T., Potts D. Field inhomogeneity correction based on gridding reconstruction for magnetic resonance imaging. IEEE Trans. Med. Imaging. 2007;26:374–384. doi: 10.1109/TMI.2006.891502. [DOI] [PubMed] [Google Scholar]
- 25.Stanisz G.J., Odrobina E.E., Pun J., Escaravage M., Graham S.J., Bronskill M.J., Henkelman R.M. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med. 2005;54:507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]