Abstract
Background
The thiomethyl group of S-adenosylmethionine is often recycled as methionine from methylthioadenosine. The corresponding pathway has been unravelled in Bacillus subtilis. However methylthioadenosine is subjected to alternative degradative pathways depending on the organism.
Results
This work uses genome in silico analysis to propose methionine salvage pathways for Klebsiella pneumoniae, Leptospira interrogans, Thermoanaerobacter tengcongensis and Xylella fastidiosa. Experiments performed with mutants of B. subtilis and Pseudomonas aeruginosa substantiate the hypotheses proposed. The enzymes that catalyze the reactions are recruited from a variety of origins. The first, ubiquitous, enzyme of the pathway, MtnA (methylthioribose-1-phosphate isomerase), belongs to a family of proteins related to eukaryotic intiation factor 2B alpha. mtnB codes for a methylthioribulose-1-phosphate dehydratase. Two reactions follow, that of an enolase and that of a phosphatase. While in B. subtilis this is performed by two distinct polypeptides, in the other organisms analyzed here an enolase-phosphatase yields 1,2-dihydroxy-3-keto-5-methylthiopentene. In the presence of dioxygen an aci-reductone dioxygenase yields the immediate precursor of methionine, ketomethylthiobutyrate. Under some conditions this enzyme produces carbon monoxide in B. subtilis, suggesting a route for a new gaseous mediator in bacteria. Ketomethylthiobutyrate is finally transaminated by an aminotransferase that exists usually as a broad specificity enzyme (often able to transaminate aromatic aminoacid keto-acid precursors or histidinol-phosphate).
Conclusion
A functional methionine salvage pathway was experimentally demonstrated, for the first time, in P. aeruginosa. Apparently, methionine salvage pathways are frequent in Bacteria (and in Eukarya), with recruitment of different polypeptides to perform the needed reactions (an ancestor of a translation initiation factor and RuBisCO, as an enolase, in some Firmicutes). Many are highly dependent on the presence of oxygen, suggesting that the ecological niche may play an important role for the existence and/or metabolic steps of the pathway, even in phylogenetically related bacteria. Further work is needed to uncover the corresponding steps when dioxygen is scarce or absent (this is important to explore the presence of the pathway in Archaea). The thermophile T. tengcongensis, that thrives in the absence of oxygen, appears to possess the pathway. It will be an interesting link to uncover the missing reactions in anaerobic environments.
Background
Availability of a huge number of genome sequences (1004 ongoing or completed genome sequencing programmes are listed at http://wit.integratedgenomics.com/GOLD/ on February 17th, 2004) now allows scientists to explore new avenues in the deciphering of metabolic pathways. While sulfur is ubiquitously used in life, it is a highly reactive atom, hence very sensitive to the oxido-reduction potential of its local environment. It is also very sensitive to dioxygen and reactive oxygen species. As a consequence, sparing processes that enable the cell to keep a sulfur atom in its final chemical and electronic environment (inside well characterized groups of atoms) often exists in cells [1]. A case in point is the fate of the ubiquitous S-adenosylmethionine molecule (AdoMet), which can donate each one of its sulfur connected groups: methyl-, aminobutyryl- (usually, after decarboxylation as aminopropyl-), and adenosyl- (ribosyl-). A vast number of AdoMet mediated methylation events continuously happen in most, if not all organisms. Methylases are present at the level of DNA metabolism, RNA metabolism and protein post-translational modifications [2-4]. They are also involved in formation of a variety of small metabolites (essential coenzymes or prosthetic groups in particular [5,6]). This group transfer results in the synthesis of S-adenosyl homocysteine (AdoHcy). The latter molecule has to be recycled or further metabolized, either after excision of the adenine moiety by hydrolysis or phosphorolysis, or by direct hydrolysis into adenosine and homocysteine. When aminopropyl- is transferred, methylthioadenosine (MTA) is a by-product. It is produced during synthesis of spermidine and spermine, and also of N-acylhomoserine lactone synthases in Gram-negative bacteria [7] and 1-aminocyclopropane-1-carboxylate (ACC) synthase in plants (and possibly in bacteria producing ethylene). Furthermore, other still unknown reactions might exist, transferring the aminopropyl-group to yet unrecognized acceptors (most likely to be nitrogen atoms). Recycling the sulfur-containing compounds S-adenosylhomocysteine and MTA is a challenge for all cells. MTA may regenerate methionine in the process. The biochemical details of the MTA to methionine pathway remains unknown in most organisms. This prompted us to make a thorough analysis of available genome sequences to try and identify some of the metabolic pathways involved in MTA recycling through genomic and functional comparisons with our recent findings [8]. The present work aims at combining comparative genomics and biochemical or genetic data to propose a list of identified and putative genes associated to the corresponding functions. A self-consistent nomenclature will be proposed in parallel, so that homologous functions in various organisms could tentatively be identified from their genome sequences.
Results
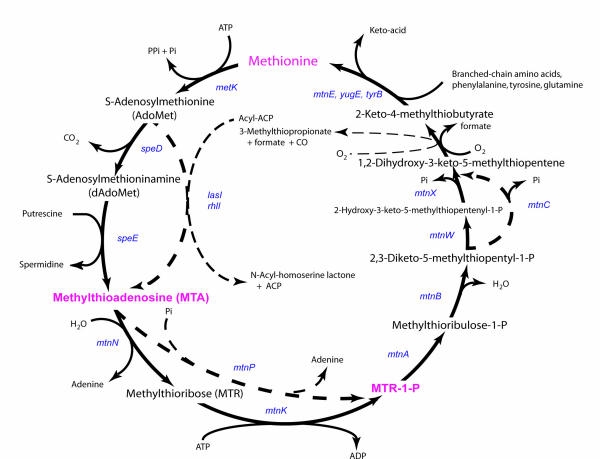
MTA can be exported from the cell, or recycled, beginning with two main alternative pathways (Figure 1). The most energy sparing one starts with MTA phosphorolysis and yields methylthioribose-1-phosphate (MTR-1-P) [9], while the second one splits this first step into two steps: the first one being hydrolysis of MTA to methylthioribose (MTR) and adenine, followed by ATP-dependent phosphorylation of MTR by MTR kinase [10,11]. In this latter case, MTR can also be excreted from the cell, especially in organisms that are not challenged by a high level of dioxygen (this happens in facultative anaerobes living in rich media, such as Escherichia coli [12]).
Figure 1.
The methionine salvage pathway. As a reference, the pathway is shown in B. subtilis (solid lines). Variants of the pathway are shown in broken lines.
MTA phosphorylase
The gene coding for MTA phosphorylase (named mtnP, see below) has been identified in many organisms, in all three kingdoms: Archaea, Bacteria and Eukarya. The activity of the enzyme has long been studied as a target to fight some kinds of tumor cells [13]. A PROSITE signature, PDOC00946, [GST]-x-G-[LIVM]-G-x-[PA]-S-x-[GSTA]-I-x(3)-E-L, is common to three classes of nucleoside phosphorylases, purine nucleosidases, uridine nucleosidases and MTA phosphorylase. A second signature, PDOC00954 [LIV]-x(3)-G-x(2)-H-x-[LIVMFY]-x(4)-[LIVMF]-x(3)-[ATV]-x(1,2)-[LIVM]-x-[ATV]-x(4)-[GN]-x(3,4)-[LIVMF](2)-x(2)-[STN]-[SA]-x-G-[GS]-[LIVM] labels some other phosphorylases (including MTA phosphorylase), from Eukarya in particular.
This family can be narrowed into three conserved domains families: COG0005, purine nucleoside phosphorylase, pfam00896, phosphorylase family 2, KOG3985, MTA phosphorylase. Interestingly, these purine nucleoside phosphorylase families share most of their sequence in amino acid residues, except for the very first residues, that appear to be made of a particular sequence in the case of MTA phosphorylase. In contrast to other purine nucleoside phosphorylases, it appears likely that the amino-terminal end of MTA phosphorylase starts very early upstream of the G [ST]G sequence that is conserved in purine nucleoside phosphorylases: the exception we find to the rule is the sequence present in Synechocystis PCC6803, but a methionine codon is placed just at the expected beginning of the protein, while translation of the upstream region is not confirmed by biochemical analysis (and it is notorious that start codons are difficult to identify in general and in cyanobacteria in particular). Immediately upstream of this first consensus sequence, a G amino acid residue is found in all known and putative MTA phosphorylases, as well as other features of the region displayed in Figure 2. It was therefore important to explore whether in Pseudomonas aeruginosa gene PA-3004 (mtnP), which has this signature, codes for MTA phosphorylase. The presence of this gene is not ubiquitous in gamma-proteobacteria, since this gene seems to be absent from Klebsiella pneumoniae (the sequence of the complete genome is not publicly available, sequences of the ongoing genome project are available at http://genome.wustl.edu/blast/client.pl).
Figure 2.

Comparison of purine and MTA phosphorylases. The beginning of MTA phosphorylases is aligned with that of purine phosphorylases extracted from phylogenetically widely diverse organisms: PSEAE Pseudomonas aeruginosa; THETE Thermoanaerobacter tengcongensis; HOMSA Homo sapiens; CAEEL Caenorhabditis elegans; SCHPO Schizosaccharomyces pombe; SYNEC Synechocystis PCC 6803; MYCTU Mycobacterium tuberculosis; METJA Methanococcus jannaschii; METTH Methanobacterium thermoautotrophicum; LEPIN Leptospira interrogans; XYLFA Xylella fastidiosa; ESCHO Escherichia coli; SACCE Saccharomyces cerevisiae; BACSU Bacillus subtilis; GEOST Geobacillus stearothermophilus; BOSTA Bos taurus. Conserved residues in both classes of enzymes are emphasized [54]. In red are shown conserved residues that are specific to one class, while conservative replacements are indicated in blue. The three residues in green indicate a region that suffered an insertion/deletion event in the course of evolution that separated the two classes. The position of the first residue of the alignment shown is displayed in blue. Note that for Synechocystis the published sequence starts earlier, but that the residue indicated is likely to be the real start of the protein.
The P. aeruginosa methionine-auxotroph PAO503, which carries a mutation in the metZ gene encoding O-succinyl-homoserine sulfhydrylase [14], grew well on minimal medium E when supplemented with either methionine, homocysteine or MTA. By contrast, a mutant derived from PAO503 by gene replacement, PAO6422 (metZ mtnP), did not grow with MTA, but still responded to methionine and homocysteine (Table 1). These results demonstrate that P. aeruginosa has an MTA recycling pathway and that MTA phosphorylase has an essential function in this pathway.
Table 1.
Growth of Pseudomonas aeruginosa mutants on various sulfur sources
| Strain | MTA | Methionine | Homocysteine |
| PAO503 metZ9011 | + | + | + |
| PAO6412 metZ9011 mtnB::Ω-Sp/Sm | - | + | + |
| PAO6422 metZ9011 mtnP::Ω-Sp/Sm | - | + | +a |
a weaker growth as compared to the other constructs.
An alignment of MTA phosphorylase and purine phosphorylase is provided in Figure 2, with sequences from Thermoanaerobacter tengcongensis (Firmicutes [15]) and P. aeruginosa (gamma-proteobacteria), as well as from an organism that is usually difficult to classify in terms of proteins, Leptospira interrogans. In addition to the position of the beginning of the protein (shorter in MTA phosphorylase) residues that are discriminating between general scavenging purine phosphorylase and MTA phosphorylase are indicated in Figure 2, around the common motif [FIMLV]-[IV]-X-G-[ST]-G-X16–22-P. Noteworthy features are the presence of conserved residues in MTA phosphorylase, that differ from those at the same position in the other phosphorylases: G upstream of the highly conserved G-[ST]-G motif and a T is present upstream of the partially conserved P residue, with a downstream aromatic residue followed by a G in MTA phosphorylase (respectively L or I and F in other phosphorylases). The product of the reaction is MTR-1-P, which undergoes further processing as detailed below.
MTA hydrolysis and methylthioribose phosphorylation
AdoMet-dependent methylases produce AdoHcy, while aminopropyl-group transfer produces MTA. It was once thought that the metabolism of AdoHcy was split in various organisms into two alternative pathways [16]: the molecule was recycled either by AdoHcy hydrolase that generates homocysteine and adenosine, or by the sequential action of AdoHcy nucleosidase that generates adenine and S-ribosylhomocysteine, followed by a S-ribosylhomocysteine-cleavage enzyme. The hydrolase has been now found in many organisms, such as Pseudomonas sp., Homo sapiens, L. interrogans, Aquifex sp. and Thermotoga maritima but it is not present in E. coli nor in Bacillus subtilis, at least with a related sequence [17].
In E. coli the nucleosidase (coded by gene pfs) has in fact a lower specificity since it hydrolyses both AdoHcy and MTA [18]. In B. subtilis it has been experimentally demonstrated that the gene initially termed yrrU has this activity [19]. The counterpart of this enzyme exists in K. pneumoniae (Figure 3). In many bacteria (including E. coli) the fate of the MTR thus produced is not known, but the molecule is found to accumulate in the external medium, suggesting the presence of an export system [12]. In B. subtilis experiments using radioactive labelling have shown that a kinase exists that transforms MTR into MTR-1-P (Figure 1). It is encoded by a gene belonging to a S-box riboswitch-controlled operon [20,21], ykrT (renamed here mtnK, see below) [22]. In contrast with the situation in E. coli, a counterpart of this gene exists in K. pneumoniae (Figure 3), consistent with the existence of a methionine salvage pathway in the latter. Interestingly, this enzyme that has been crystallized recently [23] does not belong to the ribo- or deoxyribokinase family, but, rather, to the choline/ethanolamine kinase family [24].
Figure 3.
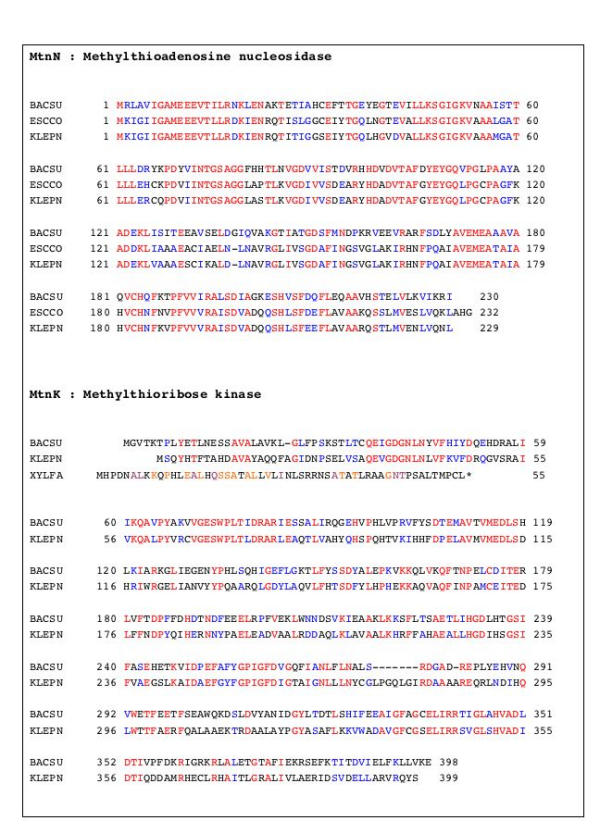
The MTA nucleosidase / MTR kinase pathway in B. subtilis and K. aeruginosa. When the methionine salvage pathway exists, while the organism does not code for a MTA phosphorylase, the combination of MTA nucleosidase and MTR kinase produce MTR-1-P from MTA (Figure 1). B. subtilis (BACSU) and K. pneumoniae (KLEPN) possess this pathway, while E. coli (ESCCO) has only the nucleosidase. Conservative replacements are shown in blue following Dayhoff's rule [54]. The short protein that may be a truncated MtnK in X. fastidiosa is shown for comparison (identities with the B. subtilis sequence in orange, and similarities in violet).
The elusive dowstream section of the methionine salvage pathway
It has long been known that MTA, once converted to adenine and MTR-1-P was able to regenerate methionine. This was found in organisms as distant as H. sapiens or K. pneumoniae. The overall reaction scheme has been unraveled by Abeles and coworkers in K. pneumoniae [25]. However, the actual enzymes or genes involved for each step have not been uncovered in totality. A gene inactivation study in B. subtilis revealed the involvement of several genes [26], but the exact nature of the enzymes needed for each step in this organism was only discovered when appropriate biochemical experiments were set up [8]. Briefly, the pathway in B. subtilis begins as follows: MTR-1-P is converted into MTRu-1-P by the isomerase encoded by gene ykrS (now mtnA, see new proposed nomenclature below) [8,22]. Remarkably, it has been observed that this gene, expressed at a level similar to that of ykrT (mtnK) [22], codes for a protein with a sequence annotated as similar to that of eukaryotic initiation factor eIF-2B (alpha subunit). Figure 4 shows a high degree of conserved residues in the corresponding alignment with genes from Firmicutes, gamma-proteobacteria, plants and humans. Interestingly this sequence is similar, especially in its carboxy-terminal end (see the Mus musculus sequence), with eIF-2B-alpha, but, as can be seen in the Figure, this similarity, although unambiguous, should perhaps not have been emphasized in genome annotations.
Figure 4.

Alignment of methylthioribose-1-phosphate isomerases. Conserved residues are indicated in red, while conservative replacements are indicated in blue. Organisms are labelled as in Figure 2, with a few additional organisms: AQUAE Aquifex aeoliqus; ARATH Arabidopsis thalina and MUSMU Mus musculus. The sequence of mouse initiation factor eIF-2B (alpha subunit) is shown for comparison. The § symbol at the beginning and the end of the K. aeruginosa sequence corresponds to regions that have not been identified from the ongoing genome programme sequences.
The next step is dehydration of MTRu-1-P by MTRu-1-P dehydratase encoded by gene ykrY (now mtnB), a member of operon ykrWXYZ, under the control of an S-box riboswitch [20,21]. The product of the reaction is the diketone 2,3-diketo-5-methylthiopentyl-1-phosphate. A counterpart of this gene can be found in many organisms (Figure 5), where it is usually associated to other genes involved in the pathway. This is the case, for example in K. pneumoniae. In P. aeruginosa, the operon aroC, PA-1682, PA-1683, PA-1684, mas http://bioinfo.hku.hk/AeruList encodes proteins similar to chorismate synthase, a metabolite transporter, MTRu-1-P dehydratase, 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase and enolase-phosphatase, respectively. Indeed, the methionine-auxotroph (metZ [14]) P. aeruginosa strain PAO503, which grew well on minimal medium E when supplemented with either methionine, homocysteine or MTA, lost the ability to use MTA when the mtnB gene (PA-1683) was inactivated by gene replacement in strain PAO6412 (metZ mtnB) (Table 1). Thus, in addition to the MTA phosphorylase, P. aeruginosa has an MTRu-1-P dehydratase which has an essential function in the methionine salvage pathway (Table 1). The MtnB sequence is a member of a large cluster of proteins that form class II aldolases (COG235, Pfam0096). Among these enzymes one finds L-ribulose-5-phosphate-4-epimerase, suggesting that recognition of the ribulose backbone is common to a variety of enzymes of the class. The N-terminal domain of these proteins form class KOG2631, which appears to overlap exactly with the dehydratase.
Figure 5.

Alignment of methylthioribulose-1-phosphate dehydratases. Important residues are indicated as in Figure 2. For A. thaliana (sequence from At5g53850) the N-terminal end of the protein is aligned. The § symbol at the beginning and the end of the K. aeruginosa sequence corresponds to regions that have not been identified from the ongoing genome programme sequences.
The dehydratase product is further metabolized by 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, encoded in B. subtilis by gene ykrW (now mtnW). Interestingly, this enzyme is highly similar to RuBisCO [8,26,27]. It produces 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate. At this stage (outcome of the enolase reaction) 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate phosphatase, encoded in B. subtilis by gene ykrX (mtnX), produces 1,2-dihydroxy-3-keto-5-methylthiopentene (Figure 1).
In other organisms, such as K. pneumoniae, the corresponding enzymes appear usually to consist of a single enolase-phosphatase, allowing the cell to bypass the last step. Interestingly, in Arabidopsis thaliana, the protein carrying the dehydratase (Figure 5) is a two-domain protein, the carboxy-terminal domain coding for the enolase-phosphatase (Figure 6). This further substantiates the proposed identification of the corresponding steps in the pathway. The enolase-phosphatase corresponds to the KOG family KOG2630. The product of the enolase-phosphatase reaction is then submitted to the action of a dioxygenase, as demonstrated in K. pneumoniae [28]. It has also been shown that dioxygen is required for the methionine salvage pathway in B. subtilis [8,26]. The sequence of the proteins is illustrated in Figure 7. The aci-reductone dioxygenase that creates 2-keto-4-methylthiobutyrate (KMTB) has counterparts in many Bacteria and Eukarya, including in mammals, belonging to family COG1791. It seems to be absent from Archaea. Interestingly Abeles and co-workers discovered that the enzyme has two activities, depending on its metal co-factor. When iron was involved, the enzyme produced KMTB, the immediate precursor of methionine, together with formate. In contrast, when nickel was involved, carbon monoxide was produced in addition to formate, together with 3-methylthiopropionate [29]. The enzyme has been crystallized [30]. In B. subtilis, using palladium salt reduction, we found that CO was likely to be produced when the cells were grown in the presence of MTR. A significant difference between the wild type and a mtnD mutant could be seen: the wild type makes more reducing gasses when MTR is the sulfur source (16.7 ppm/hour, Table 2). However, B. subtilis produced other reducing gasses as shown by the presence of a reddish-black hue in the precipitate, even in the mutant. To test for another possible source of reducing gasses we investigated reducing gas production in a yflM mutant. Its gene product, YflM, is highly similar to an NO synthase. As shown in Table 2, an yflM mutant produced somewhat less reducing gasses. The amount produced in our conditions, about 6.7 ppm/hour less than the wild type in sulfate, and 10 ppm/hour less in MTR, is however not much higher than the uncertainty of the experiment (ca 5 ppm/hour). In this background, the effect of MTR was still visible and significant as compared to growth with sulfate as sulfur source (13.4 ppm/hour). However a significant level of palladium precipitation remains in these mutants. The background level found in the absence of bacteria was of the order of 12.5 ppm/hour. Other gasses such as methanethiol and hydrogen sulfide are likely to contribute to the reducing power of the organism.
Figure 6.
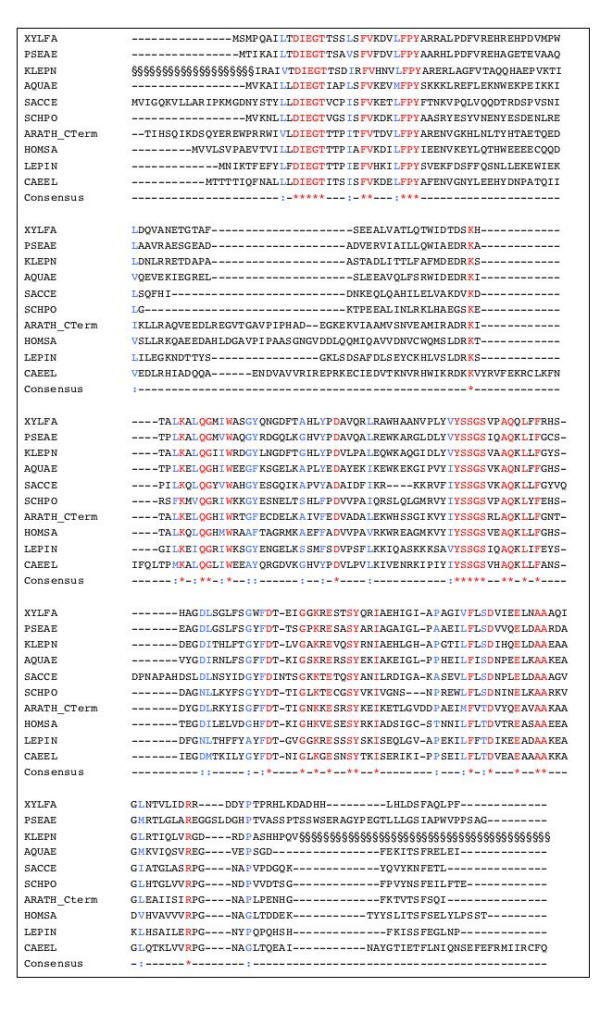
Alignment of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase-phosphatases. Important residues are indicated as in Figure 2. For A. thaliana (sequence from At5g53850) the C-terminal end of the protein is aligned. The § symbol at the beginning and the end of the K. aeruginosa sequence corresponds to regions that have not been identified from the ongoing genome programme sequences.
Figure 7.

Alignment of aci-reductone dioxygenases. Important residues are indicated as in Figure 2. The N-terminus of the protein is poorly conserved.
Table 2.
CO production by Bacillus subtilis on plates using GASTEC dosimeters.
| Strain | Sulfur source used | |
| MgSO4 | MTR | |
| WT 168 | 50.0 | 66.7 |
| BFS1853 mtnD::lacZ | 48.3 | 51.7 |
| BFS4605 yflM::lacZ | 43.3 | 56.7 |
CO production is expressed in ppm/hour accumulated during 20 hours of incubation. Background is not subtracted. A difference of 5 in the limit of the palladium metal precipitate is reproducibly visible. The values given correspond to three replicates of each experiment. Note that it is not possible to distinguish between purely black and reddish black precipitates (often due to thiol containing gasses). Background precipitation in the absence of bacteria was of the order of 12.5 ppm/hour.
When MTA is recycled, the two last steps of the methionine salvage pathway appear to be common to all organisms that grow in the presence of dioxygen. The counterpart of the dioxygenase is not known in organisms that live under anaerobic growth conditions. KMTB is transaminated to methionine. There are many analogs of the final aminotransferase, however they can more or less complement one another (as is often the case with aminoacid aminotransferases, see e.g. [31]). In B. subtilis, putative aspartate aminotransferases, the yugH, ywfG, ykrV, aspB, and patA gene products, have been cloned, expressed, and characterized for methionine regeneration activity. In this family, only YkrV (encoded by ykrV, adjacent to the ykrWXYZ operon) was able to convert KMTB to methionine, and it catalyzed the reaction only when glutamine was used as amino donor [32]. This suggests a link with nitrogen metabolism, substantiating our previous observations [33]. However the bulk of the activity resulted from transaminase YbgE, using either branched chain aminoacids or phenylalanine or tyrosine as donors could also perform a similar reaction [32]. In K. pneumoniae it is the same enzyme that makes methionine from KMTB and tyrosine from its ketoacid precursor [34].
Nomenclature
We propose the following nomenclature, taking into consideration previous names of some of the genes, as well as previous usage of names for some other groups of genes, trying to avoid naming conflicts. The acronym "mtn" (for methylthionucleoside) will be used.
Because the early steps are variable, we shall use as a mnemonic the capital letter of the enzyme's name, following Demerec's rule. Thus, mtnN will code for MTA nucleosidase, mtnK will code for MTR kinase, mtnP will code for MTA phosphorylase.
Considering that the common part of the pathway starts with MTR-1-P, we shall name the following genes sequentially: mtnA will code for MTR-1-P isomerase (ykrS in B. subtilis) and mtnB will code for MTRu-1-P dehydratase (ykrY).
In B. subtilis, the pathway is probably unusual, with mtnW (ykrW) coding for the enolase and mtnX (ykrX) for the phosphatase creating 1,2-dihydroxy-3-keto-5-methylthiopentene. In other organisms, this step is performed by an enolase-phosphatase, encoded by gene mtnC, while mtnD (ykrZ) codes for the aci-reductone dioxygenase.
Finally, mtnE codes for KMTB aminotransferase, when it exists as a specific enzyme. The pathway with this nomenclature is summarized in Figure 1.
Discussion
The beginning of the methionine salvage pathway is variable, even in phylogenetically related organisms. It can either correspond to the tandem reactions, MTA nucleosidase/MTR kinase (as in B. subtilis or K. pneumoniae) or start with MTA phosphorylase as in P. aeruginosa, many Eukarya or presumably in T. tengcongensis, a Firmicute living at high temperature in partially anaerobic conditions. The pathway can further vary at the level of enolase, with either an enolase (RuBisCO-like) and a phosphatase (in B. subtilis, B. cereus and B. anthracis, for example), or, more often, with an enolase-phosphatase as in a variety of organisms, including many Bacteria (e.g. P. aeruginosa), Plants and Animals (Figure 1). An explicit substantiation of this latter pathway can be seen in A. thaliana where gene At5g53850 codes for a protein made of two domains, similar to KOG2631 and KOG2630, with proposed activities of the Class II aldolases and enolase-phosphatases. We do not discuss here the case of Archaea because, although there seems to be a clear counterpart of the methionine salvage pathway, starting from MTA phosphorolysis [27], no clearcut biochemical data are yet available, while the last step (dioxygenase) is probably missing or altered to another type of reaction (many Archaea are strict anaerobes).
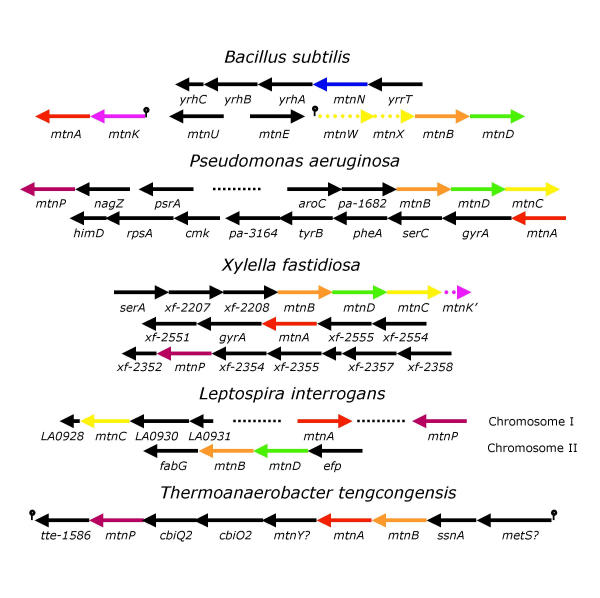
In order to further support our description of the pathway we extrapolated from the B. subtilis data to other genomes, based on chromosome neighborhood [35] (Figure 8). Here we discuss four cases: K. pneumoniae, P. aeruginosa, Xylella fastidiosa, L. interrogans and T. tengcongensis.
Figure 8.
Variety of gene organisation for the methionine salvage pathway in different bacteria. The organisation of the genes coding for the methionine salvage pathway is shown in several bacteria: B. subtilis, P. aeruginosa, X. fastidiosa, L. interrogans and T. tencongensis. The pathway is as described in Figure 1. Genes with the same function are indicated with the same color code. When a gene arrow is broken, this indicates that the function is the same, but that the gene product differs (enolase (MtnW) and phosphatase (MtnX) in B. subtilis, vs enolase-phosphatase (MtnC) in other organisms). The stem and loop symbol indicates the presence of a S-box riboswitch. In B. subtilis the enolase-phosphatase activity found in other organisms is replaced by two enzymes, a RuBisCO-like enolase and a phosphatase. In X. fastidiosa, downstream of the main operon on can see a truncated form of mtnK (broken arrow) requiring the presence of mtnP instead.
Most of the methionine salvage pathway has been uncovered in K. pneumoniae by the biochemical studies of Abeles and coworkers [25,28,29]. It was therefore interesting to identify the genes corresponding to the pathway. A complete genome sequence is known but not publicly available, while the K. pneumoniae genome programme at Washington University is still ongoing. We could however identify the genes for a complete methionine salvage pathway, which begins as in B. subtilis (MTA nucleosidase followed by MTR kinase), but differs at the level of the enolase-phosphatase step (Figures 3, 4, 5, 6).
In contrast with the behavior of K. pneumoniae, P. aeruginosa codes for an MTA phosphorylase. Gene PA-3004, located downstream from nagZ (coding for N-acetyl-D-glucosaminidase) and perhaps from transcriptional regulator gene psrA, corresponds to mtnP. Operon aroC(PA-1681)-PA-1682-PA-1683-PA-1684-masA(PA-1685) is likely to code for the core of the pathway in which PA-1682 has an unknown transport function, whereas PA-1683(mtnB), PA-1684(mtnD) and PA-1685(mtnC) correspond to the enzymatic steps described above. Finally, mtnA corresponds to gene PA-3169, possibly the first gene of a large mixed-function cluster mtnA-gyrA-serC-pheA-tyrB-aroE-cmk-rpsA-himD. This cluster is also involved in biosynthesis of aromatic molecules, and the gene tyrB coding for a wide spectrum aminotransferase is probably involved in KMTB transamination, as is its counterpart is in K. pneumoniae [34]. As a support to this analysis, both mtnP and mtnB mutants of P. aeruginosa could not grow on MTA as a sulfur source.
In X. fastidiosa a cluster of genes, presumably constituting an operon, is consistent with the existence of a complete methionine salvage pathway. This operon "serA-xf-2207-xf-2208-xf-2209-xf-2210-xf-2211-xf-2212" would code for glyceraldehyde-3-phosphate dehydrogenase (serine and one-carbon metabolism), subunits of a transporter, mtnB, mtnD, mtnC and a short fragment (N-terminus) of mtnK, respectively (Figure 3). A second gene cluster "xf-2555-xf-2554-mtnA-gyrA-xf-2551" codes for a tRNA synthetase, a protein of unknown function, mtnA, DNA gyrase and a protein of unknown function found in plants. There exists also a clear counterpart of mtnP (xf-2353, see Figure 2). This gene belongs to an operon (xf-2358->xf-2352), coding for an RNA methylase, a putative membrane protein highly conserved in cyanobacteria or bacteria associated to plants, a very short protein with similar sequence found in plants, a cell wall hydrolase, purine phosphoribosyltransferase, MtnP and a cold-shock-like protein. This operon is, at least in part, consistent with MTA and/or S-adenosylhomocysteine production and phosphorolysis. In particular, the first gene product would produce the latter molecule, while the phosphoribosyltransferase could scavenge adenine or a derivative. Thus, X. fastidiosa has all genes needed for a functional methionine salvage pathway. The truncated form of mtnK suggests that the core metabolic genes might have been obtained by horizontal gene transfer [36], and that MtnK was useless in the presence of a functional MtnP. Presence of a gene coding for spermidine synthase (xf-0143) as well as a counterpart of AdoMet decarboxylase (xf-1539) indicates that the substrate, MTA, is indeed synthesized in X. fastidiosa. The similarity between the steps of the methionine salvage pathway in K. pneumoniae, P. aeruginosa, and X. fastidiosa suggests that similar pathways will be found in many gamma-proteobacteria.
Leptospira interrogans genome annotation has been particularly difficult because the majority of the genes present in this organism are only poorly related to known genes in other organisms. Indeed, the annotated genome contains a large number of small putative genes that may not really be coding DNA sequences [37]. This prompted us to explore the presence of a methionine salvage pathway in this organism. In order to have better chances to find the genes of the pathway, we started by looking for counterparts of the first gene in the pathway, mtnA. There is a clear counterpart, LA2868, in L. interrogans (Figure 3), as an isolated gene in chromosome I. The gene for the phosphorylase with clear features (molecular footprint) of MTA phosphorylase (Figure 2), mtnP (LA4248), is also present as an isolated gene in the same chromosome. In chromosome II, operon LB294-LB293-LB292-fabG codes for translation factor EFP, MtnD, MtnB and a putative oxoacyl-ACP-reductase. The last enzyme required for the pathway is the enolase-phosphatase (MtnC): it is encoded in an operon present in chromosome I "LA0931-LA0930-LA0929-LA0928". The first gene codes for a peptide that could have a regulatory role, the following gene codes for a permease, then we find mtnC (Figure 5). The operon ends with a gene coding for a short peptide that could have features of transcriptional regulators. This indicates that L. interrogans most probably has the complete methionine salvage pathway. Interestingly, this organism has two counterparts of spermidine synthase (one on each chromosome), enzymes that should result in the production of MTA. Gene LB312, on the second chromosome could be the counterpart of speD in this genome.
Finally, we tried to explore the pathway in T. tengcongensis, because although similar to B. subtilis, this organism thrives mostly under anaerobic conditions. In T. tengcongensis the phosphorylase would be encoded in a large cluster (either continuous or split into two operons: tte-1584<-tte-1594) "ileS-ssnA-araD-tdh2-gcn3-cbiO2-cbiQ2-pnp2-tte-1586" upstream of "metC3-tte-1584". Both parts of this cluster, typical of a "sulfur island" [38], are preceded by a S-box riboswitch [20,21]. Because of the consistency of the functions encoded in this cluster it is possible that ileS has been mistakenly thought to code for isoleucine tRNA synthetase, but rather codes for methionine tRNA synthetase (both enzymes are usually extremely similar). Because annotations are often propagated by similarity only, it would be most interesting to check experimentally whether this enzyme is an authentic methionine tRNA synthetase. If this were the case, this should prompt reanalysis and annotation of the whole class of cysteine, isoleucine, leucine, methionine and valine tRNA synthetases. The second gene could code for a metal-dependent hydrolase, probably hydrolyzing C-N bonds. The third gene, in the present context, is mtnB, tdh2 could code for a metal-dependent enolase (differing from its counterpart in other organisms, hence tentatively named mtnY, but this has to be substantiated experimentally), gcn3 is mtnA, the next two genes are divalent metal transporters, followed by mtnP, while the last gene could code for a component of a transporter. Thus the operon would be "S-box-metS-ssnA-mtnB-tte-1591(mtnY)-mtnA-cbiO2-cbiQ2-mtnP-tte-1586". For this organism, it is unlikely that a dioxygenase belongs to the pathway, because it lives mostly in the absence of oxygen. Indeed, we do not find a clear counterpart of mtnD. Starting from the end of the pathway we find that a broad-spectrum aminotransferase (labelled hisC, tte-0380) belongs to a large cluster mostly devoted to divalent-metal transport and synthesis of coenzyme B12, with two unknown genes tte-0378 and tte-0379. Coenzyme B12 is often involved in pathways operating under anaerobic conditions and this should prompt further studies of the methionine salvage pathway under these conditions (T. tengcongensis, that belongs to Bacteria, might provide a missing link to explore the pathway in Archaea).
Sulfur metabolism has probably to be compartmentalized, because of the extreme reactivity of this atom towards dioxygen and radicals. In this respect, it is interesting to observe that, despite their apparent variety of origins, genes of the methionine salvage pathway have a tendency to cluster into operons (translation products of neighboring cistrons are produced in close proximity in the cell), or even fuse and give multidomain proteins (this is frequent in eukaryotes [39]). Another case of gene fusion that might be related to comparmentalization of sulfur containing molecules can be seen in Aquifex aeoliqus where the MtnA protein begins with a domain of unknown function:
MDIRTHLGINQELSGKPVKVAEGYAEVLLKTQESMKADDKGLVHGGFIFSAADYA SMLAVNHPNVVLAGANVKFLKPVKVGDEIICKARVSEDKGKKKKVLVECFKGEDKVFEGEFFTVFLID
that represents a whole COG domain, COG2050.1, PaaI. This domain has a signature for sulfur metabolite binding. This further substantiates the idea that sulfur metabolism is compartmentalized.
Finally it was interesting to note that B. subtilis, when in the presence of significant amounts of MTR, was likely to produce CO, that may act as a gaseous mediator. This is the more interesting because B. cereus and B. anthracis possess the same methionine salvage pathway as B. subtilis. Furthermore, because this pathway exists in humans, it may be relevant to the ongoing discussion about the role of CO as a neuronal or gastro-intestinal mediator [40,41]. Analysis of similar arrangements in other genomes will lead to recognition of new features of the genetics of microbial sulfur metabolism.
Conclusions
The MTA => methionine salvage pathway is present in many Bacteria. This pathway provides us with a remarkable example of enzyme recruitment in bacteria associated to their biotope. Indeed the enolase-phosphatase step in the pathway is performed by enzymes of entirely different descent, RuBisCO being the most puzzling example. In the same way it seems quite remarkable that a family of GTP binding proteins, previously shown to be involved in translation elongation, universally belong to this pathway (MtnA). It will be an interesting link to compare the reactions in anaerobic environments. This will teach us how we could modify enzyme activities in the future and reprogram biochemical processes.
Methods
Genetic and biochemical experiments
Growth conditions
The following B. subtilis strains were used in this work: the wild type strain 168 [42], BFS1853 (mtnD(ykrZ)::lacZ) and BFS4605 (yflM::lacZ). These strains have been constructed during the B. subtilis functional analysis program (http://locus.jouy.inra.fr/cgi-bin/genmic/madbase/progs/madbase.operl and http://bacillus.genome.ad.jp[26,43]). Escherichia coli strains were grown either on LB or on nutrient yeast broth (NYB) liquid or agar plates at 37°C. B. subtilis was grown in ED minimal medium: K2HPO4, 8 mM; KH2PO4, 4.4 mM; glucose, 27 mM; Na3-citrate, 0.3 mM; L-glutamine, 15 mM; L-tryptophan, 0.244 mM; ferric citrate, 33.5 μM; MgSO4, 2 mM; MgCl2, 0.61 mM; CaCl2, 49.5 μM; FeCl3, 49.9 μM; MnCl2, 5.05 μM; ZnCl2, 12.4 μM; CuCl2, 2.52 μM; CoCl2, 2.5 μM; Na2MoO4, 2.48 μM at 37°C. To assay for CO production, ED minimal medium plates were used with either a MgSO4 containing medium or a sulfur-free basal medium (where MgSO4 was replaced by MgCl2 at 2 mM concentration). In the latter case, 10 μl of the sulfur source under investigation (MTR, 200 mM stock solution) was applicated onto paper discs deposited at the center of the plate, after bacteria had been uniformly spread on the surface of the plate. All experiments were performed in accordance with the European regulation requirements concerning the contained use of Genetically Modified Organisms of Group-I (French agreement N°2735). P. aeruginosa strains were routinely grown in NYB or on nutrient agar plates at 37°C. When required, antibiotics were added to media at the following concentrations: tetracycline (Tc), 25 μg ml-1 (E. coli) or 125 μg ml-1 (P. aeruginosa); kanamycin (Km), 50 μg ml-1 (E. coli); ampicillin (Ap), 100 μg ml-1 (E. coli); spectinomycin (Sp), 25 μg ml-1 (E. coli) or 1,000 μg ml-1 (P. aeruginosa); erythromycin plus lincomycin, 1 μg ml-1 and 25 μg ml-1 (B. subtilis). To counterselect E. coli S17-1 donor cells in matings with P. aeruginosa for gene replacement, chloramphenicol was used at a concentration of 10 μg ml-1; mutant enrichment experiments were performed with tetracycline at a final concentration of 20 μg ml-1 and carbenicillin at a final concentration of 2,000 μg ml-1. Strain PAO503 and its derivatives were cultivated in NYB, washed and plated out on minimal medium E; 50 μl of methionine, homocysteine or MTA (50 mM each) were then spotted onto sterile filters placed on the bacterial lawn. Growth around the filters was scored after 18 h of incubation. The optical density (OD) of bacterial cultures was measured at 600 nm.
DNA manipulation and cloning procedures
Small- and large-scale preparations of plasmid DNA were carried out by the cetyltrimethylammonium bromide (CTAB) method [44] and with JetStar columns (Genomed, Basel, Switzerland), respectively. Chromosomal DNA was purified from P. aeruginosa with phenol:chloroform (1:1) by a method described previously [45]. Restriction enzyme digestions, PCR, ligation, agarose gel electrophoresis, and electroporation were performed using standard procedures [46,47]. For Southern blotting, chromosomal DNA was digested overnight with the appropriate enzymes, electrophoresed on a 0.8% agarose gel and transferred to a Hybond-N nylon membrane (Amersham Biosciences). Labeling of DNA probes, hybridization and detection were performed using the digoxigenin (DIG) DNA Labeling and Detection Kit (Roche) according to the manufacturer's procedures. Nucleotide sequences of PCR-derived constructs were determined on both strands with a dye terminator kit (Perkin-Elmer product 402080) and an ABI Prism 373 sequencer.
Construction of P. aeruginosa mutant strains
For the construction of strain PAO6412 (metZ9011 mtnB::Ω-Sp/Sm), the mtnB gene (= PA1683) was PCR-amplified using PAO1 genomic DNA as a template and primers PA1683-1 (5'-AAAAGAATTCTGCACCTGGAATCGCTGGG-3') and PA1683-2 (5'-AAAAGGATCCCGGTGCTCGTCGAGGAACTTG-3'), tagged with EcoRI and BamHI artificial sites (italics), respectively. The mtnB gene was interrupted by a spectinomycin-streptomycin resistance cassette (Ω-Sp/Sm) integrated as a 2-kb SmaI fragment into the SmaI site of mtnB; the resulting 3.3-kb EcoRI-XbaI fragment containing mtnB::Ω-Sp/Sm was inserted into the suicide plasmid pME3087 [48] to generate pME3880. This plasmid was mobilized into PAO503 (metZ9011) by the donor E. coli S17-1 (thi pro hsdR recA/RP4-2 Tc::Mu Km::Tn7) and chromosomally integrated with selection for tetracycline resistance. Excision of the pME3087 carrying mtnB by a second crossover was obtained by enrichment for tetracycline-sensitive cells [49]. The tetracycline-sensitive, spectinomycin-resistant clones (containing mtnB::Ω-Sp/Sm) were checked by Southern blotting (data not shown).
For the construction of strain PAO6422 (metZ9011 mtnP::Ω-Sp/Sm), the mtnP gene (= PA3004) was PCR-amplified with primers PA3004-1 (5'-AAAAGGATCCGTCGAAGGCGGTTTCGAC-3') and PA3004-2 (5'-AAAAAAGCTTCATGTGGCGATTCCCGAAGAC-3') containing artificial sites for BamHI and HindIII (italics). To generate pME3891, the 2-kb SmaI fragment containing the Ω-Sp/Sm cassette was integrated into the EcoRV site of mtnP and the resulting 3.3-kb KpnI-XbaI fragment carrying mtnP::Ω-Sp/Sm was inserted into the suicide plasmid pME3087. The resulting plasmid (pME3891) was delivered to the chromosome of PAO503 as described above, resulting in strain PAO6422, which was checked by Southern hybridization as above.
Preparation of MTR and CO assays
Methylthioribose (MTR) was prepared from MTA (Sigma, D5011) by acid hydrolysis as described by Schlenk et al. [50]. The CO production assay was carried using the GASTEC No.1DL dosimeters (Carbon Monoxide Low Range Passive Dosi-Tubes, Gastec Corporation, Japan). To detect CO production the ED minimal medium plates of 150 mm diameter were inoculated with bacteria and the dosimeter was activated by breaking the tube at its score. Plates were incubated at 37°C for 20 hrs and readings were recorded after the end of incubation period. Comparison of replicates of the same experiment (same volume of the growth medium, same temperature, same incubation time) indicated that the level of significance was better than 5 ppm. Control and MTR experiments were performed on the same day. It is not possible, however, to separate the nature of the different reducing gasses from the production of palladium metal precipitates. Replicates of the experiment on plates without bacteria gave an average level of reducing gasses of ca 12.5 ppm/h.
Computer-mediated analyses
Data were extracted from the microbial genome databases set up at http://bioinfo.hku.hk/genolist.html, using the following databases with their imbedded BLAST browsers and searching using appropriate patterns ("search patterns") when appropriate: AeruList, LeptoList, ThermaList and XylelList. For B. subtilis, the reference database was used: http://genolist.pasteur.fr/SubtiList. Analyses of domains were performed using "CD-search" at the NCBI [51]. Annotations were used with the help of SwissProt [52]. Alignments were performed using CLUSTALW and further refined manually [53].
Authors' contributions
All authors participated in the final form of the article. AS set up and performed the experiments meant to detect the presence of reducing gasses in B. subtilis and its mutants. VD performed the genetic experiments with P. aeruginosa. HA elaborated experiments of the methionine salvage pathway in B. subtilis. KM established the nomenclature and harmonized it with the Swissprot requirements. DH conceived the experiments with P. aeruginosa. AY established the biochemical protocol needed to identify the steps of the methionine salvage pathway in B. subtilis. AD performed the sequence comparisons and wrote the core of the article.
Acknowledgments
Acknowledgements
We would first like to acknowledge the contribution of the three anonymous reviewers who contributed to improve considerably the quality of this article. We wish to thank Evelyne Turlin for help in some of the experiments for monitoring reducing gasses in B. subtilis. This work benefited from support by the Innovation and Technology Fund, Hong Kong (BIOSUPPORT grant), Programme Commun de Recherche en Génie Biomédical (Swizerland) and by a grant from the Centre National de la Recherche Scientifique (Panoramix). Connection to human genes was made as a first contribution to the European Union BioSapiens programme.
Contributor Information
Agnieszka Sekowska, Email: sekowska@pasteur.fr.
Valérie Dénervaud, Email: valerie.denervaud@unifr.ch.
Hiroki Ashida, Email: hashida@bs.aist-nara.ac.jp.
Karine Michoud, Email: karine.michoud@isb-sib.ch.
Dieter Haas, Email: Dieter.Haas@imf.unil.ch.
Akiho Yokota, Email: yokota@bs.aist-nara.ac.jp.
Antoine Danchin, Email: adanchin@pasteur.fr.
References
- Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol. 2000;2:145–177. [PubMed] [Google Scholar]
- Rando RR. Chemical biology of protein isoprenylation/methylation. Biochim Biophys Acta. 1996;1300:5–16. doi: 10.1016/0005-2760(95)00233-2. [DOI] [PubMed] [Google Scholar]
- Takemura M, Kitanaka N, Kitanaka J. Signal transduction by histamine in the cerebellum and its modulation by N-methyltransferase. Cerebellum. 2003;2:39–43. doi: 10.1080/14734220309430. [DOI] [PubMed] [Google Scholar]
- Turner BM. Memorable transcription. Nat Cell Biol. 2003;5:390–393. doi: 10.1038/ncb0503-390. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc Natl Acad Sci U S A. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppenmeier U. The unique biochemistry of methanogenesis. Prog Nucleic Acid Res Mol Biol. 2002;71:223–283. doi: 10.1016/S0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- Winans SC, Bassler BL. Mob psychology. J Bacteriol. 2002;184:873–883. doi: 10.1128/jb.184.4.873-883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Saito Y, Kojima C, Kobayashi K, Ogasawara N, Yokota A. A functional link between RuBisCO-like protein of Bacillus and photosynthetic RuBisCO. Science. 2003;302:286–290. doi: 10.1126/science.1086997. [DOI] [PubMed] [Google Scholar]
- Zappia V, Oliva A, Cacciapuoti G, Galletti P, Mignucci G, Carteni-Farina M. Substrate specificity of 5'-methylthioadenosine phosphorylase from human prostate. Biochem J. 1978;175:1043–1050. doi: 10.1042/bj1751043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riscoe MK, Ferro AJ, Fitchen JH. Analogs of 5-methylthioribose, a novel class of antiprotozoal agents. Antimicrob Agents Chemother. 1988;32:1904–1906. doi: 10.1128/aac.32.12.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower PA, Alexander DB, Johnson LL, Riscoe MK. Regulation of methylthioribose kinase by methionine in Klebsiella pneumoniae. J Gen Microbiol. 1993;139 ( Pt 5):1027–1031. doi: 10.1099/00221287-139-5-1027. [DOI] [PubMed] [Google Scholar]
- Schroeder HR, Barnes CJ, Bohinski RC, Mallette MF. Biological production of 5-methylthioribose. Can J Microbiol. 1973;19:1347–1354. doi: 10.1139/m73-217. [DOI] [PubMed] [Google Scholar]
- Savarese TM, Cannistra AJ, Parks R. E., Jr., Secrist J. A., 3rd, Shortnacy AT, Montgomery JA. 5'-deoxy-5'-methylthioadenosine phosphorylase--IV. Biological activity of 2-fluoroadenine-substituted 5'-deoxy-5'-methylthioadenosine analogs. Biochem Pharmacol. 1987;36:1881–1893. doi: 10.1016/0006-2952(87)90484-9. [DOI] [PubMed] [Google Scholar]
- Foglino M, Borne F, Bally M, Ball G, Patte JC. A direct sulfhydrylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology. 1995;141:431–439. doi: 10.1099/13500872-141-2-431. [DOI] [PubMed] [Google Scholar]
- Bao Q, Tian Y, Li W, Xu Z, Xuan Z, Hu S, Dong W, Yang J, Chen Y, Xue Y, Xu Y, Lai X, Huang L, Dong X, Ma Y, Ling L, Tan H, Chen R, Wang J, Yu J, Yang H. A complete sequence of the T. tengcongensis genome. Genome Res. 2002;12:689–700. doi: 10.1101/gr.219302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RD, Duerre JA. S-adenosylhomocysteine metabolism in various species. Can J Biochem. 1975;53:312–319. doi: 10.1139/o75-044. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Burgess N, Doherty N, Kirke D, Holden MT, Linforth R, Cornell KA, Taylor AJ, Hill PJ, Williams P. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology. 2002;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure (Camb) 2001;9:941–953. doi: 10.1016/S0969-2126(01)00656-6. [DOI] [PubMed] [Google Scholar]
- Sekowska A, Danchin A. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 1999;6:255–264. doi: 10.1093/dnares/6.5.255. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Sekowska A, Mulard L, Krogh S, Tse JK, Danchin A. MtnK, methylthioribose kinase, is a starvation-induced protein in Bacillus subtilis. BMC Microbiol. 2001;1:15. doi: 10.1186/1471-2180-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SY, Yip P, Cornell KA, Riscoe MK, Howell PL. Crystallization and preliminary X-ray analysis of 5'-methylthioribose kinase from Bacillus subtilis and Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr. 2004;60:116–119. doi: 10.1107/S0907444903022042. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Hosaka K. Choline kinase from yeast. Biochim Biophys Acta. 1997;1348:63–69. doi: 10.1016/S0005-2760(97)00104-5. [DOI] [PubMed] [Google Scholar]
- Trackman PC, Abeles RH. Methionine synthesis from 5'-S-Methylthioadenosine. Resolution of enzyme activities and identification of 1-phospho-5-S methylthioribulose. J Biol Chem. 1983;258:6717–6720. [PubMed] [Google Scholar]
- Sekowska A, Danchin A. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2002;2:8. doi: 10.1186/1471-2180-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson TE, Tabita FR. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc Natl Acad Sci U S A. 2001;98:4397–4402. doi: 10.1073/pnas.081610398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RW, Abeles RH. Conversion of 5-S-methyl-5-thio-D-ribose to methionine in Klebsiella pneumoniae. Stable isotope incorporation studies of the terminal enzymatic reactions in the pathway. J Biol Chem. 1990;265:16913–16921. [PubMed] [Google Scholar]
- Wray JW, Abeles RH. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases. J Biol Chem. 1995;270:3147–3153. doi: 10.1074/jbc.270.7.3147. [DOI] [PubMed] [Google Scholar]
- Pochapsky TC, Pochapsky SS, Ju T, Mo H, Al-Mjeni F, Maroney MJ. Modeling and experiment yields the structure of acireductone dioxygenase from Klebsiella pneumoniae. Nat Struct Biol. 2002;9:966–972. doi: 10.1038/nsb863. [DOI] [PubMed] [Google Scholar]
- Gelfand DH, Steinberg RA. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol. 1977;130:429–440. doi: 10.1128/jb.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BJ, English S, Chan G, Knodel MH. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J Bacteriol. 2003;185:2418–2431. doi: 10.1128/JB.185.8.2418-2431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A, Robin S, Daudin JJ, Henaut A, Danchin A. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2001;2:RESEARCH0019. doi: 10.1186/gb-2001-2-6-research0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronn J, Wilson J, Berger BJ. Tyrosine aminotransferase catalyzes the final step of methionine recycling in Klebsiella pneumoniae. J Bacteriol. 1999;181:1739–1747. doi: 10.1128/jb.181.6.1739-1747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke P, Guerdoux-Jamet P, Chiapello H, Faroux G, Henaut C, Henaut A, Danchin A. Indigo: a World-Wide-Web review of genomes and gene functions. FEMS Microbiol Rev. 1998;22:207–227. doi: 10.1016/S0168-6445(98)00014-X. [DOI] [PubMed] [Google Scholar]
- Marques MV, da Silva AM, Gomes SL. Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa. Plasmid. 2001;45:184–199. doi: 10.1006/plas.2000.1514. [DOI] [PubMed] [Google Scholar]
- Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Sekowska A, Danchin A. Sulphur islands in the Escherichia coli genome: markers of the cell's architecture? FEBS Lett. 2000;476:8–11. doi: 10.1016/S0014-5793(00)01660-4. [DOI] [PubMed] [Google Scholar]
- Patthy L. Genome evolution and the evolution of exon-shuffling--a review. Gene. 1999;238:103–114. doi: 10.1016/S0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- Gibbons SJ, Farrugia G. The role of carbon monoxide in the gastrointestinal tract. J Physiol. 2004. [DOI] [PMC free article] [PubMed]
- Boehning D, Snyder SH. Novel neural modulators. Annu Rev Neurosci. 2003;26:105–131. doi: 10.1146/annurev.neuro.26.041002.131047. [DOI] [PubMed] [Google Scholar]
- Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sal G, Manfioletti G, Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988;16:9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper M, Ganter B, Polito MR, Haas D. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J Mol Biol. 1992;226:943–957. doi: 10.1016/0022-2836(92)91044-p. [DOI] [PubMed] [Google Scholar]
- Farinha MA, Kropinski AM. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol Lett. 1990;58:221–225. doi: 10.1111/j.1574-6968.1990.tb13982.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EG, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Voisard C, Bull CT, Keel C, Laville J, Maurhofer M, Schnider U, Défago G, Haas D. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches. In: F O'Gara, DN Dowling and B Boesten, editor. Molecular ecology of rhizosphere microorganisms. VCH Weinheim; 1994. pp. 67–89. [Google Scholar]
- Haas D, Watson J, Krieg R, Leisinger T. Isolation of an Hfr donor of Pseudomonas aeruginosa PAO by insertion of the plasmid RP1 into the tryptophan synthase gene. Mol Gen Genet. 1981;182:240–244. doi: 10.1007/BF00269664. [DOI] [PubMed] [Google Scholar]
- Schlenk F, Zydek-Cwick CR, Dainko JL. 5'-Methylthioadenosine and related compounds as precursors of S- adenosylmethionine in yeast. Biochimica et Biophysica Acta. 1973;320:357–362. doi: 10.1016/0304-4165(73)90316-4. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler GH, Mazumder R, Nikolskaya AN, Panchenko AR, Rao BS, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A, Michoud K, Rivoire C, Auchincloss AH, Coudert E, Lima T, Kersey P, Pagni M, Sigrist CJ, Lachaize C, Veuthey AL, Gasteiger E, Bairoch A. Automated annotation of microbial proteomes in SWISS-PROT. Comput Biol Chem. 2003;27:49–58. doi: 10.1016/S1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. Atlas of Protein Sequence and Structure. Vol. 5. Washington DC, National Biochemical Research Foundation; 1978. [Google Scholar]