Abstract
Background
Alzheimer's disease is multifactorial, having environmental, toxicological and genetic risk factors. Impaired folate and homocysteine metabolism has been hypothesised to increase risk. In addition to its xenobiotic-metabolising capacity, human arylamine N-acetyltransferase type-1 (NAT1) acetylates the folate catabolite para-aminobenzoylglutamate and is implicated in folate metabolism. The purpose of this study was to determine whether polymorphisms in the human NAT genes influence susceptibility to Alzheimer's disease.
Methods
Elderly individuals with and without Alzheimer's disease were genotyped at the polymorphic NAT1 (147 cases; 111 controls) and NAT2 (45 cases; 63 controls) loci by polymerase chain reaction-restriction fragment length polymorphism, and the genotype and allele frequencies were compared using the chi-squared test.
Results
Although a trend towards fast NAT2 acetylator-associated Alzheimer's disease susceptibility was indicated and the NAT1*10/1*10 genotype was observed only in cases of Alzheimer's disease (6/147, 4.1%), no significant difference in the frequency of NAT2 (p = 0.835) or NAT1 (p = 0.371) genotypes was observed between cases and controls. In addition, a novel NAT1 variant, NAT1*11B, was identified.
Conclusions
These results suggest that genetic polymorphisms in NAT1 and NAT2 do not influence susceptibility to Alzheimer's disease, although the increase in frequency of the NAT1*10 allele in Alzheimer's disease is worthy of further investigation. Due to its similarity with the NAT1*11A allele, NAT1*11B is likely to encode an enzyme with reduced NAT1 activity.
Background
Alzheimer's disease (AD) is a common multifactorial disease of the elderly, which results in progressive neurodegeneration causing severe and permanent cognitive impairment [1]. As a result of family pedigree analyses and case-control studies, multiple genetic risk factors such as mutations in the β-amyloid precursor protein (APP) [2], presenilin type-1 (PS1) [3], presenilin type-2 (PS2) [4] and apolipoprotein E (APOE) [5] genes have been identified. Genetic epidemiology studies have however, estimated that only 30% to 50% of the population risk for AD can be attributed to genetic factors [6]. This along with the low concordance rate of AD observed in monozygotic twins [7,8] suggests that environmental factors may also influence AD susceptibility. This may be the case particularly for later-onset 'sporadic' cases of the disease, which account for the majority (approximately 90%) of AD cases but show only modest familial clustering [6].
Many putative environmental risk factors for AD have been proposed. In particular, low blood levels of folate and elevated serum total homocysteine have been associated with increased risk of AD [9-11]. Differences in the level of serum homocysteine and folate were not associated with increasing duration of the symptoms of AD [10], which suggests that the observed level of these biochemical markers was not due to progression of the disease. Therefore, impaired folate and homocysteine (one-carbon) metabolism has been hypothesised as a risk factor in AD [10]. Such altered folate and homocysteine metabolism could arise as a result of genetic mutations in enzymes of folate metabolism.
The association between impaired folate and homocysteine metabolism and AD is particularly interesting in view of the putative endogenous role of the human phase II xenobiotic-metabolising enzyme arylamine N-acetyltransferase type-1 (NAT1)(and its murine homologue NAT2), in folate catabolism [12-15], and our previous demonstration of the expression of murine NAT2 in particular cell types, such as the cytoplasm and dendrites of the Purkinjie cells of the cerebellum in the adult mouse brain [16]. There has as yet been no description of the pattern of human NAT1 expression in the adult brain, although human NAT1 (the human equivalent of murine NAT2) activity has been identified in the brain early in embryonic development [17]. Human NAT1 is polymorphic [18] and inter-individual variation in NAT1 activity [19] may modulate individual folate levels. NAT1 may therefore be a potential low penetrance gene which can modify individual risk of AD.
Polymorphisms in the arylamine N-acetyltransferase type-2 (NAT2) gene, which encodes the phase II xenobiotic-metabolising isozyme NAT2, have been linked with increased susceptibility to multifactorial neurodegenerative disorders such as Parkinson's disease [20,21]. In this case the impaired ability of the individual to handle environmental xenobiotics or neurotoxins acting on the brain has been hypothesised to contribute to the development of the disease. In a similar manner human NAT2, and in addition human NAT1, in their more traditionally recognized role as phase II xenobiotic-metabolising enzymes [22], may also be modulators of AD risk as a result of chemical insult. An association study carried out by Rocha et al., [23] has previously indicated that human NAT2 may be a potential low penetrance gene in AD pathogenesis. In view of these findings we undertook a study to genotype an elderly group of individuals with and without a history of AD for major alleles at the NAT1 and NAT2 loci, to investigate the role of NAT1 and NAT2 polymorphisms in AD susceptibility.
Methods
Study populations
Genomic DNA from 148 elderly Caucasian individuals with AD and 90 elderly Caucasian individuals without AD, from the Oxford Project to Investigate Memory and Ageing (OPTIMA) cohort, was available for investigation. OPTIMA is a longitudinal study of normal, non-institutionalised elderly volunteers with good cognitive function, and elderly patients with memory problems from the Oxfordshire community (UK), and has been described in detail elsewhere [10,24]. Briefly, the OPTIMA study follows the AD course of an individual, from initial diagnosis to post mortem confirmation. Every year each subject is examined by neurological and neuropsychological tests, brain scans and biochemical analysis of blood and cerebrospinal fluid. Cognitive evaluation is undertaken using the Cambridge Examination for Mental Disorders of the Elderly [25]. This includes the Cambridge Cognitive Examination (CAMCOG), a neuropsychological test which includes elements of the Mini-Mental State Examination and assesses a broad range of cognitive functions, such as memory, language, attention, perception, praxis and thinking. A cut-off value of <80/107 CAMCOG points discriminates between demented and normal subjects. Clinical diagnosis is made according to the National Institute of Neurological and Communicative Disorders and Stroke-Alzheimer's Disease and Related Disorders Association criteria [1]. Post mortem, the brain is examined by histochemical and biochemical methods to confirm disease pathology using Consortium to Establish a Registry for Alzheimer's Disease criteria [26]. In this study the AD case individuals from the OPTIMA cohort included: 101 living patients with a clinical diagnosis of probable or possible AD, 30 patients with histologically confirmed pure AD with the absence of any additional disease pathology, 16 patients with histologically confirmed AD with the coexistence of vascular or Parkinson's disease pathology and 1 patient who demonstrated clinical symptoms of AD which were not confirmed post mortem. The non-demented control individuals from the OPTIMA cohort included: 83 living subjects with no clinically diagnosed symptoms of AD, 4 subjects with the absence of AD confirmed post mortem and 3 subjects who demonstrated no symptoms of AD which was not confirmed post mortem. The OPTIMA study had ethical approval from the central Oxford and Psychiatric Research Ethics Committees and informed consent was obtained in writing from all subjects.
Genomic DNA from 22 elderly volunteers from the Foresight Challenge cohort was also available for investigation. Foresight Challenge is a cohort of normal, non-institutionalised elderly volunteers with good cognitive function from the Oxfordshire community (UK)(non-black and Caucasian), recruited for a three-year longitudinal study aimed at further defining early markers or predictors of cognitive impairment and their relationship to the subsequent development of dementia [27]. Participants were excluded if they: (a) scored ≤80/107 points on the CAMCOG or ≤24/30 points on the Mini-Mental State Examinations [25] at the initial screening visit, (b) reported significant progressive subjective memory complaints, (c) lived in institutional care, or (d) were unable to complete the Cambridge Examination for Mental Disorders of the Elderly [25]. For the purpose of this study the individuals in the case and control groups were regarded as homogeneous groups. Subjects with other causes of dementia were excluded from the study.
Materials
All chemicals were purchased from Sigma-Aldrich Company Ltd., Merck Ltd. or BDH Laboratory Supplies, UK. Molecular biology reagents were purchased from Promega, Roche Molecular Biochemicals and New England Biolabs Inc., UK. Oligonucleotides were synthesized by Sigma-Genosys Ltd., UK.
DNA extraction
Genomic DNA from each AD case (n = 148) and control (n = 90) subject in the OPTIMA cohort, and from each control subject (n = 22) in the Foresight Challenge cohort was prepared from the buffy coat fraction of EDTA blood [28] using the Wizard® Genomic DNA Purification Kit (Promega, UK), and was stored at 4°C in TE buffer (10 mM Tris-HCl (pH 7.5), 1 mM Na2EDTA).
NAT1 genotyping
The human NAT1*3, NAT1*4, NAT1*10, NAT1*11A and NAT1*14A alleles (those unfamiliar with NAT allele nomenclature should see http://www.louisville.edu/medschool/pharmacology/NAT.html) were identified by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and allele-specific PCR (AS-PCR) step-by-step genotyping procedure [29]. To summarise, a fragment of 344 bp between nucleotides 769 and 1113 was amplified with primers N769 (5'-ACTCTGAGTGAGGTAGAAATA-3') and N1113 (5'-ACAGGCCATCTTTAGAA-3'). As a result of Mbo II sites at nucleotides 945 and 969, the alleles NAT1*3, NAT1*10 and NAT1*14A produce fragments of 176 bp, 24 bp and 144 bp on digestion with Mbo II (for NAT1*11A, as a result of a 9 bp deletion, the fragments are 176 bp, 24 bp and 135 bp). The primer N1113 introduces an additional Mbo II site into NAT1*4 such that the 144 bp fragment is cleaved to 125 bp and 19 bp. Subsequently, NAT1*3 was distinguished from NAT1*10 and NAT1*14A by an allele-specific PCR. NAT1*10 and NAT1*14A were distinguished by a further PCR and digestion with BsaO I [29]. Nucleotides which are changed from the NAT1*4 sequence (EMBL Accession Number AJ307007) are underlined.
The NAT1*14A allele (G560A, T1088A, C1095A) was confirmed and distinguished from the similar NAT1*14B allele (G560A), by further analysis using AS-PCR and nested PCR-RFLP. This analysis first amplified only NAT1*10 and NAT1*14A allele sequences, and then identified whether the G560A variation was present. AS-PCR amplification was performed in a 100 μl reaction using 1X PCR reaction buffer, 250 ng genomic DNA, 2.5 mM MgCl2 0.2 mM dNTPs, 2.5 U Taq DNA polymerase and 25 pmoles each of primers N539 (5'-TCCTAGAAGACAGCAACGACC-3') and N1110b (5'-GGCCATCTTTAAAATACATTTT-3')(with annealing temperature 54°C and a product extension of 1 minute at 72°C). Primer N1110b, which contains T1088 annealed to and amplified a 571 bp product from the NAT1*10 and NAT1*14A allele sequences only. The PCR product from the first round of amplification was diluted 1 in 20, and 1 μl used as template in a PCR with primers N539 and N714 (5'-GTGAAGCCCACCAAACAG-3'), followed by BsaO I digestion of the 175 bp amplified product [29]. NAT1*14A contains G560A; therefore no BsaO I site was generated. NAT1*10 contains G560, therefore a BsaO I site was generated resulting in fragments of 155 bp and 20 bp.
NAT2 genotyping
The human NAT2*4, NAT2*5A, NAT2*5B, NAT2*5C, NAT2*6A and NAT2*7B alleles were identified by genotyping using the PCR-RFLP method of Hickman & Sim [30], and Hickman et al. [31]. This method is more than 95% accurate at predicting the NAT2 phenotype in a Caucasian population [31]. This method of genotyping is unable to distinguish between the NAT2*4/NAT2*5B and the NAT2*5A/NAT2*5C genotypes. However, since the expected frequency of the NAT2*4 (19% to 27%) and NAT2*5B (38% to 45%) alleles in a Caucasian population is much greater than the expected frequency of the NAT2*5A (1.0% to 4.2%) and NAT2*5C (1.0% to 4.3%) alleles [32], this method is sufficient to confer an accurate genotype in greater than 99% of cases if the NAT2*4/NAT2*5B genotype is inferred. Genotyping of both NAT1 and NAT2 was carried out blind with respect to the disease status of the subjects.
Sequencing of novel NAT1 variant
PCR amplification of a 1614 nucleotide region of NAT1 for cloning and sequencing of a NAT1 variant was performed in a 100 μl reaction using 1 μg genomic DNA, 2.5 U high-fidelity Pfu DNA Polymerase (Promega, UK), 1X Pfu DNA Polymerase reaction buffer with 2 mM MgSO4, 0.2 mM dNTPs, and 25 pmoles each of primers N-438 (5'-TTGTATAAGGCTCAGCTAAAAGGG-3') and N1176 (5'-GGAATTCAACAATAAACCAACAT-3')(with annealing temperature 56°C and a product extension of 2 minutes at 72°C). Following gel purification, the amplified PCR product was A-tailed with dATP, ligated into the pGEM®-T Easy Vector (Promega, UK), and transformed into high efficiency JM109 Competent E. coli cells (Promega, UK) by heat shock. Positive clones were identified by blue-white selection on LB agar plates containing 100 μg/ml ampicillin, 0.1 mM isopropyl-β-D-thiogalactoside (Melford Laboratories Ltd., UK) and 40 μg/ml X-Gal. The identity of the cloned NAT1 sequence was confirmed by NAT1 genotyping of plasmid DNA. The cloned NAT1 inserts (two different clones for each allele) were sequenced using M13F (5'-AGGGTTTTCCCAGTCACGA-3') and M13R (5'-ACACAGGAAACAGCTATGAC-3') sequencing primers, and the NAT1-specific primer N714. Fluorescent DNA sequencing was performed by the DNA Sequencing Facility, Department of Biochemistry, University of Oxford, using an ABI 377XL Prism DNA Sequencer with ABI BigDye™ terminators (Warrington, UK). Sequencing chromatograms were analysed using Chromas version 1.45 (Conor McCarthy, Griffith University, Australia).
Statistical analysis
NAT allele and genotype frequencies for the AD case and control groups were calculated and the NAT genotype distribution for AD case and control groups was tested for Hardy-Weinberg equilibrium. Any deviations from this equilibrium were assessed using a chi-squared test. NAT allele and genotype distributions of the AD case and control groups were compared using the chi-squared test, and p values of less than 0.05 were considered to be significant. The strength of association between inheritance of a particular NAT allele or genotype and AD was assessed by calculation of crude odds ratios (OR) and 95% confidence intervals (CI).
Results
The NAT1 genotype of each individual in the Alzheimer's case group (n = 147) and the control group (n = 111) was determined and the frequency of NAT1 alleles and NAT1 genotypes is illustrated in Table 1 and Table 2 respectively. The distribution of alleles demonstrates that NAT1*4 is the most common allele in both groups (75–79%), whilst NAT1*10 is the next most frequent (15–19%). The same pattern has been observed in all Caucasian populations that have been studied [32]. The genotypes observed were determined to be in Hardy-Weinberg equilibrium.
Table 1.
NAT1 allele frequency in AD cases and controls.
| NAT1 ALLELE | AD CASES (N = 147) | CONTROLS (N = 111) | OR (95% CI) |
| 1*4 | 220 (0.75) | 176 (0.79) | 1.00 |
| 1*3 | 9 (0.03) | 7 (0.03) | 1.03 (0.38, 2.82) |
| 1*10 | 57 (0.19) | 33 (0.15) | 1.38 (0.86, 2.22) |
| 1*11A | 3 (0.01) | 2 (0.01) | 1.20 (0.20, 7.26) |
| 1*14A | 5 (0.02) | 4 (0.02) | 1.00 (0.27, 3.78) |
| TOTAL | 294 (1.00) | 222 (1.00) | - |
The number and frequency (in brackets) of NAT1 alleles in 147 AD cases and 111 controls is shown. The OR of AD to non-disease is given with 95% CI for each NAT1 allele (with NAT1*4 as the reference group).
Table 2.
NAT1 genotype frequency in AD cases and controls.
| NAT1 GENOTYPE | AD CASES (N = 147) | CONTROLS (N = 111) | EXPECTED | OR (95% CI) |
| 1*4/1*4 | 81 (0.55) | 65 (0.59) | 0.63 | 1.00 |
| 1*4/1*10 | 43 (0.29) | 33 (0.30) | 0.24 | 1.05 (0.60, 1.83) |
| 1*4/1*3 | 7 (0.05) | 7 (0.06) | 0.05 | 0.80 (0.27, 2.40) |
| 1*4/1*14A | 5 (0.03) | 4 (0.04) | 0.03 | 1.00 (0.26, 3.89) |
| 1*4/1*11A | 3 (0.02) | 2 (0.02) | 0.01 | 1.20 (0.20, 7.42) |
| 1*10/1*10 | 6 (0.04) | 0 (0) | 0.02 | - |
| 1*3/1*10 | 2 (0.01) | 0 (0) | 0.01 | - |
| Others | 0 (0) | 0 (0) | 0.01 | - |
| TOTAL | 147 (1.00) | 111 (1.00) | 1.00 | - |
The number and frequency (in brackets) of NAT1 genotypes in 147 AD cases and 111 controls is shown. Expected genotype frequency was calculated based on the allele frequency in the control group (Table 1). 'Others' includes additional NAT1 genotypes containing the alleles 1*4, 1*3, 1*10, 1*11A and 1*14A expected at low frequency which were not observed in these AD cases and controls. The OR of AD to non-disease is given with 95% CI for each NAT1 genotype (with NAT1*4/NAT1*4 as the reference group).
Although a chi-squared comparison of the two groups being investigated demonstrates that the allele and genotype frequency of the two groups does not differ significantly (χ2 = 1.84, p = 0.765 and χ2 = 6.48, p = 0.371 respectively) the number of NAT1*10 alleles is slightly elevated in the Alzheimer's patient population. Comparing the NAT1*10 homozygote genotype frequency, there are 6 out of 147 Alzheimer's cases with this genotype, whilst there are no control individuals with this genotype out of 111 investigated. Although the numbers are small, it is a finding worthy of further investigation, especially since the NAT1*10 allele may have an effect on the level of NAT1 expression [33].
NAT2 genotyping was carried out on a random subgroup of the AD cases (n = 45) and of the controls (n = 63). The frequency of NAT2 alleles and NAT2 genotypes (which were in Hardy-Weinberg equilibrium) is illustrated in Table 3 and Table 4 respectively. NAT2*5B is the most common allele in both groups (39–42%), whilst NAT2*6A is the next most frequent allele (31–36%), as seen in other Caucasian populations that have been studied [32]. A direct chi-squared comparison of the NAT2 allele and genotype frequencies in the Alzheimer's case and control groups indicates that they are not significantly different (χ2 = 1.95, p = 0.857 and χ2 = 5.76, p = 0.835 respectively). In a Caucasian population, an individual's NAT2 phenotype can be predicted quite accurately (in more than 95% of cases) by their NAT2 genotype [31]. Therefore the Alzheimer's cases and control individuals were classified as fast or slow acetylators and compared (Table 5). Although a small increase in the frequency of the NAT2 fast acetylator phenotype was observed in the AD cases (38%) compared with the controls (35%), which is in agreement with a similar study carried out in a Portuguese sporadic AD population by Rocha et al. [23], once again no significant difference between the Alzheimer's case and control group was observed (χ2 = 0.09, p = 0.761). Other studies have also demonstrated a lack of association between the NAT2 acetylator phenotype and risk of AD including those by Steventon et al. [34] and Ladero et al. [35]. However, in these cases it has been suggested that administration of therapeutic agents such as tacrine, an acetylcholinesterase inhibitor used in the treatment of AD, which is thought to be metabolised by CYP1A2 [36,37] may have affected the acetylation of sulphamethazine which was used to phenotype NAT2.
Table 3.
NAT2 allele frequency in AD cases and controls.
| NAT2 ALLELE | AD CASES (N = 45) | CONTROLS (N = 63) | OR (95% CI) |
| 2*4 | 20 (0.22) | 26 (0.21) | 1.00 |
| 2*5A | 1 (0.01) | 4 (0.03) | 0.33 (0.03, 3.14) |
| 2*5B | 35 (0.39) | 53 (0.42) | 0.86 (0.42, 1.77) |
| 2*5C | 1 (0.01) | 2 (0.02) | 0.65 (0.06, 7.69) |
| 2*6A | 32 (0.36) | 39 (0.31) | 1.07 (0.51, 2.25) |
| 2*7B | 1 (0.01) | 2 (0.02) | 0.65 (0.06, 7.69) |
| TOTAL | 90 (1.00) | 126 (1.00) | - |
The number and frequency (in brackets) of NAT2 alleles in 45 AD cases and 63 controls is shown. The OR of AD to non-disease is given with 95% CI for each NAT2 allele (with NAT2*4 as the reference group).
Table 4.
NAT2 genotype frequency in AD cases and controls.
| NAT2 GENOTYPE | AD CASES (N = 45) | CONTROLS (N = 63) | EXPECTED | OR (95% CI) |
| 2*4/2*4 | 3 (0.07) | 4 (0.06) | 0.04 | 1.00 |
| 2*4/2*5A | 1 (0.02) | 4 (0.06) | 0.01 | 0.33 (0.02, 4.74) |
| 2*4/2*5B | 6 (0.13) | 10 (0.16) | 0.17 | 0.80 (0.13, 4.87) |
| 2*4/2*6A | 6 (0.13) | 4 (0.06) | 0.13 | 2.00 (0.28, 14.20) |
| 2*4/2*7B | 1 (0.02) | 0 (0) | 0.01 | - |
| 2*5B/2*5B | 6 (0.13) | 8 (0.13) | 0.18 | 1.00 (0.16, 6.26) |
| 2*5B/2*5C | 1 (0.02) | 1 (0.02) | 0.01 | 1.33 (0.06, 31.12) |
| 2*5B/2*6A | 16 (0.36) | 25 (0.40) | 0.26 | 0.85 (0.17, 4.33) |
| 2*6A/2*6A | 5 (0.11) | 5 (0.08) | 0.10 | 1.33 (0.19, 9.31) |
| 2*5B/2*7B | 0 (0) | 1 (0.02) | 0.01 | - |
| 2*5C/2*7B | 0 (0) | 1 (0.02) | 0.00 | - |
| Others | 0 (0) | 0 (0) | 0.08 | - |
| TOTAL | 45 (1.00) | 63 (1.00) | 1.00 | - |
The number and frequency (in brackets) of NAT2 genotypes in 45 AD cases and 63 controls is shown. Expected genotype frequency was calculated based on the allele frequency in the control group (Table 3). 'Others' includes additional NAT2 genotypes containing the alleles 2*4, 2*5A, 2*5B, 2*5C, 2*6A and 2*7B expected at low frequency which were not observed in these AD cases and controls. The OR of AD to non-disease is given with 95% CI for each NAT2 genotype (with NAT2*4/NAT2*4 as the reference group).
Table 5.
NAT2 phenotype distribution in AD cases and controls.
| NAT2 PHENOTYPE | AD CASES (N = 45) | CONTROLS (N = 63) | OR (95% CI) |
| fast acetylator | 17 (0.38) | 22 (0.35) | 1.00 |
| slow acetylator | 28 (0.62) | 41 (0.65) | 0.88 (0.40, 1.96) |
| TOTAL | 45 (1.00) | 63 (1.00) | - |
The number and frequency (in brackets) of fast and slow NAT2 acetylators in 45 AD cases and 63 controls is shown. Based on their genotype, AD cases and controls were classified as either 'fast' or 'slow' NAT2 acetylators. Individuals homozygous or heterozygous for the NAT2*4 allele were assigned fast NAT2 acetylators, individuals who carried two 'slow' NAT2 alleles (NAT2*5A, NAT2*5B, NAT2*5C, NAT2*6A and NAT2*7B) in any combination were assigned slow NAT2 acetylators [31]. The OR of AD to non-disease is given with 95% CI for each NAT2 phenotype (with NAT2 fast acetylator as the reference group).
Discussion
It is hypothesized that variation in NAT1 activity may alter the risk of AD (which has been associated with low blood folate levels [9-11]), via the postulated role of NAT1 in folate metabolism. Together, the results of the present study suggest that genetic polymorphisms in NAT1 and NAT2 do not influence susceptibility to AD. It should be noted that the study presented here is preliminary and its statistical power is limited due to the relatively small number of samples analysed, particularly the study of NAT2 genotype and risk of AD. Therefore much larger case-control investigations, using more highly automated detection methods such as the LightCycler real-time PCR methods of Blömeke et al. [38] and Wikman et al. [39] will help to validate the results shown here. In order to detect a 1.5 fold increased risk of AD associated with the NAT1*10 allele, approximately 650 cases and 650 controls would need to be studied to give 80% power of achieving significance at the 5% level. Similarly, in order to detect a 1.5 fold increased risk of AD associated with the NAT2*4 allele, approximately 520 cases and 520 controls would need to be studied.
It is hypothesised that expression of a high activity NAT2 enzyme might increase the metabolic activation of environmental compounds (including neurotoxins), contributing to the neuronal tissue degeneration characteristic of AD. However, as no environmental compounds or neurotoxins currently suspected of contributing to the development of AD are known to be metabolised by the NAT enzymes, the identification of endogenous NAT substrates or NAT substrates which may be precursors of neurotoxic derivatives, will be important to strengthen the hypothesis that NAT1 and NAT2 may be risk factors for AD as a result of chemical insult.
At present, the suggestion for an endogenous role for human NAT1 has focused on the ability of human NAT1 to acetylate the folate catabolite para-aminobenzoylglutamate [12,13]. However, it is possible that NAT1 may play a role in the metabolism of other, as yet unidentified arylamines. Experiments in which the murine gene equivalent to human NAT1 (murine NAT2) has been knocked out indicate that superficially the mice are well [40]. However, in view of the distribution of murine NAT2 in the nervous system [16], it is important that subtle tests involving behaviour are carried out and the histology of the nervous system is studied in order to determine the role of murine NAT2 in the nervous system and identify whether there might be compensating factors at play in the genetically modified mice. In addition, mice over-expressing the human NAT1 gene show developmental abnormalities [41].
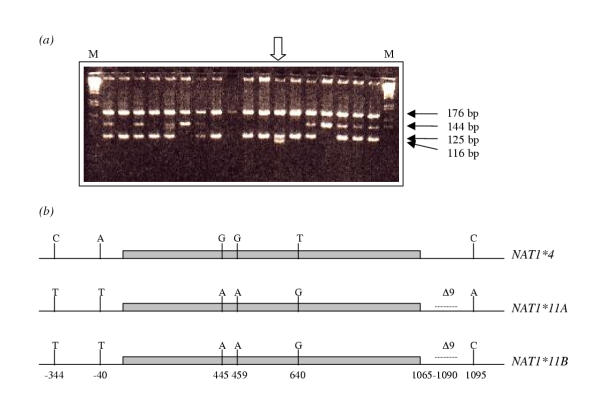
Whilst identifying the NAT1 alleles, a pattern was observed in one individual that was not consistent with any known NAT1 genotype. Only individuals with the NAT1*4 allele generate restriction fragments of 176 bp and 125 bp following PCR amplification of the region corresponding to nucleotides 769 to 1113, and digestion with the restriction enzyme Mbo II. This allele (NAT1*4) is present in the individual indicated by the open arrow in Figure 1(a). However, most other known NAT1 alleles (in which the primer N1113 does not generate an additional Mbo II restriction site) generate fragments of 176 bp and 144 bp, apart from NAT1*11, in which the corresponding fragment is 135 bp due to a 9 base pair deletion. In the case of the individual identified by the open arrow, an additional band of 116 bp was identified which was not consistent with any of the then-known alleles. To confirm whether this restriction pattern was due to the presence of a novel allele, the NAT1 alleles (a 1.6 Kb region) from the individual were cloned and sequenced. As a result, a novel allele was identified and defined as NAT1*11B (with C at 1095) due to its high sequence similarity with NAT1*11A (with A at 1095) (Figure 1(b)). NAT1*11B contains six mutations with respect to the NAT1*4 sequence: C-344T, A-40T, G445A (V149I), G459A, T640G (S214A), 9 bp deletion in the nucleotide region 1065–1090. The functional consequences of this allele are likely to be similar to that of NAT1*11A where an association with low enzymic activity has been proposed as a result of the amino acid changes V149I and S214A [42,43]. Other changes in the novel allele compared with NAT1*4 are outside the coding region (Figure 1(b)) and their effects have not yet been established. Although this allele was identified in an AD patient, it is unlikely to play a significant role in the pathogenesis of AD since it was found in only one individual.
Figure 1.
Identification of a novel NAT1 allele. (a) A 344 bp PCR product of NAT1 (amplified from genomic DNA using primers N769 and N1113, the latter introducing an additional Mbo II restriction site in NAT1*4) was digested with Mbo II and separated on a 4% (w/v) MetaPhor® agarose (FMC BioProducts, UK)/0.5X TBE gel, against a 1 kb DNA Ladder (Gibco BRL, UK)(Lane M). Detectable NAT1*4 fragments are 176 bp and 125 bp. Fragments of other known alleles are 176 bp and 144 bp (NAT1*3, NAT1*10, NAT1*14A) or 176 bp and 135 bp (NAT1*11A). The individual marked with an open arrow had a novel restriction fragment (116 bp) which did not correspond to any known allelic combination. (b) Both NAT1 alleles of the individual marked with an open arrow were sequenced across the coding and 5' and 3' flanking regions and the sequence of the novel allele (NAT1*11B) is illustrated in comparison with the most common Caucasian NAT1 allele, NAT1*4 (EMBL Accession Number AJ307007) and the most similar NAT1 allele, NAT1*11A. NAT1*11B contains sequence variations identical to those of NAT1*11A (C-344T, A-40T, G445A, G459A, T640G and a 9 base deletion (Δ) in the nucleotide range 1065–1090, compared with the NAT1*4 sequence), but contains a cytosine at nucleotide 1095. The 870 bp coding region of NAT1 is shaded grey, with 438 nucleotides upstream and 306 nucleotides downstream also shown.
Conclusions
In conclusion, the study presented here, which to our knowledge includes the first known investigation of the association between NAT1 genotype and susceptibility to AD, suggests that genetic polymorphisms in NAT1 and NAT2 do not influence susceptibility to AD. However, the inclusion of SNP analysis at positions 1088 and 1095 in a multi-variant analysis would be warranted based on the present studies since there is an indication that NAT 1*10 is elevated in AD.
Competing interests
None declared.
Authors' contributions
NJ carried out the NAT1 and NAT2 genotyping and NAT1 sequence analysis, performed the statistical analysis and drafted the manuscript. PB and VJ participated in the NAT1 genotyping. MB participated in the design of the study and provided study samples. ES conceived of the study, and participated in its design and coordination. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We are very grateful to The Wellcome Trust, Bristol-Myers Squibb and the MRC (Foresight Challenge Grant) for generous financial support. We thank Prof. A.D. Smith and his staff at OPTIMA for their co-operation as well as the individuals who provided the samples used in this study.
Contributor Information
Nichola Johnson, Email: nichola.johnson@icr.ac.uk.
Peter Bell, Email: peter.bell@pharm.ox.ac.uk.
Vesna Jonovska, Email: vesna.jonovska@pharm.ox.ac.uk.
Marc Budge, Email: marc.budge@act.gov.au.
Edith Sim, Email: edith.sim@pharm.ox.ac.uk.
References
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Force under the auspices of Department of Health and Human Services Task Force on Alzheimer's disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KA, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer's disease locus on chromosome 1. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- St George-Hyslop PH. Molecular genetics of Alzheimer's disease. Biological Psychiatry. 2000;47:183–199. doi: 10.1016/S0006-3223(99)00301-7. [DOI] [PubMed] [Google Scholar]
- Nee LE, Eldridge R, Sunderland T, Thomas CB, Katz D, Thompson KE, Weingartner H, Weiss H, Julian C, Cohen R. Dementia of the Alzheimer type: clinical and family study of 22 twin pairs. Neurology. 1987;37:359–363. doi: 10.1212/wnl.37.3.359. [DOI] [PubMed] [Google Scholar]
- Small GW, Leuchter AF, Mandelkern MA, La Rue A, Okonek A, Lufkin RB, Jarvik LF, Matsuyama SS, Bondareff W. Clinical, neuroimaging, and environmental risk differences in monozygotic female twins appearing discordant for dementia of the Alzheimer type. Arch Neurol. 1993;50:209–219. doi: 10.1001/archneur.1993.00540020085022. [DOI] [PubMed] [Google Scholar]
- Joosten E, Lesaffre E, Riezler R, Ghekiere V, Deremaeker L, Pelemans W, Dejaeger E. Is metabolic evidence for vitamin B-12 and folate deficiency more frequent in elderly patients with Alzheimer's disease? J Gerontol A Biol Sci Med Sci. 1997;52:M76–M79. doi: 10.1093/gerona/52a.2.m76. [DOI] [PubMed] [Google Scholar]
- Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- Snowdon DA, Tully CL, Smith CD, Riley KP, Markesbery WR. Serum folate and the severity of atropy of the neocortex in Alzheimer disease: findings from the Nun Study. Am J Clin Nutr. 2000;71:993–998. doi: 10.1093/ajcn/71.4.993. [DOI] [PubMed] [Google Scholar]
- Ward A, Summers MJ, Sim E. Purification of recombinant human N-acetyltransferase type 1 (NAT1) expressed in E. coli and characterisation of its potential role in folate metabolism. Biochem Pharmacol. 1995;49:1759–1767. doi: 10.1016/0006-2952(95)00087-G. [DOI] [PubMed] [Google Scholar]
- Minchin RF. Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem J. 1995;307:1–3. doi: 10.1042/bj3070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Rodgers L, Levy GN, Weber WW. Substrate selectivity of mouse N-acetyltransferases 1, 2 and 3 expressed in COS-1 cells. Drug Metabolism and Disposition. 1998;26:502–505. [PubMed] [Google Scholar]
- Payton M, Smelt V, Upton A, Sim E. A method for genotyping murine arylamine N-acetyltransferase type 2 (NAT2): a gene expressed in preimplantation embryonic stem cells encoding an enzyme acetylating the folate catabolite p-aminobenzoylglutamate. Biochem Pharmacol. 1999;58:779–785. doi: 10.1016/S0006-2952(99)00171-9. [DOI] [PubMed] [Google Scholar]
- Stanley LA, Copp AJ, Pope J, Rolls S, Smelt V, Perry VH, Sim E. Immunochemical detection of arylamine N-acetyltransferase during mouse embryonic development and in adult mouse brain. Teratology. 1998;58:174–182. doi: 10.1002/(SICI)1096-9926(199811)58:5<174::AID-TERA3>3.3.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Pacifici GM, Bencini C, Rane A. Acetyltransferase in humans: Development and tissue distribution. Pharmacology. 1986;32:283–291. doi: 10.1159/000138181. [DOI] [PubMed] [Google Scholar]
- Vatsis KP, Weber WW. Structural heterogeneity of Caucasian N-acetyltransferase at the NAT1 gene locus. Arch Biochem Biophys. 1993;301:71–76. doi: 10.1006/abbi.1993.1116. [DOI] [PubMed] [Google Scholar]
- Weber WW, Vatsis KP. Individual variability in p-aminobenzoic acid N-acetylation by human N-acetyltransferase (NAT1) or peripheral blood. Pharmacogenetics. 1993;3:209–212. doi: 10.1097/00008571-199308000-00006. [DOI] [PubMed] [Google Scholar]
- Bandmann O, Vaughan J, Holmans P, Marsden CD, Wood NW. Association of slow acetylator genotype for N-acetyltransferase 2 with familial Parkinson's disease. Lancet. 1997;350:1136–1139. doi: 10.1016/S0140-6736(97)03495-8. [DOI] [PubMed] [Google Scholar]
- Agúndez JA, Jimenez-Jimenez FJ, Luengo A, Molina JA, Orti-Pareja M, Vazquez A, Ramos F, Duarte J, Coria F, Ladero JM, Alvarez-Cermeno JC, Benitez J. Slow allotypic variants of the NAT2 gene and susceptibility to early-onset Parkinson's disease. Neurology. 1998;51:1587–1592. doi: 10.1212/wnl.51.6.1587. [DOI] [PubMed] [Google Scholar]
- Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol Rev. 1985;37:25–79. [PubMed] [Google Scholar]
- Rocha L, Garcia C, de Mendonca A, Gil JP, Bishop DT, Lechner MC. N-acetyltransferase (NAT2) genotype and susceptibility to sporadic Alzheimer's disease. Pharmacogenetics. 1999;9:9–15. [PubMed] [Google Scholar]
- Jobst KA, Smith AD, Szatmari M, Molyneux A, Esiri ME, King E, Smith A, Jaskowski A, McDonald B, Wald N. Detection in life of confirmed Alzheimer's disease using a simple measurement of medial temporal lobe atropy by computed tomography. Lancet. 1992;340:1179–1183. doi: 10.1016/0140-6736(92)92890-R. [DOI] [PubMed] [Google Scholar]
- Roth M, Tym E, Mountjoy CQ, Huppert FA, Hendrie H, Verma S, Goddard R. CAMDEX. A standardised instrument for the diagnosis of mental disorder in the elderly with special reference to the early detection of dementia. B J Psychiatry. 1986;149:698–709. doi: 10.1192/bjp.149.6.698. [DOI] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Budge M, Johnston C, Hogervorst E, de Jager C, Milwain E, Iversen SD, Barnetson L, King E, Smith AD. Plasma total homocysteine and cognitive performance in a volunteer elderly population. Ann NY Acad Sci. 2000;903:407–410. doi: 10.1111/j.1749-6632.2000.tb06392.x. [DOI] [PubMed] [Google Scholar]
- Ward A, Hickman D, Gordon JW, Sim E. Arylamine N-acetyltransferase in human red blood cells. Biochem Pharmacol. 1992;44:1099–1104. doi: 10.1016/0006-2952(92)90373-Q. [DOI] [PubMed] [Google Scholar]
- Payton MA, Sim E. Genotyping human arylamine N-acetyltransferase type 1 (NAT1). The identification of two novel allelic variants. Biochem Pharmacol. 1998;55:361–366. doi: 10.1016/S0006-2952(97)00478-4. [DOI] [PubMed] [Google Scholar]
- Hickman D, Sim E. N-acetyltransferase polymorphism. Comparison of phenotype and genotype in humans. Biochem Pharmacol. 1991;42:1007–1014. doi: 10.1016/0006-2952(91)90282-A. [DOI] [PubMed] [Google Scholar]
- Hickman D, Risch A, Camilleri JP, Sim E. Genotyping human polymorphic arylamine N-acetyltransferase: identification of new slow allotypic variants. Pharmacogenetics. 1992;2:217–22. doi: 10.1097/00008571-199210000-00004. [DOI] [PubMed] [Google Scholar]
- Upton A, Johnson N, Sandy J, Sim E. Arylamine N-acetyltransferases – of mice, men and microorganisms. Trends Pharmacol Sci. 2001;22:140–146. doi: 10.1016/S0165-6147(00)01639-4. [DOI] [PubMed] [Google Scholar]
- Smelt VA, Mardon HJ, Sim E. Placental expression of arylamine N-acetyltransferases: evidence for linkage disequilibium between NAT1*10 and NAT2*4 alleles of the two human arylamine N-acetyltransferase loci NAT1 and NAT2. Pharmacol Toxicol. 1998;83:149–157. doi: 10.1111/j.1600-0773.1998.tb01461.x. [DOI] [PubMed] [Google Scholar]
- Steventon GB, Heafield MT, Waring RH, Williams AC, Sturman S, Green M. Xenobiotic metabolism in Alzheimer's disease. Neurology. 1990;40:1095–1098. doi: 10.1212/wnl.40.7.1095. [DOI] [PubMed] [Google Scholar]
- Ladero JM, Barquero MS, Coria F, Molina JA, Jimenez-Jimenez FJ, Benitez J. Acetylator polymorphism in Alzheimer's disease. Eur J Med. 1993;2:281–283. [PubMed] [Google Scholar]
- Madden S, Woolf TF, Pool WF, Park BP. An investigation into the formation of stable, protein-reactive and cytotoxic metabolites from tacrine in vitro. Studies with human and rat liver microsomes. Biochem Pharmacol. 1993;46:13–20. doi: 10.1016/0006-2952(93)90342-T. [DOI] [PubMed] [Google Scholar]
- Spaldin V, Madden S, Pool WF, Woolf TF, Park BF. The effect of enzyme inhibition on the metabolism and activation of tacrine by human liver microsomes. Br J Clin Pharmacol. 1994;38:15–22. doi: 10.1111/j.1365-2125.1994.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blömeke B, Sieben S, Spotter D, Landt O, Merk HF. Identification of N-acetyltransferase 2 genotypes by continuous monitoring of fluorogenic hybridisation probes. Anal Biochem. 1999;275:93–97. doi: 10.1006/abio.1999.4288. [DOI] [PubMed] [Google Scholar]
- Wikman H, Thiel S, Jager B, Schmezer P, Spiegelhalder B, Edler L, Dienemann H, Kayser K, Schulz V, Drings P, Bartsch H, Risch A. Relevance of N-acetyltransferase 1 and 2 (NAT1, NAT2) genetic polymorphisms in non-small cell lung cancer susceptibility. Pharmacogenetics. 2001;11:157–168. doi: 10.1097/00008571-200103000-00006. [DOI] [PubMed] [Google Scholar]
- Cornish VA, Pinter KA, Boukouvala S, Johnson N, Labrousse C, Payton M, Priddle H, Smith AJH, Sim E. Generation and analysis of mice with a targeted disruption of the arylamine N-acetyltransferase type 2 gene. Pharmacogenomics J. 2003;3:169–177. doi: 10.1038/sj.tpj.6500170. [DOI] [PubMed] [Google Scholar]
- Sim E, Pinter K, Mushtaq A, Upton A, Bhakta S, Noble M. Arylamine N-acetyltransferases: a pharmacogenomic approach to drug metabolism and endogenous function. Biochemical Society Transactions. 2003;31:615–619. doi: 10.1042/bst0310615. [DOI] [PubMed] [Google Scholar]
- Risch A, Smelt V, Lane D, Stanley L, van der Slot W, Ward A, Sim E. Arylamine N-acetyltransferase in erythrocytes of cystic fibrosis patients. Pharmacol Toxicol. 1996;78:235–240. doi: 10.1111/j.1600-0773.1996.tb00211.x. [DOI] [PubMed] [Google Scholar]
- Hughes NC, Janezic SA, McQueen KL, Jewett MA, Castranio T, Bell DA, Grant DM. Identification and characterisation of variant alleles of human acetyltransferase NAT1 with defective function using p-aminosalicylate as an in-vivo and in-vitro probe. Pharmacogenetics. 1998;8:55–66. doi: 10.1097/00008571-199802000-00008. [DOI] [PubMed] [Google Scholar]